Abstract
The evolution of tight coupling between the circadian system and redox homeostasis of the cell has been proposed to coincide roughly with the appearance of the first aerobic organisms, around 3 billion years ago. The rhythmic production of oxygen and its effect on core metabolism are thought to have exerted selective pressure for the temporal segregation of numerous metabolic pathways. Until recently, the only evidence for such coupling came from studies showing circadian cycles in the abundance of various redox metabolites, with many arguing that these oscillations are simply an output from the transcription/translation-feedback loop (TTFL). The recent discovery that the peroxiredoxin (PRX) proteins exhibit circadian cycles in their oxidation status, even in the absence of transcription, demonstrated the existence of autonomous oscillations in the redox status of the cell. The PRXs are a family of cellular thiol peroxidases whose abundance and high reaction rate make them the major cellular sink for cellular peroxides. Interestingly, as part of the normal catalytic cycle, PRXs become inactivated by their own substrate via over-oxidation of the catalytic residue, with the inactivated form of the enzyme displaying circadian accumulation. Here, we describe the biochemical properties of the PRX system, with particular emphasis on the features important for the experimental analysis of these enzymes. We will also present a detailed protocol for measuring PRX over-oxidation across circadian time in adherent cell cultures, red blood cells and fruit flies (Drosophila melanogaster), providing practical suggestions for ensuring consistency and reproducibility of the results.
1. Introduction: circadian and redox coupling in the cell
The intimate link between circadian rhythmicity and cellular redox state has been suggested to date back to the Great Oxidation Event that occurred approximately 3 billion years ago (Crowe et al., 2014), facilitating the subsequent development of aerobic metabolism (Konhauser et al., 2012). Rising oxygen levels, attributed to the ability of photosynthetic bacteria to use water as the main electron donor, are thought to have created a strong selective pressure on anaerobes to evolve defence systems to deal with this harsh and unprecedented oxidizing environment. Therefore driven by the day-night cycle, diurnal rhythms in oxygen or reactive oxygen species (ROS) of biological origin, as well as ROS generated directly by UV radiation, could have potentially driven the co-evolution of circadian and redox systems so that processes sensitive to ROS would anticipate, and be buffered against, harmful oxidative stress.
Evidence for circadian oscillations in redox pathways were documented in the early days of chronobiology, during the pre-genomic era, and circadian variation in assorted redox parameters over several days were consistently observed (Brody & Harris, 1973). Before any of the ‘clock genes’ were discovered, rhythms in NADP+:NADPH ratio, as well as rhythms of adenine nucleotides abundance, were demonstrated in plant seedlings kept in constant darkness (Wagner & Frosch, 1974). Several studies performed in rodents demonstrated that key redox couples such as glutathione and NADP+:NADPH ratio display diurnal cycles in their oxidation status in the liver (Kaminsky et al., 1984; Farooqui & Ahmed, 1984). Even if these oscillations might be driven partially by feeding, glutathione levels in platelets kept in vitro (Scheer et al., 2011; Radha, Hill, Rao, & White, 1985) and in blood serum (Blanco et al., 2007) also display diurnal variation, implying that the redox balance of the blood resonates with cycles of day and night. Moreover, a recent study in rodents demonstrated that circadian redox oscillations control neuron excitability in the suprachiasmatic nuclei (SCN) by regulating multiple potassium (K+) channels (Wang et al., 2012).
The first evidence for coupling of the redox and circadian transcriptional/translational feedback loop (TTFL) came from a biochemical study demonstrating that the DNA-binding affinity of CLOCK/NPAS2:BMAL1 – the main transcriptional activators of the TTFL – are regulated by the redox state of the adenine dinucleotide coenzymes NAD(P)H in vitro (Rutter, 2001). However, these experiments were performed in a purified system and used concentrations of NAD(P)H in the millimolar range, thus leaving unanswered questions about the relevance of these findings in vivo. Nonetheless, there are a number of examples that show direct interaction between the redox state and circadian gene regulation. For example, heme-binding, and thus activity of the nuclear receptor REV-ERBβ, is governed by a redox-sensitive cysteine (Gupta & Ragsdale, 2011). Further afield, experiments performed in zebrafish Z3 cells demonstrate that light induces hydrogen peroxide (H2O2) release, which in turn drives the rhythmic transcription of clock genes (Hirayama, Cho, & Sassone-Corsi, 2007). In addition, this was recapitulated by application of a single H2O2 bolus, showing that H2O2 is the signal transducer in the light entrainment of zebrafish. Redox regulation of clock components has also been demonstrated in the cyanobacterium Synechococcus elongatus in which the Kai system operates as the major oscillator (Ivleva, Bramlett, Lindahl, & Golden, 2005). The light sensitive protein ldpA, which acts as a redox sensor by means of two iron-sulfur clusters, was shown to interact with the core clock machinery in a redox-regulated manner, thereby adjusting the organism’s period length. It has been suggested that this mechanism acts to fine tune the core oscillation by relaying nutritional (i.e. redox) cues (Ivleva et al., 2005).
Reciprocally, some redox components have been shown to be regulated directly by the TTFL. A well-established example is the NAD+-producing enzyme nicotinamide phosphoribosyltransferase (NAMPT), involved in the NAD+-salvage pathway. The rhythmic transcription of this enzyme results in the rhythmic output of NAD+, which subsequently has the potential to feed back to the TTFL either indirectly, by modulating the activity of key regulator enzymes such as the deacetylase SIRT1, or directly via redox sensitive transcription factors, although the latter lacks in vivo confirmation (Asher et al., 2008; Ramsey, Yoshino, Brace, Abrassart et al, 2009). In addition, rhythmic NAD+ production has the potential to generate rhythmic NADPH levels, since one of the major sources of NADPH is the enzymatic conversion of NADH to NADPH, catalysed by mitochondrial nicotinamide nucleotide transhydrogenase (NNT) and cytosolic NAD kinases (NADK) enzymes (Circu & Aw, 2010).
Furthermore, genetic studies have demonstrated that the ablation of core clock components affects the antioxidant capacity of the cell. For example, the per01 mutant of Drosophila melanogaster exhibits impaired, but circadian, period-luciferase rhythms (Plautz et al., 1997) and compromised antioxidant defence (Krishnan, Davis, & Giebultowicz, 2008). Similarly, in mammals behaviourally arrhythmic Bmal1−/− mice display increased accumulation of ROS and premature aging (Kondratov, 2006) and neurodegenerative change (Musiek et al., 2013). Given that not all ‘clock gene’ mutants exhibit such phenotypes, such effects could be secondary to non-circadian activities of Bmal1 as a transcription factor, beyond its role in the clockwork. Moreover, glutathione synthesis has recently been shown to be under direct TTFL control through the rhythmic transcription of the rate-limiting enzyme glutamate cysteine ligase in flies (Beaver et al., 2012). Rhythmic synthesis of redox metabolites does not, however, necessarily imply rhythms in the ratio of their reduced and oxidised forms, which more likely depends on functional metabolic oscillations. This therefore demonstrates an inherent difficulty in teasing apart cause and effect in the interaction between the TTFL and redox oscillations, since the two processes are no doubt intertwined at the molecular and biochemical levels.
As summarised above, research into circadian redox rhythms has mainly relied on assaying various redox metabolites across circadian time. However, a significant difficulty in performing such analyses lies in the labile nature of many redox-active metabolites, and the post-lysis artifacts that can result from the uncontrolled release of various cellular enzymes and redox-active small molecules, as well as the introduction of molecular oxygen. In addition, in cells it is expected that co-factors such as NADPH are generated and act locally, predominantly in cellular micro-domains rather than at the whole-cell level, which further complicates analysis (Stangherlin & Reddy, 2013). Last, but not least, it is difficult to tease apart the relative contributions of transcriptional and redox oscillations since these are normally tightly coupled in the cell (Reddy & Rey, 2014). In this regard, the finding of autonomous circadian rhythms in the overoxidation of the peroxiredoxin proteins marked an exciting discovery in the circadian field and presented researchers with the first biochemical readout of an underlying redox oscillator (Edgar et al., 2012; O’Neill & Reddy, 2011; O’Neill et al., 2011).
Using antiserum directed against the over-oxidised peroxiredoxins (PRX), we were able to demonstrate that two non-transcriptional eukaryotic systems display robust circadian rhythms in PRX oxidation (O’Neill et al., 2011; O’Neill & Reddy, 2011). These oscillations exhibit all of the key characteristics of circadian clocks, namely, persistence in constant conditions, entrainment by external stimuli, and temperature compensation. Importantly, in contrast to the divergent nature of TTFL components across phyla, PRX redox oscillations are conserved in all domains of life, including metazoans, plants, fungi, bacteria and even archaea (Edgar et al., 2012). The deep phylogenic conservation of PRX oscillations, together with the fact that they likely evolved to counteract oxidative stress, suggested that redox-based non-transcriptional time keeping mechanisms might be as ancient as the first aerobic organisms. Importantly, ‘clock’ mutant organisms exhibit PRX oxidation cycles, and conversely, PRX mutant animals retain rhythmic gene expression. However, in both cases the rhythms seem less robust, displaying perturbations in their phase and amplitude (Edgar et al., 2012). This suggested that although the two systems might be individually dispensable for the maintenance of rhythmicity, in the intact cell these oscillations are coupled to generate one unambiguous cellular timekeeping mechanism, adapted to the particular organism’s physiological needs and to its environmental niche.
2. The biochemical properties of the peroxiredoxin system
PRXs are ubiquitous antioxidant proteins that comprise a highly conserved family found in almost all aerobic organisms from Archaea to humans (Flohé & Harris, 2007). Most species possess more than one PRX enzyme, while the mammalian system expresses six different PRXs (PRX1-6). The importance of this family of peroxidases is underlined by the fact that they are present in virtually every cellular compartment, with PRX 1,2 and 6 being exclusively nucleocytoplasmic, PRX3 located in the mitochondrion, PRX4 in the endoplasmic reticulum (ER) and PRX5 being present in the mitochondrion, peroxisomes and cytosol (Park, Lee, Lee, & Kang, 2014). Initially identified through their ability to protect proteins from oxidative damage by ROS generated in the presence of dithiothreitol (DTT) (Rhee, Woo, Kil, & Bae, 2012), it is now known that this class of peroxidases offers essential antioxidant support to the cell and is responsible for detoxifying various organic peroxides, including hydrogen peroxide (H2O2) (Randall et al., 2013) and peroxinitrite (OONO−) (Bryk, Griffin, & Nathan, 2000).
2.1 The catalytic cycle of peroxiredoxins
Peroxiredoxins differ from other cellular peroxidases in the structure of their catalytic center, which does not rely on cofactors such as metals, heme, or selenocysteine, but rather employs a highly reactive cysteine residue, termed the peroxidatic cysteine (CysP). The first step of the catalytic cycle involves the oxidation of this residue to cysteine sulphenic acid (–SOH). The fate of this highly reactive intermediate forms the basis upon which PRXs are classified into three major groups. In the ‘typical’ and ‘atypical’ 2-Cys PRXs, the oxidized CysP condenses with another conserved residue, termed the resolving cysteine (CysR), which resides on a different PRX monomer in typical 2-Cys enzymes, or the same subunit in the atypical class (Wood, Schröder, Robin Harris, & Poole, 2003). In the 1-Cys PRX group, once oxidized, the redox-active cysteine residue can be regenerated into the active thiolate form by small thiol reductants such as glutathione (Manevich et al., 2004). The typical 2-Cys PRX (mammalian Prx1-4) initially exist as non-covalent dimers, in a head-to-tail configuration (i.e. the CysP of one faces the CysR of the another monomer). Oxidation then causes the formation of a disulphide-linked dimer, which can be visualised by non-reducing SDS-PAGE. The dimer is regenerated into its active form by the thioredoxin system, using NADPH as electron donor (Figure 1).
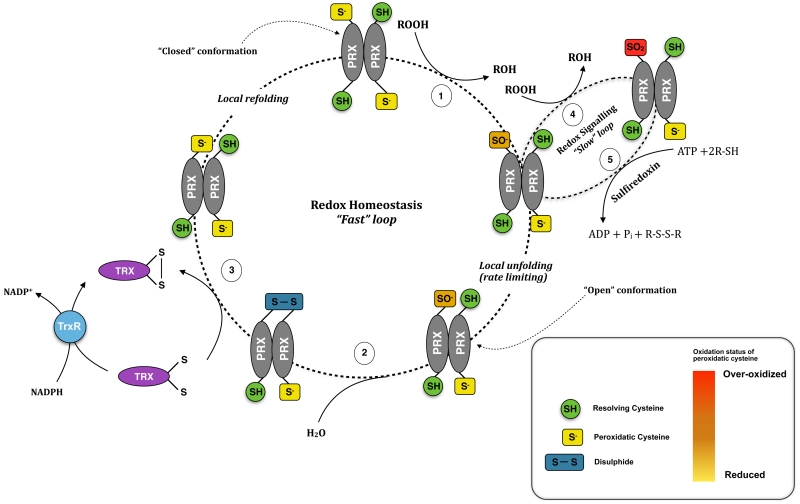
Figure 1.
The catalytic cycle of 2-Cys Peroxiredoxins. In vivo, PRX monomers associate into obligate dimers in head-to-tail manner via non-covalent interactions. In the reduced conformation (“fully folded conformation”), the CysP and CysR reside 13Å apart. In the first step (1), oxidation of the peroxidatic cysteine (thiolate form shown) by an incoming hydroperoxide (ROOH) leads to the formation of sulphenic acid (–SOH). In order to interact, the CysP and CysR have to be brought in close proximity, which is accomplished by local unfolding of the active site loop. In the second step (2), the sulphenic acid is resolved by reversible condensation with CysR to form intra-molecular disulfide bond. (3) The active form of the enzyme is subsequently regenerated by the thioredoxin system in a series of reactions involving thioredoxin (Trx), thioredoxin reductase (TrxR) and NADPH as electron donor. (4) Since the fully folded (FF) conformation of 2-Cys Prxs is more stable compared to the locally unfolded (LU) conformation, the longer-lived sulphenic acid can be further oxidised by a second peroxide molecule to sulphinic acid (SO2H), and perhaps sulphonic acid (SO3H, see figure 2). In most proteins, these forms are considered to be irreversible. However, the sulphinic acid form of PRXs have been shown to be resolved by the only known cellular sulphinic acid reductase, sulfiredoxin, in an ATP-dependent reaction (5).
Initially, the physiological relevance of PRXs as cellular peroxidases was challenged by studies demonstrating that the rate of the reaction with H2O2 was ≈105 M−1s−1 compared to ≈107-108 M−1s−1 for catalase (CAT) and glutathione peroxidase (GRX). More recent studies, however, showed that PRXs display kinetics comparable to CAT and GRX (Cox et al., 2009a; Peskin et al., 2013; 2007). The high reaction rates, together with the fact that PRXs are among the most abundant cellular proteins (around 1% of the total protein in most mammalian cells), implies that they are the major H2O2 sink in the cell (Randall et al., 2013). The initial underestimation of PRX reaction rates was due to the fact that activity measurements were done indirectly by assaying the oxidation of NADPH, compounded by a lack of mechanistic explanation for the extreme reactivity of CysP at that time. However, it was subsequently shown that the regeneration of the enzyme by the thioredoxin system is the rate-limiting step in catalytic cycles (Poynton & Hampton, 2014). Moreover, recent structural investigations revealed that four conserved amino acids in the active site of all 2-Cys PRXs not only decrease the pKa of the CysP thiol (thereby making it more nucleophilic in its thiolate form), but also activate the peroxide substrate and stabilize the transition state during catalysis (Hall, Nelson, Poole, & Karplus, 2011; Wood et al., 2003).
2.2 Oxidative inactivation and oligomerization
One intriguing feature of PRXs that differentiate them from other cellular peroxidases is that the oxidised intermediate can undergo further oxidation to sulphinic acid (–SO2H) and subsequently to sulphonic acid (–SO3H), collectively termed overoxidation here (Figure 2). These overoxidation states of the active site lead to the deactivation of PRX peroxidase activity, and have been shown to have functional significance for downstream signalling and chaperone activity (Wood, 2003; Yang, 2002). The mechanism of inactivation of the enzyme is explained through the existence of two conformations: Fully folded (FF) and locally unfolded (LU).
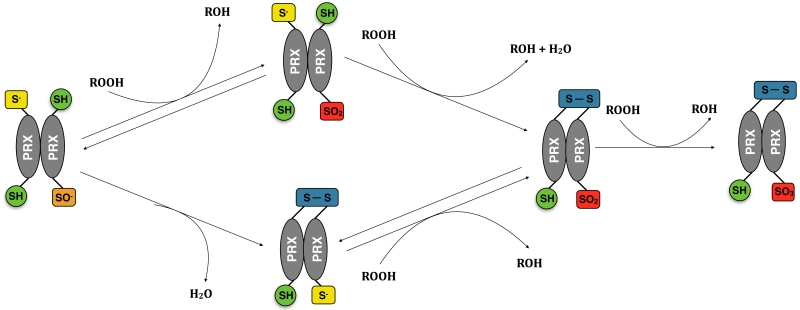
Figure 2.
Routes to formation of over-oxidised dimers. An oxidised PRX dimer could either form a disulphide bond via condensation reaction between CysP and CysR (1) or be further oxidised to sulphinic acid (2). Since a dimer possesses a second pair of CysP and CysR, it could undergo another catalytic cycle and form an over-oxidised dimer. The over-oxidized dimer could form by further oxidation of a disulfide-linked dimer (3), or by the formation of a disulfide bond between an over-oxidised monomer and a reduced monomer (4). The over-oxidised dimer could in turn be oxidised to form a sulphonic acid (5), which is regarded as irreversibly oxidised, and is recycled via proteosomal degradation. For clarity, in step 3 we have omitted the first oxidation reaction forming sulphenic acid and the reverse reaction, involving the reduction of the oxidized species by the thioredoxin system.
In the FF conformation the CysP and CysR are positioned 13Å apart. Therefore, the formation of a disulphide bond requires structural rearrangement to bring the two residues in close proximity. While the oxidised CysP is still in the active site, it can react with a second H2O2 molecule thereby forming sulfinic acid (Barranco-Medina, Lázaro, & Dietz, 2009). Thus, disulphide bond formation and overoxidation are in continuous competition for determining the fate of the sulphenic acid (–SOH) intermediate. In fact, the only known enzymes to react faster with H2O2 than sulphenic acids are the cellular peroxidases (Peskin et al., 2013). Interestingly, it was suggested that overoxidation is a characteristic selected for during evolution since it occurs predominantly in eukaryotic PRXs due to a feature of the C-terminal tail, which stabilises the FF conformation and retards the rate of unfolding (Wood, 2003).
Initially considered as a ‘dead-end’ form of the enzyme, the sulphinic acid form is now known to be recycled through an ATP-dependent reaction by the reductase sulfiredoxin (Figure 2). However, owing to the slow kinetics of the reduction reaction, over-oxidised forms can persist in the cell for several hours (Woo, 2003). The structural peculiarity of the C-terminal tail, together with the slow rates of regeneration, led some authors to suggest that the controlled inactivation of the enzyme by its own substrate allows H2O2 to act as a second messenger in signal transduction, which has been described as the “flood-gate hypothesis” (Wood et al., 2003). In support of this hypothesis, circadian cycles of overoxidation of mitochondrial PRX3 have been demonstrated to be essential for adrenal steroidogenesis (Kil et al., 2012).
PRX dimers associate via non-covalent interactions into higher order oligomers both in vivo and in vitro (Barranco-Medina et al., 2009; Wood et al., 2003; Wood, Poole, Hantgan, & Karplus, 2002), with the predominant oligomeric species being a decamer possessing putative chaperone activity. Investigations into the dynamics of oligomer formation demonstrated that decamer assembly is governed by the redox status of the monomers, with the fully reduced and overoxidised form associated with oligomer stabilization, whereas the oxidized form promotes disassembly (Figure 3). This is further supported by structural studies showing that the reduced and overoxidised dimers adopt a similar conformation, while the disulphide-linked dimer displays marked differences (Figure 4).
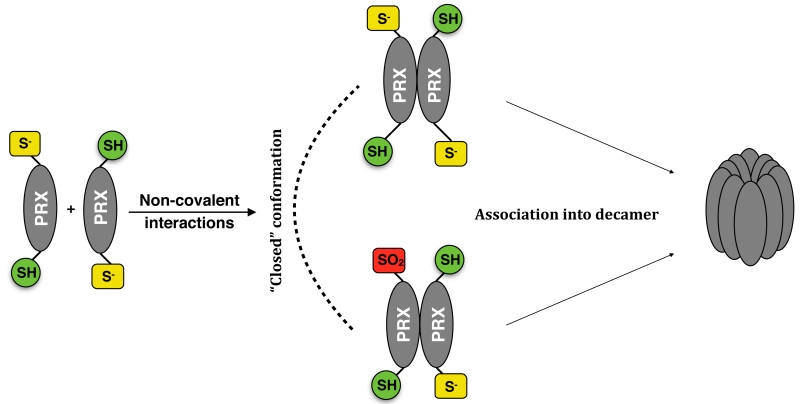
Figure 3.
Peroxiredoxin oligomerization. In addition to dimers, PRXs can form higher order oligomers. The predominant high-molecular species is a decamer, which is proposed to have chaperone function. Structural studies indicate that the decamer is stabilized by dimers in the fully folded conformation – i.e. it is made formed from reduced or overoxidized dimers.
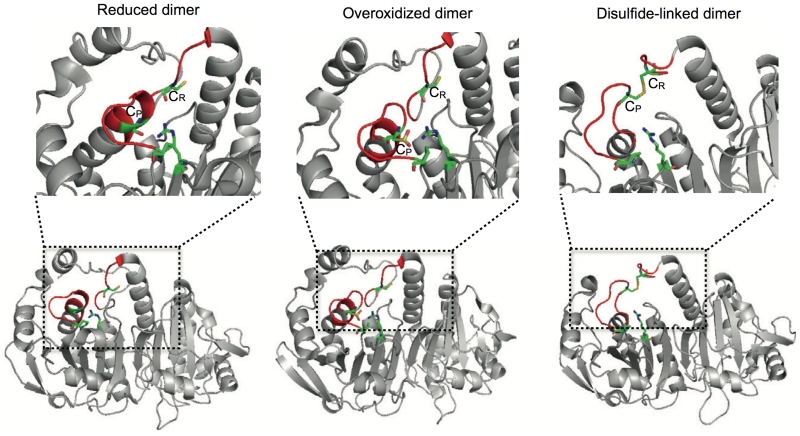
Figure 4.
The structure of the 2-Cys PRX dimer is governed by the oxidation status of the active site cysteine. Lower panel. Reduced (PDB entry 2Z9S), over-oxidized (PDB entry 1QMV) and disulfide-linked (PDB entry 1QQ2) dimers represented in ribbon diagrams. The reduced and over-oxidized dimers assume almost identical conformations, with the active site loop (shown in red) in the fully folded state. Notice that the active site loop in the disulfide-linked dimer is partially unfolded to allow the CR-CP interaction. Top panel. The active sites of the dimers - CysP,CysR and the stabilizing Arg128 (Arg127 for 1QQ2) and Pro45 (Pro44 for 1QQ2) are shown in “sticks” representation, the regions in the active site which undergo significant structural rearrangements to allow disulphide bond formation are coloured in red.
Insights into the molecular mechanism allowed the construction of a model to describe oligomer dynamics (Hall et al., 2011). Briefly, reduced non-covalent dimers in the fully folded conformation associate via non-covalent interactions into a decamer. Oxidation results in structural rearrangements in the dimer, which adopts a locally unfolded conformation, thereby interfering with the dimer-dimer interactions and promoting dissociation of the decamer. However, if the levels of H2O2 are sufficiently high, overoxidation of CysP effectively out-competes disulphide bond formation and stabilizes the decamer. Similar to the floodgate hypothesis, some authors have proposed that when H2O2 levels exceed the antioxidant capacity of the cell, overoxidation acts as a molecular switch that activates the chaperone-like properties of PRXs at the expense of the peroxidase function. The dimer and the decamer are not the only oligomeric forms that PRXs adopt; tetramers and higher order oligomers have also been observed (Barranco-Medina et al., 2009; Hall et al., 2011; Lee et al., 2007).
3. Analysis of PRX Redox Oscillations
The protocols presented below will describe a straightforward method for the measurement of the accumulation of over-oxidised PRX species across circadian time in cultured adherent cells, erythrocytes and flies (Drosophila melanogaster).
3.1 Methodology and important considerations
Owing to their extreme reactivity, PRXs are prone to artifactual oxidation during lysis. There are several methods described in the literature to limit post-lytic oxidation of PRXs and the method we find to be most efficacious is a derivative of the protocol first described by Peskin and colleagues (Cox, Winterbourn, & Hampton, 2010; Peskin et al., 2013). For determination of absolute PRX over-oxidation there are three major steps: (1) all buffers to be used are pre-cleared from adventitious H2O2 by the addition of catalase, (2) the cells are pre-incubated with alkylating reagent to block all free cysteine residues and (3) lysis is performed with low volume of buffer in order to retain relatively high PRX concentration.
The most commonly used alkylating reagents are N-ethylmaleimide (NEM) and iodoacetamide (IAM). The former displays higher reaction rates, less pH dependence and higher specificity, and is thus regarded as the most effective alkylating reagent (Zander, Nikhil, & Bardwell, 1998). PRX alkylation represents one of the greatest ironies in redox biology. Despite their extremely high reactivity with peroxides, PRXs display very low reaction rates with alkylating reagents and even when cells are lysed in the presence of NEM, artifactual oxidation can still occur (Cox, Peskin, Paton, Winterbourn, & Hampton, 2009b; Peskin et al., 2007). This could in part be due to the fact that NEM reacts less effectively with buried or partially exposed thiols (Zander et al., 1998). An essential step to ensure complete alkylation, therefore, is the pre-incubation of cells with alkylating buffer prior to lysis (Cox et al., 2010). It has to be noted that alkylation is not absolutely essential when measuring relative levels of overoxidation across time, since the characteristic overoxidation pattern at different time points would not be affected assuming that lysis conditions remain constant. We have found that excessive handling of the samples such as pre-incubation with NEM can lead to greater variability across time points and thereby result in inconsistent extraction efficiency, especially when working with red blood cells. Moreover, pre-incubation is often impractical when working with whole tissue samples. In order to minimize experimental error we therefore may omit the alkylation step from our protocol depending on the context.
Another method to limit artifactual oxidation includes lysing cells under completely denaturing conditions, for example, in the presence of 2% sodium dodecyl sulfate (SDS), 6 M guanidium chloride, 8 M urea, or strong acids such as trichloroacetic acid (TCA), termed acid quenching (Zander et al., 1998). Cell lysis with TCA in particular has three principal advantages: (1) it leads to a decrease in pH below the pKa of thiols thereby ensuring that all free thiols are protonated and thus not available for reaction; (2) it fully denatures proteins ensuring that enzymatic reduction/oxidation or disulphide exchange will not occur; and (3) metabolites such as H2O2 remain in the supernatant, thus allowing the re-suspension of the protein pellet in H2O2-free buffer. We have employed this method, similarly to others (Cox et al., 2010), but have found that it is not well-suited for processing complex cell/tissue extracts consistently, mainly due to the fact that re-suspension of the resulting protein pellet requires excessive processing of the samples and high detergent concentration. Direct lysis using high SDS, guanidium chloride or urea concentrations are accompanied by other technical issues (protein quantification, protein resolubilization and long-term storage, respectively). It would be desirable if such a denaturing lysis method could be optimized for circadian time course experiments.
When performing a protein time course it is essential that consistency be maintained throughout the protocol. For example, it is important to maintain consistency when re-suspending RBCs to ensure that an equal amount of cells is taken each time (see Protocol). When working with proteins, the time the sample spends at room temperature, and in contact with air, has to be minimized as much as possible. Therefore, the use of flash-freezing procedures is highly recommended when preparing the samples for storage. In our experience with circadian time courses, it is desirable to keep the number of experimentalists to a minimum (ideally one) in order to ensure consistency in sample handling. For obvious reasons this becomes impractical for time courses of several days duration and it is thus important to ensure the agreed protocol is followed without deviation.
Although DTT is routinely employed as a reducing agent in protein biochemistry it should be used with caution and avoided as a constituent of lysis buffers since it is known to catalyze the reduction of molecular oxygen to superoxide in the presence of trace metal ions via Fenton chemistry (Kim, Rhee, & Stadtman, 1985). This chemical cycle results in the continued formation of additional H2O2 post-lysis unless steps are taken to prevent it. Moreover, common metal hexadentate chelators, such as ethylenediaminetetraacetic acid (EDTA), an often-used constituent of lysis buffers, potentiate this reaction. Indeed, the concentration of trace metals is kept negligible in vivo by means of metal storage proteins and metal-binding enzymes, however, upon cell lysis, proteins are denatured and the metal cofactors are released in the form that can participate in Fenton reactions, thereby introducing artifactual oxidation of the sample. This becomes even more important when working with tissues. For instance, the hemolymph of Drosophila melanogaster is a rich source of trace metals. For analysis of redox biochemistry, it is therefore strongly recommended that octadentate ligands, such as diethylene triamine pentaacetic acid (DTPA), are employed in place of EDTA/EGTA, so that any transition metal ions released during lysis are no longer accessible.
We also find that incubation of cell/tissue extracts with standard reductants under non-denaturing conditions cannot be assumed to result in complete reduction of PRX disulphides, possibly due to poor solvent accessibility of this bond in the fully folded protein. For this reason we find it preferable to reduce samples at the final step, following protein determination, when they are diluted into denaturing sample buffer immediately prior to heating samples for SDS-PAGE. For biochemical characterization of PRX activity however, it is generally desirable to perform SDS-PAGE under both reducing and non-reducing conditions (Figure 5).
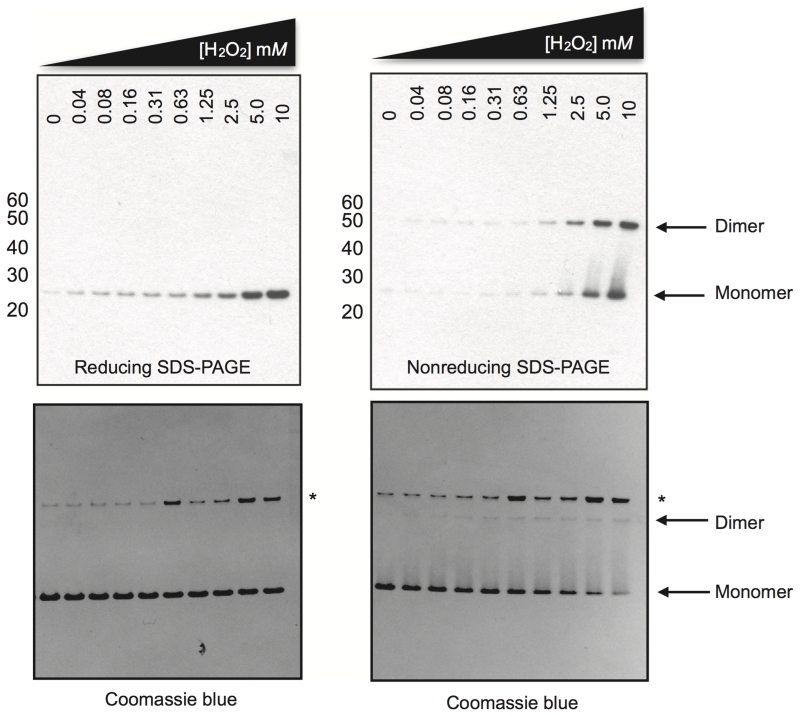
Figure 5.
dPRX1/Jafrac1 behaves like a classic 2-cys peroxiredoxin. Drosophila PRX1/Jafrac1 was expressed, purified and H2O2-treated as in (Peskin et al., 2013) with the following modifications: 50 mM TCEP was used for the initial reduction step prior to desalting, 5 μM recombinant protein was used in a total volume of 50 μL for each concentration, and the incubation time with H2O2 prior to catalase (marked with an asterisk on the SDS-PAGE) addition and NEM treatment was 15 minutes. PRX identity and oxidation state was confirmed by mass spectrometry, antibody activity against the over-oxidized recombinant protein was confirmed by immunopreciptation/mass spectrometry. Marker used: MagicMark XP (Life Technologies).
3.2 Protocol
3.2.1 Sample preparation
Cultured cells
Lysis buffer:
50 mM Tris-HCl pH 7.4
150 mM NaCl
0.20% lithium dodecyl sulfate (LDS) or sodium dodecyl sulphate (SDS)
0.5% sodium deoxycholate
1% Triton X-100
5 mM diethylenetriamine-pentaacetic acid (DTPA)
+ cOmplete Ultra Protease Inhibitor cocktail (Roche).
*All buffers should be pre-treated with 10μg/mL catalase, which is subsequently removed by passage through an Amicon Ultra-15 10K filter (Millipore).
At each time point, one plate is removed from the incubator.
-
Aspirate the medium from each well and wash once with room temperature phosphate-buffered saline (PBS).
Or
(Optional – for determination of absolute PRX oxidation status) Aspirate the medium from each well and replace with ice cold phosphate-buffered saline (PBS) containing 10 mM NEM (freshly diluted into ice cold PBS from 0.5M stock in 100% ethanol). Incubate on ice for 15 minutes.
Aspirate the PBS and replace with ice-cold Lysis buffer. Swirl gently to cover the wells and incubate on ice for 20 minutes. Use cell scrapers to detach cellular debris if necessary and transfer lysate to a new tube.
The lysate is clarified by centrifugation at 21,000 × g for 15 minutes at 4°C in a refrigerated tabletop centrifuge.
When de-frosting, incubate on ice for 30 - 40 minutes to ensure samples are thoroughly defrosted, remove an aliquot and to it add 4xLDS sample buffer (NuPAGE Novex, Life Technologies) to give 1.5X final concentration. For reducing conditions, TCEP should be included and used at a final concentration of 50 mM (diluted from 0.5 M stock, Sigma 646547). Ensure samples are completely mixed.
Transfer the LDS samples to a shaking heatblock at 70°C, shaking at 800 rpm for exactly 10 minutes.
Allow cooling at room temperature for 10 minutes before loading on an SDS-PAGE gel.
Red blood cells
Krebs Buffer: Krebs-Henseleit Buffer (Sigma K3753) supplemented with 2.1g/l sodium bicarbonate, 0.373 g/l calcium chloride dehydrate, 25ug/ml each Pen/Strep, 0.1% BSA. pH is adjusted to 7.4 with NaOH/HCl and osmolarity to 290 mOsm with NaCl.
Red blood cells are isolated from whole blood freshly taken from a human subject using the following protocol:
Note: Warm all solutions to room temperature. Perform steps 1-7 in a tissue culture hood, taking care to maintain best practice as regards aseptic technique. Blood collected from donors who have not been recently screened for transmissible blood born diseases should be treated as potentially hazardous, and the appropriate steps taken to minimize any risk of infection.
Add 5mL Histopaque-1007 to a 15-mL conical centrifuge tube.
Carefully layer 5 mL of whole blood onto the upper gradient of the tube from step 1.
Centrifuge at 700 × g for 15 minutes at room temperature. Carefully remove centrifuge tubes. Aspirate and discard fluid up to the surface of the RBC pellet.
Wash the RBC pellet twice with 10ml PBS, centrifuging at 700 × g for 5 minutes each time. Make up final pellet to 9 ml with Krebs Buffer.
Add 500 μl of anti-CD15 Dynabeads (Life Technologies) resuspended in Krebs Buffer, and place on roller at room temperature for 15 minutes.
Use magnet to pull out Dynabeads. Transfer RBCs to a new tube.
100 μl of RBC suspension are dispensed in sterile PCR tubes, closed, and then tranferred to a thermal cycler (e.g. at 37°C) for the time course.
At each time point, tubes are removed from the PCR machine. RBCs will have pelleted at the bottom of the tube and are re-suspended by pipetting up and down four times (it is important to maintain consistency when re-suspending to ensure that an equal number of cells is sampled at each time point).
Remove exactly 50 μl and transfer it into pre-labelled microcentrifuge tubes containing 650 μl 2X-LDS sample buffer (Life Technologies) supplemented with 5 mM DTPA. Pipette up and down four times and keep at room temperature for 15 minutes.
Place the LDS samples in the heatblock for exactly 10 minutes at 70°C, shaking at 800 rpm. It is important to maintain consistency - do not leave at 70°C for longer than 10 minutes.
Flash-freeze and store samples at −80°C.
When de-frosting, incubate at room temperature for 30 minutes to ensure samples are thoroughly defrosted. For reducing conditions, remove an aliquot from each sample and add reductant to desired concentration then transfer to shaking heatblock at 70°C, shaking at 800 rpm for exactly 10 minutes
Allow cooling at room temperature for 10 minutes before loading on an SDS-PAGE gel.
Drosophila melanogaster
Lysis buffer:
50 mM Tris-HCl pH 7.4
150 mM NaCl,
0.20% lithium dodecyl sulfate (LDS)
0.5. sodium deoxycholate
1% Triton X-100
5 mM diethylenetriamine-pentaacetic acid (DTPA)
+ cOmplete Ultra Protease Inhibitor cocktail (Roche).
*All buffers should be pre-treated with 10 μg/mL catalase, which is subsequently removed by passage through an Amicon Ultra-15 10K filter (Millipore).
At each time point, take one culture vial from the incubator. Flash-freeze and harvest the tissue of interest (e.g. heads, bodies).
Homogenise the tissue on ice in ice-cold Lysis buffer. For optimal extraction, use a motorised homogeniser (e.g. Kontes pellet pestles cordless motor).
The lysate is clarified by centrifugation at 21,000 × g for 15 minutes at 4°C in a refrigerated tabletop centrifuge. Remove supernatant. Clarify supernatant again by centrifugation at 21,000 × g for 15 minutes at 4°C.
Flash-freeze and store samples at −80°C.
When de-frosting, incubate on ice for 30 - 40 minutes to ensure samples are thoroughly defrosted and dilute with 4×LDS sample buffer (NuPAGE Novex, Life Technologies) to a final 1.5× concentration. For reducing conditions, TCEP should be included and used at a final concentration of 50 mM (diluted from 0.5 M stock, Sigma 646547). Ensure samples are completely mixed.
Transfer the LDS samples to shaking heatblock at 70°C, shaking at 800 rpm for exactly 10 minutes.
Allow cooling at room temperature for 10 minutes before loading on an SDS-PAGE gel.
3.2.2 Gel electrophoresis and protein transfer to nitrocellulose membranes for blotting
Before loading on a SDS-PAGE gel, samples are diluted with LDS sample buffer (NuPAGE Novex, Life Technologies) to a final 1.5× concentration. We use 4–12% Bis- Tris gradient gels (NuPAGE Novex, Life Technologies) and run them using non-reducing MES-SDS running buffer (Life Technologies) at 200V constant for 42 minutes. For reducing gels, samples are diluted with LDS sample buffer containing TCEP to give 50 mM final concentration. To ensure that reduced samples are not oxidized during the electrophoresis, 500 μL NuPAGE® Antioxidant (Life Technologies) are added to the running buffer. Other reducing reagents often used in redox biology include the thiol reducing agent dithiothreitol (DTT) and β-mercaptoethanol (βME). We have tested these compounds for their ability to reduce PRX dimers and found 50 mM TCEP to be the most appropriate for the this application since it is resistant to air oxidation and does not reduce transition metal ions. After electrophoresis, the proteins are transferred to nitrocellulose membranes using the iBlot system (Life Technologies) with a standard (P3, 7 min) protocol.
3.2.3 Immunoblotting
For measuring PRX overoxidation we use the commercially available anti-PRX-SO2/3 antibody (Abcam, ab16830). This antibody has been raised against a 9 amino acid sequence, which mimics the over-oxidised active site of 2-Cys PRXs (mammalian Prx1-4) (Woo, 2003). Some investigators have argued that the sulphonic acid modification is an artifact resulting from air oxidation of the samples with time. This does not affect our analysis in any way due to the fact that the antibody has been shown to recognize both forms with equal affinity (Woo, 2003). Since, PRX3 and PRX4 are slightly larger than PRX1 and PRX2, the PRX-SO2/3 monomer appears as a doublet under non-reducing conditions, provided that protein levels are not too high, in which case the two bands merge and appear as one thick band. This also depends on the relative expression levels of different PRXs in the particular cell type. For example, in erythrocytes, where PRX2 is much more highly expressed than the other PRXs, the monomer appears as a single band.
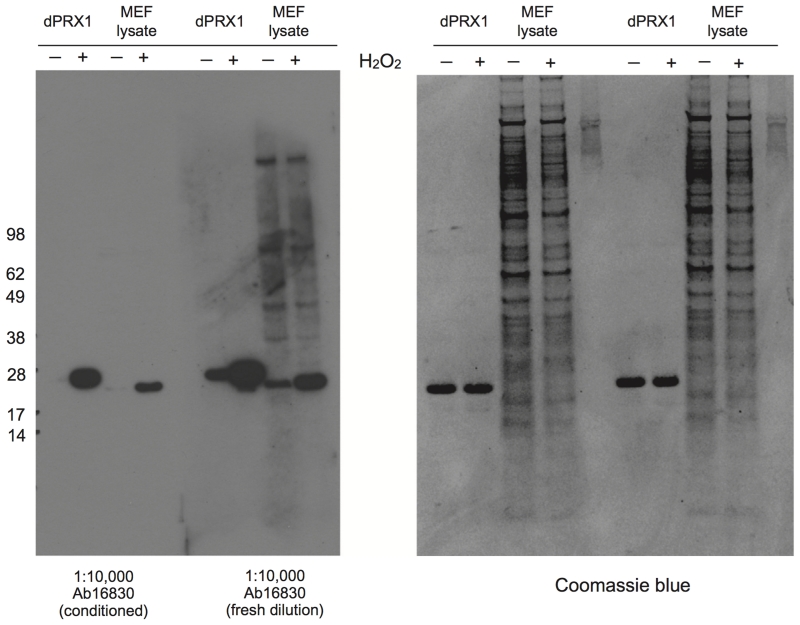
A critical step for improving the quality of the immunoblots is “conditioning” of the polyclonal anti-PRX-SO2/3 antiserum in order to deplete for antibodies that recognize other features of the active site epitope, and are therefore less specific for the oxidative modification in question (see Figure 6 and 7). To condition the commercial antiserum, first make a ‘dummy’ nitrocellulose blot from lysates that are going to be blotted subsequently (e.g. for human cells, use human cell lysate; for Drosophila, use head or body lysate). Then, make up a 1:10,000 dilution of ab16830 + 0.02% sodium azide in blocking buffer and incubate it with the dummy blot, overnight at 4°C on a shaking table. This should be repeated twice more. This antiserum dilution can be reused multiple times and can be stored for several months at 4°C. We further validated this observation by generating a new antiserum against overoxidised PRX-SO2/3 using a more stringent affinity purification and depletion strategy (for details see Figure 8).
Figure 6.
PRX-SO2/3 antibody conditioning is essential for specific epitope recognition and reduced background. 5 μM purified recombinant Drosophila PRX1/Jafrac1 in PBS + 0.1 mM DTPA was treated with 5 mM H2O2 or vehicle for 15 minutes at room temperature prior to reducing SDS-PAGE, in order to generate recombinant positive and negative blotting controls. To generate similar controls in cell lysate mouse embryonic fibroblasts (MEFs) were treated with 2 mM H2O2 or vehicle for 30 minutes prior to treatment with NEM treatment and then lysis, as described in Sample preparation. Reducing SDS-PAGE, immunoblotting and antibody conditioning were performed as described in this document. Note that antibody conditioning reduces background, removes all non-specific banding, and results in immunoblots that recognize over-oxidised PRX exclusively. Marker used: SeeBlue® Plus2 (Life Technologies).
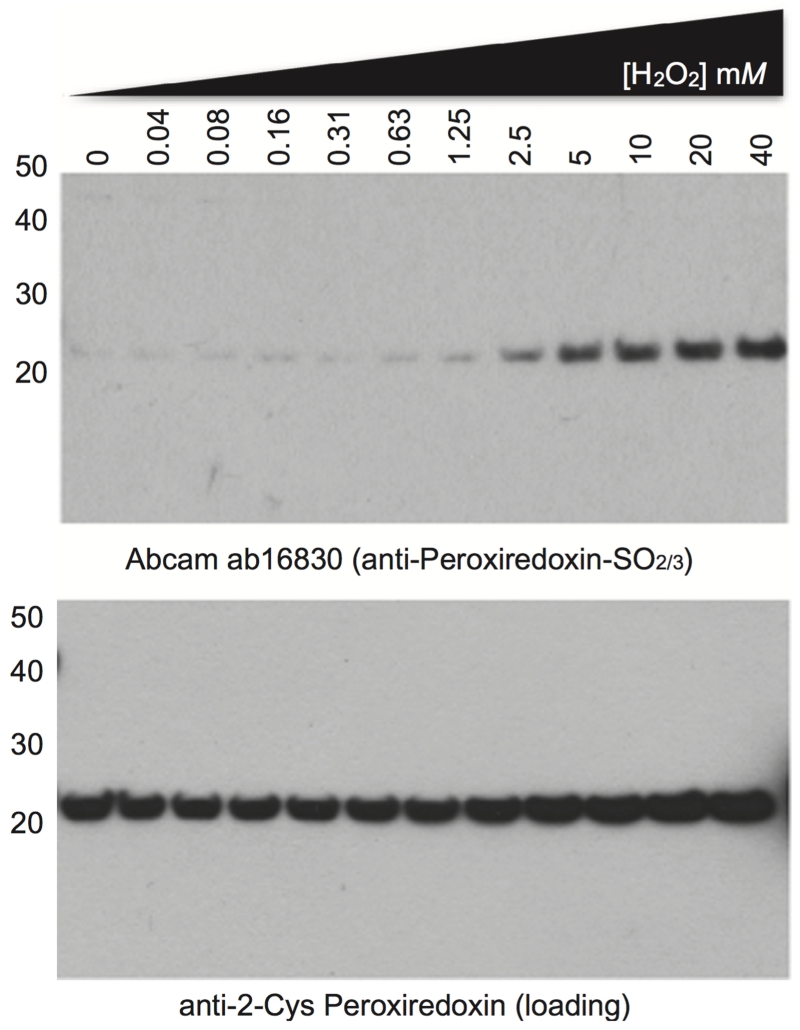
Figure 7.
Antibody specificity can be readily confirmed in Drosophila extract using an in vitro assay for PRX oxidation. Drosophila were lysed and clarified as described in this document. The supernatant was then transferred to a new tube and reduced with 50 mM TCEP for 1 hour at 4°C with gentle agitation. Excess reductant was removed by desalting into PBS+0.1 mM DTPA as described in (Peskin et al. 2013), to give a final concentration of 5 mg/ml total protein. H2O2 treatment were also performed as in Peskin et al. (Peskin et al., 2013), with the exception that a total volume of 50 μL extract was used for each incubation, the incubation time with H2O2 prior to catalase addition and NEM treatment was 15 minutes, and 50 mM TCEP was used for reducing SDS-PAGE. Note that the faint PRX over-oxidation signal in the untreated lane (0 mM) relative to higher H2O2 concentrations (>2.5 mM) indicates that only a small proportion of PRX is over-oxidised under normal conditions in vivo relative to the total PRX pool, consistent with its dynamic redox cycle. Marker used: MagicMark XP (Life Technologies).
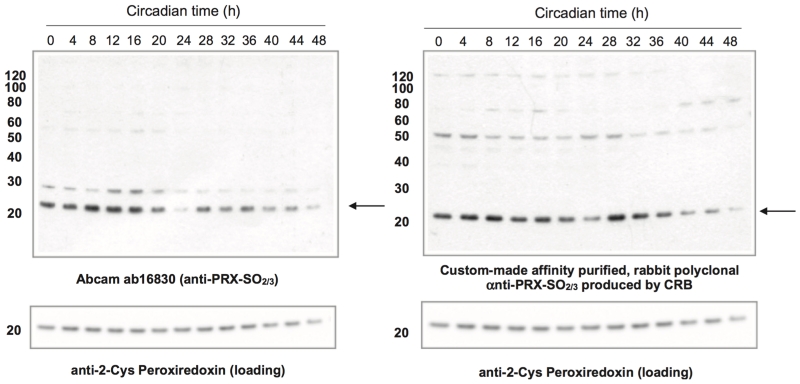
Figure 8.
Peroxiredoxin oxidation cycles in fruit flies. Two independently generated polyclonal antisera against over-oxidised peroxiredoxin (PRX-SO2/3) demonstrate circadian pattern in the over-oxidation of PRX in head lysates of Drosophila melanogaster, sampled under constant dark (DD) conditions (the 2nd and 3rd DD cycles are shown). The custom-made antiserum was generated by Cambridge Research Biochemical (CRB) using a ‘double purification’ approach: The crude serum was first depleted of non-sulfonylated epitopes using a peptide containing serine (–OH) in its (i.e. DFTFVSPTEI) followed by retrieval of antigen-specific antibodies against the antigen peptide (sulfonylated sequence, DFTFV[C-SO3]PTEI). The membranes were re-probed with anti-2-Cys PRX antibody (1:1,000 dilution) as a loading control. Abcam ab16830 and CRB anti-PRX-SO2/3 were used at 1:10, 000 and 1:1,000 dilution, respectively. Marker used: MagicMark XP (Life Technologies).
The nitrocellulose membrane is washed briefly in Tris-buffered saline/0.05% Tween-20 (TBST), and then blocked in 0.5% (w/w) BSA/non-fat dried milk (Marvel) dissolved in TBST for 1h at room temperature. We have determined that centrifugal clarification or syringe filtration (0.45 μM) of the blocking buffer prior to use significantly reduces background, presumably by removing undissolved particles.
Membranes are then incubated with primary antibody diluted in blocking buffer overnight at 4°C.
The following day, the membranes are washed three times for 5 minutes in TBST and then incubated with HRP-conjugated secondary antibody for 1 hour at room temperature.
The membranes are then washed four times for 10 minutes in TBST at room temperature.
Chemiluminescence detection is performed using the Luminata Forte Western Blot reagent (Millipore) for 3-5 minutes at room temperature, although specific exposure times will of course vary dependent on sample type.
Recommended antibody dilutions are provided in Table 1.
Table 1.
Working dilutions of the antibodies used in the protocol
| Antibody | Working dilution |
|---|---|
| Mouse anti–β–actin (Santa Cruz, sc–47778) | 1:5000 in 0.5% BSA/milk |
| Rabbit anti–Peroxiredoxin–SO2/3 (Abeam, ab 16830) | 1:10,000 in 0.5% BSA/milk |
| Mouse anti–2–Cys Peroxiredoxin (Abeam, ab 16765) | 1:1000 in 0.5% BSA/milk |
All antibody dilutions are made up in blocking buffer with 0.02% sodium azide and incubated with a blot overnight at 4 °C on a shaking table.
3.2.4 Normalization and controls
Normalization of PRX levels is crucial for accurate interpretation of the results. The most commonly used method in our laboratory is the re-probing of membranes with anti-β-actin monoclonal antibody. In organisms where no suitable loading control is present (e.g. archaea species), protein levels can be normalized by staining a parallel gel with Coomassie blue. This method is less sensitive but serves as a reasonable alternative. Finally, normalization could be performed with the recently developed anti-2-Cys PRX antibody (Abcam, ab16765), which detects total levels of most 2-Cys PRXs. Normalization with this antibody requires reducing conditions so that a single monomeric band is present (Figure 7 and 8).
When blotting for PRX overoxidation in a new organism or novel context, blotting conditions may need to be optimized empirically. As such, it is essential to run the appropriate positive and negative controls alongside the sample(s) of interest in order to demonstrate antiserum specificity. See Supplementary Figure S7 and S8 of Edgar et al. for an example (Edgar et al., 2012).
3.2.5 Alternative methods for the measuring of PRX overoxidation
Several other methods for measuring PRX overoxidation have been described in the literature. Although somewhat old-fashioned, 2D-PAGE gels still represent one of the most sensitive ways for measuring PRX overoxidation. However, there are two problems associated with this method: (1) each sample has to be run on a separate gel which increases the sample variability and (2) the analysis might be perplexed by isoelectric point (pI) shifts due to other modifications (e.g. PRXs are phosphorylated, glutathionalylated etc.). Cox and colleagues have described an immunoblotting method for measuring PRX overoxidation using antibodies against individual PRX proteins (Cox et al., 2009a; 2010). This method relies on the fact that upon lysis in the absence of alkylating reagents, all reduced PRXs will oxidise and form dimers whereas over-oxidised PRXs will remain as monomers, as their CysP is not available to participate in dimer formation. Using this method they were able to show accumulation of over-oxidised PRX1 and PRX2 monomers in Jurkat cells treated with exogenous H2O2(Cox et al., 2009a). The results were validated with the anti-PRX-SO2/3 antibody. Although this method provides a way of monitoring overoxidation of specific PRXs, the analysis is limited to monomeric species.
Another method involves using the alkylation reagent (iodoacetyl)amino-stilbene-2,2’-disulfonic acid (AMS) (Poynton & Hampton, 2014). Owing to its rather bulky nature (it adds approximately 500 Da per reduced cysteine) this agent could be used to separate oxidized and reduced species using non-reducing SDS-PAGE. Addition of one AMS molecule to the available CysP of the over-oxidised dimer causes it to run with higher apparent mass than the disulfide-linked dimer, whereas the reduced monomer (two AMS molecules) appears larger than the over-oxidised monomer (one AMS molecule). Finally, overoxidation has also been observed by mass spectrometry. Similar to 2D-PAGE gels, however, this method is also limited by the run-to-run variability (Peskin et al., 2013).
3.3 Example: Circadian cycles of PRX oxidation in fruit flies
Drosophila tissue (approx. 40 heads per replicate, per time-point) was homogenised on ice in 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.2% LDS, 0.5% sodium deoxycholate, 1% Triton X-100, 5 mM DTPA + protease inhibitors (Roche). The lysate was clarified for 15 minutes at 4°C at 21,000 × g, the supernatant removed and further clarified for 15 minutes at 4°C at 21,000 × g. Total protein in each sample was quantified using Coomassie staining of SDS-PAGE gels. Blots were performed as described in the protocol above and “conditioned” antiserum was employed for the Abcam (ab16830) antiserum (it was pre-incubated with Drosophila head lysate on nitrocellulose membranes, or re-used at least three times to reduce non-specific banding patterns on the immunoblots) (Figure 8).
Acknowledgements
ABR acknowledges funding from the Wellcome Trust (100333/Z/12/Z), the European Research Council (ERC Starting Grant No. 281348, MetaCLOCK), EMBO Young Investigators Programme and the Lister Institute of Preventative Medicine. JSO’N is supported by the Medical Research Council (MC_UP_1201/4) and the Wellcome Trust (093734/Z/10/Z). GR is supported by an EMBO Long-term Fellowship.
References
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, et al. SIRT1 Regulates Circadian Clock Gene Expression through PER2 Deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Barranco-Medina S, Lázaro J-J, Dietz K-J. The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Letters. 2009;583(12):1809–1816. doi: 10.1016/j.febslet.2009.05.029. [DOI] [PubMed] [Google Scholar]
- Beaver LM, Klichko VI, Chow ES, Kotwica-Rolinska J, Williamson M, Orr WC, et al. Circadian Regulation of Glutathione Levels and Biosynthesis in Drosophila melanogaster. PLoS ONE. 2012;7(11):e50454. doi: 10.1371/journal.pone.0050454.s003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco RA, Ziegler TR, Carlson BA, Cheng P-Y, Park Y, Cotsonis GA, et al. Diurnal variation in glutathione and cysteine redox states in human plasma. The American Journal of Clinical Nutrition. 2007;86(4):1016–1023. doi: 10.1093/ajcn/86.4.1016. [DOI] [PubMed] [Google Scholar]
- Brody S, Harris S. Circadian rhythms in neurospora: spatial differences in pyridine nucleotide levels. Science. 1973;180(4085):498–500. doi: 10.1126/science.180.4085.498. [DOI] [PubMed] [Google Scholar]
- Bryk R, Griffin P, Nathan C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature. 2000;407(6801):211–215. doi: 10.1038/35025109. [DOI] [PubMed] [Google Scholar]
- Circu ML, Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radical Biology and Medicine. 2010;48(6):749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AG, Pearson AG, Pullar JM, Jönsson TJ, Lowther WT, Winterbourn CC, Hampton MB. Mitochondrial peroxiredoxin 3 is more resilient to hyperoxidation than cytoplasmic peroxiredoxins. Biochemical Journal. 2009a;421(1):51–58. doi: 10.1016/S0969-2126(00)00147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AG, Peskin AV, Paton LN, Winterbourn CC, Hampton MB. Redox Potential and Peroxide Reactivity of Human Peroxiredoxin 3. Biochemistry. 2009b;48(27):6495–6501. doi: 10.1021/bi900558g. [DOI] [PubMed] [Google Scholar]
- Cox AG, Winterbourn CC, Hampton MB. Measuring the Redox State of Cellular Peroxiredoxins by Immunoblotting. Thiol Redox Transitions in Cell Signaling, Part B. 1st ed. Vol. 474. Elsevier Inc.; 2010. pp. 51–66. [DOI] [PubMed] [Google Scholar]
- Crowe SA, Døssing LN, Beukes NJ, Bau M, Kruger SJ, Frei R, Canfield DE. Atmospheric oxygenation three billion years ago. Nature. 2014;501(7468):535–538. doi: 10.1038/nature12426. [DOI] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485(7399):459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui MY, Ahmed AE. Circadian periodicity of tissue glutathione and its relationship with lipid peroxidation in rats. Life Sci. 1984;34(24):2413–2418. doi: 10.1016/0024-3205(84)90430-2. [DOI] [PubMed] [Google Scholar]
- Flohé L, Harris JR. Peroxiredoxin Systems: Structures and Fuctions. Vol. 44. Springer; Netherlands: 2007. Series: Suncellular Biochemistry. [Google Scholar]
- Gupta N, Ragsdale SW. Thiol-disulfide Redox Dependence of Heme Binding and Heme Ligand Switching in Nuclear Hormone Receptor Rev-erb. Journal of Biological Chemistry. 2011;286(6):4392–4403. doi: 10.1074/jbc.M110.193466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Nelson K, Poole LB, Karplus PA. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxidants & Redox Signaling. 2011;15(3):795–815. doi: 10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J, Cho S, Sassone-Corsi P. Circadian control by the reduction/oxidation pathway: catalase represses light-dependent clock gene expression in the zebrafish. Proceedings of the National Academy of Sciences. 2007;104(40):15747–15752. doi: 10.1073/pnas.0705614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: a component of the circadian clock senses redox state of the cell. The EMBO Journal. 2005;24(6):1202–1210. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky YG, Kosenko EA, Kondrashova MN. Analysis of the circadian rhythm in energy metabolism of rat liver. International journal of Biochemistry. 1984;16(6):629–639. doi: 10.1016/0020-711x(84)90032-6. [DOI] [PubMed] [Google Scholar]
- Kil IS, Lee SK, Ryu KW, Woo HA, Hu M-C, Bae SH, Rhee SG. Feedback Control of Adrenal Steroidogenesis via H. Molecular Cell. 2012;46(5):584–594. doi: 10.1016/j.molcel.2012.05.030. [DOI] [PubMed] [Google Scholar]
- Kim K, Rhee SG, Stadtman ER. Nonenzymatic cleavage of proteins by reactive oxygen species generated by dithiothreitol and iron. Journal of Biological Chemistry. 1985;260(29):15394–15397. [PubMed] [Google Scholar]
- Kondratov RV. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes & Development. 2006;20(14):1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konhauser KO, Lalonde SV, Planavsky NJ, Pecoits E, Lyons TW, Mojzsis SJ, et al. Aerobic bacterial pyrite oxidation and acid rock drainage during the Great Oxidation Event. Nature. 2012;478(7369):369–373. doi: 10.1038/nature10511. [DOI] [PubMed] [Google Scholar]
- Krishnan N, Davis AJ, Giebultowicz JM. Circadian regulation of response to oxidative stress in Drosophila melanogaster. Biochemical and Biophysical Research Communications. 2008;374(2):299–303. doi: 10.1016/j.bbrc.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Choi KS, Riddell J, Ip C, Ghosh D, Park JH, Park YM. Human Peroxiredoxin 1 and 2 Are Not Duplicate Proteins: THE UNIQUE PRESENCE OF CYS83 IN Prx1 UNDERSCORES THE STRUCTURAL AND FUNCTIONAL DIFFERENCES BETWEEN Prx1 AND Prx2. Journal of Biological Chemistry. 2007;282(30):22011–22022. doi: 10.1074/jbc.M610330200. [DOI] [PubMed] [Google Scholar]
- Manevich Y, Feinstein SI, Fisher AB. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with ΠGST. Proceedings of the National Academy of Sciences. 2004;101(11):3780–3785. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiek ES, Lim MM, Yang G, Bauer AQ, Qi L, Lee Y, et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. Journal of Clinical Investigation. 2013;123(12):5389–5400. doi: 10.1172/JCI70317DS1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469(7331):498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget F-Y, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469(7331):554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee S, Lee S, Kang SW. 2-Cys Peroxiredoxins: Emerging Hubs Determining Redox Dependency of Mammalian Signaling Networks. International Journal of Cell Biology. 2014;2014(44):1–10. doi: 10.1042/BJ20130030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskin AV, Dickerhof N, Poynton RA, Paton LN, Pace PE, Hampton MB, Winterbourn CC. Hyperoxidation of Peroxiredoxins 2 and 3: RATE CONSTANTS FOR THE REACTIONS OF THE SULFENIC ACID OF THE PEROXIDATIC CYSTEINE. Journal of Biological Chemistry. 2013;288(20):14170–14177. doi: 10.1074/jbc.M113.460881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskin AV, Low FM, Paton LN, Maghzal GJ, Hampton MB, Winterbourn CC. The High Reactivity of Peroxiredoxin 2 with H2O2 Is Not Reflected in Its Reaction with Other Oxidants and Thiol Reagents. Journal of Biological Chemistry. 2007;282(16):11885–11892. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms. 1997;12(3):204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- Poynton RA, Hampton MB. Peroxiredoxins as biomarkers of oxidative stress. Biochimica et Biophysica Acta. BBA - General Subjects. 2014;1840(2):906–912. doi: 10.1016/j.bbagen.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Radha E, Hill TD, Rao GH, White JG. Glutathione levels in human platelets display a circadian rhythm in vitro. Thrombosis Research. 1985;40(6):823–831. doi: 10.1016/0049-3848(85)90319-6. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009 doi: 10.1126/science.1171641. al, E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall LM, Ferrer-Sueta G, Denicola A. Peroxiredoxins as Preferential Targets in H. Hydrogen Peroxide and cell signaling, Part B. 1st ed. Vol. 527. Elsevier Inc.; 2013. pp. 41–63. [DOI] [PubMed] [Google Scholar]
- Reddy AB, Rey G. Metabolic and Nontranscriptional Circadian Clocks: Eukaryotes. Annual Review of Biochemistry. 2014;83(1):165–189. doi: 10.1146/annurev-biochem-060713-035623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SG, Woo HA, Kil IS, Bae SH. Peroxiredoxin Functions as a Peroxidase and a Regulator and Sensor of Local Peroxides. Journal of Biological Chemistry. 2012;287(7):4403–4410. doi: 10.1074/jbc.R111.283432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J. Regulation of Clock and NPAS2 DNA Binding by the Redox State of NAD Cofactors. Science. 2001;293(5529):510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- Scheer FAJL, Michelson AD, Frelinger AL, Evoniuk H, Kelly EE, McCarthy M, et al. The Human Endogenous Circadian System Causes Greatest Platelet Activation during the Biological Morning Independent of Behaviors. PLoS ONE. 2011;6(9):e24549. doi: 10.1371/journal.pone.0024549.t001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangherlin A, Reddy AB. Regulation of circadian clocks by redox homeostasis. Journal of Biological Chemistry. 2013 doi: 10.1074/jbc.R113.457564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E, Frosch S. Cycles in plants. Journal of Interdisiplinary Cycle Research. 1974;5(3-4):231–239. doi: 10.1080/09291017409359428. [DOI] [Google Scholar]
- Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, Coleman TP, et al. Circadian Rhythm of Redox State Regulates Excitability in Suprachiasmatic Nucleus Neurons. Science. 2012;337(6096):839–842. doi: 10.1126/science.1222826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HA, Kang S, Kim HK, Yang K, Chae HZ, Rhee SG. Reversible Oxidation of the Active Site Cysteine of Peroxiredoxins to Cysteine Sulfinic Acid: IMMUNOBLOT DETECTION WITH ANTIBODIES SPECIFIC FOR THE HYPEROXIDIZED CYSTEINE-CONTAINING SEQUENCE. Journal of Biological Chemistry. 2003;278(48):47361–47364. doi: 10.1074/jbc.C300428200. [DOI] [PubMed] [Google Scholar]
- Wood ZA. Peroxiredoxin Evolution and the Regulation of Hydrogen Peroxide Signaling. Science. 2003;300(5619):650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- Wood ZA, Poole LB, Hantgan RR, Karplus PA. Dimers to Doughnuts: Redox-Sensitive Oligomerization of 2-Cysteine Peroxiredoxins †,‡. Biochemistry. 2002;41(17):5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- Wood ZA, Schröder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends in Biochemical Sciences. 2003;28(1):32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- Yang KS. Inactivation of Human Peroxiredoxin I during Catalysis as the Result of the Oxidation of the Catalytic Site Cysteine to Cysteine-sulfinic Acid. Journal of Biological Chemistry. 2002;277(41):38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- Zander T, Nikhil P, Bardwell J. Methods in Enzymalogy. Vol. 290. Academic Press; 1998. Disulfide Bond Catalysis in E. coli. [DOI] [PubMed] [Google Scholar]