Abstract
Mono-ubiquitination of Fancd2 is essential for repairing DNA inter-strand crosslinks (ICLs) but the underlying mechanisms are unclear. The Fan1 nuclease, also required for ICL repair, is recruited to ICLs by ubiquitinated (Ub)-Fancd2. This could in principle explain how Ub-Fancd2 promotes ICL repair, but we show recruitment of Fan1 by Ub-Fancd2 is dispensable for ICL repair. Instead Fan1 recruitment – and activity –restrains DNA replication fork progression, and prevent chromosome abnormalities from occurring, when DNA replication forks stall. Accordingly, Fan1 nuclease-defective knockin mice are cancer-prone. Moreover, we show that a Fan1 variant in high-risk pancreatic cancers abolishes recruitment by Ub-Fancd2, and causes genetic instability without affecting ICL repair. Therefore, Fan1 recruitment enables processing of stalled forks that is essential for genome stability and health.
ICLs block DNA replication. Defects in repairing ICLs are implicated in Fanconi anemia (FA), a rare autosomal recessive disease typified by hypersensitivity to ICL-inducing agents such as mitomycin-C (MMC) or diepoxybutane (DEB), exaggerated G2 arrest and increased chromosome abnormalities after exposure to these agents (1). FA is caused by mutations in any of the nineteen Fanc proteins, comprising the FA network (2). The mono-ubiquitination of Fancd2 at Lysine-561 is essential for ICL repair, but the underlying mechanisms are unclear (2, 3). The Fan1 nuclease is recruited to ICLs in S-phase via the interaction between its ubiquitin-binding (UBZ) domain and ubiquitinated (Ub-) Fancd2 (4-7). However, mutations in Fan1 do not cause FA as might be expected, but rather lead to karyomegalic interstitial nephritis (KIN) (8), suggesting that Fan1 and Fancd2 play distinct roles in ICL repair. Therefore, the functional significance of the Fan1/Ub-Fancd2 interaction is unclear, and we took several approaches to address this problem.
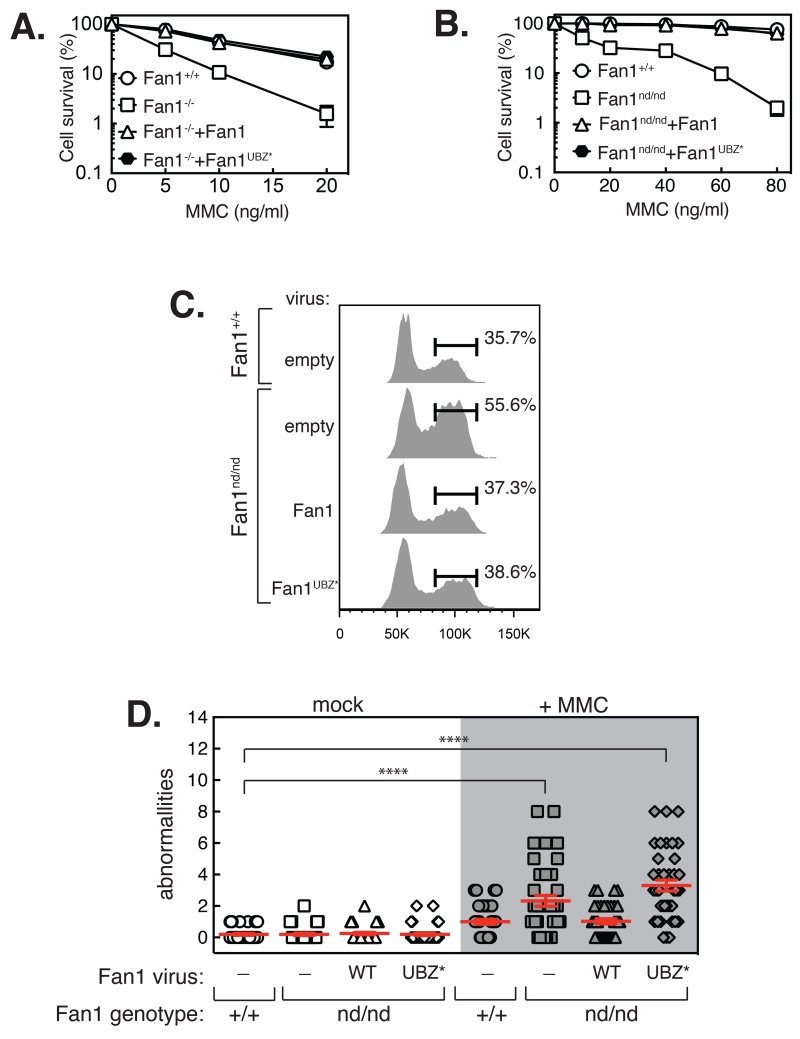
First we tested the ability of UBZ-mutated Fan1 (UBZ*) to recue the MMC sensitivity of human U2OS Fan1−/− cells (9). Mutating two conserved residues (C44A+C47A) in the Fan1 UBZ domain that abolishes foci (4) fully rescued the MMC hypersensitivity of Fan1−/− cells (Fig. 1A; Fig. S1A). Embryonic fibroblasts (MEFs) from Fan1 nuclease-defective (nd) mice harboring a deletion of the last 41 amino acids of Fan1, including key catalytic residues (Figs. S2, S3; mice described in Supplemental Information), were hypersensitive to MMC (Fig. 1B). The murine Fan1 UBZ* mutant fused to GFP, expressed at roughly 4-times the level of endogenous Fan1, failed to form foci (Figs. S1B-D) but rescued the MMC hypersensitivity of Fan1nd/nd MEFs (Fig. 1B). These data suggest that the recruitment of Fan1 by Ub-Fancd2 is dispensable for ICL repair. Furthermore, we found that the UBZ-mutated Fan1 fully rescued the exaggerated G2 arrest seen in Fan1nd/nd MEFs (Fig. 1C), but failed to rescue the increase in chromosome abnormalities (radial structures and chromatid breaks) induced by MMC (Fig. 1D).
Fig. 1. Fan1 UBZ domain is not required for efficient ICL repair.
A. U2OS Fan1−/− cells stably expressing GFP-tagged Fan1 or Fan1 C44A+C47A (UBZ*), exposed to the concentrations of MMC indicated, were subject to clonogenic survival assays. For each cell population, viability of untreated cells is defined as 100%. B. Clonogenic survival analysis of Fan1nd/nd MEFs stably expressing GFP-tagged murine Fan1 or Fan1 UBZ* mutant. Wild-type MEFs were used as control. C. FACS analysis of DNA content in Fan1nd/nd MEFs stably expressing GFP- Fan1 or GFP-Fan1 UBZ* mutant after exposure to MMC for 24 h. Wild-type MEFs (Fan1+/+) with empty vector were used as control. D. The number of chromosome abnormalities per metaphase in spreads of MEFs exposed to MMC. Data in A and B are represented as mean ± SD. In D., significance was calculated using one-way ANOVA (****, p < 0.0001) followed by Bonferroni’s Multiple Comparison Test.
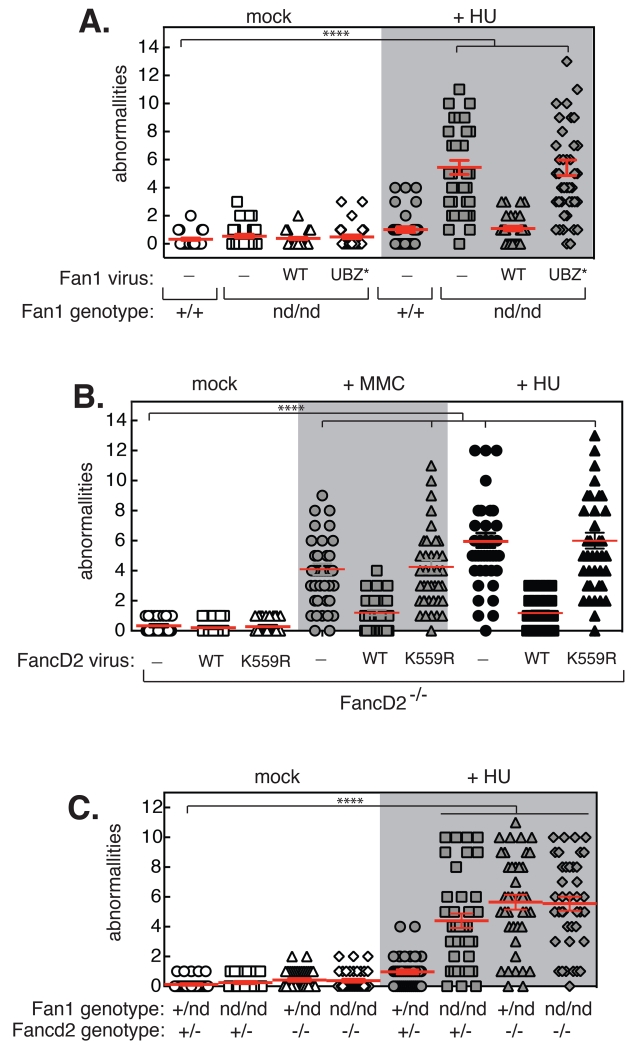
Fan1nd/nd MEFs also showed a high level of chromosome abnormalities in response to fork-stalling agents such as methymethanesulfonate (MMS; Fig. S4) and hydroxyurea (HU; Fig. 2A) which cause base alkylation and nucleotide depletion, respectively. The increase in chromosome abnormalities in Fan1nd/nd MEFs induced by HU were not reversed by the Fan1 UBZ* mutant (Fig. 2A). These data indicate Fan1 nuclease activity and interaction with Ub-Fancd2 prevent chromosome abnormalities at stalled forks independent of ICL repair. This is consistent with Fancd2 null cells showing high levels of chromosome abnormalities after exposure to HU (10), but the requirement of ubiquitination was not investigated. We found that the non-ubiquitinatable mouse Fancd2 K559R mutant was unable to rescue the high levels of chromosome abnormalities induced by MMC or HU in Fancd2−/− MEFs (Figs. 2B, S5A), consistent with the idea that interaction of Fan1 with Ub-Fancd2 is required for this response. However, Ub-Fancd2 and Fan1 may prevent chromosome abnormalities at stalled forks by different mechanisms. To address this point we crossed Fan1nd/nd mice with Fancd2−/− mice to generate double mutants. As shown in Fig. 2C, the level of HU-induced chromosome abnormalities in MEFs from Fan1nd/nd Fancd2−/− double mutant mice was not higher than in the respective single mutants, suggesting that Fan1 and Fancd2 are epistatic in this respect. Therefore the Fan1/Ub-Fancd2 interaction, and Fan1 nuclease activity, is required for preventing chromosome abnormalities when forks stall, and this role is independent of ICL repair.
Fig. 2. Fan1 nuclease and Ub-Fancd2 interaction prevent chromosome abnormalities at stalled forks.
After exposure of cells to HU or MMC as indicated, metaphase spreads were prepared and the number of chromosome abnormalities (radial strucutres and chromatid breaks) was quantitated. The following cells were analysed: A. Fan1+/+ MEFs infected with empty virus, and Fan1nd/nd MEFs complemented with GFP-tagged Fan1 wild-type (WT), Fan1 UBZ* mutant or empty virus. B. Fancd2−/− MEFs infected with empty virus, or virus expressing mCherry-Fancd2 or mCherry-Fancd2 K559R. C. MEFs from littermate mice of the genotypes indicated. Statistical significance was calculated using one-way ANOVA (****, p < 0.0001) followed by Bonferroni’s Multiple Comparison Test.
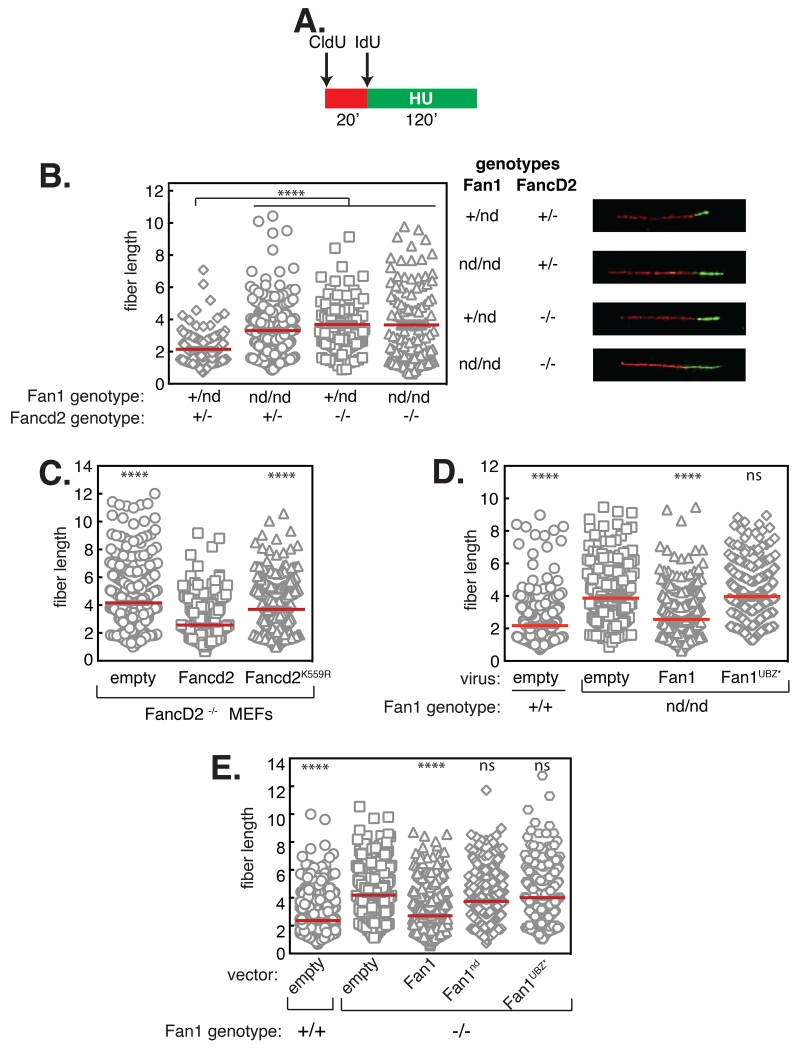
As well as preventing chromosome abnormalities, Fancd2 has been implicated in restraining progression of forks that stall after exposure to HU (11). We set out to test if Fan1 plays a similar role using DNA fiber analysis. Primary MEFs were pulsed with CldU followed by IdU with or without HU, and IdU track length was measured (Fig. 3A). Fancd2−/− MEFs had substantially longer IdU tracks in HU than wild type MEFs (11), and a similar effect was seen in Fan1nd/nd MEFs (Fig. 3B). Tracks from Fan1nd/nd Fancd2−/− double mutant MEFs were not longer than in the respective single mutant cells, implying epistasis (Fig. 3B). No differences in track length were observed in the absence of HU (Fig. S5B). Consistent with the fiber data, we observed more DNA synthesis in Fan1nd/nd MEFs exposed to HU than in Fan1+/nd MEFs, judged by FACS analysis of EdU incorporation (Fig. S6). Thus, Fancd2 and Fan1 restrain the progression of stalled forks, and we next tested if their interaction is required. Fancd2−/− MEFs expressing the Fancd2 K559R mutant had substantially longer IdU tracks in HU than cells expressing Fancd2 (Fig. 3C). Furthermore, we found that the Fan1 UBZ* mutant failed to restore normal track length in HU-treated Fan1nd/nd MEFs, in contrast with wild-type Fan1 (Fig. 3D). Accordingly, U2OS Fan1−/− cells had longer replication tracks in HU than parental cells and neither the Fan1 UBZ* mutant nor the nuclease-dead mutant rescued this defect (Fig. 3E). Together these data show that the Fan1/Ub-Fancd2 interaction restrains the progression of stalled forks, a function that also requires Fan1 nuclease activity.
Fig. 3. Fan1 nuclease activity Ub-Fancd2 interaction control progression of stalled forks.
A. Fiber labeling protocol. Primary MEFs were pulsed with CldU followed by IdU with HU, and IdU track length was measured. B-D. Dot plots of ldU replication tracks in MEFs of the genotypes indicated. In B. representative CldU and IdU replication tracks are shown (right panels). Cells were infected with viruses expressing the Fancd2 (C) or Fan1 (D) proteins indicated. E. Dot plot of IdU replication tracks in U2OS cells, and U2OS Fan1−/− cells stably stably transfected with the vectors indicated. “nd” nuclease-defective”. Red lines represent mean track length. Statistical significance was calculated using one-way ANOVA (****, p < 0.0001) followed by Bonferroni’s Multiple Comparison Test.
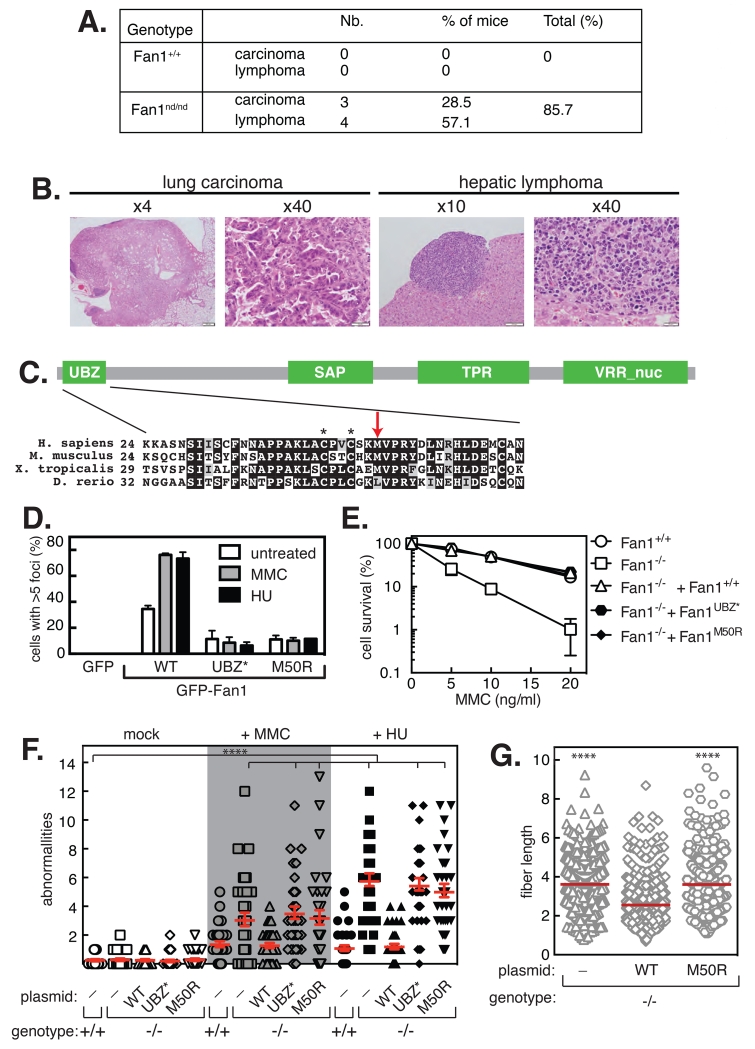
Bi-allelic mutations in Fancd2, or genes that promote Fancd2 ubiquitination, cause FA, typified by cancer predisposition (1, 2). We speculated therefore that Fan1 defects might also cause cancers. No tumors were observed in Fan1nd/nd mice at 6 months (data not shown), but by 20 months around 85% of Fan1nd/nd mice developed cancers; no malignancies were evident in age-matched Fan1+/+ mice (Fig. 4A). Pulmonary carcinomas, epithelial-type cancers similar to those reported in FA patients and Fancd2−/− mice (12), were observed in 28% of the mice, while 57% developed lymphoma (Fig 4A,B). These data suggest that Fan1 nuclease activity is a tumor suppressor, at least in mice.
Fig. 4. Fan1 nuclease activity and interaction with Ub-Fancd2 prevents cancers.
A. Incidence of carcinoma and lymphoma in Fan1+/+ and Fan1nd/nd mice at approximately 20 months of age revealed by genotype-blind whole-body pathological analyses. B. Representative images of pulmonary carcinoma and hepatic lymphoma from Fan1nd/nd mice. C. Alignment of the UBZ domain of FAN1. Identical residues black, similar residues grey. Asterisks denote C44 and C47 residues. Red arrow denotes M50. D. U2OS Fan1−/− cells stably expressing the proteins indicated were treated, or not, with MMC or HU and the proportion of cells with > 5 GFP–FAN1 foci were counted. E. Clonogenic survival analysis of U2OS Fan1−/− stably expressing the GFP-tagged Fan1 proteins indicated, after exposure to the indicated concentrations of MMC for 18 h. F. Chromosome abnormalities in U2OS Fan1−/− cells, stably expressing the proteins indicated, after treatment with MMC or HU. G. Dot plots of IdU replication tracks in U2OS Fan1−/− cells, stably expressing the GFP-Fan1 proteins indicated, after exposure to HU. “WT”, wild-type. In D, E. data are represented as mean ± SD. In F and G, significance was calculated using one-way ANOVA (****, p < 0.0001) followed by Bonferroni’s Multiple Comparison Test.
As suppression of chromosome abnormalities by Fan1 requires interaction with Ub-Fancd2 as well as Fan1 nuclease activity, we postulated that mutations in the human Fan1 UBZ domain might cause cancers. Whole exome sequencing recently identified a recurrent germline Fan1 variant, M50R, occurring at a relatively high frequency in high-risk pancreatic cancers (13). The M50R Fan1 variant, which co-segregates with pancreatic cancer in two separate families, is a strong candidate pancreatic cancer predisposition gene. M50 lies in the UBZ domain of Fan1 (Fig. 4C). Similar to the UBZ* mutation (C44A+C47A), the Fan1 M50R mutation abolished Fan1 foci but rescued the MMC sensitivity of U2OS Fan1−/− cells (Figs. 4D,E). The M50R mutant failed, however, to prevent chromosome abnormalities induced by HU or MMC in Fan1−/− cells (Fig. 4F). Moreover, expression of wild-type Fan1 in Fan1−/− cells restored normal track length in HU, but the Fan1 M50R mutant failed to do so (Fig. 4G). Therefore, the Fan1 M50R variant associated with high-risk pancreatic cancers causes unrestrained replication fork progression and chromosomal instability known to drive carcinogenesis.
In this study we made the unexpected finding that although Ub-Fancd2 recruits Fan1 to ICL-blocked replication forks, this is not required for ICL repair judged by MMC sensitivity and G2 arrest. Instead Fan1 recruitment it is vital for protective responses when forks stall even in the absence of DNA crosslinks. Cells defective in Fan1 recruitment, or activity, show a high frequency of chromosome abnormalities and increased fork rate, when forks are forced to stall. The mechanisms underlying these defects are not yet clear but cells depleted of the HLTF tranlocase or RAD51 recombinase, which both drive fork reversal, show longer replication tracks in HU similar to Fan1-defective cells (14, 15). Therefore the Fan1 activity might promote fork reversal but this remains to be tested. It is not yet clear if the chromosome abnormalities seen after fork stalling in Fan1-defective cells are related to the increased fork speed, or if they arise independently. It seems counter-intuitive, perhaps, that a nuclease activity is required to prevent chromosome breaks at stalled forks. One potential explanation is that Fan1 cleaves stalled forks in a way that enables replication to resume after fork stalling, consistent with a recent report that Fan1 promotes replication fork recovery (16). Failure of Fan1-mediated fork processing may result in the persistence of structures that are cleaved inappropriately by other nucleases leading to forks breaking in a way that is refractory to repair.
Our observations that Fan1 nuclease activity and interaction with Ub-Fancd2 prevent cancers prompt future investigations as to whether cancer predisposition associated with FA might be caused by defective fork processing as opposed to defective ICL repair. Identifying a separation of function Fan1 mutant affecting ICL repair but not stalled fork processing would be valuable for these efforts. Besides pancreatic cancer, germline mutations in Fan1 have been identified in colon cancer (17). Loss of heterozygosity (LOH) has not been observed in tumours from the M50R carriers, or in Fan1-muated colon cancers (13, 17). Epigenetic inactivation of Fan1, haplo-insufficiency, or dominant negative effects may provide explanations but these ideas remain to be investigated. KIN caused by biallelic Fan1 mutations is very rare disease but early onset cancers were reported in two affected families (17). These reports, together with the present study, are consistent with Fan1 acting as a tumor suppressor with multiple roles in genome maintenance vital for preventing human diseases.
Supplementary Material
Acknowledgements
We are grateful to James Hastie, Hilary MacLauchlan and the Antibody Production Team at Division of Signal Transduction Therapy (DSTT) and the DNA Sequencing Service at SLS, University of Dundee. We thank CIRRU staff, especially Lorraine Malone and Carol Clacher for help with animal husbandry, and Gail Gilmour and Elaine Forsyth for help with genotyping. We thank Lynn Oxford and Lynn Stevenson (Veterinary Diagnostic Services, University of Glasgow) for support with tissue processing. We thank Alan d’Andrea for Fancd2−/− mice. We are grateful to Laura Feeney for critical reading of this manuscript. We thank Ashton Connor, Alyssa Smith, Steven Gallinger and George Zogopoulos for communicating data prior to publication. CL is the recipient of a Marie Curie Intra-European fellowship. AM was supported by Wellcome Trust Investigator Award WT096598MA to JJB. This study was supported by the Medical Research Council and the pharmaceutical companies supporting the DSTT (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck KgaA, Janssen Pharmaceutica and Pfizer) associated with MRC PPU.
References
- 1.Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Higuera I, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 4.MacKay C, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kratz K, et al. Deficiency of FANCD2-Associated Nuclease KIAA1018/FAN1 Sensitizes Cells to Interstrand Crosslinking Agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 6.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 7.Smogorzewska A, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou W, et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet. 2012;44:910–915. doi: 10.1038/ng.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munoz IM, Szyniarowski P, Toth R, Rouse J, Lachaud C. Improved genome editing in human cell lines using the CRISPR method. PLoS One. 2014;9:e109752. doi: 10.1371/journal.pone.0109752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lossaint G, et al. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol Cell. 2013;51:678–690. doi: 10.1016/j.molcel.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 12.Houghtaling S, et al. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith AL, et al. Candidate DNA repair susceptibility genes identified by exome sequencing in high-risk pancreatic cancer. Cancer Lett. 2015 doi: 10.1016/j.canlet.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zellweger R, et al. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol. 2015;208:563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kile AC, et al. HLTF’s Ancient HIRAN Domain Binds 3′ DNA Ends to Drive Replication Fork Reversal. Mol Cell. 2015;58:1090–1100. doi: 10.1016/j.molcel.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhury I, Stroik DR, Sobeck A. FANCD2-controlled chromatin access of the Fanconi-associated nuclease FAN1 is crucial for the recovery of stalled replication forks. Mol Cell Biol. 2014;34:3939–3954. doi: 10.1128/MCB.00457-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segui N, et al. Germline Mutations in FAN1 Cause Hereditary Colorectal Cancer by Impairing DNA Repair. Gastroenterology. 2015 doi: 10.1053/j.gastro.2015.05.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.