Abstract
We report scM&T-seq, a method for parallel single-cell genome-wide methylome and transcriptome sequencing, allowing discovery of associations between transcriptional and epigenetic variation. Profiling of 61 mouse embryonic stem cells confirmed known links between DNA methylation and transcription. Notably, the method reveals novel associations between heterogeneously methylated distal regulatory elements and transcription of key pluripotency genes.
Multi-parameter sequencing-based analysis of single cells offers a powerful tool to dissect relationships between epigenetic, genomic and transcriptional heterogeneity1. Recent advances have enabled single-cell genome-wide or reduced-representation bisulfite sequencing (scBS-seq, scRRBS2-4), allowing exploration of intercellular heterogeneity of DNA methylation5, 6. We and others have recently described methods for parallel genome and transcriptome sequencing within single cells7, 8. Importantly our method, G&T-seq, utilizes physical separation of RNA and DNA allowing bisulfite conversion of DNA without affecting the transcriptome. We now apply scBS-seq to genomic DNA purified according to the G&T-seq protocol to generate methylomes and transcriptomes from the same single cells (Fig. 1a, Supplementary Fig. 1). Parallel-profiling using scM&T-seq will enable detailed study of the complex relationship between DNA methylation and transcription in heterogeneous cell populations9, 10 and may be used to provide multi-dimensional information in clinical contexts where material is severely limited (e.g. in vitro fertilisation).
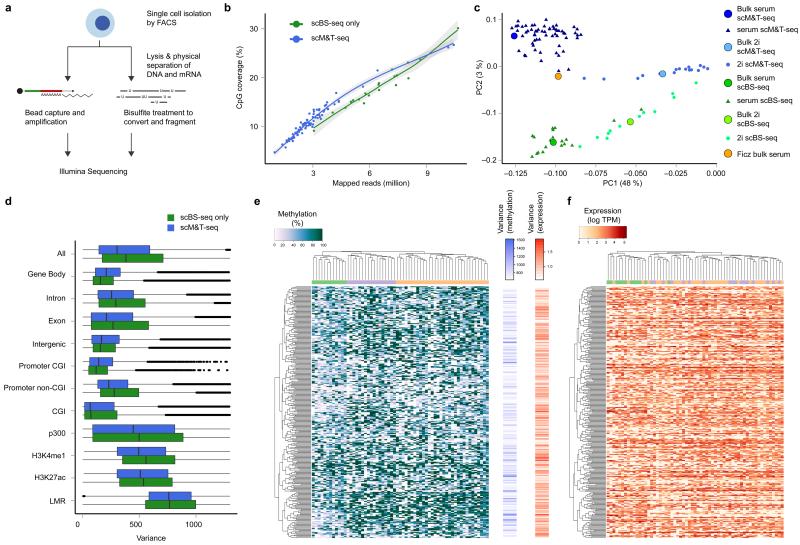
Figure 1. Quality control and global methylation and transcriptome patterns identified in serum ESCs profiled using scM&T-seq.
a) Schematic overview of the scM&T-seq protocol. b) CpG coverage of single cells as a function of the number of mapped sequencing reads. Green: stand-alone scBS-seq3, Blue: scM&T-seq. c) Joint principal component analysis of the methylomes (gene body methylation) of 61 serum ESCs (dark blue) and 16 2i ESCs (light blue) obtained using scM&T-seq, as well as 20 serum ESCs (green) and 12 2i ESCs (yellow) sequenced using stand-alone scBS-seq3. The solid circles correspond to synthetic bulk datasets form the same cells. For comparison, we also included a bulk serum ESC DNA methylation dataset16 (orange). Cell type explained a substantially larger proportion of variance (PC1, 48%) than protocol (PC2, 3%). d) Comparison of epigenetic heterogeneity in different genomic context, either considering 61 serum ESCs obtained using scM&T-seq (blue), or 20 serum ESCs sequenced using stand-alone scBS-seq3 (green). e, f) Clustering analysis of transcriptome and methylation data from 61 serum ESCs, considering gene body methylation (e) and gene expression (f) for the 300 most heterogeneous genes (based on gene body methylation). The order of genes was taken from an individual clustering analysis based on gene body methylation whereas cells were clustered separately either using DNA methylation or expression data, and coloured by methylation cluster. The bar plots in the center show the heterogeneity in DNA methylation (left) and gene expression (right).
To demonstrate the potential of the method, we applied scM&T-seq to mouse embryonic stem cells (ESCs). In the presence of serum, these cells are a metastable population with stochastic switching between transcriptional states11-12. This transcriptional heterogeneity has been linked to the differentiation potential of ESCs, with NANOG-low cells having an increased propensity to differentiate13 and elevated expression of differentiation markers12, 14, 15. Experiments in sorted populations of cells have also linked transcriptional and epigenetic heterogeneity by demonstrating differences in DNA methylation between transcriptional states, such as gains in DNA methylation in NANOG-low and REX1-low (REX1 also known as ZFP42) cells11, 16. The development of single-cell techniques has allowed the transcriptional heterogeneity of ESCs to be studied at unprecedented detail, revealing a complex population structure and multiple sources of variation17, 18. Using scBS-seq, we have also demonstrated DNA methylation heterogeneity in ESCs at the single-cell level3. To further investigate the link between epigenetic and transcriptional heterogeneity in ESCs, we performed scM&T-seq on 76 individual serum ESCs and 16 ESCs grown in “2i” media, which induces genome-wide DNA hypomethylation16.
We obtained an average of 2.7 million (M) scRNA-seq reads per cell, and excluded cells with fewer than 2 M mapped reads (Supplementary Table 1). We have previously shown that the scRNA-seq data generated by the G&T-seq method is of comparable quality to that generated using stand-alone Smart-seq27. In ESCs passing scRNA-seq QC, we detected transcripts from between 4,000 and 8,000 genes exceeding 1 transcript per million (TPM), consistent with previous measurements made using the method (see Supplementary Fig. 2 for additional scRNA-seq quality metrics).
To assess the quality of the scBS-seq data, we compared the resulting single-cell methylomes with published data from 20 serum and 12 2i ESCs for which stand-alone scBS-seq was performed3. Sequencing of the scBS-seq libraries was performed at relatively low depth (an average of 11.1 M reads), with an average of 3.15 M genomic reads mapped per cell (Supplementary Table 1). We excluded cells with a mapping efficiency of less than 7%, or a bisulfite conversion efficiency less than 95% (as estimated by non-CpG methylation). Cells passing these QC steps had a mean mapping efficiency of 15.6% (compared to a mean of 17.2% for single ESCs by stand-alone scBS-seq3, Supplementary Table 1, Supplementary Fig. 3). The low mappibility is not due to foreign DNA, as negative controls aligned less than 2% but can be explained by high primer contamination (Supplementary Fig. 3). Due to reduced sequencing depth, methylome coverage in scM&T-seq libraries was lower. However, genome-wide CpG coverage at matched sequencing depth was consistent across protocols (Fig. 1b; for additional quality metrics, including analysis of representation bias in different contexts see Supplementary Fig. 3) and we found that scM&T-seq covered a large proportion of sites in different genomic contexts with sufficient frequency to enable the analysis of epigenome heterogeneity across cells (Supplementary Fig. 4 and 5). To evaluate the potential coverage of scM&T-seq we sequenced a randomly chosen subset of four libraries at increased depth (mean of 25.9 M raw reads), which yielded a CpG coverage in line with the previous method (4.5 M compared to 3.6 M after 20.2 M raw reads for ESCs in stand-alone scBS-seq). Saturation depth was not reached in these four cells (mean duplication rate of 25.5%), meaning that additional sequencing would yield greater coverage as demonstrated previously3. As additional validation, we assessed the discrimination of serum and 2i ESCs by both stand-alone scBS-seq and scM&T-seq, finding a similar degree of separation that was consistent with bulk datasets published previously16 (Fig. 1c), with similar conclusions when using a joint hierarchical clustering across all cells (Supplementary Fig. 6). Notably, the difference between protocols and biological batches had a substantially smaller effect (PC2, 3% variance) than cell type differences (PC1, 48% variance), and by combining data across cells, we found that both protocols yield genome-wide methylation profiles that accurately recapitulate bulk methylation profiles in the same cell type (Supplementary Fig. 7). Finally, we compared estimates of methylation heterogeneity in different genomic contexts, again finding good agreement between protocols (Fig. 1d). Taken together, these analyses provide confidence that the parallel scM&T-seq method yields results that are in agreement with data from stand-alone scBS-seq.
For subsequent analyses, we focused on serum ESCs only since transcription and DNA methylation are uncoupled in 2i ESCs16, 19. A comparison of the principal components derived from the two data types - gene body methylation and gene expression- revealed that the global sources of variation were partially linked (Supplementary Fig. 8 and 9). However, a hierarchical clustering analysis of gene body methylation and gene expression for the 300 most variable genes (based on DNA methylation variance; for alternatives see Supplementary Fig. 10) revealed distinct clustering of cells when using either source of information (Fig. 1e,f). This suggests that global methylome and transcriptome profiles yield complementary, but distinct, aspects of cell state. This is also consistent with previous observations that the transcriptome and methylome are partially uncoupled in serum ESCs16.
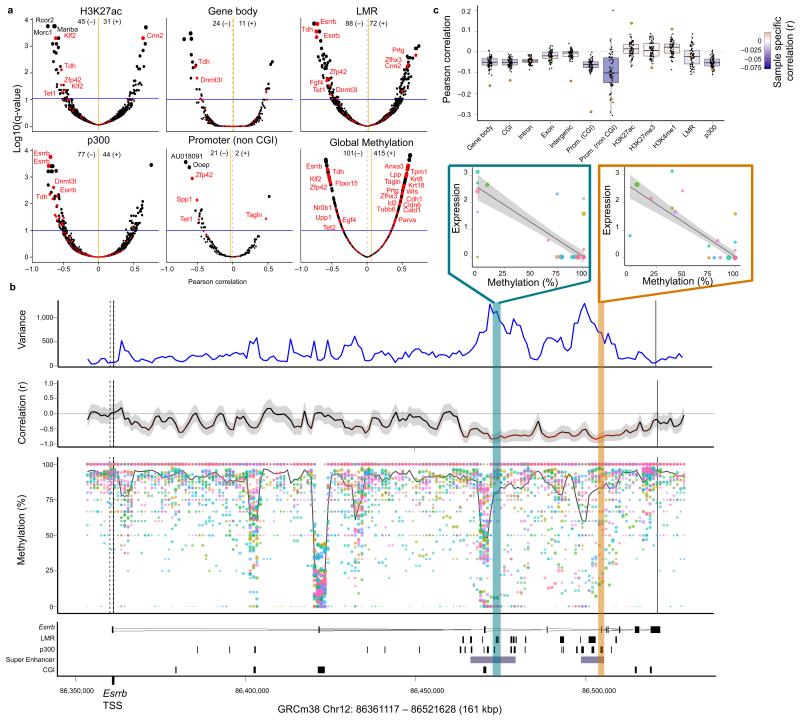
Next, we tested for associations between expression of individual genes and DNA methylation variation at several genomic contexts (Methods; Supplementary Table 2), identifying a total of 1,493 associations (FDR < 10%; see Fig. 2a, Supplementary Table 3 and 4), which were robust when using a bootstrapping approach to subsample the set of cells (Supplementary Fig. 11). We found both positive and negative associations, highlighting the complexity of interactions between the methylome and transcriptome9, 10. While methylation of non-CGI promoters is known to be associated with transcriptional repression, the role of enhancer methylation is less clear. Accordingly, negative correlations between DNA methylation and gene expression were predominant for non-CGI promoters, while distal regulatory elements including low methylated regions20 (LMRs) had a more even balance of positive and negative associations (Fig. 2a, Supplementary Fig. 12 and 13). Interestingly, associated genes were enriched for known pluripotency and differentiation genes18 (FDR < 1%, Fisher’s exact test; Supplementary Table 5). Our results provide the first evidence that heterogeneous methylation of distal regulatory elements (e.g. LMRs) accompanies heterogeneous expression of key pluripotency factors in stem cell populations6, 21. As an example, Figure 2b shows the association map of Esrrb, a known hub gene in pluripotency networks22 whose expression negatively correlates with the methylation of several LMR and p300 sites overlapping ‘super enhancers’ in the genomic neighbourhood23. We also found 516 genes whose expression correlated with the overall methylation level (FDR < 10%), indicating substantial links between transcriptional heterogeneity and global methylation levels (Fig. 2a).
Figure 2. Genome-wide associations between methylation and transcriptional heterogeneity in mouse ESCs.
a) Volcano plots of correlation coefficients (Pearson r2) from association tests between gene expression heterogeneity of individual genes and DNA methylation heterogeneity in alternative genomic contexts. Shown is the correlation coefficient for every gene (x-axis) versus the adjusted p-value (using Benjamini-Hochberg correction; y-axis). The size of dots corresponds to the adjusted p-value. A set of 86 known pluripotency and differentiation genes18 are highlighted in red. The blue horizontal line corresponds to the FDR 10% significance threshold. The total number of significant positive (+) and negative (−) correlations (FDR < 10%) for each annotation is shown in the header of each panel. The orange vertical bar corresponds to the average correlation coefficient across all genes for a given context. b) Representative zoom-in view for the gene Esrrb. From bottom to top, shown is: the annotation of the Esrrb locus with LMR, p300, super enhancer and CGI sites indicated; the estimated methylation rate of 3kb windows for each cell with the size of dots representing the CpG coverage and the solid line indicating the weighted mean methylation rate across all cells; the correlation between the methylation rate and Esrrb expression for each region coloured by the strength of the correlation and with the shaded area corresponding to the 95% confidence interval of the correlation coefficient; and the estimated weighted DNA methylation variance between cells. The top two scatter plots depict the association between DNA methylation at a p300 region (yellow) and an LMR (blue) and Esrrb expression. c) Gene-specific association analysis, assessing correlations between DNA methylation in different genomic contexts and gene expression in individual cells. For each annotation, shown are box plots of methylation-expression correlations for all variable genes in single cells, with the correlation obtained from matched RNA-seq and BS-seq of a bulk cell population superimposed16 (orange circles).
In addition to between-cell analyses, scM&T-seq can also be used to correlate the methylome and transcriptome between genes in individual cells (Fig. 2c). We found that correlation between methylation and gene expression varied substantially between cells but was consistent in direction with matched RNA-seq and BS-seq data from a population of cells16. Again, this attests to scM&T-seq being sufficiently accurate to reliably study epigenome-transcriptome linkages. Our results also point to the possibility of heterogeneity between cells in the degree of coupling between the methylome and the transcriptome. Although we have ruled out obvious confounding factors such as average methylation rate and sequence coverage (Supplementary Fig. 14 and 15), more data will be required to understand possible technical components in these linkages.
Our work demonstrates that parallel profiling of the methylome and transcriptome from the same single cell is feasible, obtaining data of similar quality to methods profiling either feature in isolation. For the first time, scM&T-seq allows the relationship between DNA methylation and expression to be studied at specific genes in single cells. We have confirmed a negative association between non-CGI promoter methylation and transcription in single cells and identified both positive and negative associations at distal regulatory regions. The expression levels of many pluripotency factors, e.g. Esrrb, were found to be negatively associated with DNA methylation, suggesting that an important mechanistic component of fluctuating pluripotency in serum ESCs is epigenetic heterogeneity. Finally, we demonstrate that the strength of the connection between methylome and transcriptome can vary from cell to cell. scM&T-seq is a powerful approach to interrogate the poorly understood connectivity between transcriptional and DNA methylation heterogeneity in single cells and provides the potential to identify factors that regulate this relationship.
Online Methods
Sample collection & Single-cell sequencing
E14 ESCs were cultured in serum and LIF or 2i media as described previously16. Single cells were collected by FACS following ToPro-3 and Hoechst 33342 staining to select for live cells with low DNA content (i.e. G0 or G1 phase cells). Cells were collected in RLT plus lysis buffer (Qiagen) containing 1 U/μl SUPERase-In (Ambion) and processed using the G&T-seq protocol7, but following physical separation of mRNA and genomic DNA from single cells, the DNA was eluted into 10 μl of H2O.
Single-cell bisulfite libraries were then prepared as previously described3 but with the following modifications. Conversion was carried out using EZ Methylation Direct bisulfite reagent (Zymo) on purified DNA in the presence of AMPure XP beads (Beckman Coulter) following G&T-seq. Purification and desulphonation of converted DNA was performed with magnetic beads (Zymo) on a Bravo Workstation (Agilent), eluting into the mastermix for the first strand synthesis. Primers for first and second strand synthesis contained a 3′-random hexamer and biotin capture of first strand products was omitted, however an extra 0.8× AMPure XP purification was performed between second strand synthesis and PCR. Each pre-PCR AMPure XP purification was carried out using a Bravo Workstation. To avoid batch effects all libraries were prepared in parallel in a 96 well plate. Purified scBS-seq libraries were sequenced in pools of 16-20 per lane of an Illumina HiSeq2000 using 125-bp paired-end reads.
RNA sequencing libraries were prepared from the single-cell cDNA libraries using the Nextera XT kit (Illumina) as per the manufacturer's instructions but using one-fifth volumes. Multiplexed library pools were sequenced on one lane of an Illumina HiSeq2000 generating 125-bp paired-end reads.
Sequence data processing and raw data analysis
BS-seq read alignment
Sequencing data was processed as previously described3, with small modifications. Briefly, raw sequence reads were trimmed to remove the first 6 base pairs (the 6N random priming portion of the reads), adapter contamination and poor-quality base calls using Trim Galore (v0.3.8, parameters: --clip_r1 6 (or 9) --clip_r2 6 (or 9). Trimmed reads were aligned in single-end mode to the GRCm38 mouse genome assembly using Bismark24 (v0.13.1, parameters: --bowtie2 --non-directional). Methylation calls were extracted after duplicate alignments had been removed. (Note: due to multiple rounds of random priming with oligo 1, the single-cell bisulfite libraries are non-directional).
RNA-seq read alignment and gene expression quantification
GSNAP25 (version 2014.02.28) was used to align all RNA-seq libraries onto the mouse genome assembly GRCm38 (with the --use-splicing option). For computing the transcriptome raw read count table, an aligned read was counted towards a gene if it overlaps with any exonic region of that gene. To normalize transcriptome counts for library size, library size estimates obtained from DESeq226 were used. For computing the transcriptome TPM table (Transcripts Per Million table), the output from cufflinks27 (with the --frag-bias-correct --compatible-hits-norm --multi-read-correct option) were normalized to TPM values. Ensembl annotation version 75 was used whenever gene annotations were required.
BS-seq and RNA-seq quality assessment
We included four negative controls (empty wells) in the library preparation procedure, to exclude the possibility of DNA or RNA contamination. Single-cell BS-seq libraries from negative controls had < 2% mapping efficiency (% raw sequencing reads aligned), and scRNA-seq libraries from these samples had an alignment rate of less than 1%.
Single-cell BS-seq libraries with low alignment rates (< 7% raw sequencing reads aligned), or poor bisulfite conversion < 95% (based on Bismark CHH and CHG methylation estimates), were excluded. Out of a total of 92 single-cell libraries, 81 passed this quality filter.
To identify low quality scRNA-seq libraries, we required a minimum of 2 M mapped reads. Four serum and 2 2i ESCs were excluded based on this criterion (Supplementary Table 1).
Of the 92 single cell samples, 75 (61 serum ESCs, 14 2i ESCs, 81.5%) passed quality assessment for both their methylome and transcriptome sequencing. Complete QC data for both scRNA-seq and scBS-seq are provided as Supplementary Table 1.
Statistical analyses
Clustering analyses
PCA analysis in Figure 1c) was performed jointly on gene-body methylation of 12 2i and 20 serum cells profiled by stand-alone scBS-seq3, 61 serum and 16 2i cells profiled by scM&T-seq, as well as a bulk BS-seq sample16 and single-cell bulk methylation rates corresponding to genome-wide averages.
DNA methylation-gene expression association analysis
For association analyses, gene expression levels were considered on a logarithmic scale, using log10 normalized TPM counts (see above). Binary single-base pair CpG methylation states were estimated by the ratio of methylated read counts to total read counts. The methylation rate in different genomic contexts, such as gene-body, promotor, or enhancer annotations, were estimated as the mean CpG methylation rate within the region defined by the context (Supplementary Table 2). Following the approach of Smallwood et. al.3, weighted arithmetic mean and variance estimates were obtained for each context and cell, thereby accounting for differences in CpG coverage between cells.
For correlation analysis, genes with low expression levels or low expression and methylation variability between cells were discarded, following the rational of independent filtering28. First, a minimum expression level (at least 10 TPM counts) in at least 10% of all cells was required. From these, the 7,500 most variable genes were considered for analysis. Second, methylated regions were required to be covered by at least one read in at least 50% of all cells. For association tests, all possible relationships between genes and methylated regions within 10 kbp of the gene (upstream and downstream of gene start or stop) were considered. Association tests were based on weighted Pearson correlation coefficient, thereby accounting for differences in CpG coverage between cells. Precisely, let e be a vector with expression rates of cells for a particular gene, m be methylation rates of the associated region, and w be weights corresponding to the number of covered CpGs within the region. Then the weighted Pearson correlation cor(e, m; w) between gene-expression e and methylation m is:
Here, cov(x, y; w) is the weighted covariance
and m(x; w) the weighted arithmetic mean:
Two-sided Student’s t-tests were performed to test for non-zero correlation, and p-values were adjusted for multiple testing for each context using the Benjamini-Hochberg procedure. For the zoom-in plot in Fig. 2b, we considered a sliding window approach (3kb sized windows, step-size 1kb) to estimate the methylation rate in consecutive regions. Each region was tested for association with gene expression, again using weighted correlation coefficients as defined above.
For correlating methylation and expression of a single cell across genes (Fig. 2c), we filtered genes in the same way as described above, and used again the weighted Pearson correlation to test for associations.
R version 3.1.2 was used for all the analysis. The corresponding source code is available on Github (https://github.com/PMBio/scMT-seq). SeqMonk version 0.30 was used to compute methylation rates and CpG coverage for different regions (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/). Ensembl annotation version 75 was used whenever gene annotations were required.
Supplementary Material
Acknowledgements
We thank A. Kolodziejczyk and S. A. Teichmann for providing a list of 86 ESC pluripotency and differentiation genes18. We thank W. Haerty for his supervision and valuable advice to T.X.H. We thank the Welcome Trust Sanger Institute sequencing pipeline team for assistance with Illumina sequencing. We thank the members of the Sanger-EBI single-cell genomics centre for general advice. W.R. is supported by BBSRC, Wellcome Trust, and EU, G.K. is supported by BBSRC, MRC, and EU, H.J.L. is supported by EU NoE EpiGeneSys and O.S. is supported by EMBL, Wellcome Trust, EU.
Footnotes
Accession codes scRNA and scBS-sequencing data from all 92 ESCs and 4 negative controls are available under the accession number GSE74535. This study reuses some data from Smallwood et. al.3, which has previously been deposited under accession GSE56879.
Competing Financial Interests Statement W.R. is a consultant and shareholder of Cambridge Epigenetix. All other authors declare no competing financial interests.
References
- 1.Shapiro E, Biezuner T, Linnarsson S. Nature reviews. Genetics. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 2.Guo H, et al. Genome research. 2013;23:2126–2135. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smallwood SA, et al. Nature methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farlik M, et al. Cell Rep. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 6.Yan L, et al. Nature structural & molecular biology. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 7.Macaulay IC, et al. Nature methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 8.Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A. Nat Biotech. 2015;33:285–289. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schubeler D. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA. Nature reviews. Genetics. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 11.Singer ZS, et al. Mol Cell. 2014;55:319–331. doi: 10.1016/j.molcel.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalmar T, et al. PLoS biology. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers I, et al. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 14.Singh AM, Hamazaki T, Hankowski KE, Terada N. Stem cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- 15.Torres-Padilla ME, Chambers I. Development. 2014;141:2173–2181. doi: 10.1242/dev.102624. [DOI] [PubMed] [Google Scholar]
- 16.Ficz G, et al. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein Allon M., et al. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolodziejczyk AA, et al. Cell Stem Cell. 2015;17:471–485. doi: 10.1016/j.stem.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habibi E, et al. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Stadler MB, et al. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 21.Lee HJ, Hore TA, Reik W. Cell Stem Cell. 2014;14:710–719. doi: 10.1016/j.stem.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papp B, Plath K. The EMBO journal. 2012;31:4255–4257. doi: 10.1038/emboj.2012.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whyte WA, et al. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Online methods references
- 24.Krueger F, Andrews SR. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu TD, Nacu S. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, et al. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourgon R, Gentleman R, Huber W. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.