Supplemental Digital Content is Available in the Text.
Key Words: HIV, antibodies, neutralization, diversity, epitopes
Abstract
Background:
Highly potent broadly neutralizing monoclonal antibodies (bNAbs) have been obtained from individuals infected by HIV-1 group M variants. We analyzed the cross-group neutralization potency of these bNAbs toward non-M primary isolates (PI).
Material and Methods:
The sensitivity to neutralization was analyzed in a neutralization assay using TZM-bl cells. Twenty-three bNAbs were used, including reagents targeting the CD4-binding site, the N160 glycan-V1/V2 site, the N332 glycan-V3 site, the membrane proximal external region of gp41, and complex epitopes spanning both env subunits. Two bispecific antibodies that combine the inhibitory activity of an anti-CD4 with that of PG9 or PG16 bNAbs were included in the study (PG9-iMab and PG16-iMab).
Results:
Cross-group neutralization was observed only with the bNAbs targeting the N160 glycan-V1/V2 site. Four group O PIs, 1 group N PI, and the group P PI were neutralized by PG9 and/or PG16 or PGT145 at low concentrations (0.04–9.39 μg/mL). None of the non-M PIs was neutralized by the bNAbs targeting other regions at the highest concentration tested, except 10E8 that neutralized weakly 2 group N PIs and 35O22 that neutralized 1 group O PI. The bispecific bNAbs neutralized very efficiently all the non-M PIs with IC50 below 1 μg/mL, except 2 group O strains.
Conclusion:
The N160 glycan-V1/V2 site is the most conserved neutralizing site within the 4 groups of HIV-1. This makes it an interesting target for the development of HIV vaccine immunogens. The corresponding bNAbs may be useful for immunotherapeutic strategies in patients infected by non-M variants.
INTRODUCTION
Through technological advances associating B-cell clonal amplification, antibody cloning in expression vectors, and high-throughput neutralization assays, many extremely potent and broad donor-derived human monoclonal neutralizing antibodies (bNAbs) directed to the human immunodeficiency virus type 1 (HIV-1) were discovered over the past 5 years (for reviews, see Refs. 1–4). They target sites of vulnerability on the HIV-1 envelope glycoproteins: 3 within the external glycoprotein gp120 corresponding to the CD4-binding site (CD4bs), a V1/V2-glycan-dependent site (N160 glycan-V1/V2), and a V3-glycan-dependent site (N332 glycan-V3), 1 corresponding to the membrane proximal external region (MPER) of the transmembrane glycoprotein gp41, and more recently described, epitopes located at the gp120-gp41 interface.5–8 When passively transferred, the bNAbs can protect against Simian HIV infection in nonhuman primates at low concentrations.9,10 In addition, they can also suppress established infection in humanized mice and macaques.11–13 After the identification of this new generation of bNAbs, both their breadth and potency were still improved by either structure-based gene modifications14,15 or creation of bispecific antibodies that combine the HIV-1 inhibitory activity of an anti-human CD4 with that of anti-gp120 bNAbs (BibNAbs).16
All the data described above were obtained using group M HIV-1 viruses. The identification of antigenic targets of the bNAbs that would be conserved among highly divergent HIV-1 variants, such as variants related to groups N, O, and P, might be of additional value to inform HIV-1 vaccine design. Indeed, a high conservation of a neutralizing antigenic site would suggest its major role in the biology of the HIV-1 species and, therefore, its potential importance as a vaccine component. We recently presented data with some evidence for conservation of the N160 glycan-V1/V2 site within the different groups.17 The aim of this study was to extend the previous analysis to a larger panel of non-M viruses and to the most potent bNAbs described today, including BibNAbs.
METHODS
Virus Isolates
Sixteen HIV-1 primary isolates (PIs) related to groups O, N, and P were used (Table 1). There were 12 group O viruses, 2 group N viruses, 1 group P virus, and 1 recombinant M/O virus. The entire env region of the recombinant M/O virus was related to group M. These viruses were isolated from blood samples collected between 1994 and 2011, either in Cameroon or in France (see references in Table 1). As shown in Figure 1, the PIs were representative of the non-M diversity because they did not cluster in specific branches of the tree. The sequence divergence among the viruses used in the study is shown in Table S1 (see Supplemental Digital Content 1, http://links.lww.com/QAI/A753). Virus stocks were prepared by passaging isolates only once or twice on phytohemagglutinin-stimulated peripheral blood mononuclear cells from a HIV-negative blood donor.
TABLE 1.
Characteristics of the HIV-1 Strains Related to Groups O, N, and P, Used in the Study
FIGURE 1.
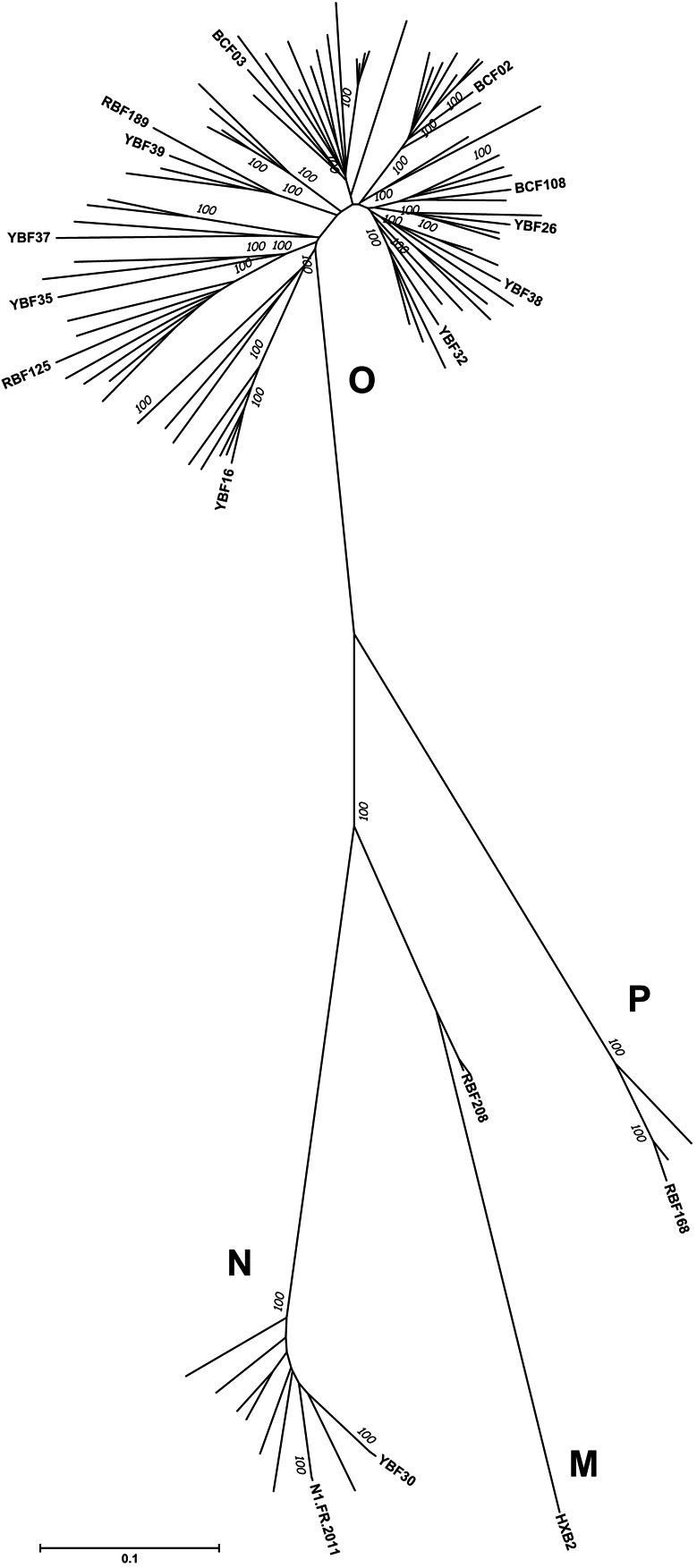
Phylogenetic analysis of env sequences. The 16 env sequences of the PIs included in the study were aligned with 72 env sequences from non-M variants that were available in the Los Alamos database. The evolutionary history was inferred using the Neighbor-Joining method.34 The optimal tree with the sum of branch length = 752,171,464 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches.35 The tree is drawn to scale with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Tamura-Nei method36 and are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). All ambiguous positions were removed for each sequence pair. There were a total of 3165 positions in the final data set. Evolutionary analyses were conducted in MEGA6.37
Neutralizing Antibodies and Soluble CD4
Twenty-three neutralizing antibodies were used (Table 2). Eight human monoclonal bNAbs target the CD4bs: VRC01, VRC03, 3BNC117, and 5 clonal variants of the bNAb NIH45-46G54W.14 Four bNAbs target the N160 glycan-V1/V2 site: PGT145, PG9, PG16, and the modified PG9-PG16 RSH.15 Three bNAbs target the N332 glycan-V3 site: PGT121, PGT128, and 10-1074. Three bNAbs target the MPER: 2F5, 4E10, and 10E8. In addition, we used 2 human monoclonal bNAbs 8ANC195 and 35O22 that target the interface of both env subunits5,8 and the single-domain llama antibody JM4sdAb that targets an epitope involving elements of both coreceptor-binding sites and CD4bs on gp120.24
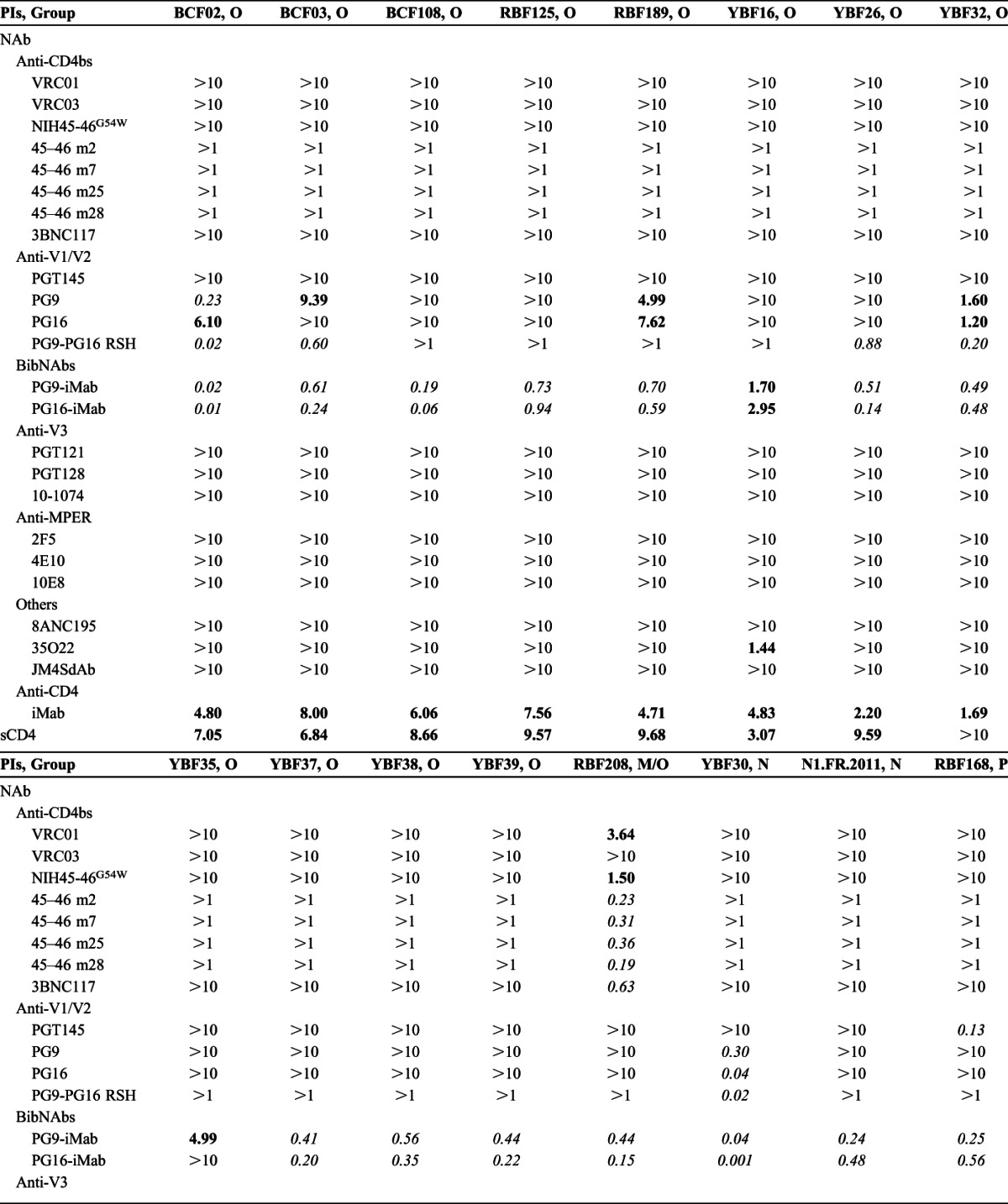
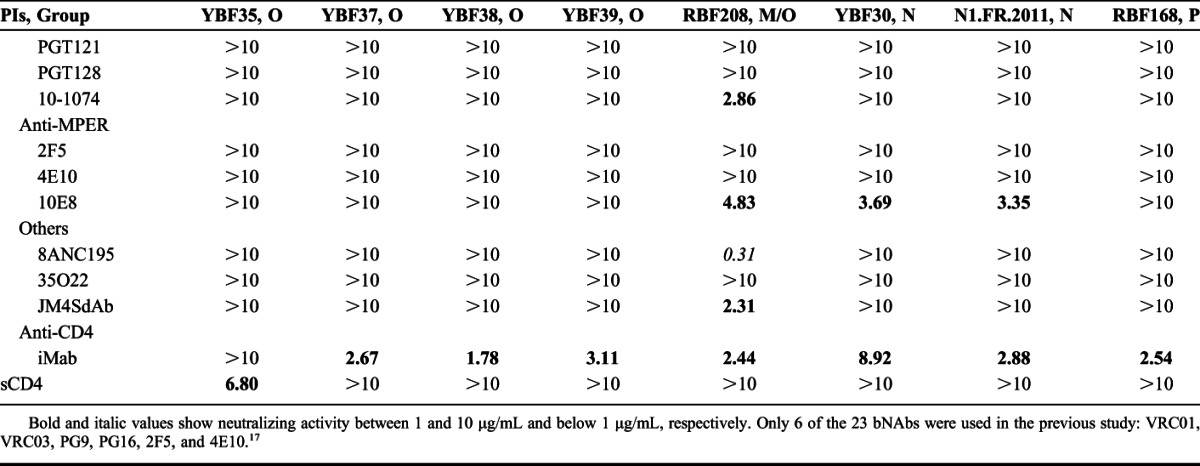
TABLE 2.
Sensitivity to Neutralization (IC50) of 16 Non-M PIs
Two BibNAbs, PG9-iMab, and PG16-iMab, that exhibit exceptional breadth and potency toward HIV-1 group M isolates, were analyzed.16 They are bispecific antibodies that combine the HIV-1 inhibitory activity of ibalizumab (iMab), a humanized Mab directed to domain 2 of human CD4, with that of PG9 or PG16. They were constructed by engraftment of the single-chain variable fragment (scFv) of PG9 or PG16 at the N-terminus of the heavy chain of iMab by a flexible linker. The parental anti-CD4 iMab, whose both safety and anti-HIV-1 efficacy have been established in clinical trials, was used as control.25 The sensitivity to soluble CD4 (sCD4; Progenics Pharmaceuticals, Tarrytown, NY) was also analyzed.
Neutralization Assay
Sensitivity to bNAbs or sCD4 of the PIs was assessed in TZM-bl cells as previously described.17 Briefly, virus stocks were diluted to 10,000 TCID50/mL. Aliquots of 50 μL corresponding to 500 TCID50 were then incubated for 1 hour at 37°C with 50 μL of 3-fold serial dilutions of each reagent (starting at 10 μg/mL for all the reagents, except at 1 μg/mL for PG9-PG16 RSH and the clonal variants of NIH45-46 G54W). The virus–antibody mixture was then used to infect 10,000 TZM-bl cells in the presence of 30 μg/mL DEAE-dextran. Infection levels were determined after 48 hours by measuring the luciferase activity of cell lysates using the Bright-Glo luciferase assay (Promega, Madison, WI) and a Centro LB 960 luminometer (Berthold Technologies, Bad Wildbad, Germany). The results were expressed as the mean values of the assays performed in duplicate. IC50 values were defined as the bNAb concentration required to reduce the relative luminescence units by 50%.
RESULTS
None of the bNAbs targeting the CD4bs or the N332 glycan-V3 site was capable of neutralizing the non-M viruses (Table 2). Similarly, 8ANC195 and JM4SdAb did not neutralize any of the non-M viruses, at least at concentration below 10 μg/mL. 35O22 neutralized only 1 group O strain (IC50 = 1.44 μg/mL). Among the anti-MPER bNAbs, no neutralization was detected for 2F5 and 4E10, and 10E8 showed a moderate neutralizing activity only toward the 2 group N viruses (IC50 = 3.35–3.69 μg/mL). In contrast, cross-group neutralization was observed with the bNAbs, targeting the N160 glycan-V1/V2 site. PG9 and PG16 neutralized 3 group O PIs (IC50 = 0.23–7.62 μg/mL), and a fourth group O strain (BCF03) was neutralized weakly by PG9 only (IC50 = 9.39 μg/mL). Both PG9 and PG16 were highly efficient toward 1 N strain (YBF30; IC50 = 0.30 and 0.04 μg/mL, respectively), but the second N strain, N1.FR.2011, appeared resistant. The resistance of the N1.FR.2011 strain could be attributed to an N/K mutation at position 160 (Fig. 2). PG9-PG16 RSH is a chimeric antibody in which 3 key amino acids from the PG16 light chain were introduced in PG9.15 Logically, the potency of the modified PG9-PG16 RSH bNAb toward the PG9/PG16-sensitive strains was enhanced, at least clearly for 4 of them (BCF02, BCF03, YBF32, and YBF30; Table 2), and the YBF26 strain which was resistant to PG9 and PG16 (IC50 >10 μg/mL) became sensitive to the modified PG9-PG16 RSH (IC50 = 0.88 μg/mL). Although the group P prototype strain RBF168 was resistant to neutralization by PG9 and PG16, it was sensitive to PGT145 (IC50 = 0.13 μg/mL). All together, these data strongly suggest that the neutralizing epitopes located in the N160 glycan-V1/V2 site are conserved within all groups of the HIV-1 species, at least at some degree. Not surprisingly, the most sensitive strain to the various bNAbs was the M/O recombinant RBF208 because of the nature of its env gene that is related to group M.
FIGURE 2.
Conservation of amino acids involved in antibody binding epitopes. An alignment of the env protein sequences of the non-M viruses used in the study is depicted, with dashes representing gaps introduced to improve the alignment. HXB2 sequence is shown as reference. Amino acids are colored based on their physicochemical properties. The logo plots denote the conservation of individual amino acids, with the height of each letter indicating the proportion of sequences that contain the residue at that site. Contact residues of VRC01 (blue), JM4SdAb (yellow), and MPER bAbs (brown) are highlighted below the alignment. The Y symbols indicate the positions of the glycans associated with antibody neutralizing activity (see colors in the inserted legend to identify the corresponding bNAbs).
The BibNAbs PG9-iMab and PG16-iMab neutralized very efficiently all the non-M PIs with IC50 below 1 μg/mL, except 2 group O strains, YBF16 and YBF35, which were neutralized at IC50 between 1 and 10 μg/mL or by PG9-iMab only, respectively (Table 2). Ibalizumab alone neutralized all the non-M viruses except 1 group O strain (YBF35), but at higher IC50 (1.78–8.92 μg/mL). When comparing the BibNAbs with the parental antibodies, the median IC50 values were 0.47 and 0.23 μg/mL for PG9-iMab and PG16-iMab, respectively, compared to 3.91 μg/mL for iMab and above 10 μg/mL for PG9 and PG16. The greater potency of PG9-iMab and PG16-iMab compared with the parental antibodies was observed for both the PG9-sensitive and PG16-sensitive and the PG9-resistant and PG16-resistant viruses (Fig. 3A). Indeed, the 5 dual-sensitive viruses (BCF02, BCF03, RBF189, YBF32, and YBF30) were neutralized approximately 10-fold more potently by the BibNAbs than by PG9 or PG16. The data suggest that the enhanced potency was not simply due to the additive effects of the parental antibodies. The most sensitive viruses to PG9-iMab and PG16-iMab were also the most sensitive to PG9 or PG16 (Fig. 3A), suggesting that the high potency of the BibNAbs was mediated by the gp120-binding activity of PG9 and PG16 scFvs. The improved activity of PG9-iMab and PG16-iMab is illustrated also in Figure 3B, where viral neutralization coverage as a function of increasing concentrations is shown. More than 80% viral coverage was achieved by PG9-iMab and PG16-iMab at IC50 below 1 μg/mL, whereas this coverage was achieved at approximately 8 μg/mL for iMab alone while PG9 or PG16 alone neutralized less than 35% of the non-M PIs at 10 μg/mL.
FIGURE 3.
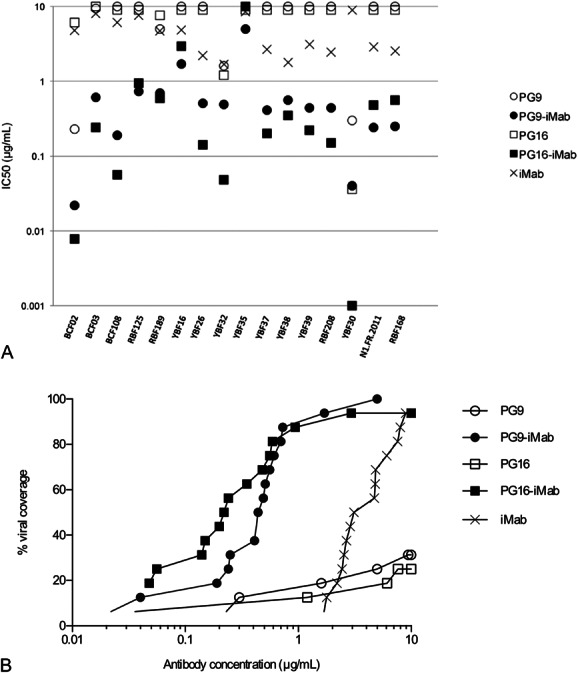
Neutralization breadth and potency of PG9-iMab and PG16-iMab, and parental Mabs against the panel of non-M viruses. A, Comparison of potency. For each virus, IC50 are represented with a closed circle for PG9-iMab, an open circle for PG9, a closed square for PG16-iMab, an open square for PG16, and a cross for iMab. B, Percent viral coverage achieved by PG9-iMab, PG16-iMab, PG9, PG16, and iMab. Cumulative frequency distribution of IC50 values of the antibodies tested against the 16 viruses. Symbols as above.
Soluble CD4 neutralized 8 of the 12 group O strains, but none of the N or P variants at 10 μg/mL (Table 2).
DISCUSSION
HIV-1 non-M variants are mainly endemic in Cameroon, where the prevalence has been stable over the last 10 years, and sporadic cases were reported outside this region, particularly in France.26 In contrast to the prevalence of HIV-1M that has progressed exponentially worldwide, the non-M viruses did not spread and their prevalence remains stable for reasons that are still greatly unknown, even in Cameroon.26 As such, they do not seem as a major public health problem. However, because of their distant genetic relatedness to HIV-1, they represent a relevant tool to study conserved biological properties. We, and others previously, have shown that sera from HIV-1–infected individuals may share cross-neutralizing activities against groups M, N, O, and P PIs.17,27–29 The identification of antigenic targets of bNAbs that would be conserved among highly divergent HIV-1 variants could be informative for HIV vaccine design. Taking the opportunity to analyze the sensitivity of a panel of rare non-M PIs to the most potent bNAbs described today, we confirm and expand the impact of our previous findings that suggested some degree of conservation of the PG9/PG16 epitopes within these divergent groups.17 Indeed, 17 of the 23 bNAbs that we used in this study were not included in our previous work.17 One limitation of our findings might be the representativeness of the panel of non-M strains because we included less than 20 PIs. However, because of the scarcity of non-M PIs, we consider that it represents a relatively large panel. In addition, the phylogenetic analysis that included all the env gene sequences from non-M variants available in the Los Alamos database revealed that our isolates did not cluster in particular branches. It suggests that the neutralization properties that we describe may be representative of the entire non-M population.
We provide data showing that variants related to groups O, N, and P are neutralized by bNAbs, targeting the N160 glycan-V1/V2 site, either by PG9 and PG16 for groups O and N viruses or by PGT145 for the single–group P isolate. Although the bNAbs directed to the CD4bs or to the N332 glycan-V3 site that we used were selected among the most potent reagents that exist up to now, we did not detect cross-neutralizing activity involving these 2 major gp120 antigenic sites. In view of the lack of effect of antibodies to the CD4b, we analyzed the sensitivity of the viruses to neutralization by soluble CD4. Only approximately half of them were neutralized by sCD4, suggesting a lower sensitivity to sCD4 of non-M viruses also. The 2 bNAbs 8ANC195 and 35O22 that target epitopes located at the gp120-gp41 interface had no cross-neutralization potential, except 35O22 which neutralized a single–group O isolate (YB16) at an IC50 of 1.44 μg/mL. JM4SdAb, that targets an epitope involving elements of both coreceptor-binding sites and CD4bs on gp120, did not neutralize the non-M viruses. Except a moderate neutralizing activity of 10E8 toward 2 group N isolates, we did not detect cross-neutralizing activity of the anti-MPER bNAbs. Therefore our data indicate that the N160 glycan-V1/V2 antigenic site is the most conserved neutralizing site within the 4 groups of HIV-1, and as such is a valuable site to include in vaccine candidates. Concordant data were reported very recently by Barbian et al30 when analyzing the neutralization properties of Simian immunodeficiency virus infecting chimpanzees and gorillas. Complementary to this observation, it is interesting to note that antibodies to the V1/V2 region of multiple HIV-1 subtypes were identified as a correlate of decreased risk of HIV-1 infection in the RV144 HIV-1 efficacy trial.31 It is fascinating that a single domain would share 2 apparently opposite properties. Its high variability, confined to the loop V1 and V2 regions, contributes in evading antibody-mediated neutralization. In contrast, its conservation, in the 4-stranded β-sheet domain,32 makes it a target of both cross-group neutralizing antibodies and antibodies that may be associated to a successful vaccine outcome.
We tried to identify a structural explanation for the observed cross-neutralization. Several specific amino acids at some positions have been identified as critical for binding of the V1/V2 bNAbs. The core epitopes of PG9, PG16, and PGT145 contain 2 essential glycosylation sites N156 and N160 and also a lysine-rich motif in strand C of V1V2.32,33 YBF30 which was the most sensitive to neutralization by PG9 and PG16 was the single non-M isolate harboring these 2 N-glycosylation sites and the K-rich motif (Fig. 2). In contrast, YBF35 which was the most resistant to PG9 and PG16, including PG9-iMab and PG16-iMab, did not harbor N156 nor the K-rich motif. The resistance of the N1.FR.2011 strain, albeit it possessed both the N156 and the K-rich motif, could be attributed to an N/K mutation at position 160. Among the other non-M viruses, all harbored both the K-rich motif and the N160 (except 1, RBF125), but all lacked the N156. Despite this common feature, some of them were sensitive to PG9 and PG16, whereas others were not. Thus, variations in other amino acid positions in this region or in other Env regions must also be involved to confer the antigenicity of the corresponding epitopes. Specifically dedicated studies should be conducted to better identify the molecular determinants of sensitivity or resistance to the V1/V2 bNAbs. Similar studies involving other bNAbs could be also informative. Indeed, for instance, all the non-M viruses were resistant to PGT128 and 8ANC195, albeit the relevant N-glycosylation sites N332 and N234/N276 were present in some or most of the isolates, respectively, and the 2 N isolates were sensitive to 10E8, albeit 2 mutations, N673S for YBF30 and S678T for N1.FR.2011, were present in the core epitope (Fig. 2).
The 2 BibNAbs, PG9-iMab and PG16-iMab, exhibited exceptional breadth and potency toward a panel of diverse HIV-1 group M viruses.16 Our data show that they are also highly potent toward highly divergent strains related to groups O, N, and P, even toward those that were not neutralized by PG9 or PG16 alone. In addition, the PG9/PG16-sensitive viruses were neutralized approximately 10-fold more potently by PG9-iMab and PG16-iMAb than by iMab or PG9 and PG16 alone. It has been suggested that the improvement in neutralization potency of the BibNAbs could be due to a synergistic effect through an increase in PG9-scFv or PG16-scFv concentration at the location where HIV-1 penetrates into its target cell.16 The fact that the most sensitive viruses to PG9-iMab and PG16-iMab in our study were those that were the most sensitive to PG9 or PG16 suggests also that the high potency of the BibNAbs would be mediated by the gp120-binding activity of PG9 and PG16 scFvs.
The recent developments in the field of HIV monoclonal antibodies have led to the idea that the clinical potential of bNAbs or BibNAbs would deserve to be considered for immunoprophylaxis and immunotherapy. Ongoing or future clinical trials should inform on their efficacy and their safety. In addition to bring an additional basic knowledge on the neutralizing epitopes that might be useful for vaccine designers, our study suggests that patients exposed to or infected by the rare non-M variants of HIV-1 could benefit also from these new strategies.
ACKNOWLEDGMENTS
The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl cells from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc.; VRC01 and VRC03 from Dr. John Mascola; 10E8 and 35O22 from Dr. Mark Connors. The authors thank Sylvie Brunet for her excellent technical support and Marie Leoz for previous work on isolation and characterization of the viral strains.
Footnotes
Supported by the Agence Nationale de Recherche sur le SIDA et les Hépatites (ANRS, Paris, France). M. Morgand was supported by a doctoral fellowship from the ANRS. M. Bouvin-Pley was supported by doctoral fellowships from the Région Centre and Sidaction (France).
Presented in part as a poster presentation at the “AIDS vaccine 2013” meeting, October 7–9, 2013, Barcelona, Spain.
The authors have no conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.jaids.com).
REFERENCES
- 1.Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat Rev Immunol. 2012;13:693–701. [DOI] [PubMed] [Google Scholar]
- 2.Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol Rev. 2013;254:225–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein F, Mouquet H, Dosenovic P, et al. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West AP, Jr, Scharf L, Scheid JF, et al. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156:633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scharf L, Scheid JF, Lee JH, et al. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep. 2014;7:785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner C, lee JH, Sliepen K, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falkowska E, Le KM, Ramos A, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang J, Kang BH, Pancera M, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515:138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moldt B, Rakasz EG, Schultz N, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A. 2012;109:18921–18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shingai M, Donau OK, Plishka RJ, et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211:2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horwitz JA, Halper-Stromberg A, Mouquet H, et al. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc Natl Acad Sci U S A. 2013;110:16538–16543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barouch DH, Whitney JB, Moldt B, et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shingai M, Nishimura Y, Klein F, et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503:277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diskin R, Klein F, Horwitz JA, et al. Restricting HIV-1 pathways for escape using rationally designed anti-HIV-1 antibodies. J Exp Med. 2013;210:1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pancera M, Shahzad-ul-Hussan S, Doria-Rose NA, et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol. 2013;20:804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pace CS, Song R, Ochsenbauer C, et al. Bispecific antibodies directed to CD4 domain 2 and HIV envelope exhibit exceptional breadth and picomolar potency against HIV-1. Proc Natl Acad Sci U S A. 2013;110:13540–13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braibant M, Gong EY, Plantier JC, et al. Cross-group neutralization of HIV-1 and evidence for conservation of the PG9/PG16 epitopes within divergent groups. AIDS. 2013;27:1239–1244. [DOI] [PubMed] [Google Scholar]
- 18.Roques P, Robertson DL, Souquiere S, et al. Phylogenetic analysis of 49 newly derived HIV-1 group O strains: high viral diversity but no group M-like subtype structure. Virology. 2002;302:25–273. [DOI] [PubMed] [Google Scholar]
- 19.Depatureaux A, Leoz M, De Oliveira F, et al. Specific diagnosis and follow-up of HIV-1 group O infection: RES-O data. Med Mal Infect. 2010;40:669–676. [DOI] [PubMed] [Google Scholar]
- 20.Plantier JC, Leoz M, Dickerson JE, et al. A new human immunodeficiency virus derived from gorillas. Nat Med. 2009;15:871–872. [DOI] [PubMed] [Google Scholar]
- 21.Simon F, Mauclere P, Roques P, et al. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4:1032–1037. [DOI] [PubMed] [Google Scholar]
- 22.Delaugerre C, De Oliveira F, Lascoux-Combe C, et al. HIV-1 group N: travelling beyond Cameroon. Lancet. 2011;378:1894. [DOI] [PubMed] [Google Scholar]
- 23.Vessiere A, Rousset D, Kfutwah A, et al. Diagnosis and monitoring of HIV-1 group O-infected patients in Cameroun. J Acquir Immune Defic Syndr. 2010;53:107–110. [DOI] [PubMed] [Google Scholar]
- 24.Matz J, Kessler P, Bouchet J, et al. Straightforward selection of broadly neutralizing single-domain antibodies targeting the conserved CD4 and coreceptor binding sites of HIV-1 gp120. J Virol. 2013;87:1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson JM, Kuritzkes DR, Godovsky E, et al. Safety, pharmacokinetics, and antiretroviral activity of multiple doses of ibalizumab (formerly TNX-335), an anti-CD4 monoclonal antibody, in human immunodeficiency virus type 1-infected adults. Antimicrob Agents Chemother. 2009;53:450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mourez T, Simon F, Plantier JC. Non-M variants of human immunodeficiency virus type 1. Clin Microbiol Rev. 2013;26:448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nyambi PN, Nkengasong J, Peeters M, et al. Reduced capacity of antibodies from patients infected with human immunodeficiency virus type 1 (HIV-1) group O to neutralize primary isolates of HIV-1 group M viruses. J Infect Dis. 1995;172:1228–1237. [DOI] [PubMed] [Google Scholar]
- 28.Beirnaert E, Nyambi P, Willems B, et al. Identification and characterization of sera from HIV-infected individuals with broad cross-neutralizing activity against group M (env clade A-H) and group O primary isolates. J Med Virol. 2000;62:14–24. [PubMed] [Google Scholar]
- 29.Ferrantelli F, Kitabwalla M, Rasmussen RA, et al. Potent cross-group neutralization of primary human immunodeficiency virus isolates with monoclonal antibodies: implications for acquired immunodeficiency syndrome vaccine. J Infect Dis. 2004;189:71–74. [DOI] [PubMed] [Google Scholar]
- 30.Barbian HJ, Decker JM, Bibollet-Ruche F, et al. Neutralization properties of simian immunodeficiency viruses infecting chimpanzees and gorillas. MBio. 2015;6:e00296–e00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zolla-Pazner S, deCamp A, Gilbert PB, et al. Vaccine-induced IgG antibodies to V1V2 regions of multiple HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PLoS One. 2014;9:e87572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mc Lellan JS, Pancera M, Carrico C, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doria-Rose NA, Georgiev I, O'Dell S, et al. A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. J Virol. 2012;86:8319–8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. [DOI] [PubMed] [Google Scholar]
- 35.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. [DOI] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]



