Abstract
BACKGROUND
Black women are at greater risk for peripartum cardiomyopathy (PPCM). The guanine nucleotide-binding proteins beta-3 subunit (GNB3) has a polymorphism C825T. The GNB3 TT genotype more prevalent in blacks is associated with poorer outcomes. We evaluated GNB3 genotype and myocardial recovery in PPCM.
METHODS AND RESULTS
97 women with PPCM were enrolled and genotyped for the GNB3 T/C polymorphism. Left ventricular ejection fraction (LVEF) was assessed by echocardiography at entry, 6 and 12 months postpartum. LVEF over time in subjects with the GNB3 TT genotype was compared to those with the C allele overall and in black and white subsets. The cohort was 30% black, age 30 + 6, LVEF 0.34 + 0.10 at entry 31+25 days post-partum. The % GNB3 genotype for TT/CT/CC =23/41/36 and differed markedly by race (Blacks= 52/38/10 versus Whites=10/44/46, p<0.001). In subjects with the TT genotype, LVEF at entry was lower (TT= 0.31+ 0.09; CT+CC= 0.35+0.09, p=0.054) and this difference increased at 6 (TT=0.45+ 0.15; CT+CC= 0.53+0.08, p=0.002), and 12 months (TT=0.45+ 0.15; CT+CC= 0.56+ 0.07, p<0.001.). The difference in LVEF at 12 months by genotype was most pronounced in blacks (12 months LVEF for GNB3 TT=0.39+ 0.16; versus CT+CC= 0.53+ 0.09, p=0.02) but evident in whites (TT=0.50++0.11; CT+CC= 0.56+0.06, p=0.04).
CONCLUSIONS
The GNB3 TT genotype was associated with lower LVEF at 6 and 12 months in women with PPCM, and this was particularly evident in blacks. Racial differences in the prevalence and impact of GNB3 TT may contribute to poorer outcomes in black women with PPCM.
Keywords: peripartum, polymorphism genetics, cardiomyopathy, recovery, remodeling heart failure, pregnancy
Peripartum cardiomyopathy (PPCM) is a rare complication of pregnancy which remains a significant cause of maternal morbidity and mortality. The prevalence in the Unites States is approximately 1 in 2000 to 1 in 4000 live births (1–3), the incidence varies based on patient geography, patient ethnicity, and the challenges of global population based studies (4–6). While the exact etiology remains uncertain, several hypotheses have been proposed, with immunologic/inflammatory, vascular and genetic factors potentially playing significant roles (7–9).
The prevalence of PPCM is highest globally in Haiti and parts of Africa and in the United States among blacks, suggesting that African genomic ancestry may be a risk factor for development of PPCM (2). Clinical outcomes in women with PPCM vary substantially. Recovery of left ventricular function appears to be poorer among blacks than in comparable white cohorts (10,11). However, whether genomic variation underlies apparent racial differences in clinical outcomes in PPCM has not been previously investigated.
The guanine nucleotide-binding protein beta-3 subunit (GNB3) has a polymorphism of exon-10, C825T. The T allele is associated with enhanced alpha2-adrenergic receptor intracellular signaling (12), and an increased risk of hypertension (13,14), low plasma renin (15), and potentially cardiac remodeling (16). The T allele has a much higher prevalence in black cohorts than in whites (17). Roughly 50% of black subjects have the GNB3 TT genotype, compared to only 10% of whites. In the genetic sub-study of AHeFT, the African American Heart Failure Trial, blacks with the GNB3 TT genotype had the poorest outcome on placebo and received the greatest benefit from therapy (18). Despite clear racial differences in outcomes with PPCM, the impact of GNB3 TT genotype on myocardial recovery has not been previously examined.
The Peripartum Cardiomyopathy Network (PCN) was formed as a thirty center collaborative group to facilitate research in this disorder and recently completed the Investigations of Pregnancy Associated Cardiomyopathy (IPAC) study. IPAC was a multi-center prospective investigation of the genetic and clinical predictors of outcomes for PPCM patients in North America and reported significantly less myocardial recovery among black women with PPCM when compared to whites (11). Given the distinct differences in allele frequency for the GNB3 C825T polymorphism between black and white cohorts and the impact of GNB3 TT genotype in black subjects in AHeFT, we examined the relationship of the GNB3 genotype to myocardial recovery in the IPAC cohort.
METHODS
Cohort
Ninety seven women with newly diagnosed PPCM were enrolled within the first thirteen weeks postpartum at 30 centers (appendix) between December 2009 and September 2012. All women were at least 18 years of age and had no previous history of cardiac disease, an estimated clinical LVEF of < 0.45 at the time of enrollment, and an evaluation consistent with idiopathic non-ischemic cardiomyopathy. Women with significant valvular disease, coronary disease (>50% stenosis of a major epicardial vessel or a positive non-invasive study), evidence of ongoing bacterial septicemia (positive blood cultures), ongoing drug or alcohol abuse, history of chemotherapy or chest radiation with 5 years of enrollment, or a history of a previous cardiomyopathy were excluded.
Protocol
The study protocol was approved by the institutional review boards at all participating centers, and informed consent was obtained from all subjects. At the time of enrollment demographic information including self-designated race, previous clinical evaluation and current medical therapy were recorded. Women were followed until one year post-partum. All hospitalizations and major cardiac events including death, cardiac transplantation, or implantation of a left ventricular assist device (LVAD) were recorded.
Left Ventricular Function
All subjects had an echocardiogram to assess left ventricular ejection fraction at the time of enrollment which was repeated at 6 and 12 months post-partum. In addition, women enrolled early (within the six weeks postpartum, n=65) had a repeat assessment of left ventricular function at 2 months. Echocardiograms were reviewed by a core laboratory at the University of Pittsburgh for assessment of ventricular volumes and calculation of ejection fraction. The left ventricular (LV) volumes and EF were assessed by biplane Simpson’s rule using manual tracing of digital images. Left ventricular end-diastolic diameter (LVEDD) was assessed in the parasternal long-axis view. A subset of echocardiograms obtained were not available for assessment by the core laboratory due to format (22 of 310, 7%) and for these studies the left ventricular ejection fraction calculated locally was used.
DNA isolation and genotype assays
Peripheral blood was obtained from subjects at the time of entry and shipped overnight at room temperature to the core laboratory (University of Pittsburgh). DNA was isolated from peripheral blood by leukocyte centrifugation and cell lysis using the PureGene DNA purification kit (Gentra Systems, Minneapolis, Minnesota). The guanine nucleotide binding protein (G-protein) beta polypeptide-3 (GNB3) position 825 C/T polymorphism was assessed using a TaqMan SNP Genotyping Assay (Applied Biosystems, Inc., Foster City, California) with tagged primers (reporter 1 tagged dye ¼ VIC; reporter 2 tagged dye ¼ FAM). Context sequence for the GNB3 825 C/T polymorphism was as follows: AGAGCATCATCTGCGGCATCACGTC[C/T] GTGGCCTTCTCCCTCAGTGGCCG CC. Products were read using the Applied Biosystems 7000 (ABI).
Statistical and Genetic Analysis
All statistical analyses were performed in SPSS v22. Continuous variables (eg. age, blood pressure) are reported as means and standard deviations. Student t tests and Fisher exact tests were used to compare subject characteristics by genotype. Non-normally distributed variables (eg. days postpartum, gravida and para) are summarized as median and 25th and 75th percentiles, and compared by non-parametric Wilcoxon rank sum tests. The impact of GNB3 genotype on LVEF during recovery (defined as LVEF at 12 months) was first examined by one way ANOVA with Tukey’s post hoc tests for pair wise comparisons. Since we had LVEF measured at multiple time points, we performed a repeated measures ANOVA (using the repeated measures under GLM option). LVEF at baseline, 6 months and 12 months were entered as repeated measures outcome, genotype (TT vs. C allele) and LVEDD were entered as factor and covariate respectively. In multivariable models, LVEDD and race are significant predictors of LVEF recovery in this cohort (11). Finally, we used ANCOVA to evaluate the impact of genotype on LVEF over time adjusting for these two variables. Since allele and genotype frequencies for this genotype differ significantly between whites and blacks, race-specific analyses were done in white and black subsets. These analyses compared TT genotype carriers to C allele (TC and CC genotype subjects combined) carriers in hierarchical linear regression models. Hierarchical linear regression is a specific analytical approach in which predictors are entered successively in each step. The regression model built at each step is related but separate from the immediately previous and subsequent models. This allows for comparison of related regression models within a single framework. This technique helps to quantify the predictive power of a variable while accounting for other covariates in the model. LVEDD was entered in the first step and genotypes were entered in the second step.
The Kaplan-Meier method was used to estimate survival free from events (cardiac transplantation or the need for mechanical circulatory support) with days postpartum as the time interval (starting at delivery). Event free survival of subjects with the GNB3 TT genotype was compared to those with the C allele by the exact log-rank test.
RESULTS
Sample Characteristics
Of the cohort of 97 women, 30% (n= 29) were black, 65% were white (n=63), and 5% other (n=5). Seventy percent were NYHA Class II–III, and over 80% of patients were on ACE inhibitor and beta blocker therapy. Baseline LVEF for these patients was 0.34± 0.09. For the entire cohort women were enrolled at a median of 24 days post-partum (25th percentile, 75th percentile= 11.5, 51 days). Sixty five women were enrolled early (range 0 to 42 days post-partum, median 14 days) while 32 were enrolled late (range 43 to 95 days, median 59).
Genetic analysis of GNB3 C635T in IPAC
For the GNB3 T/C polymorphism, 22 (23%) subjects were TT genotype, 40 (41%) TC, and 35 (36%) CC. Genotype differed markedly by race. In black women, 15 (52%) were GNB3 TT, 11 (38%) TC and 3 (10%) CC, while in white subjects TT/CT/CC= 6(10%) / 28(44%) / 29 (46%). The SNP was in Hardy Weinberg Equilibrium in whites (χ2 = 0.013, P = 0.91), in blacks (χ2 = 0.21, P = 0.65) and in the combined race cohort (χ2 = 2.49, P = 0.11).
The clinical characteristics for the cohort overall and by GNB3 genotype are listed in the Table. Comparing subjects with the GNB3 TT genotype to those with the C allele (TC and CC combined), there was no significant difference in age, New York Heart Association (NYHA) class, heart rate, body mass index (BMI), gravida or para. There was no significant difference in days postpartum to study entry by GNB3 genotype overall (GNB3 TT genotype median=34.5 days (25th percentile, 75th percentile= 14, 63), C allele=24 days (10, 49), p=0.17). In addition there was no significant difference in days postpartum to entry by GNB3 genotype either within the subset of black women (GNB3 TT subjects median 51 days (22, 72) vs C allele median=33.5 days (22, 54), p=0.53) or in the subset of white women (GNB3TT median=21 day (6.5, 42) vs C allele median=19 days (9, 45), p=0.76). As expected, there were a significantly higher percentage of blacks subjects in the TT subset compared to those with the C allele (68% versus 19%, p<0.001). The mean systolic and diastolic blood pressures were 4 points higher in GNB3 TT subjects but both failed to reach statistical significance.
TABLE.
Clinical Characteristics by GNB3 Genotype
| All (N=97) |
TT (N=22) |
TC+CC (N=75) |
p value* | |
|---|---|---|---|---|
| Age (years) | 30 ± 6 | 28 ± 7 | 30 ± 6 | 0.15 |
| Gravida | 2 (1,4) | 2.5 (1,4) | 2 (1,4) | 0.93 |
| Para | 2 (1,3) | 3 (1,3) | 2 (1,3) | 0.38 |
| Race, % Black | 30 | 68 | 19 | <0.001 |
| Days post-Partum | 24 (11,51) | 34.5 (14,63) | 24 (10,49) | 0.17 |
| BMI | 29 ± 7 | 30 ± 7 | 29 ± 8 | 0.42 |
| NYHA Class (% I/II/III/IV) | 12/45/25/18 | 18/27/36/18 | 11/51/21/17 | 0.22 |
| HR | 86 ± 16 | 86 ± 15 | 86 ± 17 | 0.94 |
| BP systolic | 112 ± 17 | 115 ± 19 | 111 ± 17 | 0.35 |
| BP diastolic | 70 ± 13 | 73 ± 13 | 69 ± 13 | 0.20 |
| ACE Inhibitor (%) | 81 | 91 | 79 | 0.16 |
| Aldosterone receptor antagonist | 27 | 32 | 25 | 0.36 |
| Beta Blocker (%) | 89 | 96 | 87 | 0.23 |
Comparison of women with the GNB3 TT genotype to those with the GNB3 C allele. BMI=body mass index, HR = heart rate, BP= blood pressure, NYHA=New York Heart Association, ACE=angiotensin converting enzyme. For gravida, para and days post-partum values listed are the median (with 25th and 75th percentiles). All other values are mean and standard deviation. There was a significantly higher percentage of black women in the subset with the GNB3 TT genotype (68%) than in the subset with the C allele (19%), p<0.001. No other significant differences.
There was no significant difference in the % of subjects on ACE inhibitors at study entry by genotype (TABLE: GNB3 TT subjects 91%, C allele=79%, p=0.16). The predominant ACE inhibitor utilized was lisinopril which was used by 62% of subjects overall, and there was no significant difference in the median daily dose by genotype at entry (TT =10 mg/day (25th, 75th=5, 17.5), C allele=5 mg/day (3, 10), p=0.39. The ACE inhibitor dose was slightly higher among GNB3 TT subjects at 12 months postpartum (TT=10 mg/day (5, 20), C allele=5mg/day (2.5, 10), p=0.03). There was no significant difference in the % of subjects on beta blockers at study entry by genotype (TABLE: GNB3 TT subjects 96%, C allele=87%, p=0.23). The predominant beta blocker utilized was carvedilol which was used by 69% of subjects overall and there was no significant difference in the median daily dose by genotype either at entry (TT=12.5 mg/day (6.25, 12.5), C allele=12.5mg/day (6.25, 25), p=0.20) or at 12 months postpartum (TT=12.5mg/day (9.375, 37.5), C allele=25 mg/day (20, 50), p=0.10).
For analysis of LVEF over time by all three GNB3 genotypes, LVEF did not differ by genotype at entry but did by an overall ANOVA at 6 (p=0.007) and 12 months (p<0.001) (Figure 1A). Subjects with the GNB3 TT genotype demonstrated a significantly lower LVEF by from GNB3 CT subjects at 6 (p=0.007) and 12 months (p=0.001) and from GNB3 CC subjects at 6 (p=0.02) and 12 months (p=0.008). Tukey’s post hoc analysis confirmed subjects with the GNB3 TT genotype demonstrated a significantly lower LVEF from CT subjects at 6 (p=0.009) and 12 months, (p<0.0001) and from CC subjects at 6 (p=0.02) and 12 months (p=0.004). However, the mean LVEF for TC and CC subjects was similar and did not differ at either 6 month (p=0.61) or 12 month (p=0.22) follow up. The association between myocardial recovery (defined as LVEF at 12 months) and GNB3 genotypes persisted after adjustment for race and LVEDD (F= 7.04, df = 2, P = 0.002). Pairwise contrasts showed that TT genotype carriers had significantly lower adjusted LVEF than CT (P = 0.002) and CC (0.005) groups. Given their similarities in terms of mean LVEF over time, CC and TC genotype carriers were combined into a single “C allele” group and compared to GNB3 TT subjects
Figure 1. LVEF over time (entry, 6 and 12 months post-partum) by GNB3 Genotype.
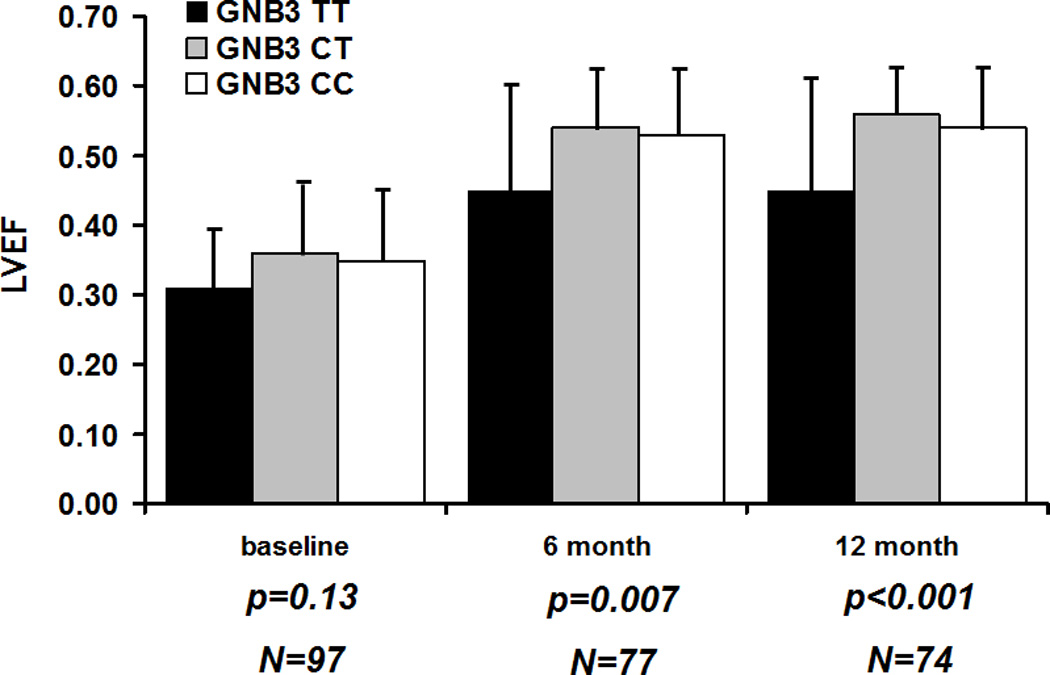
A. Comparison of three genotype classes: Women homozygous for GNB3 T (black bars), heterozygous women (gray bars) and those homozygous of the C allele (white bars). Lines extending from bars represent standard deviations. P values represent an analysis of variance (ANOVA) of an overall difference between the three genotype classes. No significant difference at entry (p value=0.13) but a significantly lower mean LVEF for GNB3 TT subjects at 6 (p=0.007) and 12 months (p<0.001).
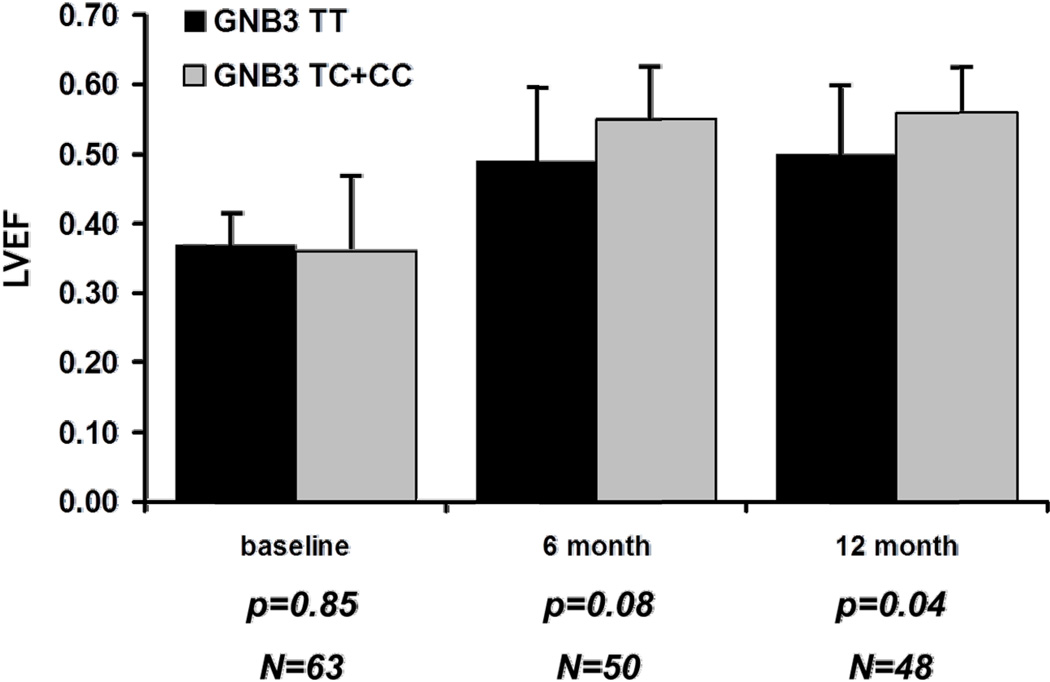
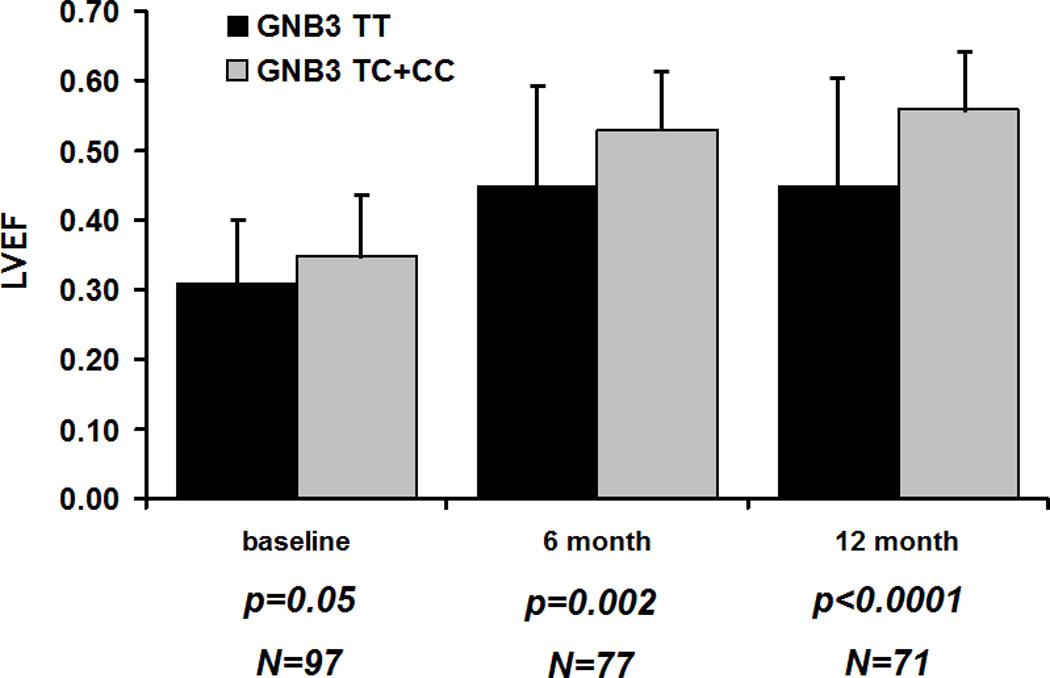
B. Same comparison but combining heterozygous subjects and those homozygous for the C allele into one group. Subjects homozygous for GNB3 T (black bars) compared to subjects with the C allele (gray bars). Not statistically significant at entry (p value=0.054) but a significantly lower mean LVEF for GNB3 TT subjects at 6 (p=0.002) and 12 months (p<0.0001).
The predominant difference in LVEF by genotype was between subjects homozygous for the GNB3 T allele (GNB3 TT) compared to subjects with the GNB3 C allele. When comparing LVEF over time specifically between subjects with the GNB3 TT genotype versus those with the C allele, subjects enrolled early post-partum had similar mean LVEF regardless of genotype (TT= 0.35± 0.07; C allele= 0.35±0.09, p=0.98), however significant differences were apparent at 2 months (TT=0.38± 0.15; C allele= 0.44± 0.10, p=0.02), 6 months (TT=0.45± 0.15; C allele= 0.53± 0.08, p=0.002), and 12 months (TT=0.45± 0.15; C allele= 0.56± 0.07, p<0.0001.). For the entire cohort at the time of entry (combining women enrolled early and late), the mean LVEF for GNB3 TT subjects was slightly lower at entry though this failed to reach significance (p=0.054). The differences in LVEF by GNB3 genotype became more pronounced at 6 months and 12 months (Figure 1B). These differences persisted when the model was further adjusted for race and baseline LVEDD at 6 months (P = 0.011) and 12 months (P <0.00001). With repeated measures data assuming four time points (entry, 2 months, 6 months, 12 months) with genotype (TT vs. C allele carriers) and LVEDD as predictors, results showed that TT genotype was a significant negative predictor of LVEF recovery over time (F= 15.3; P <0.0001).
In subset analysis by race, black women with the GNB3 TT genotype had a significantly lower mean LVEF at entry compared to C allele carriers (0.28±0.09 vs. 0.35± 0.08; p=0.04), and this difference increased to14 EF units at 12 months (12 months LVEF TT=0.39± 0.16; C allele= 0.53± 0.09, p=0.02, Figure 2A). In white women, the mean LVEF for GNB3 TT subjects was similar to C allele carriers at entry (GNB3 TT=0.37± 0.040; C allele=36± 0.10; p = 0.85), but significantly lower at 12 months (0.50 ± 0.11 vs. 0.56 ± 0.06; p= 0.04, Figure 2B). In hierarchical regression models adjusted for baseline LVEDD, presence of the TT genotype was associated with 6.2% lower LVEF recovery in blacks (β (SE) = −6.2 (2.8); P = 0.03) and the genotype explained only 10% of variation in this race. In whites the TT genotype corresponded to a 9.7% lower LVEF recovery (β (SE) = −9.7 (4.4); P = 0.04) and explained 53% of variation.
Figure 2. Subset analysis by race for LVEF over time by GNB3 Genotype.
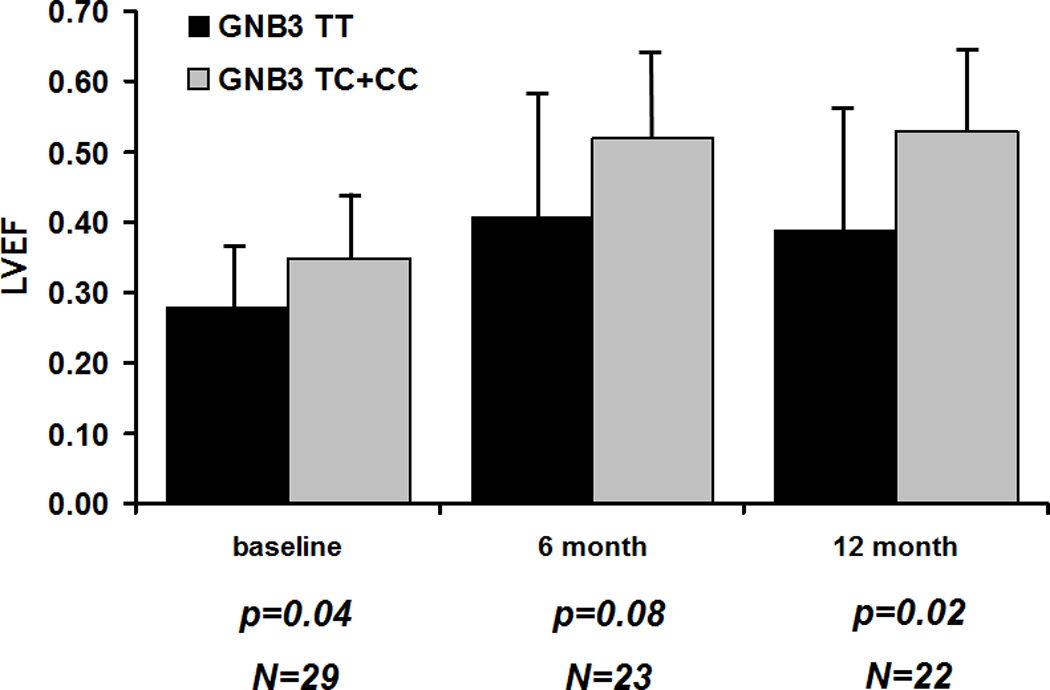
A. Black Women (n=29): Women homozygous for GNB3 TT (black bars) compared to women carrying the C allele (heterozygous subjects and women homozygous for the C allele combined, gray bars). Significantly lower LVEF for GNB3 TT subjects at entry (p value=0.04), which increases to 14 EF units by 12 months (p=0.02).
B. White women (n=63): Women homozygous for GNB3 TT (black bars) with subjects carrying the C allele (gray bars). No significant difference in LVEF at entry (p value=0.04), but a significantly lower LVEF in women with the GNB3 TT genotype by 12 months (p=0.04).
Overall 71% of women recovered to a final LVEF of ≥0.50. Recovery to this degree was significantly less likely overall in GNB3 TT subjects than in those with the GNB3 C allele (TT=48% versus C allele=78%, p=0.01). Subset analysis by race revealed similar trends in both white women (final LVEF ≥ 0.50 for GNB3 TT=50% versus C allele=81%, p=0.12) and black women (TT=43% versus C allele=77%, p=0.12). In addition, analysis of NYHA functional class by GNB3 genotype revealed no significant difference at study entry (Table, p=0.22) but a significant difference at 12 month follow up with higher NYHA class in GNB3 TT subjects (% NYHA class I/II/III/IV at 12 months GNB3 TT subject=35/41/18/6 versus the C allele=79/17/2/2, p=0.004).
Event Free survival
Among the 97 women in IPAC there were few events. Two women died and 4 received LVADs prior to one year postpartum. Of the subjects who received LVADs, 2 women subsequently died and one was transplanted. No women recovered on LVAD and the remaining LVAD subject remained on support at one year. Analysis of event free survival revealed that the overall transplant free survival was 95% at 12 months, while the LVAD free survival was 93%. There was no significant difference by GNB3 genotype in either the LVAD free survival at one year post-partum (GNB3 TT versus C allele= 96% (95% confidence interval=100% to 87%) versus 92% (99% to 86%), p=0.44) nor transplant free survival: TT=96% (100% to 87%) versus C allele 95% (100% to 89%), p=0.86.
DISCUSSION
While genetic predispositions have been increasingly recognized as a risk factor for development of PPCM (19, 20), the role genomic background plays in racial differences in myocardial recovery remains largely unexplored. In this study from IPAC, women with PPCM with the GNB3 TT genotype demonstrated a significantly lower LVEF at 6 and 12 months postpartum. While this genotype clearly impacts both whites and blacks, the greater impact and higher frequency of the GNB3 TT homozygote among blacks appears to contribute to the lower LVEF evident in black women with PPCM at 12 months postpartum.
This common GNB3 polymorphism, although functionally silent, is in 100% linkage disequilibrium with a splicing variant of exon 9 (GNB3s) and the T allele is always co-inherited with the truncated subunit. This truncated subunit enhances alpha adrenergic activation (21–22), and the GNB3 825T allele, which is much more prevalent in African Americans (17), has been associated with low renin hypertension (15). In IPAC, GNB3 TT subjects did not demonstrate significant systolic or diastolic hypertension, although an association with hypertension may have been masked by the use of both ACE inhibitors and beta blockers in the majority of subjects.
The effect of GNB3 genotype on outcomes in African Americans with chronic heart failure has been examined in a genetic sub-study of AHeFT (18). Subjects with the GNB3 TT genotype on placebo in AHeFT demonstrated poorer hospitalization-free survival than subjects with the C allele. This was reversed by treatment with a fixed dose combination of isosorbide dinitrate and hydralazine (FDC I/H) suggesting the GNB3 genotype influences the impact of pharmacotherapy in African American with chronic heart failure. Of note while beta blockers and ACE inhibitors were widely utilized in IPAC, FDC I/H received limited use. Whether use of FDC I/H could have improved myocardial recovery in IPAC subjects with the GNB3 TT genotype remains to be determined.
The impact of genotype on cardiac remodeling and LVEF in patients with chronic heart failure was examined in AHeFT and several small studies. Of note, genotypes related to the renin angiotensin aldosterone system (23), nitric oxide synthesis (24), and the adrenergic receptors (25) have been explored as potentially modulating remodeling and the impact of therapy. In contrast to its impact on event free survival, GNB3 TT genotype did not impact remodeling in AHeFT (18). In addition to the work in AHeFT, the GNB3 T allele has been associated with and in increase in arrhythmic events in chronic heart failure (26). Modulation of GNB3 has been associated with reverse remodeling after cardiac resynchronization therapy (27). However, no previous analysis has evaluated the impact of GNB3 on myocardial recovery in recent onset cardiomyopathy, and it is possible that factors influencing left ventricular recovery in PPCM are distinct from those impacting remodeling in more chronic left ventricular dysfunction.
The impact of the GNB3 T allele on myocardial recovery in IPAC was evident by 2 months postpartum and may involve a response to injury which reflects differences in vascular tone and increased afterload. Despite previous work linking GNB3 T to the increase risk of hypertension, blood pressure was not significantly different by genotype in IPAC. Black subjects with the GNB3 TT genotype had a greater heart rate response to the cold pressor test of sympathetic activation (28), suggesting the GNB3 polymorphism may influence vascular tone through alterations of autonomic function. Metabolic stress testing revealed an association between the GNB3 T allele and poorer peak oxygen consumption and altered heart rate variability, further suggesting that the T allele may be associated with autonomic modulation and cardiovascular stress response (29).
This study is limited by the small number of patients which diminishes the power to firmly establish a genetic association between GNB3T allele and LV recovery in patients with PPCM. This is particularly evident in subset analysis by race, given the small number of African American women per genotype class. Therapy with ACE inhibitors and beta blockers was used at all centers, but was not standardized or optimized by protocol, and variation in treatment by site may have contributed to the heterogeneity of outcomes. The association of GNB3 TT with less myocardial recovery may reflect its higher prevalence in blacks and its role as a nonspecific marker of African genomic heritage, rather than a true functional impact of the truncated subunit itself. However, the strong impact of the GNB3 TT genotype within the black subset itself argues that its impact is not just as a marker of race.
CONCLUSION
This is the first study to establish a relationship between difference in genetic background and LV remodeling and recovery in women with PPCM. Black women are more predisposed to PPCM and less likely to recover. GNB3 TT genotype, more prevalent in black women, was associated with lower LVEF at 6 and 12 months postpartum. Women presenting with PPCM with this specific genotype may be at higher risk for chronic cardiomyopathy. The relationship between this specific genotype, adrenergic signaling and the potential for genotype targeted therapy requires further investigation.
Supplementary Material
Clinical Perspective
The ability to predict recovery of left ventricular (LV) function after a patient develops cardiomyopathy remains a significant challenge to clinicians. Clinical predictors have been examined, including patient factors, serological factors, and factors on cardiac imaging such as echocardiographic features. These have been explored in patients with both ischemic and non ischemic cardiomyopathy. There are also some studies that have subsequently examined genetic factors that might help predict recovery of LV function as well. This is the first study to explore genetic factors that might help predict LV recovery in women with peripartum cardiomyopathy (PPCM). The particular gene explored (GNB3 C825T) is associated with hypertension and also with African American background, both of which are associated with the development of PPCM. The ability to predict recovery of LV function in patients with PPCM remains clinically very relevant, and the role of patient genetic profile might also help further our understanding of the physiology underlying the development of PPCM. Patient genotype may potentially help clinicians better individualize the patient discussions regarding the ramifications of the cardiomyoptahy, and also help potentially tailor medical therapy individually to women with PPCM.
Footnotes
Disclosures
None.
References
- 1.Sliwa K, Hilfiker-Kleiner D, Petrie MC, Mebazaa A, Pieske B, Buchmann E, Regitz-Zagrosek V, Schaufelberger M, Tavazzi L, van Veldhuisen DJ, Watkins H, Shah AJ, Seferovic PM, Elkayam U, Pankuweit S, Papp Z, Mouquet F, McMurray JJ Heart Failure Association of the European Society of Cardiology Working Group on Peripartum Cardiomyopathy. Current state of knowledge on aetiology, diagnosis, management, and therapy of peripartum cardiomyopathy: a position statement from the Heart Failure Association of the European Society of Cardiology Working Group on peripartum cardiomyopathy. Eur J Heart Fail. 2010;12:767–778. doi: 10.1093/eurjhf/hfq120. [DOI] [PubMed] [Google Scholar]
- 2.Elkayam U. Clinical Characteristics of Peripartum Cardiomyopathy in the United States: Diagnosis, Prognosis, and Management. J Am Coll Cardiol. 2011;58:659–670. doi: 10.1016/j.jacc.2011.03.047. [DOI] [PubMed] [Google Scholar]
- 3.Capriola, Michael. Peripartum cardiomyopathy: a review. Int J Womens Health. 2013;5:1–8. doi: 10.2147/IJWH.S37137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gentry MB, Dias JK, Luis A, Patel R, Thornton J, Reed GL. African-American women have a higher risk for developing peripartum cardiomyopathy. J Am Coll Cardiol. 2010;55:654–659. doi: 10.1016/j.jacc.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moser M, Brown CM, Rose CH, Garovic VD. Hypertension in pregnancy: is it time for a new approach to treatment? J Hypertens. 2012;30:1092–1100. doi: 10.1097/HJH.0b013e3283536319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruz M, Briller J, Hibbard J. Update on Peripartum Cardiomyopathy. Obstet Gynecol Clin N Am. 2010;37:283–303. doi: 10.1016/j.ogc.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Ansari AA, Fett JD, Carraway RE, Mayne AE, Onlamoon N, Sundstrom JB. Autoimmune mechanisms as the basis for human peripartum cardiomyopathy. Clin Rev Allergy Immunol. 2002;23:301–324. doi: 10.1385/CRIAI:23:3:301. [DOI] [PubMed] [Google Scholar]
- 8.Bultmann BD, Klingel K, Nabauer M, Wallwiener D, Kandolf R. High prevalence of viral genomes and inflammation in peripartum cardiomyopathy. Am J Obstet Gynecol. 2005;193:363–365. doi: 10.1016/j.ajog.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 9.Lamparter S, Pankuweit S, Maisch B. Clinical and immunologic characteristics in peripartum cardiomyopathy. Int J Cardiol. 2007;118:14–20. doi: 10.1016/j.ijcard.2006.04.090. [DOI] [PubMed] [Google Scholar]
- 10.Goland S, Modi K, Hatamizadeh P, Elkayam U. Differences in clinical profile of African-American women with peripartum cardiomyopathy in the United States. J Card Fail. 2013;19:214–218. doi: 10.1016/j.cardfail.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 11.McNamara DM, Elkayam U, Alharethi R, Damp J, Hsich E, Ewald G, Modi K, Alexis JD, Ramani GV, Semigran MJ, Haythe J, Markham DW, Marek J, Gorcsan J, 3rd, Wu WC, Lin Y, Halder I, Pisarcik J, Cooper LT, Fett JD IPAC Investigators. Clinical Outcomes for Peripartum Cardiomyopathy in North America: Results of the IPAC Study (Investigations of Pregnancy-Associated Cardiomyopathy) J Am Coll Cardiol. 2015;66:905–914. doi: 10.1016/j.jacc.2015.06.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meirhaeghe A, Bauters C, Helbecque N, Hamon M, McFadden E, Lablanche JM, Bertrand M, Amouyel P. The human G-protein beta3 subunit C825T polymorphism is associated with coronary artery vasoconstriction. Eur Heart J. 2001;22:845–848. doi: 10.1053/euhj.2000.2400. [DOI] [PubMed] [Google Scholar]
- 13.Siffert W, Rosskopf D, Siffert G, Busch S, Moritz A, Erbel R, Sharma AM, Ritz E, Wichmann HE, Jakobs KH, Horsthemke B. Association of a human G-protein b3 subunit variant with hypertension. Nat Genet. 1998;18:45–48. doi: 10.1038/ng0198-45. [DOI] [PubMed] [Google Scholar]
- 14.Siffert W. G-protein b3 subunit 825T allele and hypertension. Curr Hypertens Rep. 2003;5:47–53. doi: 10.1007/s11906-003-0010-4. [DOI] [PubMed] [Google Scholar]
- 15.Schunkert H, Hense HW, Döring A, Riegger GA, Siffert W. Association between a polymorphism in the G-protein b3 subunit gene and lower renin and elevated diastolic blood pressure levels. Hypertension. 1998;32:510–513. doi: 10.1161/01.hyp.32.3.510. [DOI] [PubMed] [Google Scholar]
- 16.Schmitz B, De Maria R, Gatsios D, Chrysanthakopoulou T, Landolina M, Gasparini M, Campolo J, Parolini M, Sanzo A, Galimberti P, Bianchi M, Lenders M, Brand E, Parodi O, Lunati M, Brand SM. Identification of genetic markers for treatment success in heart failure patients: insight from cardiac resynchronization therapy. Circ Cardiovasc Genet. 2014;7:760–770. doi: 10.1161/CIRCGENETICS.113.000384. [DOI] [PubMed] [Google Scholar]
- 17.Siffert W, Forster P, Jöckel KH, Mvere DA, Brinkmann B, Naber C, Crookes R, Du P, Heyns A, Epplen JT, Fridey J, Freedman BI, Müller N, Stolke D, Sharma AM, Al Moutaery K, Grosse-Wilde H, Buerbaum B, Ehrlich T, Ahmad HR, Horsthemke B, Du Toit ED, Tiilikainen A, Ge J, Wang Y, Rosskopf D. Worldwide ethnic distribution of the G-protein b3 subunit 825T allele and its association with obesity in Caucasian, Chinese and black African individuals. J Am Soc Nephrol. 1999;10:1921–1930. doi: 10.1681/ASN.V1091921. [DOI] [PubMed] [Google Scholar]
- 18.McNamara DM, Taylor AL, Tam SW, Worcel M, Yancy CW, Hanley-Yanez K, Cohn JN, Feldman AM. G-protein beta-3 subunit genotype predicts enhanced benefit of fixed-dose isosorbide dinitrate and hydralazine: results of A-HeFT. JACC Heart Fail. 2014;2:551–557. doi: 10.1016/j.jchf.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 19.Cemin R, Janardhananc R, Donazzana L, Davesb M. Peripartum Cardiomyopathy: Moving Towards a More Central Role of Genetics. Current Cardiology Reviews. 2013;9:179–184. doi: 10.2174/1573403X113099990029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Tintelen JP, Pieper PG, Van Spaendonck-Zwarts KY, Van Den Berg MP. Pregnancy, cardiomyopathies, and genetics. Cardiovasc Res. 2014;15:571–578. doi: 10.1093/cvr/cvu014. [DOI] [PubMed] [Google Scholar]
- 21.Rosskopf D, Manthey, Siffert W. Identification and ethnic distribution of major haplotypes in the gene GNB3 encoding the G-protein b3 subunit. Pharmacogenetics. 2002;12:209–220. doi: 10.1097/00008571-200204000-00005. [DOI] [PubMed] [Google Scholar]
- 22.Baumgart D, Naber C, Haude M, Oldenburg O, Erbel R, Heusch G, Siffert W. G protein beta3 subunit 825T allele and enhanced coronary vasoconstriction on alpha(2)-adrenoceptor activation. Circ Res. 1999;85:965–969. doi: 10.1161/01.res.85.10.965. [DOI] [PubMed] [Google Scholar]
- 23.McNamara DM, Tam SW, Sabolinski ML, Tobelmann P, Janosko K, Taylor AL, Cohn JN, Feldman AM, Worcel M. Aldosterone Synthase Promoter Polymorphism Predicts Outcome in African Americans With Heart Failure: Results From the A-HeFT Trial. J Am Coll Cardiol. 2006;48:1277–1282. doi: 10.1016/j.jacc.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 24.McNamara DM, Tam SW, Sabolinski ML, Tobelmann P, Janosko K, Venkitachalam L, Ofili E, Yancy C, Feldman AM, Ghali JK, Taylor AL, Cohn JN, Worcel M. Endothelial Nitric Oxide Synthase (NOS3) Polymorphisms in African Americans With Heart Failure: Results From the A-HeFT Trial. J Cardiac Fail. 2009;15:191–198. doi: 10.1016/j.cardfail.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 25.Terra SG, Hamilton KK, Pauly DF, Lee CR, Patterson JH, Adams KF, Schofield RS, Belgado BS, Hill JA, Aranda JM, Yarandi HN, Johnson JA. B1-Adrenergic receptor polymorphisms and left ventricular remodeling changes in response to b-blocker therapy. Pharmacogenetics and Genomics. 2005;15:227–234. doi: 10.1097/01213011-200504000-00006. [DOI] [PubMed] [Google Scholar]
- 26.Chemello D, Rohde LE, Santos KG, Silvello D, Goldraich L, Pimentel M, Rosa PR, Zimerman L, Clausell N. Genetic polymorphisms of the adrenergic system and implantable cardioverter-defibrillator therapies in patients with heart failure. Europace. 2010;12:686–691. doi: 10.1093/europace/euq040. [DOI] [PubMed] [Google Scholar]
- 27.Schmitz B, De Maria R, Gatsios D, Chrysanthakopoulou T, Landolina M, Gasparini M, Campolo J, Parolini M, Sanzo A, Galimberti P, Bianchi M, Lenders M, Brand E, Parodi O, Lunati M, Brand SM. Identification of Genetic Markers for Treatment Success in Heart Failure Patients: Insight From Cardiac Resynchronization Therapy. Circ Cardiovasc Genet. 2014;7:760–770. doi: 10.1161/CIRCGENETICS.113.000384. [DOI] [PubMed] [Google Scholar]
- 28.Kurnik D, Friedman EA, Muszkat M, Sofowora GG, Xie HG, Dupont WD, Wood AJ, Stein CM. Genetic variants in the α2C-adrenoceptor and G-protein contribute to ethnic differences in cardiovascular stress responses. Pharmacogenet Genomics. 2008;18:743–750. doi: 10.1097/FPC.0b013e3282fee5a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faruque MU, Millis RM, Dunston GM, Kwagyan J, Bond V, Jr, Rotimi CN, Davis T, Christie R, Campbell AL. Association of GNB3 C825T Polymorphism with Peak Oxygen Consumption. Int J Sports Med. 2009;30:315–319. doi: 10.1055/s-0029-1202259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.