Introduction
Ion channels are the most rapid of all signaling entities. Minor fluctuations in the actions of regulatory signals can switch the conductance state of a single channel so as to influence the transmembrane movement of millions of ions per second (Clapham, 2001). This is a particularly impressive example of signal amplification. The conductance of the particular Cl− channel that we study, ClC3, is activated by CaMKII (Robinson et al., 2004). ClC3 is also regulated by one member of the inositol phosphate signaling family, Ins(3,4,5,6)P4 (Fig. 1), which inhibits Cl− conductance through this channel (Mitchell et al., 2008). The cellular levels of Ins(3,4,5,6)P4 are dynamically regulated by receptor-initiated control over the activities of a multifunctional kinase/phosphotransferase (ITPK1) that interconverts Ins(3,4,5,6)P4 with Ins(1,3,4,5,6)P5 (Fig. 1). It is this enzyme, and its biological significance, that are the focus for this review.
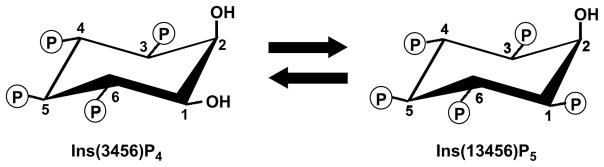
Fig. 1.
The structures of Ins(3,4,5,6)P4 and Ins(1,3,4,5,6)P5. The myo-inositol building block is a ring structure made from six carbon groups, each of which has a free hydroxyl. The hydroxyl attached to the 2-carbon is axial (perpendicular) to the plane of the ring, and the remaining hydroxyls are equatorial (i.e. approximately in the same plane as the ring). The carbons are numbered in an anticlockwise direction when the ring is viewed from above. Substitution of four of the hydroxyls with phosphates at positions 3, 4, 5 and 6 produces inositol 3,4,5,6-tetrakisphosphate. The standard abbreviation for this polyphosphate – Ins(3,4,5,6)P4 – therefore reflects the recognition by inositol phosphate nomenclature of the number of phosphate groups (denoted by the subscript), as well as their positions around the inositol ring.
Ins(3,4,5,6)P4 regulates Cl− channel conductance
Whole-cell electrophysiological analysis has been used to demonstrate that Ins(3,4,5,6)P4 is a concentration-dependent inhibitor of a CaMKII-activated Cl− conductance that is located in the plasma membrane (Ho et al., 2001; Mitchell et al., 2008; Xie et al., 1996, 1998). At least in mammalian cells, the inhibition of Cl− channel conductance by Ins(3,4,5,6)P4 is an exquisitely specific regulatory process; it is not imitated by any of the many other inositol phosphates that exist inside cells (Ho et al., 2001; Ho and Shears, 2002; Xie et al., 1996).
In secretory epithelia, the major role for Cl− channels is to sustain salt and fluid secretion (Petersen, 1992), so Ins(3,4,5,6)P4 is now viewed as playing a key regulatory role in this important biological process. Ins(3,4,5,6)P4 has also been reported to regulate cell growth through an effect on Cl− fluxes in a plant model, namely, the apex of the pollen tube from lily and tobacco (Zonia et al., 2002). The recent identification of ClC3 as the channel that Ins(3,4,5,6)P4 regulates, at least in mammalian cells (Mitchell et al., 2008), has greatly expanded the biological repertoire of this inositol phosphate. For example, ClC3 is responsible for the Ins(3,4,5,6)P4-regulated Cl− conductance in hippocampal neurones (Mitchell et al., 2008), which is thought to contribute to the overall regulation of the synaptic efficacy in generating action potentials (Wang et al., 2006). Long-term changes in synaptic efficacy comprise a cellular basis for information storage and memory formation (Bliss and Collingridge, 1993). Thus, Ins(3,4,5,6)P4 is a molecule that has the potential to affect neuronal development. It therefore seems pertinent that Ins(3,4,5,6)P4 has also previously been suggested to have the characteristics of a “memory molecule”, because its relatively slow rate of metabolism permits its physiological effects to long outlast the duration of the stimulus that initially prompts intracellular Ins(3,4,5,6)P4 to accumulate (Ho and Shears, 2002). ClC3 is known to have many other roles, including tumor cell migration (Mao et al., 2008), bone remodeling (Okamoto et al., 2008), apoptosis (Claud et al., 2008), insulin secretion (Barg et al., 2001), and inflammatory responses (Moreland et al., 2006). We can now anticipate that Ins(3,4,5,6)P4 might also regulate these processes.
Some of the newly appreciated functions for Ins(3,4,5,6)P4 arise because ClC3 is not only present in the plasma membrane, but in addition this ion channel resides in intracellular vesicles such as insulin granules (Barg et al., 2001) and the early endosomal compartment (Gentzsch et al., 2003; Hara-Chikuma et al., 2005; Mitchell et al., 2008; Stobrawa et al., 2001; Zhao et al., 2007). In these intracellular compartments, considerable ClC3 driven Cl− flux occurs even in the apparent absence of CaMKII activation (Mitchell et al., 2008). It has been proposed that Cl− influx into these vesicles provides the charge-neutralization without which the electrogenic H+-ATPase would not be capable of acidifying the vesicle interior (Hara-Chikuma et al., 2005; Weylandt et al., 2007). A recent complication for the latter hypothesis is the determination that two close relatives of ClC3, namely, ClC4 and ClC5, are actually nCl−/H+ antiporters (n > 1) (Picollo and Pusch, 2005; Scheel et al., 2005). As yet, there is no direct evidence that ClC3 is also a transporter rather than a channel. In fact, a recent study (Lisal and Maduke, 2008) has rationalized how the ClC family might actually include both channels and transporters. It is even possible that a ClC protein might functionally switch between the two modes; this could, for example, be mediated by CaMKII-dependent phosphorylation, or perhaps by the association of regulatory proteins (Wang et al., 2006). Irrespective of the mechanisms involved, there is no doubt that ClC3 contributes to endosomal acidification (Jentsch, 2008). Indeed, when a cell-permeant analogue of Ins(3,4,5,6)P4 was added to cells so as to inhibit ClC3, the pH of certain vesicular sub-compartments became more alkaline (Mitchell et al., 2008; Renström et al., 2002). What is the biological significance of this regulation of intra-vesicular pH? With regards to insulin granules, it has been proposed that their intraluminal acidification is a priming process, without which they become less competent to fuse with the plasma membrane and release their cargo (Barg et al., 2001). In support of this idea, we have shown that alkalinization of insulin granules by Ins(3,4,5,6)P4 has the effect of reducing insulin secretion from pancreatic β-cells (Renström et al., 2002). This has previously led us (Renström et al., 2002) to consider what might be the pathological consequences of a persistently activated pancreatic Ins(3,4,5,6)P4 signal, such as that which would inevitably (see Section 4) accompany sustained glucose-dependent activation of PLC (Trimble et al., 1987). Perhaps in some individuals, such abnormally elevated Ins(3,4,5,6)P4 levels contribute to the hyperglycemia-dependent refractoriness of β-cells (Meyer et al., 2002) which typifies type 2 diabetes etiology (Kilpatrick and Robertson, 1998).
In many other cell types, the acidification of the intracellular vesicles by H+-ATPases serves other important functions, including modulation of certain ligand–protein interactions during endocytosis, enzyme targeting, H+-coupled uptake of small molecules (such as neurotransmitters), and optimization of proteolytic activities of, for example, prohormone processing enzymes (Nishi and Forgac, 2002). It appears that we have only scratched the surface of our understanding of the biological importance of Ins(3,4,5,6)P4. Unfortunately, we have not yet determined the mechanism by which Ins(3,4,5,6)P4 regulates ClC3.
Besides ClC3, are there other species of Cl− channels that might be inhibited by Ins(3,4,5,6)P4? The proteins that are most closely related to ClC3 are ClC4 and ClC5, but neither of these appear to be regulated by Ins(3,4,5,6)P4 (Mitchell et al., 2008). We have to move outside this protein family, to another group of entirely different proteins, the so-called ClCA family, in order to find any evidence for other Cl− channels being regulated by Ins(3,4,5,6)P4 (Ismailov et al., 1996). In the latter study, recombinant bovine tracheal CLCA (bCLCA1) was expressed in Xenopus oocytes, and then membrane fragments were prepared and incorporated into lipid bilayers. However, it is rather puzzling that Ins(3,4,5,6)P4 inhibits bCLCA1 at low nanomolar concentrations (Ismailov et al., 1996), far below the micromolar levels observed even in resting cells (Section 4). It is difficult to avoid the conclusion that, if these observations are biologically relevant, bCLCA1 must be constitutively inhibited by the levels of Ins(3,4,5,6)P4 that prevail even in non-stimulated cells. There is no additional information to indicate that these observations might have any other regulatory context. Another confounding issue is that the CLCA family have been reported to exhibit pharmacological properties (inhibition by dithiothreitol (Fuller et al., 2005)) and electrophysiological parameters (13–30 pS unitary conductance (Fuller et al., 2005)) that do not match those of the native Ins(3,4,5,6)4-inhibited Cl− current (insensitivity to dithiothreitol and 1–2 pS unitary conductance (Ho et al., 2001)). Moreover, there are good reasons to believe that the CLCA proteins, some of which are clearly secreted from cells, do not form chloride channels per se, but instead have other functions (Jentsch and Günther, 1996; Mundhenk et al., 2006). Thus, it is difficult to understand what could be the biological relevance of the apparent interaction of Ins(3,4,5,6)P4 with bCLCA1.
The pathway of Ins(3,4,5,6)P4 synthesis
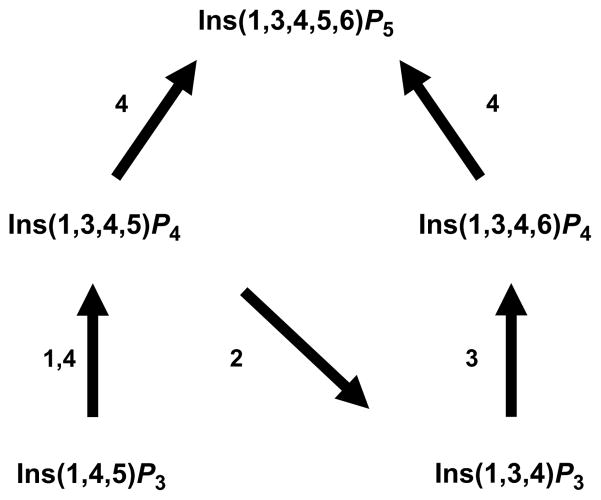
The idea that de novo synthesis of Ins(3,4,5,6)P4 occurs by dephosphorylation of Ins(1,3,4,5,6)P5 (Fig. 1) was first proposed by our laboratory in 1990 (Menniti et al., 1990), although more than a decade passed before we were able to experimentally validate this hypothesis (Chamberlain et al., 2007; Ho et al., 2002). As for Ins(1,3,4,5,6)P5 itself, there are two routes by which this molecule is synthesized from Ins(1,4,5)P3 (Fig. 2). The more protracted of these, Ins(1,4,5)P3 -> Ins(1,3,4,5)P4 -> Ins(1,3,4)P3 -> Ins(1,3,4,6)P4 -> Ins(1,3,4,5,6)P5, was the first to be discovered around 20 years ago (Hunyady et al., 1988; McConnell et al., 1991; Menniti et al., 1990; Shears, 1989; Stephens et al., 1988) and this involves ITPK1. Subsequently, a more direct route of Ins(1,3,4,5,6)P5 synthesis was discovered, following the identification and molecular cloning of an enzyme (IPMK/IPK2) that harbors both 3- and 6-kinase activities and, therefore, can single handedly phosphorylate Ins(1,4,5)P3 to Ins(1,3,4,5,6)P5 (Odom et al., 2000; Saiardi et al., 1999, 2000). The latter route to Ins(1,3,4,5,6)P5 is generally believed to be the more evolutionarily ancient of the two pathways; its origin is now thought to even predate the evolution of Ins(1,4,5)P3 as a Ca2+-mobilizing signal (Irvine and Schell, 2001; Schell et al., 1999; York, 2006). In fact, some organisms (yeasts and insects) do not possess an Itpk1 gene, yet they still synthesize adequate quantities of inositol phosphates. In higher animals, which possess both pathways of Ins(1,3,4,5,6)P5 (and hence also InsP6) synthesis, ITPK1 may have a multitasking capability, contributing both to the pathway of InsP6 synthesis (a metabolic function) and regulation of Ins(3,4,5,6)P4 levels (a signaling function). However, the relative contributions of ITPK1-dependent and -independent pathways for the synthesis of Ins(1,3,4,5,6)P5 (Fig. 2) and InsP6 are still being debated (Alcazar-Roman and Wente, 2008; Leyman et al., 2007; Verbsky et al., 2005). Perhaps this varies between cell types.
Fig. 2.
The metabolic link between Ins(1,4,5)P3 and Ins(1,3,4,5,6)P5 in animal cells. The figure shows the quantitatively most important reactions in animal cells that link Ins(1,4,5)P3 to Ins(1,3,4,5,6)P5. Numbers in the figure refer to various enzymes as follows: 1. Ins(1,4,5)P3 3-kinase (EC 2.7.1.127); 2. Ins(1,4,5)P3/Ins(1,3,4,5)P4 5-phosphatase (EC 3.1.3.56); 3. ITPK1 (EC 2.7.1.134); 4. Inositol phosphate multikinase (EC 2.7.1.151); note that kinetic data have led to it being questioned whether IPMK can contribute significantly to Ins(1,3,4,5)P4 synthesis de novo in animal cells (Chang et al., 2002).
Interestingly, plants contain multiple homologues of ITPK1 (Josefsen et al., 2007; Shi et al., 2003; Stiles et al., 2008; Sweetman et al., 2007; Wilson and Majerus, 1997). Rice has six such genes (Suzuki et al., 2007). The catalytic efficiencies of the different plant ITPK isoforms can differ by up to two orders of magnitude (Stiles et al., 2008). There is good evidence of an important and ubiquitous role for at least some of these ITPK isoforms in the synthesis of higher inositol phosphates. For example, a maize mutant has been identified which has a defective ITPK gene which results in decreased levels of InsP6 (Shi et al., 2003). Perhaps there are tissue specific differences in the expression of the various plant isoforms that might preferentially fulfill either metabolic functions (synthesis of InsP6) or signaling functions (synthesis of Ins(3,4,5,6)P4).
The contributions that ITPK makes to Ins(1,3,4,5,6)P5/InsP6 synthesis in plants involve metabolic pathways that are independent of both PLC and Ins(1,4,5)P3; these pathways do not exist in animal cells (Brearley and Hanke,1996; Stiles et al., 2008; Sweetman et al., 2007). For example, the phosphorylation of Ins(3,4,6)P3 to Ins(1,3,4,6)P4 is likely a plant-specific step in the Ins(1,3,4,5,6)P5/InsP6 synthetic pathway (Brearley and Hanke, 1996; Stiles et al., 2008).
Receptor-dependent regulation of Ins(3,4,5,6)P4 levels by ITPK1
The inhibition of CaMKII-activated Cl− conductance by Ins(3,4,5,6)P4 shows an IC50 value of approximately 5 μM (Mitchell et al., 2008; Xie et al., 1996). This is a significant observation because it demonstrates that the actions of Ins(3,4,5,6)P4 occur within a physiologically relevant concentration range: cellular levels of Ins(3,4,5,6)P4 are around 1 μM in resting cells, and they increase to the 5–10 μM range whenever PLC is activated (Ho and Shears, 2002). It is worth emphasizing this point: in all animal cells, irrespective of the mechanism by which PLC is activated, there is an accompanying elevation of Ins(3,4,5,6)P4 concentration (Barker et al., 1992; Li et al., 1992; Menniti et al., 1990; Wong et al., 1992). This obligatory connection between Ins(3,4,5,6)P4 levels and the PLC-dependent production of Ins(1,4,5)P3 is a vital component of this entire signaling system. Clearly, mass-action effects can explain why elevations in Ins(1,4,5)P3 levels inevitably lead to accompanying increases in some of the downstream metabolites (Fig. 2). In some cells, this metabolic domino effect may “knock-on” as far as Ins(3,4,5,6)P4. However, in most cases we do not believe that this is the major mechanism by which Ins(3,4,5,6)P4 levels are regulated. We initially came to this conclusion after it was demonstrated that Ins(3,4,5,6)P4 and Ins(1,3,4,5,6)P5 belong to a metabolic pool that is separate from that of Ins(1,4,5)P3 and its more closely related metabolites (Menniti et al., 1990; Wong et al., 1992). In other words, in the short-term, the metabolic pool of Ins(3,4,5,6)P4 is somewhat insulated from changes in Ins(1,4,5)P3 concentrations. This phenomenon is clearly seen during short-term radiolabelling of cells with [3H]inositol; the Ins(1,4,5)P3 pool becomes saturated with the radiolabel several days faster than does Ins(3,4,5,6)P4 (Menniti et al., 1990). Furthermore, it has been observed that a receptor-dependent increase in Ins(3,4,5,6)P4 levels can take place in parallel with a decrease in Ins(1,3,4,5,6)P5 levels (Menniti et al., 1990). The latter phenomenon clearly does not reflect a mass-action effect. Instead, there is receptor-dependent activation of Ins(1,3,4,5,6)P5 dephosphorylation to Ins(3,4,5,6)P4 by ITPK1.
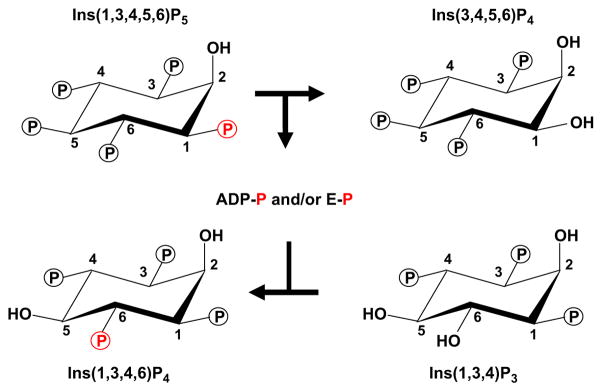
In 2000, we (Yang and Shears, 2000) determined that ITPK1 is also an active Ins(3,4,5,6)P4 1-kinase. Later, we discovered that ITPK1 also synthesizes Ins(3,4,5,6)P4 from Ins(1,3,4,5,6)P5 through a unique phosphotransferase activity (Chamberlain et al., 2007; Ho et al., 2002). It was this elucidation of the ADP-dependent phosphotransferase activity of mammalian ITPK1, an unprecedented phenomenon in the inositol phosphate field (Chamberlain et al., 2007; Ho et al., 2002), that uncovered the molecular mechanism by which Ins(3,4,5,6)P4 levels are coupled to receptor-regulated PLC activity. One important factor in this process (Fig. 3) is the tenacity with which ITPK1 binds adenine nucleotide; crystallographic data show that less than 10% of the nucleotide is solvent exposed (Chamberlain et al., 2007; Miller et al., 2005). In its ADP-bound form, ITPK1 dephosphorylates Ins(1,3,4,5,6)P5 to Ins(3,4,5,6)P4 (Fig. 3). The Ins(3,4,5,6)P4 is released to the bulk phase in exchange for Ins(1,3,4)P3, but the nucleotide – now ATP – remains bound. In other words, the inorganic phosphate that is removed from Ins(1,3,4,5,6)P5 is not released. Instead, it is fated to be passed on to the newly bound Ins(1,3,4)P3, thereby phosphorylating it to Ins(1,3,4,6)P4, which the active site then exchanges for a new molecule of Ins(1,3,4,5,6)P5 (Fig. 3), and the entire phosphotransferase cycle is repeated. Importantly, the rate at which Ins(1,3,4,5,6)P5 is dephosphorylated to Ins(3,4,5,6)P4 has been shown to be stimulated as the concentration of phosphate acceptor – Ins(1,3,4)P3 – is increased (Ho et al., 2002). In turn, the cellular levels of Ins(1,3,4)P3 – a metabolite of Ins(1,4,5)P3 – mirror both the intensity and the duration of receptor-activated PLC activity (Batty et al., 1998; Batty and Downes, 1994). In other words, the degree of PLC activity sets Ins(1,3,4)P3 levels, which controls Ins(3,4,5,6)P4 synthesis. This is the molecular basis for the integration of inositol phosphate signaling pathways via human ITPK1.
Fig. 3.
The phosphotransferase activity of ITPK1. The graphic illustrates the proposed enzymatic reactions by which the 1-phosphate on Ins(1,3,4,5,6)P5 (coloured red) is transferred to Ins(1,3,4)P3. The evidence for this reaction pathway came from HPLC analysis of the reaction products following the metabolism of [1-32P]-Ins(1,3,4,5,6)P5 by ITPK1 (Chamberlain et al., 2007). It has not yet been established whether or not a phosphoryl-enzyme (E–P) intermediate is involved, but this is a likely possibility.
The reaction mechanisms for these phosphotransferase reactions (Fig. 3) have not yet been established. Crystal structures have been obtained for human ITPK1 (Chamberlain et al., 2007) and an amoeboid homologue of ITPK1 (Miller et al., 2005), but unfortunately substrate-bound crystals have not yet been isolated, which has contributed to uncertainties concerning the reaction mechanisms. It has been proposed (Miller and Hurley, 2004) that the phosphorylation of Ins(1,3,4)P3 involves “in-line” transfer of the γ-phosphate from ATP directly to the inositol phosphate, i.e., without the participation of a phospho-enzyme intermediate. However, recently obtained preliminary data suggest that a phospho-histidine intermediate may accumulate when ITPK1 phosphorylates substrate (Majerus et al., 2008).
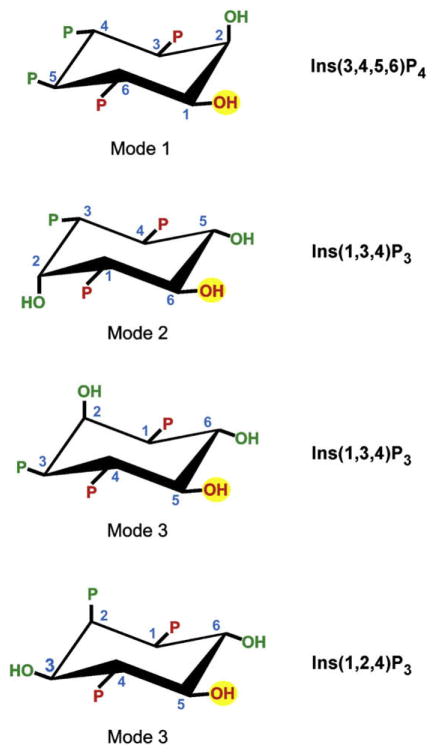
In the absence of structural information on enzyme–ligand interactions, we (Ho et al., 2002; Riley et al., 2006) have put forward a proposal which is based on the long-standing observation (Wilcox et al., 1994) that some inositol phosphates may interact with the binding sites of receptors and enzymes in more than one orientation (i.e. “mode”), enabling one inositol phosphate to mimic another by presenting to the docking site some key recognition features. Thus, we have proposed that ITPK1 uses three different binding modes (Ho et al., 2002) (Fig. 4): Mode 1 binding (permitting 1-kinase activity) was designated for Ins(3,4,5,6)P4. We further proposed that Ins(1,3,4)P3 could itself bind to the active site in two different orientations (Fig. 4), designated mode 2 (permitting 6-kinase activity) and mode 3 (in an effort to explain why Ins(1,3,4)P3 can also be phosphorylated at the 5-position (Abdullah et al., 1992; Shears, 1989)). Our three mode binding model has the advantage of accounting for the 5-hydroxyl phosphorylation of the non-physiological substrate, Ins(1,2,4)P3 (Adelt et al., 2001). Note that Miller et al. (2005) also advocate the same three different binding “modes” that we have put forward (Ho et al., 2002). However, the model of Miller et al. (2005) differs from ours in several key respects. First, they propose only a single binding mode for Ins(1,3,4)P3 (equivalent to our “mode 2”) with both the 5- and 6-hydroxyls being close enough to the γ-phosphate of ATP that either can be phosphorylated. A more provocative aspect of the model of Miller et al. (2005) is the proposal that, for the amoeboid ITPK1 at least, ligand specificity is not significantly affected by either the hydroxyl groups, or by their orientation (i.e. axial vs equatorial), or by the stereochemistry at any of the six stereogenic centers of the inositol ring. The amoeboid ITPK1 is certainly the most promiscuous of the enzymes that metabolize inositol phosphates (Field et al., 2000; Miller et al., 2005), but it would be a truly exceptional enzyme if it lacked all stereochemical specificity. In any case, we have shown that this is absolutely not the case for human ITPK1 (Chamberlain et al., 2007; Riley et al., 2006). We have clearly shown that the determinants of ligand binding include the two-dimensional arrangement of phosphates and hydroxyls around the inositol ring, and also the three-dimensional stereochemistry at each position of the ring. This stereochemically based model is, perhaps, most clearly vindicated by the empirical demonstration that Ins(1,4,5,6)P4 is not a substrate for ITPK1 (Riley et al., 2006), in contrast to the prediction that arose out of the model of Miller et al. (2005).
Fig. 4.
A model for the structural determinants of ligand specificity for mamalian ITPK1. The figure depicts our proposal (Ho et al., 2002; Riley et al., 2006) that there are three modes of binding of inositol phosphates to mamalian ITPK1. It can be illuminating to consider these different binding modes (i.e., “1”, “2” and “3”) as permitting 1-kinase, 6-kinase and 5-kinase activities, respectively. These phosphorylation sites are marked with a yellow circle. Three groups in Ins(3,4,5,6)P4, Ins(1,3,4)P3 and Ins(1,2,4)P3 (coloured red) are conserved in all three of these proposed binding modes. We have previously noted that these groups by themselves are insufficient to designate substrate specificity, so we have proposed a combinatorial recognition model in which some of the additional groups (coloured green) contribute to ligand recognition, but in a mode-specific manner.
In our model, three groups around the inositol ring (groups coloured red in Fig. 4), are common to each binding mode, but by themselves, these are insufficient to fully define ligand specificity, since all three groups are also present on Ins(1,4)P2, which is not a substrate (Ho et al., 2002; Ongusaha et al., 1998). We therefore proposed that ligand recognition was combinatorial in nature, with some groups on the inositol ring only contributing to specificity in just one or two of the three proposed substrate-binding modes (Ho et al., 2002). A possible molecular basis for this model for ligand binding is that there are some rigid regions of the active site, which could then be used in multiple binding modes, whereas others might be more flexible (for mode-specific binding).
The extent to which receptor-activation of PLC leads to an elevation in cellular Ins(3,4,5,6)P4 levels can be influenced by alterations in the degree of ITPK1 expression. This has been shown using T84 cells in which ITPK1 was over-expressed by about 2-fold, leading to an approximate doubling of the increase in Ins(3,4,5,6)P4 levels that were observed during PLC activation (Ho et al., 2002). A similar conclusion arose from a comparison of Ins(3,4,5,6)P4 levels in immortalized murine tracheal cells derived from wild-type and cftr (−/−) mice (Yang et al., 2006). In the latter study, the cells from the cftr (−/−) mice expressed less Itpk1, and so less Ins(3,4,5,6)P4 was formed in these cells (Yang et al., 2006). Such observations may be of clinical interest in the treatment of the cystic fibrosis (CF) condition. For example, cell-permeant antagonists of Ins(3,4,5,6)P4 are being developed for the purpose of enhancing the activity of Ca2+-activated Cl− channels and thereby improving salt and fluid secretion from CF individuals (Rudolf et al., 2003). We (Yang et al., 2006) have previously proposed that the degree of improvement in Cl− secretion that can be elicited by Ins(3,4,5,6)P4 antagonists would be expected to be influenced by the extent to which endogenous Ins(3,4,5,6)P4 inhibits the Cl− channels, which in turn depends upon the Ins(3,4,5,6)P4 concentration (Ho et al., 2001). Therefore, those CF individuals with the higher levels of ITPK1 expression (and hence higher levels of Ins(3,4,5,6)P4) potentially stand to benefit the most from a therapy based on Ins(3,4,5,6)P4 antagonists, should an appropriate drug eventually become available. Conversely, those CF individuals with the lower levels of ITPK1 in airway cells would synthesize less Ins(3,4,5,6)P4 following activation of PLC. This is a significant point because another potential therapy for CF is the inhalation of purinergic agonists in order to activate airway PLC, mobilize Ca2+, and activate Ca2+-activated Cl− channels (Ho and Shears, 2002). Unfortunately, PLC activation also elevates Ins(3,4,5,6)P4 levels, thereby blocking the very channels the therapy is designed to activate. However, patients with less ITPK1, and therefore less Ins(3,4,5,6)P4, might be expected to receive greater benefit from the inhalation of purinergic agonists. We (Yang et al., 2006) have proposed that clinical trials of these candidate pharmacological approaches to CF should be correlated with ITPK1 expression profiling in order to detect responsive patient subgroups.
ITPK1 is not a protein kinase
A pair of publications from Majerus’ laboratory concluded that human ITPK1 is not just an inositol phosphate kinase, but additionally acts as a protein kinase (Sun et al., 2002; Wilson et al., 2001). Several apparent substrates were identified, including the following transcription factors: IκBα, c-jun and ATF-2. This was an unexpected observation since ITPK1 has no recognizable protein kinase domains (Wilson et al., 2001). In one of their studies, Majerus and colleagues (Sun et al., 2002) also heterologously expressed recombinant ITPK1 in sf21 cells and then incubated the enzyme with [32P]-ATP; ITPK1 became phosphorylated on serine and tyrosine residues (Sun et al., 2002). This phenomenon was interpreted as evidence of autophosphorylation (Sun et al., 2002). Others (Qin et al., 2005) have suggested this itself to be direct evidence that ITPK1 is a protein kinase. However, this is not a strong argument; many ATP-binding proteins that are not protein kinases still undergo autophosphorylation (Hunter, 1995). “Autophosphorylation” of ITPK1 can also be the result of contaminating protein kinase activity. Indeed, we (Qian et al., 2005) subsequently demonstrated that the protein kinase activity that is associated with recombinant ITPK1 produced in insect cells is, in fact, a persistent contaminant; once free of this impurity, ITPK1 showed no protein kinase activity, even though its inositol phosphate kinase activity was near identical to preparations produced by the Majerus laboratory. Furthermore, recombinant ITPK1 that is expressed in Escherichia coli does not “autophosphorylate” on serine or tyrosine (Majerus et al., 2008). This particular difference in behaviour in the enzyme, which depends upon which expression system is used, can be explained by phosphorylation (rather than “autophosphorylation”) by a protein kinase that contaminates preparations of ITPK1 that are expressed in the insect cells but not in the bacteria. Finally, when Majerus and colleagues (Miller et al., 2005) described the crystal structure of an amoeboid homologue of ITPK1, they were unable “to model binding of a peptide substrate” into the constricted, “small-molecule modifying” active site. There was no alternative means by which a protein could get close enough to the tightly embedded ATP. A similar conclusion that ITPK1 cannot be a protein kinase can be drawn from crystal structure of the human enzyme (Chamberlain et al., 2007).
Summary
The synthesis and the metabolism of inositol 3,4,5,6-tetrakisphosphate (Ins(3,4,5,6)P4) are the responsibility of a single multifunctional kinase/phosphotransferase, ITPK1. This enzyme dynamically couples the cellular levels of Ins(3,4,5,6)P4 to the receptor-dependent hydrolysis of inositol lipids by phospholipase C. This is a biologically significant event because Ins(3,4,5,6)P4 regulates the conductance of a specialized class of chloride ion channels, which regulate many cellular functions including epithelial salt and fluid secretion, synaptic efficacy, bone remodelling, tumor cell migration, insulin release from pancreatic β-cells, and inflammatory responses. This review assesses the current state of our knowledge of this versatile and ubiquitous signalling cascade.
References
- Abdullah M, Hughes PJ, Craxton A, Gigg R, Desai T, Marecek JF, et al. Purification and characterization of inositol 1,3,4-trisphosphate 5/6-kinase from rat liver using an inositol hexakisphosphate affinity column. J Biol Chem. 1992;267:22340–5. [PubMed] [Google Scholar]
- Adelt S, Plettenburg O, Dallmann G, Ritter FP, Shears SB, Altenbach H-J, et al. Regiospecific phosphohydrolases from dictyostelium as tools for the chemoenzymatic synthesis of the enantiomers D-myo-inositol 1,2,4-trisphosphate and D-myo-inositol 2,3,6-trisphosphate: non-physiological, potential analogues of biologically active D-myo-inositol 1,3,4-trisphosphate. Bioorg Med Chem Lett. 2001;11:2705–8. doi: 10.1016/s0960-894x(01)00536-4. [DOI] [PubMed] [Google Scholar]
- Alcazar-Roman AR, Wente SR. Inositol polyphosphates: a new frontier for regulating gene expression. Chromosoma. 2008;117:1–13. doi: 10.1007/s00412-007-0126-4. [DOI] [PubMed] [Google Scholar]
- Barg S, Huang P, Eliasson L, Nelson DJ, Obermüller S, Rorsman P, et al. Priming of insulin granules for exocytosis by granular chloride uptake and acidification. J Cell Sci. 2001;114:2145–54. doi: 10.1242/jcs.114.11.2145. [DOI] [PubMed] [Google Scholar]
- Barker CJ, Wong NS, Maccallum SM, Hunt PA, Michell RH, Kirk CJ. The interrelationships of the inositol phosphates formed in WRK-1 stimulated rat mammary tumour cells. Biochem J. 1992;286:469–74. doi: 10.1042/bj2860469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty IH, Currie RA, Downes CP. Evidence for a model of integrated inositol phospholipid pools implies an essential role for lipid transport in the maintenance of receptor-mediated phospholipase C activity in 1321N1 cells. Biochem J. 1998;330:1069–77. doi: 10.1042/bj3301069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty IH, Downes CP. The inhibition of phosphoinositide synthesis and muscarinic-receptor-mediated phospholipase C activity by Li+ as secondary selective consequences of inositol depletion in 1321N1 cells. Biochem J. 1994;297:529–37. doi: 10.1042/bj2970529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–9. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Brearley CA, Hanke DE. Metabolic evidence for the order of addition of individual phosphate esters to the myo-inositol moiety of inositol hexakisphosphate in the duckweed Spirodela polrhiza L. Biochem J. 1996;314:227–33. doi: 10.1042/bj3140227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain PP, Qian X, Stiles AR, Cho J, Jones DH, Lesley SA, et al. Integration of inositol phosphate signaling pathways via human ITPK1. J Biol Chem. 2007;282:28117–25. doi: 10.1074/jbc.M703121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S-C, Miller AL, Feng Y, Wente SR, Majerus PW. The human homologue of the rat inositol phosphate multikinase is an inositol 1,3,4,6-tetrakisphosphate 5-kinase. J Biol Chem. 2002;277:43836–43. doi: 10.1074/jbc.M206134200. [DOI] [PubMed] [Google Scholar]
- Clapham D. How to lose your hippocampus by working on chloride channels. Neuron. 2001;29:1–6. doi: 10.1016/s0896-6273(01)00172-6. [DOI] [PubMed] [Google Scholar]
- Claud EC, Lu J, Wang XQ, Abe M, Petrof EO, Sun J, et al. Platelet-activating factor induced chloride channel activation is associated with intracellular acidosis and apoptosis of intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1191–200. doi: 10.1152/ajpgi.00318.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J, Wilson MP, Mai Z, Majerus PW, Samuelson J. An Entamoeba histolytica inositol 1,3,4-trisphosphate 5/6-kinase has a novel 3-kinase activity. Mol Biochem Parasitol. 2000;108:119–23. doi: 10.1016/s0166-6851(00)00197-3. [DOI] [PubMed] [Google Scholar]
- Fuller CM, Kovacs G, Anderson SJ, Benos DJ. The CLCAs: proteins with ionchannel, cell adhesion and tumor suppressor functions. Adv Exp Med Biol. 2005;558:83–102. [Google Scholar]
- Gentzsch M, Cui L, Mengos A, Chang XB, Chen JH, Riordan JR. The PDZ-binding chloride channel ClC-3B localizes to the Golgi and associates with cystic fibrosis transmembrane conductance regulator-interacting PDZ proteins. J Biol Chem. 2003;278:6440–9. doi: 10.1074/jbc.M211050200. [DOI] [PubMed] [Google Scholar]
- Hara-Chikuma M, Yang B, Sonawane ND, Sasaki S, Uchida S, Verkman AS. ClC-3 chloride channels facilitate endosomal acidification and chloride accumulation. J Biol Chem. 2005;280:1241–7. doi: 10.1074/jbc.M407030200. [DOI] [PubMed] [Google Scholar]
- Ho MWY, Yang X, Carew MA, Zhang T, Hua L, Kwon Y-U, et al. Regulation of Ins(3456)P4 signaling by a reversible kinase/phosphatase. Curr Biol. 2002;12:477–82. doi: 10.1016/s0960-9822(02)00713-3. [DOI] [PubMed] [Google Scholar]
- Ho MWY, Kaetzel MA, Armstrong DL, Shears SB. Regulation of a human chloride channel: a paradigm for integrating input from calcium, CaMKII and Ins(3,4,5,6)P4. J Biol Chem. 2001;276:18673–80. doi: 10.1074/jbc.M101128200. [DOI] [PubMed] [Google Scholar]
- Ho MWY, Shears SB. Regulation of calcium-activated chloride channels by inositol 3,4,5,6-tetrakisphosphate. In: Fuller CM, editor. Current Topics in Membranes. London: Academic Press; 2002. pp. 345–63. [Google Scholar]
- Hunter T. When is a lipid kinase not a lipid kinase? When it is a protein kinase Cell. 1995;83:1–4. doi: 10.1016/0092-8674(95)90225-2. [DOI] [PubMed] [Google Scholar]
- Hunyady L, Baukal AJ, Guillemette G, Balla T, Catt KJ. Metabolism of inositol-1,3,4,6-tetrakisphosphate to inositol pentakisphosphate in adrenal glomerulosa cells. Biochem Biophys Res Commun. 1988;157:1247–52. doi: 10.1016/s0006-291x(88)81008-8. [DOI] [PubMed] [Google Scholar]
- Irvine RF, Schell M. Back in the water: the return of the inositol phosphates. Nat Rev Mol Cell Biol. 2001;2:327–38. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Ismailov Fuller CM, II, Berdiev BK, Shlyonsky VG, Benos DJ, Barrett KE. A biologic function for an “orphan” messenger: D-myo-inositol 3,4,5,6-tetrakisphosphate selectively blocks epithelial calcium-activated chloride current. Proc Nat Acad Sci USA. 1996;93:10505–9. doi: 10.1073/pnas.93.19.10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch TJ. CLC chloride channels and transporters: from genes to protein structure, pathology and physiology. Crit Rev Biochem Mol Biol. 2008;43:3–36. doi: 10.1080/10409230701829110. [DOI] [PubMed] [Google Scholar]
- Jentsch TJ, Günther W. Chloride channels: an emerging molecular picture. Bioessays. 1996;19:117–126. doi: 10.1002/bies.950190206. [DOI] [PubMed] [Google Scholar]
- Josefsen L, Bohn L, Sorensen MB, Rasmussen SK. Characterization of a multifunctional inositol phosphate kinase from rice and barley belonging to the ATP-grasp superfamily. Gene. 2007;397:114–25. doi: 10.1016/j.gene.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Kilpatrick ED, Robertson RP. Differentiation between glucose-induced desensitization of insulin secretion and b-cell exhaustion in the HIT-T15 cell line. Diabetes. 1998;47:606–11. doi: 10.2337/diabetes.47.4.606. [DOI] [PubMed] [Google Scholar]
- Leyman A, Pouillon V, Bostan A, Schurmans S, Erneux C, Pesesse X. The absence of expression of the three isoenzymes of the inositol 1,4,5-trisphosphate 3-kinase does not prevent the formation of inositol pentakisphosphate and hexakisphosphate in mouse embryonic fibroblasts. Cell Signalling. 2007;19:1497–504. doi: 10.1016/j.cellsig.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Li G, Pralong W-F, Pittet D, Mayr GW, Schlegel W, Woolheim CB. Inositol tetrakisphosphate isomers and elevation of cytosolic calcium in vasopressin-stimulated insulin-secreting RINm5F cells. J Biol Chem. 1992;267:4349–56. [PubMed] [Google Scholar]
- Lisal J, Maduke M. The ClC-0 chloride channel is a ‘broken’ Cl(−)/H(+) antiporter. Nat Struct Mol Biol. 2008;15:805–10. doi: 10.1038/nsmb.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerus PW, Zou J, Marjanovic J, Kisseleva MV, Wilson MP. The role of inositol signaling in the control of apoptosis. Adv Enzym Regul. 2008;48:10–7. doi: 10.1016/j.advenzreg.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Chen L, Xu B, Wang L, Li H, Guo J, et al. Suppression of ClC-3 channel expression reduces migration of nasopharyngeal carcinoma cells. Biochem Pharmacol. 2008;75:1706–16. doi: 10.1016/j.bcp.2008.01.008. [DOI] [PubMed] [Google Scholar]
- McConnell FM, Stephens LR, Shears SB. Multiple isomers of inositol pentakisphosphate in Epstein–Barr virus transformed T5-1 lymphocytes. Identification of inositol 1,3,4,5,6-pentakisphosphate, D-inositol 1,2,4,5,6-pentakisphosphate and L-inositol 1,2,4,5,6-pentakisphosphate. Biochem J. 1991;280:323–9. doi: 10.1042/bj2800323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menniti FS, Oliver KG, Nogimori K, Obie JF, Shears SB, Putney JW., Jr Origins of myo-inositol tetrakisphosphates in agonist-stimulated rat pancreatoma cells. Stimulation by bombesin of myo-inositol 1,3,4,5,6-pentakisphosphate breakdown to myo-inositol 3,4,5,6-tetrakisphosphate. J Biol Chem. 1990;265:11167–76. [PubMed] [Google Scholar]
- Meyer J, Sturis J, Katschinski M, Arnold R, Göke B, Byrne MM. Acute hyperglycemia alters the ability of the normal b-cell to sense and respond to glucose. Am J Physiol Endocrinol Metab. 2002;282:E917–22. doi: 10.1152/ajpendo.00427.2001. [DOI] [PubMed] [Google Scholar]
- Miller GJ, Hurley JH. Crystal structure of the catalytic core of inositol 1,4,5-trisphosphate 3-kinase. Mol Cell. 2004;15:703–11. doi: 10.1016/j.molcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Miller GJ, Wilson MP, Majerus PW, Hurley JH. Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase. Mol Cell. 2005;18:201–12. doi: 10.1016/j.molcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Mitchell J, Wang X, Zhang G, Gentzsch M, Nelson DJ, Shears SB. An expanded biological repertoire for Ins(3, 4,56)P4 through its modulation of ClC-3 function. Curr Biol. 2008;18:1600–5. doi: 10.1016/j.cub.2008.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland JG, Davis AP, Bailey G, Nauseef WM, Lamb FS. Anion channels, including ClC-3, are required for normal neutrophil oxidative function, phagocytosis, and transendothelial migration. J Biol Chem. 2006;281:12277–88. doi: 10.1074/jbc.M511030200. [DOI] [PubMed] [Google Scholar]
- Mundhenk L, Alfalah M, Elble RC, Pauli BU, Naim HY, Gruber AD. Both cleavage products of the mCLCA3 protein are secreted soluble proteins. J Biol Chem. 2006;281:30072–80. doi: 10.1074/jbc.M606489200. [DOI] [PubMed] [Google Scholar]
- Nishi T, Forgac M. The vacuolar (H+)-ATPases – nature’s most versatile proton pumps. Nature Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–9. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Okamoto F, Kajiya H, Toh K, Uchida S, Yoshikawa M, Sasaki S, et al. Intracellular ClC-3 chloride channels promote bone resorption in vitro through organelle acidification in mouse osteoclasts. Am J Physiol Cell Physiol. 2008;294:C693–701. doi: 10.1152/ajpcell.00251.2007. [DOI] [PubMed] [Google Scholar]
- Ongusaha PP, Hughes PJ, Davey J, Michell RH. Inositol hexakisphosphate in Schizosaccharomyces pombe: synthesis from Ins(1,4,5)P3 and osmotic regulation. Biochem J. 1998;335:671–9. doi: 10.1042/bj3350671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen OH. Stimulus-secretion coupling: cytoplasmic calcium signals and the control of ion channels in exocrine acinar cells. J Physiol (London) 1992;448:1–51. doi: 10.1113/jphysiol.1992.sp019028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picollo A, Pusch M. Chloride/proton antiporter activity of mammalian CLC proteins ClC-4 and ClC-5. Nature. 2005;436:420–3. doi: 10.1038/nature03720. [DOI] [PubMed] [Google Scholar]
- Qian X, Mitchell J, Wei SJ, Williams J, Petrovich RM, Shears SB. The Ins(1,3,4)P3 5/6-kinase/Ins(3,4,5,6)P4 1-kinase is not a protein kinase. Biochem J. 2005;389:389–95. doi: 10.1042/BJ20050297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin ZX, Chen QJ, Tong Z, Wang XC. The Arabidopsis inositol 1,3,4-trisphosphate 5/6 kinase, AtItpk-1, is involved in plant photomorphogenesis under red light conditions, possibly via interaction with COP9 signalosome. Plant Physiol Biochem. 2005;43:947–54. doi: 10.1016/j.plaphy.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Renström E, Ivarsson R, Shears SB. Ins(3,4,5,6)P4 inhibits insulin granule acidification and fusogenic potential. J Biol Chem. 2002;277:26717–20. doi: 10.1074/jbc.C200314200. [DOI] [PubMed] [Google Scholar]
- Riley AM, Deleu S, Qian X, Mitchell J, Chung SK, Adelt S, et al. On the contribution of stereochemistry to human ITPK1 specificity: Ins(1,4,5,6)P4 is not a physiologic substrate. FEBS Lett. 2006;580:324–30. doi: 10.1016/j.febslet.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Robinson NC, Huang P, Kaetzel MA, Lamb FS, Nelson DJ. Identification of an N-terminal amino acid of the CLC-3 chloride channel critical in phosphorylation-dependent activation of a CaMKII-activated chloride current. J Physiol. 2004;556:353–68. doi: 10.1113/jphysiol.2003.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf MT, Dinkel C, Traynor-Kaplan AE, Schultz C. Antagonists of myo-inositol 3,4,5,6-tetrakisphosphate allow repeated epithelial chloride secretion. Bioorgan Med Chem. 2003;11:3315–29. doi: 10.1016/s0968-0896(03)00188-3. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett. 2000;468:28–32. doi: 10.1016/s0014-5793(00)01194-7. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Erdjument-Bromage H, Snowman A, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–6. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- Scheel O, Zdebik AA, Lourdel S, Jentsch TJ. Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature. 2005;436:424–7. doi: 10.1038/nature03860. [DOI] [PubMed] [Google Scholar]
- Schell MJ, Letcher AJ, Brearley CA, Biber J, Murer H, Irvine RF. PiUS (Pi uptake stimulator) is an inositol hexakisphosphate kinase. FEBS Lett. 1999;461:169–72. doi: 10.1016/s0014-5793(99)01462-3. [DOI] [PubMed] [Google Scholar]
- Shears SB. The pathway of myo-inositol 1,3,4-trisphosphate phosphorylation in liver. Identification of myo-inositol 1,3,4-trisphosphate 6-kinase, myo-inositol 1,3,4-trisphosphate 5-kinase, and myo-inositol 1,3,4,6-tetrakisphosphate 5-kinase. J Biol Chem. 1989;264:19879–86. [PubMed] [Google Scholar]
- Shi J, Wang H, Wu Y, Hazebroek J, Meeley RB, Ertl DS. The maize low-phytic acid mutant Ipa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 2003;131:507–15. doi: 10.1104/pp.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens LR, Hawkins PT, Barker CJ, Downes CP. Synthesis of myo-inositol 1,3,4,5,6-pentakisphosphate from inositol phosphates generated by receptor activation. Biochem J. 1988;253:721–33. doi: 10.1042/bj2530721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles AR, Qian X, Shears SB, Grabau EA. Metabolic and signaling properties of an Itpk gene family in Glycine max. FEBS Lett. 2008;582:1853–8. doi: 10.1016/j.febslet.2008.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobrawa SM, Breiderhoff T, Takamori S, Engel D, Schweizer M, Zdebik AA, et al. Disruption of ClC-3, a chloride channel expressed on synaptic vesicles, leads to a loss of hippocampus. Neuron. 2001;29:185–96. doi: 10.1016/s0896-6273(01)00189-1. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wilson MP, Majerus PW. Inositol 1,3,4-trisphosphate 5/6-kinase associates with the COP9 signalosome by binding to CSN1. J Biol Chem. 2002;277:45759–64. doi: 10.1074/jbc.M208709200. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Tanaka K, Kuwano M, Yoshida KT. Expression pattern of inositol phosphate-related enzymes in rice (Oryza sativa L): implications for the phytic acid biosynthetic pathway. Gene. 2007;405:55–64. doi: 10.1016/j.gene.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Sweetman D, Stavridou I, Johnson S, Green P, Caddick SE, Brearley CA. Arabidopsis thaliana inositol 1,3,4-trisphosphate 5/6-kinase 4 (AtITPK4) is an outlier to a family of ATP-grasp fold proteins from Arabidopsis. FEBS Lett. 2007;581:4165–71. doi: 10.1016/j.febslet.2007.07.046. [DOI] [PubMed] [Google Scholar]
- Trimble ER, Bruzzone R, Meehan CJ, Biden TJ. Rapid increases in inositol 1,4,5-trisphosphate, inositol 1,3,4,5-tetrakisphosphate and cytosolic free Ca2+ in agonist-stimulated pancreatic acini of the rat. Biochem J. 1987;242:289–92. doi: 10.1042/bj2420289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky JW, Chang SC, Wilson MP, Mochizuki Y, Majerus PW. The pathway for the production of inositol hexakisphosphate in human cells. J Biol Chem. 2005;280:1911–20. doi: 10.1074/jbc.M411528200. [DOI] [PubMed] [Google Scholar]
- Wang XQ, Deriy LV, Foss S, Huang P, Lamb FS, Kaetzel MA, et al. CLC-3 channels modulate excitatory synaptic transmission in hippocampal neurons. Neuron. 2006;52:321–33. doi: 10.1016/j.neuron.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Weylandt KH, Nebrig M, Jansen-Rosseck N, Amey JS, Carmena D, Wiedenmann B, et al. ClC-3 expression enhances etoposide resistance by increasing acidification of the late endocytic compartment. Mol Cancer Ther. 2007;6:979–86. doi: 10.1158/1535-7163.MCT-06-0475. [DOI] [PubMed] [Google Scholar]
- Wilcox RA, Safrany ST, Lampe D, Mills SJ, Nahorski SR, Potter BVL. Modification at C2 of myo-inositol 1,4,5-trisphosphate produces inositol trisphosphates and tetrakisphosphates with potent biological activities. Eur J Biochem. 1994;223:115–24. doi: 10.1111/j.1432-1033.1994.tb18972.x. [DOI] [PubMed] [Google Scholar]
- Wilson MP, Majerus PW. Characterization of a cDNA encoding Arabidopsis thaliana inositol 1,3,4-trisphosphate 5/6-kinase. Biochem Biophys Res Commun. 1997;232:678–81. doi: 10.1006/bbrc.1997.6355. [DOI] [PubMed] [Google Scholar]
- Wilson MP, Sun Y, Cao L, Majerus PW. Inositol 1,3,4-trisphosphate 5/6-kinase is a protein kinase that phosphorylates the transcription factors c-jun and ATF-2. J Biol Chem. 2001;276:40998–1004. doi: 10.1074/jbc.M106605200. [DOI] [PubMed] [Google Scholar]
- Wong NS, Barker CJ, Morris AJ, Craxton A, Kirk CJ, Michell RH. The inositol phosphates of WRK1 rat mammary tumour cells. Biochem J. 1992;286:459–68. doi: 10.1042/bj2860459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Kaetzel MA, Bruzik KS, Dedman JR, Shears SB, Nelson DJ. Inositol 3,4,5,6-tetrakisphosphate inhibits the calmodulin-dependent protein kinase II-activated chloride conductance inT84 colonic epithelial cells. J Biol Chem. 1996;271:14092–7. doi: 10.1074/jbc.271.24.14092. [DOI] [PubMed] [Google Scholar]
- Xie W, Solomons KRH, Freeman S, Kaetzel MA, Bruzik KS, Nelson DJ, et al. Regulation of Ca2+-dependent Cl− conductance in T84 cells: cross-talk between Ins(3,4,5,6)P4 and protein phosphatases. J Physiol (London) 1998;510:661–73. doi: 10.1111/j.1469-7793.1998.661bj.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Reece J, Gabriel SE, Shears SB. Apical localization of ITPK1 enhances its ability to be a modifier gene product in a murine tracheal cell model of cystic fibrosis. J Cell Sci. 2006;119:1320–8. doi: 10.1242/jcs.02836. [DOI] [PubMed] [Google Scholar]
- Yang X, Shears SB. Multitasking in signal transduction by a promiscuous human Ins(3,4,5,6)P4 1-Kinase/Ins(1,3,4)P3 5/6-kinase. Biochem J. 2000;351:551–5. [PMC free article] [PubMed] [Google Scholar]
- York JD. Regulation of nuclear processes by inositol polyphosphates. Biochim Biophys Acta. 2006;1761:552–9. doi: 10.1016/j.bbalip.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Li X, Hao J, Winston JH, Weinman SA. The ClC-3 chloride transport protein traffics through the plasma membrane via interaction of an N-terminal dileucine cluster with clathrin. J Biol Chem. 2007;282:29022–31. doi: 10.1074/jbc.M703506200. [DOI] [PubMed] [Google Scholar]
- Zonia L, Cordeiro S, Tupý J, Feijó JA. Oscillatory chloride efflux at the pollen tube apex has a role in growth and cell volume regulation and is targeted by inositol 3,4,5,6-tetrakisphosphate. Plant Cell. 2002;14:2233–49. [PubMed] [Google Scholar]