Abstract
Background
Approximately 10 % of the Mycobacterium tuberculosis genome is made up of two families of genes that are poorly characterized due to their high GC content and highly repetitive nature. The PE and PPE families are typified by their highly conserved N-terminal domains that incorporate proline-glutamate (PE) and proline-proline-glutamate (PPE) signature motifs. They are hypothesised to be important virulence factors involved with host-pathogen interactions, but their high genetic variability and complexity of analysis means they are typically disregarded in genome studies.
Results
To elucidate the structure of these genes, 518 genomes from a diverse international collection of clinical isolates were de novo assembled. A further 21 reference M. tuberculosis complex genomes and long read sequence data were used to validate the approach. SNP analysis revealed that variation in the majority of the 168 pe/ppe genes studied was consistent with lineage. Several recombination hotspots were identified, notably pe_pgrs3 and pe_pgrs17. Evidence of positive selection was revealed in 65 pe/ppe genes, including epitopes potentially binding to major histocompatibility complex molecules.
Conclusions
This, the first comprehensive study of the pe and ppe genes, provides important insight into M. tuberculosis diversity and has significant implications for vaccine development.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2467-y) contains supplementary material, which is available to authorized users.
Background
Tuberculosis disease (TB) is a major global public health problem, with control becoming difficult due to increasing drug resistance and in some populations HIV co-infection [1]. The available vaccine, Bacillus Calmette–Guérin (BCG), has limited efficacy and recent attempts to develop more effective protective vaccines have not been successful [2]. TB is caused by bacteria of the Mycobacterium tuberculosis complex, which have low overall genetic diversity and a striking clonal population structure. M. tuberculosis sensu stricto consists of seven lineages, including four that are predominant; 1 Indo-Oceanic, 2 East-Asian including Beijing, 3 East-African-Indian, 4 Euro-American [3]. These lineages are postulated to have differential impacts on pathogenesis, disease outcome and vaccine efficacy [4–7]. For example, modern lineages, such as Beijing and Euro-American Haarlem strains exhibit more virulent phenotypes compared to ancient lineages, such as East African Indian8. Whilst some genetic differences between lineages have been identified3, the molecular mechanisms responsible for differences in pathogenesis and virulence remain largely unknown [8].
Two groups of proteins, the PE and PPE families have been implicated in immune evasion and virulence [9]. Members of the pe/ppe gene families are characterized by the presence of proline-glutamate (PE) and proline-proline-glutamate (PPE) signature motifs near the N-terminus of their gene products [10]. The pe (99 loci) and ppe (69) gene families constitute ~7–10 % of the coding potential of M. tuberculosis and are scattered throughout the genome [9]. The families can be subdivided based on similarities in their N-terminal regions [11]. Many of the pe and ppe gene products are predicted to be localised to the cell membrane or secreted including those in the PE_PGRS domain containing subgroup and the PPE_MPTR domain containing subgroup [12, 13]. It has been speculated that these proteins may play a role in virulence [14]. Pe/ppe genes are differentially expressed during infection [15] and some PE/PPE proteins have been shown to elicit immune responses by the host [14, 16] and there is evidence that the PGRS domain can inhibit antigen processing [16, 17].
Whilst pe_pgrs and ppe_mptr genes represent some of the most variable M. tuberculosis regions, some members of the pe/ppe family are conserved across strains and species, therefore implying different functional roles. Only the protein structures of PE25 and PPE41 have been characterised [18], and in lieu of experimental and functional work, insights into their function and interaction partners must come from in silico analysis of large-scale ‘omics data. However, due to the repetitive nature and high GC content genetic variation in the pe/ppe genes, it has been difficult to characterize them using traditional mapping approaches, leading to their systematic exclusion from analysis [18]. There have been conflicting studies reporting either high or little or no sequence divergence [19–21], but studies have been limited by the number of genes and diversity of strains analysed.
There is a need to fully characterize pe/ppe family sequence diversity across strain-types to provide better understanding of these genes and their possible role in virulence and immune evasion. The availability of high throughput short sequencing technologies has revolutionized the study of M. tuberculosis genetic diversity. In an attempt to characterize these elusive genes we have performed whole genome assembly on next generation sequence data with a high depth of coverage across the pe/ppe gene regions from 518 clinical and experimental isolates. These isolates represent the four major lineages, each with known informative barcoding SNPs [3]. The approach was validated by examination of 21 reference genomes from established databases (www.tbdb.org; www.ebi.ac.uk), including 2 new strains with complete genomes sequenced using long read Pacific Bioscience (PacBio) technology [22, 23].
Results
Assembly of M. tuberculosis genomes
Conventional alignment-based analysis approaches have been of limited use in analysis of highly repetitive loci, including the pe/ppe genes. Here, we de novo assembled the genomes of 518 samples from 9 different countries covering the four main lineages (1 (n = 42), 2 (n = 38), 3 (n = 53), and 4 (n = 385)), with high sequence coverage in pe/ppe genes (mean 233-fold, range 100–1544) (Additional file 1: Tables S1 and S2). For each sample, at least 120 of the 168 pe/ppe genes were fully assembled and at least 90 % assembled for the remaining 48 genes (Additional file 1: Table S3). This level of assembly quality ensured low levels of assembly fragmentation and minimised poor gene characterization. Subsequent analysis involving manual inspection or re-mapping of reads to the assemblies using REAPR software, revealed all genes (168 pe/ppe; 3,654 other genes; 2,820 with an assigned function) to be of high quality (median REAPR score of 1 across all bases, reflecting high levels of accuracy in genome assemblies). A further 21 independent complete reference genomes representing all four lineages (Additional file 1: Table S1), were aligned against H37Rv to call variants, and used to further validate the results found in the assembled dataset.
Variant detection and population genetic analysis
A total of 50,539 genome-wide SNPs were identified by comparing the 518 assembled genomes to the H37Rv (lineage 4, Euro-American T) reference strain. Of these, 5,853 (11.6 %) SNPs were located within pe/ppe regions, with greater density than the rest of the genome (median SNPs per kb: ppe/pe = 12.9, non-ppe/pe = 9.1, Wilcoxon P < 2.2 × 10−14). In the 257 Malawi samples, our assembly procedure revealed 3,467 additional SNP variants genome-wide (1,438 (41.5 %) SNPs in 72 pe/ppe genes) compared to the standard approach of aligning short reads to the H3Rv reference. Of the 50,539 SNPs inferred from the assemblies, the majority (45,681, 90.3 %) were located in coding regions from all genes and consisted of 28,235 (61.8 %) non-synonymous SNPs and 17,446 (38.2 %) synonymous SNPs. This observation is in agreement with the higher abundance of non-synonymous mutations reported in the literature [19]. A large number of rare variants (i.e. present in only one isolate) were observed in all lineages, indicative of purifying selection and population expansion described by others [24]. The peaks in the spectrum represent a number of SNPs that are fixed in all isolates from sub-lineages (Additional file 2: Figure S1).
The ratio of non-synonymous to synonymous mutations was similar in pe/ppe and other genes (median: pe/ppe genes = 1.65, other genes = 1.75, Wilcoxon P = 0.68). The density of non-synonymous mutations was 2.98 times greater in pe/ppe genes compared to others (pe/ppe genes: 1 every 3933 bp, other genes: 1 every 11,706 bp, Wilcoxon P < 0.0001), consistent with another report [25]. When analysed by sub-family we observed the greatest ratio of densities in the pe_pgrs genes (pe_pgrs 3.89) compared to the other types (ppe 1.75, pe (non-pe_pgrs) 1.80), similar to that reported previously [25]. The nucleotide diversity (π) was ~2-fold greater in the pe/ppe genes (median: pe/ppe genes 2.7 × 10−4, other genes 1.4 × 10−4, Wilcoxon P < 1.4 × 10−10). Although estimates of genetic diversity may be influenced by sampling bias, nucleotide diversity varied by lineage, being greater in lineage 1 (Indo-Oceanic median: pe/ppe 1.7 × 10−4, other 9.0 × 10−5) and lower in lineage 2 (East-Asian median: pe/ppe 7.3 × 10−5, other 0) (Additional file 1: Table S2), all consistent with previous work [3]. Loci identified as being highly diverse (π > 0.003, top 0.2 %, Table 1, Fig. 1), included 5 pe/ppe genes (pe_pgrs3, pe_pgrs4, ppe57, ppe59 and ppe60), and 3 others (Rv0030, Rv0095c and lppB). The diversity per gene was compared to those from 21 complete reference genomes, and peaks were observed at Rv0095c, pe_pgrs3, pe_pgrs4, ppe57 and ppe60, independently supporting five out of the eight loci identified in the 518 global samples (Additional file 3: Figure S2).
Table 1.
Loci that are highly diverse, with recombination, or under selective pressure
| Gene | Locus | No. SNPs | Diversity π | phi p-value | phi p-valuea | dN/dS (w) | No. sitesb | Lineage specific phi |
|---|---|---|---|---|---|---|---|---|
| Rv0030 | Rv0030 | 3 | 0.0033 | 1.000 | 1.000 | - | 0 | - |
| Rv0095c | Rv0095c | 10 | 0.0059 | 0.005 | 0.021 | 10.13 | 3 | - |
| Rv0182c | sigG | 3 | 0.0003 | 0.046 | 0.046 | - | 0 | - |
| Rv0278c | pe_pgrs3 | 130 | 0.0193 | <0.001 | <0.001 | 10.5 | 49 | 1,3,4 |
| Rv0279c | pe_pgrs4 | 49 | 0.0035 | 0.001 | 0.419 | 10.5 | 20 | - |
| Rv0282 | eccA3 | 5 | 0.0005 | 0.007 | 0.210 | 9.697 | 6 | - |
| Rv0850 | Rv0850 | 2 | 0.0031 | 1.000 | 1.000 | 9.264 | 4 | - |
| Rv0978c | pe_pgrs17 | 9 | 0.0005 | 0.003 | 1.000 | 10.495 | 9 | - |
| Rv1148c | Rv1148c | 18 | 0.0022 | <0.001 | 0.015 | 10.492 | 5 | 4 |
| Rv1793 | esxN | 6 | 0.0023 | 0.034 | 0.159 | 9.694 | 2 | 4 |
| Rv1945 | Rv1945 | 18 | 0.0010 | <0.001 | 0.026 | 10.433 | 5 | - |
| Rv2048c | pks12 | 80 | 0.0008 | <0.001 | 0.012 | 10.5 | 79 | 4 |
| Rv2543 | lppA | 8 | 0.0015 | 0.006 | 0.002 | 10.036 | 5 | 4 |
| Rv2544 | lppB | 60 | 0.0123 | <0.001 | <0.001 | 5.336 | 33 | 1,2,4 |
| Rv3425 | ppe57 | 31 | 0.0154 | 0.431 | 1.000 | 10.5 | 21 | - |
| Rv3429 | ppe59 | 19 | 0.0041 | <0.001 | 0.084 | 10.419 | 29 | 4 |
| Rv3466 | Rv3466 | 6 | 0.0010 | 0.004 | 0.373 | 7.757 | 3 | - |
| Rv3478 | ppe60 | 105 | 0.0061 | <0.001 | 0.004 | 7.502 | 54 | 4 |
| Rv3619c | esxV | 3 | 0.0022 | 0.025 | 1.000 | 10.391 | 2 | - |
π nucleotide diversity, phi recombination, NS not significant
aafter removing sites under selection
bnumber of sites under selection using the Bayes Empirical Bayes method
Bolded refers to π > 0.003 or phi p-value < 0.05
Fig. 1.
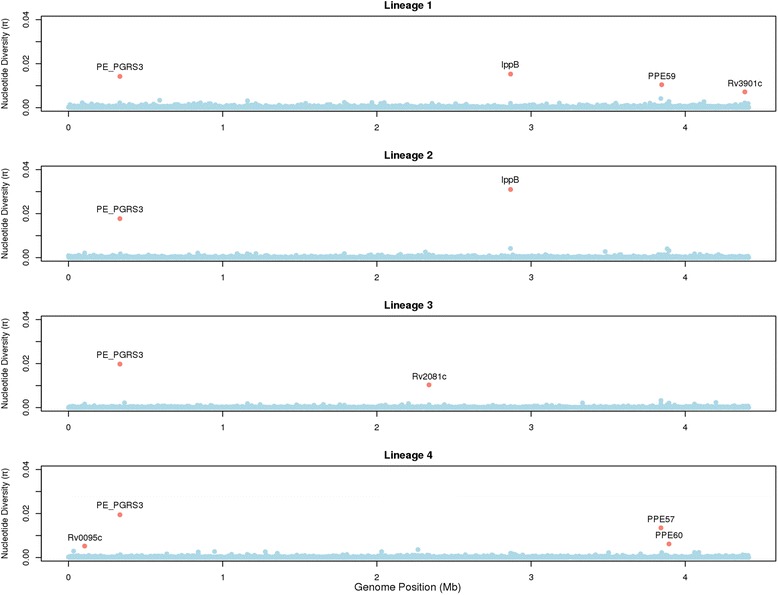
Nucleotide diversity (π) across the genome by lineage; genes with high diversity (π > 0.003) are highlighted. The pe_pgrs3 gene appears to have high nucleotide diversity in all lineages. Some lineage-specific hotspots are seen in lineage 1 (ppe59 and Rv3901c), lineage 3 (Rv2081c) and lineage 4 (ppe57 and ppe60)
Phylogenetics
To examine the link between genetic variation and lineage, a phylogenetic tree was constructed using the 50,539 SNPs. It revealed clustering by lineage, thereby further validating the quality of the assembled genomes (Additional file 4: Figure S3). However, a similar analysis using 5,853 pe/ppe specific SNP positions led to a tree with lineage 2 being split into two distinct clades, surrounded by lineage 4 strains (Fig. 2a). Subsequent analysis using SNP-based population differentiation FST and site-specific log likelihood scores approaches (Additional file 5: Figure S4) revealed that the pe_pgrs3 gene (genomic position 333 kb, lineage 2 – 104 SNPs differentiating) was predominantly responsible for the ambiguity. Removal of the 281 SNPs in the pe_pgrs3 gene led to a pe/ppe-based tree that clustered by lineage (Fig. 2b), very similar in topology to that based on the genome-wide SNPs (Additional file 4: Figure S3). This demonstrated that a core set of pe/ppe SNPs appears to be lineage specific, and further analysis revealed a set of 87 (1.4 %) SNPs (66 non-synonymous) that were lineage specific, potentially forming the basis of a lineage-specific molecular barcode (Additional file 1: Table S4). None of these 87 mutations were present in M. bovis (GCA_000195835) or M. africanum (NC_015758.1) sequences, and therefore robust as M. tuberculosis lineage-specific markers. Using only the pe_pgrs3 SNPs led to a tree with two large clades (Additional file 6: Figure S5), one containing H37Rv and strains with similar sequence, and the other consistent with isolates similar to M323 and 18b strains (Additional file 1: Table S1b), both undergoing recent sequencing using PacBio long read technology. The M323, 18b and similarly clustered assembled samples have a pe_pgrs3 gene with conserved regions at both 3′ and 5′ ends, surrounding a highly similar hypervariable core. A different hypervariable core is present in H37Rv and similarly clustered assemblies, which interestingly is also present in the pe_pgrs4 gene of 18b, and recombination is a potential explanation.
Fig. 2.
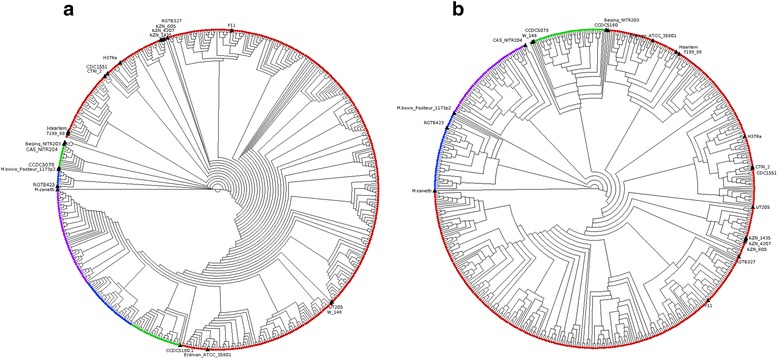
Phylogenetic tree constructed with SNPs. a pe/ppe genes (5,404 SNPs, 10 % of the genome); the pe_pgrs3 gene was identified as having SNPs leading to the lineages not perfectly clustering, potential evidence of recombination affecting these loci. b pe/ppe genes excluding pe_pgrs3 (5,572 SNPs, ~10 % of the genome). Clear clustering according to lineage can be seen (Lineage 1 (Indo-Oceanic, green), lineage 2 (East-Asian (Beijing), blue), lineage 3 (East-African-Indian, purple), lineage 4 (Euro-American, red)). Reference genomes are labelled. M. canetti is annotated in cyan
Recombination detection
Although it has been thought that M. tuberculosis undergoes little or no homologous recombination, PE_PGRS and PPE_MPTR families contain long domains comprised of series of tandem repeats, giving them a higher propensity to undergo recombination. There is published evidence of intra-chromosomal cross-over ahead of a few loci [9], including pe_pgrs3, pe_pgrs4, and ppe1 [26]. We hypothesized that recombination may be the reason for the observed high genetic diversity and distortion in the pe/ppe tree. We applied the pairwise homoplasy index (phi) method [27] genome-wide to establish if there was any evidence of recombination in pe_pgrs3 and other loci (Fig. 3). The method calculates a p-value (phi P) of observing the sequence data under the null hypothesis of no recombination. The analysis revealed 16 genes with potential recombination events (phiP < 0.05) present across all lineages: 5 in pe/ppe genes (pe_pgrs3, pe_pgrs4, pe_pgrs17, ppe59 and ppe60), and 11 others (Rv0095, sigG, eccA3, Rv1148, esxN, Rv1945, pks12, lppA, lppB, Rv3466 and esxV).
Fig. 3.
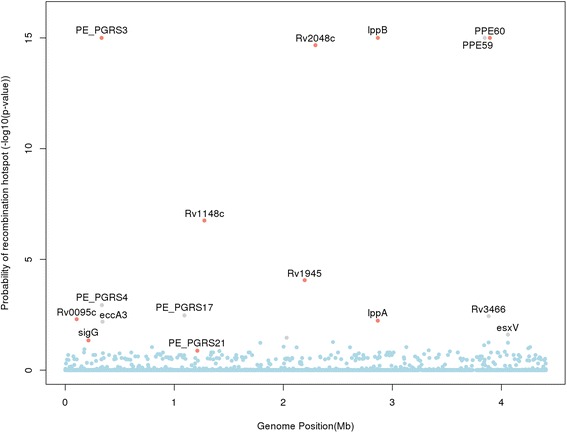
Evidence of recombination at a gene level. A Manhattan plot showing genes that are likely to be recombination hotspots. The (−log10) p-value for recombination is plotted against genome position. All genes with p-values <0.05 are labelled. Genes labelled in colour grey (eccA3, pe_pgrs4, pe_pgrs17, ppe59, Rv3466 and esxV) become statistically non-significant after removing sites under selection
It could be expected that the vast majority of any genomic recombination events are intra-lineage and that these events will pass unnoticed by other analyses, especially in studies of small sample size. Lineage-specific hotspots were also present (Additional file 7: Figure S6), including possible pathogenicity factors lppA/lppB in lineage 2 (Beijing) and pe_pgrs3 in lineage 4. An analysis of the 21 complete reference genomes revealed an overall high degree of concordance of the homoplasy phi statistic with the assembled data, with six recombination peaks in common (Rv0095c, pe_pgrs3, pe_pgrs4, pe_pgrs17, Rv1148c and Rv1945) (Additional file 8: Figure S7). Together, these results provide evidence for recombination.
Detecting selection pressure
It is possible that recombination and population expansion [24] could have introduced not only the observed increased diversity in the pe/ppe genes, but contributed to an excess of non-synonymous mutation diversity in general; especially in genes expected to be under positive or diversifying selection such as the cell wall component genes [24]. Proteins in contact with the host proteome could be under pressure to change their amino acid sequence in order to avoid detection or unfavourable interaction with the host immune system. We decided to investigate the role of selection in the pe/ppe genes compared to other categories of genes. The distribution of dN/dS values (denoted ω, = 1 neutral evolution, >1 positive selection, <1 purifying selection), calculated for each gene across all sites and branches of the phylogenetic tree, was similar between pe/ppe and other genes (median ω: pe/ppe genes 0.81, other genes 0.73; Wilcoxon P = 0.16). These values are broadly similar to those previously reported on much lower numbers of samples and pe/ppe genes [25]. The genes were further divided into functional Clusters of Orthologous Groups (COG) categories [28]. Higher median ω values were observed in genes associated with signal transduction mechanisms (median = 0.95), perhaps due to their contact with the host, and the lowest values found in genes associated with RNA processing and modification (median = 0.38) (Additional file 9: Figure S8).
In most genes it would be expected that only a small subset of sites would undergo positive selection and so calculation of a single ω value over all sites in the gene may dilute an effect. For example, this is possible in pe/ppe genes where there is less variation in the N- compared to the C- terminus [21]. We therefore used a likelihood ratio based approach that accounts for the variability of ω between sites. After implementation, we detected a greater proportion of pe/ppe loci under positive selection compared to other genes (ω > 1 and P < 0.05: pe/ppe genes 65 (39 %) vs. other genes 590 (15 %)). This observation remained consistent when the non-pe/ppe genes were subdivided into functional categories (P-values for evidence of ω >1, Wilcoxon P <0.001) (Fig. 4). Using the COG categories, the genes associated with cell motility and the pe/ppe genes again showed greater evidence of significant positive selection (Additional file 10: Figure S9). All genes annotated as possible recombination hotspots were identified as being under positive selection, except Rv0182c. To localize the specific polymorphisms under selection we applied the Bayes Empirical Bayes (BEB) method [29], and identified a small number of sites in each gene (median (range): pe/ppe genes 0 (0–60), other genes 0 (0–48), P = 1.2 × 10−10). In total 99 pe/ppe genes had sites under positive selection, including ten genes with selection at more than ten sites (Additional file 1: Table S5). For 1,106 non-pe/ppe genes, only 37 had ten or more sites under positive selection. The proportion of segregating sites under positive selection (Sp/Ss) per gene was higher in the pe/ppe loci compared to others (pe/ppe genes 0.04, other genes 0.00, Wilcoxon P = 2.58 × 10−7). There was a correlation between the number of positively selected and segregating sites (Pearson’s r, pe/ppe 0.81, and other genes 0.32).
Fig. 4.
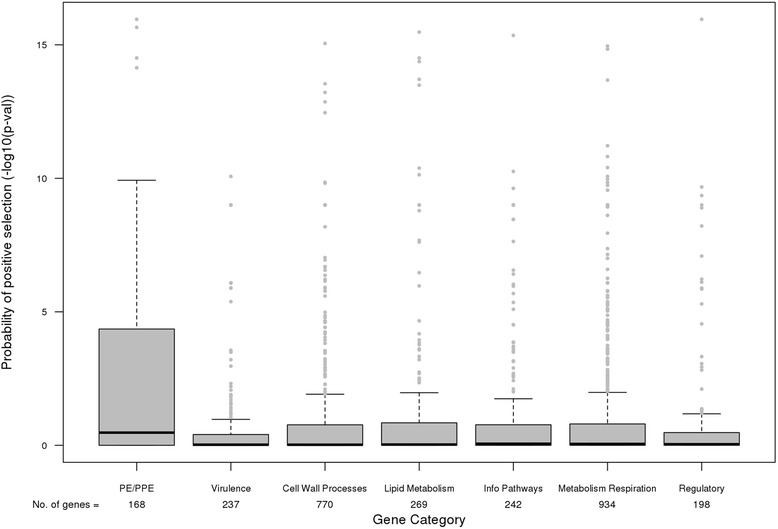
Evidence of positive selection between the pe/ppe and other genes by functional annotation. Distributions of (−log10) p-values for positive selection (evidence of ω >1) across the pe/ppe and other genes by functional annotation
We considered the 3,686 sites in the 1,106 non-pe/ppe genes with some evidence of positive selection (ω > 1). These sites were compared to a list of drug resistance-conferring mutations (www.tbdb.org), which because of a survival advantage may be expected to be under positive selection. Eighteen drug resistance markers were found, including in inhA (I21T, S94A, I194T, P251A; associated with the drug isoniazid), katG (S315T; isoniazid), gyrA (A90V; fluoroquinolones), rpoB (P45L, rifampicin), rpoL (K43R; rifampicin), and ponA1 (P631S; rifampicin). Other regions of interest included rodA (T336S) involved in cell wall processes and required for survival in primary murine macrophages, and pks6 (V504L) involved in lipid metabolism and in vitro growth. Repeating the recombination detection analysis after removing the sites under positive selection identified by the BEB method, revealed six genes that lost their statistical significance (phiP > 0.05, eccA3, pe_pgrs4, pe_pgrs17, ppe59, Rv3466 and esxV), leaving 10 as crossover hotspots (Fig. 3). Given that variation in these genes is not caused by positive selection it is highly likely that recombination hotspots are indeed present at these ten loci. The proportion of sites under selection was high for lppA (7 %) and lppB (43 %) loci. The BEB method identified 38 codons in lppA/B at which ω > 1, with almost all the related mutations present in lineage 2 (East-Asian) samples. None of these codons were in previously described conserved positions [30], implying that the core function of the protein was not disturbed, and the mutations may contribute to antigenic variation.
Selection on epitopes
Epitopes potentially binding to major histocompatibility complex molecules were predicted in all PE/PPE proteins using the netMHCpan software (Additional file 1: Table S6). The number of epitopes varied by pe/ppe gene (median 45, range 0 – 455). Some pe/ppe sites identified as being under selection using the BEB approach did overlap with regions predicted to be epitopes. In particular, for 10 genes (pe6, pe_pgrs26, pe18, pe_pgrs49, pe_pgrs60, ppe27, ppe57, ppe59, ppe60 and ppe65), more than 20 % of predicted epitopes had sites under positive selection (Additional file 1: Table S6).
Discussion
Members of the PE/PPE family of proteins have been found to trigger innate immune responses, are targets of the adaptive immune system, and potentially a rich source of diagnostic and vaccine antigens. As large ‘omic studies in M. tuberculosis have often excluded pe/ppe genes from analysis (e.g. [3]), the understanding of their function and diversity is poor compared to other loci. Assessing diversity across M. tuberculosis strain types is critical, as lineages may vary in propensity to transmit and cause disease. By applying a de novo assembly approach, we were able to characterize accurately nearly all 168 pe/ppe genes in 518 isolates with high genomic coverage, representing lineages 1 (Indo-Oceanic), 2 (East-Asian), 3 (East-African-Indian) and 4 (Euro-American). After identifying ~50 k genome-wide SNPs from whole genome alignments, we confirmed that pe/ppe genes, especially the pe_pgrs family, have a high density of non-synonymous mutations compared to other M. tuberculosis loci. This observation is consistent with their involvement in antigenic variation and immune evasion, where proteins that are directly exposed to host immune surveillance tend to show higher levels of polymorphism. A lower degree of polymorphism in the ppe genes (compared to pe_pgrs) is likely to reflect a strong functional constraint of the PPE proteins.
Using all SNPs in a phylogenetic analysis, we observed clustering by M. tuberculosis lineage and therefore consistency with other published topologies [3, 31]. There was evidence of lineage specific pe/ppe repertories, with a very similar phylogeny being attained by restricting analysis to all polymorphisms in 167 PE/PPE genes (excluding pe_pgrs3), as well as a derived subset of 87 informative SNPs. The pe_pgrs3 gene had high nucleotide diversity across all lineages, was not lineage informative, and is likely to be have been subject to recombination in lineages 1, 3 and 4. Both M. bovis and M. canetti contain two genes annotated as orthologues of pe_pgrs3, providing further evidence towards the propensity of this region to undergo genomic rearrangements. Interestingly the positioning of the M. tuberculosis reference strains in the pe/ppe gene phylogenetic tree was altered; some strains clustering near the M. canetti and ancestral strains while some of the known virulent reference strains were positioned at a further distance. Further study is needed to elucidate this effect. Other recombination and diversity hotspots included lppB (lineages 1, 2, and 4) and ppe60 (lineage 4) genes, both known to have undergone homologous recombination. LppB (and lppA) are non-essential exported lipoproteins that are unique to pathogenic mycobacteria and may encode antigens [32, 33]. The lppA/B SNPs driving this effect were found mostly in lineage 2 (Beijing) strains, and seemed to be conferring a selective advantage. The role of lppA/B proteins on virulence should be investigated further. Although, pe_pgrs17, whose protein is in contact with the host immune system [34], was identified as a recombination hotspot, this observation may be confounded by positive selection. However, recombination has been described in pe_pgrs17, with large numbers of SNPs and indels in the pe_pgrs17 and pe_pgrs18 pair observed across the different lineages, potentially arising from gene conversion events [35]. We can rule out the results being confounded due to a sampling frame that included different geographical regions, as there was strongest clustering by lineage and not geographical source.
Across all M. tuberculosis genomes there was evidence that most genes were undergoing purifying selection pressures (dN/dS < 1). However, the pe/ppe genes were most likely to be under positive selection (dN/dS > 1), consistent with some PE/PPE proteins providing antigenic variation. It is possible the dN/dS ratios may be underestimated, as the methodology is more appropriate to divergent species and not for comparisons within a population [25]. Further, the signatures from very localised regions of selection within a gene may be diluted by surrounding genetic variation. A site-specific analysis confirmed the results from the gene-based dN/dS. Whilst the majority of the sixty-five genes identified as being under positive selection had only a single positively selected site, a disproportionate number of pe_pgrs genes had multiple positively selected sites. A potential limitation of this analysis is the time dependence of dN/dS for closely related bacterial genomes. This leads to possible over-estimation of the dN/dS and difficulties in interpretation when comparing the strength of selection between genes, genomes or populations over very short time-scales [36]. The power of the dN/dS statistic to detect positive selection is reduced when samples come from a single population [25]. In addition to the site under selection, multiple neighbouring and linked sites may show evidence of selection due to hitchhiking effects.
Our findings provide potential insights into the use of PE/PPE proteins as vaccine components. The high levels of polymorphism observed and the lineage-specific nature in certain members of these protein families could limit their effectiveness. A PE/PPE protein that displays higher sequence conservation across many strains may be a more effective vaccine candidate. For example, the highly immunogenic PE_PGRS62 protein has been considered as a vaccine target [37], and as only one of the 14 non-synonymous mutations observed was lineage specific, it may have broad strain coverage. However, one roadblock is the limited immunogenicity data available at the pe/ppe epitope level. It has been found that human T-cell epitopes are highly conserved in the M. tuberculosis complex [38], and like others [25] we found many epitopes predicted in PE/PPE proteins. Our analysis revealed a number of pe/ppe genes with a high proportion of epitopes potentially subjected to diversifying or positive selection. As these epitopes may be used by M. tuberculosis to evade the host immune system they would be relevant for TB vaccination strategies.
A cohesive understanding of the function of the 168 PE/PPE family of proteins remains elusive. By analysing SNP variation in 518 samples across the four main M. tuberculosis lineages we identified pe/ppe genes that are highly diverse, recombination hotspots and under positive selection. Such analyses can assist with prioritising candidates for functional studies, potentially leading to TB control measures, such as vaccines, diagnostics and drugs.
Conclusions
Human tuberculosis poses a major burden on health services worldwide. There is a need to understand the complex interactions between the human host and bacterial pathogen so that new control measures, such as vaccines and drugs, can be developed. Recent technological advances have allowed large-scale studies to determine the genetic signatures of strain-types or ancestral lineages and drug resistance outcomes. Despite this advance, some highly variable regions of the genome are often excluded [39, 40]. This includes the pe/ppe gene family, whose members are thought to interact with the human immune system, but little is still known of their diversity and function. Here we present the first comprehensive study of the genetic diversity of the 168 pe/ppe genes. We find most genes vary in a lineage specific manner, consistent with strain-specific repertories. However, there were exceptions to this pattern, with evidence of some genes undergoing genetic cross-over events. Further, by looking for the genes under selective pressure genome-wide, we found enrichment in the number of pe/ppe genes undergoing positive selection. Overall, our work highlights the importance of pe/ppe genes, describes their suitability as vaccine candidates, and provides the basis for further exploration of the proteins involved in the host immune system and pathogen interactions.
Methods
The raw sequencing fastq files for 518 M. tuberculosis samples with more than 100-fold genomic coverage were sourced from the PolyTB [41], rapid TB [42] and global drug resistance (Coll F, McNerney R, Hill-Cawthorn G et al. Whole genome association analysis of a global collection of Mycobacterium tuberculosis clinical isolates gives new insight into drug resistance, Submitted) projects (Additional file 1: Table S1a). A list of ENA accession numbers is available for download (http://pathogenseq.lshtm.ac.uk/ppe). Lineages were inferred using robust barcoding SNPs [3]. Lineages 1, 2, 3 and 4 were represented with 42, 38, 53 and 385 samples from each respectively (Additional file 1: Table S2). A separate set of twenty-one samples representing lineages 1 to 4 with complete or near complete genomes were used for validation (Additional file 1: Table S1b). In particular, all analyses performed on the main 518 samples were also applied to the validation dataset in an attempt to confirm signals and potentially rule out spurious findings. Assembly of all short reads was performed using MaSuRCA, SGA, Velvet and SPAdes [43–46] software, run in paired end mode with default and recommended parameters, across multiple k-mer values ranging from 31 to 91. The final Velvet run was implemented with a k-mer value of 63. Quast [47] software was used to extract assembly quality metrics using the H37Rv strain (Gene bank: AL123456) as the reference. The Samtools rmdup utility [48] was used to remove duplicates from each sample’s BAM file, and picard SamToFastq (http://broadinstitute.github.io/picard/) was used to convert the BAM files to fastq format. IMAGE software [49] was used to close gaps from the contigs produced by Velvet. After running IMAGE for 3 iterations using a k-mer size of 55, the number of pe/ppe genes assembled increased for all samples, especially in high coverage samples. The majority (range: 78–98 %) of gaps were closed within 3 iterations, which provided a threshold to justify the compromise between runtime and gaps closed in new contigs (fasta format). REAPR software was used to assess the quality of the assemblies, and calculates a quality score per base (http://www.sanger.ac.uk/science/tools/reapr reapr). The final assemblies are available for download (http://pathogenseq.lshtm.ac.uk/ppe). The pe/ppe and other genes were called by aligning the assemblies to the well annotated H37Rv genome. The 50,539 SNPs genome-wide were identified using nucmer [50] with H37Rv as the reference genome. To assess the robustness of the aligned sequences and resulting SNPs and analyses, we also mapped samples to a Mycobacterium africanum lineage reference (GCA_000253355.1), but observed no major differences from those using H37Rv (lineage 4). Phylogenetic data (alignments, phylogenetic trees) are deposited in Dryad (http://datadryad.org/).
The alignments of the genotypes for the 50,539 SNPs formed the basis of the majority of population genetic analyses, except where stated otherwise. SNP locations at which more than 10 % of the genotypes were missing were excluded from analyses. Other missing data was kept in the multiple alignments and was processed according to the default settings of the analysis software applied. Indels were identified by nucmer but were not analysed in this study. Regions where multiple contigs overlapped or where no contigs mapped to were annotated as missing data. FastTree [51] software employing the generalised time-reversible model was used to produce the final phylogenetic trees. The trees included the ancient M. canettii strain (NC_019950.1). The FST measure was calculated for each SNP to identify markers with complete between-lineage allele differentiation (FST >0.99). Similarly, the ancestral reconstructed sequence for the lineage-defining node in the phylogenetic tree was compared with its closest ancestral node, and the SNP differences derived. Nucleotide diversity (π) and the number of segregating sites were calculated using variscan software applied to sequence alignments [52]. To test for recombination we used the pairwise homoplasy index (phi) statistic calculated in sliding windows, as implemented in Phipack software [27]. The non-synonymous to synonymous ratio was calculated using PAML software [53]. To discover the effect of positive selection on the pe/ppe genes compared to all other genes, codeml was used to fit a number of models to the data using a maximum likelihood approach. This is generally thought to be more robust than counting methods. A dN/dS (ω) value was calculated per gene across all positions and all branches of the phylogenetic tree. For each gene, we then performed a likelihood ratio test using PAML software to assess evidence of positive selection, which compared two models: (a) variable selective pressure but no positive selection (0 < ω < 1) (M8a) and (b) variable selective pressure with positive selection (M8) (ω > 1). The test statistic has a χ2 (1 degree of freedom) distribution, and the resulting p-value reflects the likelihood of positive selection acting on a gene. To localize the specific polymorphisms under selection we applied the Bayes Empirical Bayes (BEB) method [29]. The proportion of segregating sites under positive selection (Sp/Ss) was calculated using the results from variscan and BEB. Epitopes were predicted using netMHCpan [54] using HLA alleles previously suggested [21].
No ethical approvals were required for this study.
Availability of supporting data
The list of raw sequence data accession numbers for the ENA short read archive, final assemblies and links to the phylogenetic data (alignments, phylogenetic trees) in Dryad can be found in http://pathogenseq.lshtm.ac.uk/ppe.
Acknowledgements
JP is supported by a Biotechnology and Biological Sciences Research Council UK PhD studentship. FC was the recipient of a Bloomsbury Research Fund PhD studentship. SLS receives funding from the South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation (UID 86539). AP is supported by the KAUST faculty baseline research fund (KAUST-BRF). TGC receives funding from the Medical Research Council UK (grant numbers MR/K000551/1, MR/M01360X/1, MR/N010469/1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional files
a) The samples used for the assembly (*Malawi [55, 56], Netherlands [57], Pakistan [58], Portugal [59]) and b) the 21 reference strains. Table S2. Lineage, sequence coverage and polymorphism. π nucleotide diversity; Lineage 1 Indo-Oceanic; Lineage 2 East-Asian (Beijing); Lineage 3 East-African-Indian; Lineage 4 Euro-American. Table S3. Completeness of pe/ppe gene assemblies. Table S4. List of 87 pe/ppe lineage specific-markers. S synonymous, NS non-synonymous, * genes bolded if there are sites under selection using the Bayes Empirical Bayes method; Lineage 1 Indo-Oceanic; Lineage 2 East-Asian (Beijing); Lineage 3 East-African-Indian; Lineage 4 Euro-American. Table S5. Genes with more than 10 sites under selective pressure (dN/dS (ω) >1). Table S6. Epitopes. * identified using netMHCpan, ** epitopes that had sites under positive selection according to the Bayes Empirical Bayes (BEB) method. (DOCX 70 kb)
Allele frequency spectra for each lineage by synonymous (blue) and non-synonymous (red) mutations. The peaks at intermediate allele frequencies include sub-lineage defining SNPs (Lineage 1 Indo-Oceanic; Lineage 2 East-Asian (Beijing); Lineage 3 East-African-Indian; Lineage 4 Euro-American). (TIF 207 kb)
Gene-based nucleotide diversity (π) for the 21 reference genomes. All genes with high nucleotide diversity (π > 0.0075) are labelled. (TIF 148 kb)
Phylogenetic tree constructed using 50,540 genome-wide SNPs. Clear clustering according to lineage can be seen (Lineage 1 (Indo-Oceanic, green), lineage 2 (East-Asian (Beijing), blue), lineage 3 (East-African-Indian, purple), lineage 4 (Euro-American, red)). Reference genomes are labelled. M. canetti is annotated in cyan. (TIF 69 kb)
Identifying sites leading to differences in tree topologies based on all SNPs (Additional file 4: Figure S3a) and only those from pe/ppe genes (Additional file 4: Figure S3b). The Δ Site wise log likelihood score (Δ SSLS) is calculated for each SNP in the pe/ppe gene alignments. Negative differences indicate SNP positions favouring the pe/ppe tree. SNPs in pe_pgrs3, ppe57 and ppe60 produce strong phylogenetic signals supporting the pe/ppe tree. (TIF 113 kb)
Phylogenetic tree created using only SNPs from pe_pgrs3. No clear clustering by lineage is observed. However there are two major clades, one consistent with H37Rv (bottom-left). (TIF 126 kb)
Lineage-specific recombination hotspots. Manhattan plots showing genes that are likely to be recombination hotspots in each lineage (Lineage 1 Indo-Oceanic; Lineage 2 East-Asian (Beijing); Lineage 3 East-African-Indian; Lineage 4 Euro-American). The (−log10) p-value for the phi statistic is plotted against genome position. All genes with p-values < 0.05 are labelled. (TIF 147 kb)
Evidence of recombination at a gene level in the 21 reference genomes. A Manhattan plot showing genes that are likely to be recombination hotspots. The (−log10) p-value for the phi statistic is plotted against genome position. Genes with p-values less than 0.05 are shown. (TIF 120 kb)
Selection dN/dS values for each gene within Clusters of Orthologous Groups (COG*) categories. *ppe/N = pe/ppe genes annotated as COG category N, * COG categories: A RNA processing and modification, B Chromatin Structure and dynamics, C Energy production and conversion, D Cell cycle control and mitosis, E Amino Acid metabolism and transport, F Nucleotide metabolism and transport, G Carbohydrate metabolism and transport, H Coenzyme metabolism, I Lipid metabolism, J Translation, K Transcription, L Replication and repair, M Cell wall/membrane/envelope biogenesis, N Cell motility, O Post-translational modification, protein turnover, chaperone functions, P Inorganic ion transport and metabolism, Q Secondary Structure, T Signal Transduction, U Intracellular trafficking and secretion, Y Nuclear structure, Z Cytoskeleton, R General Functional Prediction only, S Function Unknown. (TIF 124 kb)
Non-neutral evolution for genes within Clusters of Orthologous Groups (COG*) categories. Boxplots are constructed using (-log10) p-values of non-neutral evolution for each gene. *ppe/N = pe/ppe genes annotated as COG category N, * COG categories: A RNA processing and modification, B Chromatin Structure and dynamics, C Energy production and conversion, D Cell cycle control and mitosis, E Amino Acid metabolism and transport, F Nucleotide metabolism and transport, G Carbohydrate metabolism and transport, H Coenzyme metabolism, I Lipid metabolism, J Translation, K Transcription, L Replication and repair, M Cell wall/membrane/envelope biogenesis, N Cell motility, O Post-translational modification, protein turnover, chaperone functions, P Inorganic ion transport and metabolism, Q Secondary Structure, T Signal Transduction, U Intracellular trafficking and secretion, Y Nuclear structure, Z Cytoskeleton, R General Functional Prediction only, S Function Unknown. (TIF 127 kb)
Footnotes
Ruth McNerney and Taane G Clark are joint authors
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JEP, FC, RM, and TGC conceived the project. IB, RMA, RW, SLS, NCGvP, JRG, ACC, AA, TBB, SC, KD, LG, RH, ZH, AM, DM, SP, JP, IP, PS, EdOS, EMS, PDvH, MV, and AP contributed isolates, sequencing resources and/or reagents. JEP, FC, SC, MLH and TGC contributed towards data analysis. JEP, RM and TGC wrote the first version of the manuscript, with all other authors contributing to the final version. All authors have read and approve of the final version of the manuscript.
Contributor Information
Jody E. Phelan, Email: jody.phelan@lshtm.ac.uk
Francesc Coll, Email: francesc.coll@lshtm.ac.uk.
Indra Bergval, Email: i.bergval@kit.nl.
Richard M. Anthony, Email: r.anthony@kit.nl
Rob Warren, Email: rw1@sun.ac.za.
Samantha L. Sampson, Email: ssampson@sun.ac.za
Nicolaas C. Gey van Pittius, Email: ngvp@sun.ac.za
Judith R. Glynn, Email: Judith.glynn@lshtm.ac.uk
Amelia C. Crampin, Email: mia.crampin@lshtm.ac.uk
Adriana Alves, Email: geral@ispup.up.pt.
Theolis Barbosa Bessa, Email: theolis@gmail.com.
Susana Campino, Email: susana.campino@lshtm.ac.uk.
Keertan Dheda, Email: keertan.dheda@uct.ac.za.
Louis Grandjean, Email: Louis.Grandjean@lshtm.ac.uk.
Rumina Hasan, Email: rumina.hasan@aku.edu.
Zahra Hasan, Email: zahra.hasan@aku.edu.
Anabela Miranda, Email: bsp1am@hotmail.com.
David Moore, Email: david.moore@lshtm.ac.uk.
Stefan Panaiotov, Email: spanaiotov@yahoo.com.
Joao Perdigao, Email: jrlperdigao@gmail.com.
Isabel Portugal, Email: isabel.portugal@ff.ul.pt.
Patricia Sheen, Email: patricia.sheen@gmail.com.
Erivelton de Oliveira Sousa, Email: oliveira.lacenbahia@gmail.com.
Elizabeth M. Streicher, Email: lizma@sun.ac.za
Paul D. van Helden, Email: pvh@sun.ac.za
Miguel Viveiros, Email: MViveiros@ihmt.unl.pt.
Martin L. Hibberd, Email: martin.hibberd@lshtm.ac.uk
Arnab Pain, Email: arnab.pain@kaust.edu.sa.
Ruth McNerney, Email: ruth.mcnerney@gmail.com.
Taane G. Clark, Email: taane.clark@lshtm.ac.uk
References
- 1.World Health Organization . Global Tuberculosis Report 2014. 2014. [Google Scholar]
- 2.Wilkie MEM, McShane H. TB vaccine development: where are we and why is it so difficult? Thorax. 2015;70:299–301. doi: 10.1136/thoraxjnl-2014-205202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, Viveiros M, Portugal I, Pain A, Martin N, Clark TG. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014;5:4812. [DOI] [PMC free article] [PubMed]
- 4.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7:328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 5.de Jong BC, Hill PC, Aiken A, Awine T, Antonio M, Adetifa IM, Jackson-Sillah DJ, Fox A, Deriemer K, Gagneux S, Borgdorff MW, McAdam KPWJ, Corrah T, Small PM, Adegbola RA. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J Infect Dis. 2008;198:1037–43. [DOI] [PMC free article] [PubMed]
- 6.Caws M, Thwaites G, Dunstan S, Hawn TR, Lan NTN, Thuong NTT, Stepniewska K, Huyen MNT, Bang ND, Loc TH, Gagneux S, van Soolingen D, Kremer K, van der Sande M, Small P, Anh PTH, Chinh NT, Quy HT, Duyen NTH, Tho DQ, Hieu NT, Torok E, Hien TT, Dung NH, Nhu NTQ, Duy PM, van Vinh CN, Farrar J. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 2008;4:e1000034. doi: 10.1371/journal.ppat.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ordway DJ, Shang S, Henao-Tamayo M, Obregon-Henao A, Nold L, Caraway M, Shanley CA, Basaraba RJ, Duncan CG, Orme IM. Mycobacterium bovis BCG-mediated protection against W-Beijing strains of Mycobacterium tuberculosis is diminished concomitant with the emergence of regulatory T cells. Clin Vaccine Immunol. 2011;18:1527–35. [DOI] [PMC free article] [PubMed]
- 8.Niemann S, Supply P. Diversity and evolution of Mycobacterium tuberculosis: moving to whole-genome-based approaches. Cold Spring Harb Perspect Med. 2014;4:a021188. doi: 10.1101/cshperspect.a021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fishbein S, van Wyk N, Warren RM, Sampson SL. Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol Microbiol. 2015. [DOI] [PubMed]
- 10.Akhter Y, Ehebauer MT, Mukhopadhyay S, Hasnain SE. The PE/PPE multigene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: perhaps more? Biochimie. 2012;94:110–116. doi: 10.1016/j.biochi.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 11.van Pittius NC G, Sampson SL, Lee H, Kim Y, van Helden PD, Warren RM. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol. 2006;6:95. doi: 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bottai D, Di Luca M, Majlessi L, Frigui W, Simeone R, Sayes F, Bitter W, Brennan MJ, Leclerc C, Batoni G, Campa M, Brosch R, Esin S. Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol Microbiol. 2012;83:1195–209. [DOI] [PubMed]
- 13.Majlessi L, Prados-Rosales R, Casadevall A, Brosch R. Release of mycobacterial antigens. Immunol Rev. 2015;264:25–45. doi: 10.1111/imr.12251. [DOI] [PubMed] [Google Scholar]
- 14.Delogu G, Cole ST, Brosch R. Handbook of Tuberculosis: Molecular Biology and Biochemistryitle. 2008. The PE and PPE Protein Families of Mycobacterium Tuberculosis; pp. 131–150. [Google Scholar]
- 15.Mohareer K, Tundup S, Hasnain SE. Transcriptional regulation of Mycobacterium tuberculosis PE/PPE genes: a molecular switch to virulence? J Mol Microbiol Biotechnol. 2011;21:97–109. doi: 10.1159/000329489. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Dong D, Tang S, Chen X, Gao Q. PPE38 of Mycobacterium marinum triggers the cross-talk of multiple pathways involved in the host response, as revealed by subcellular quantitative proteomics. J Proteome Res. 2013;12:2055–2066. doi: 10.1021/pr301017e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh KK, Zhang X, Patibandla AS, Chien P, Laal S. Antigens of Mycobacterium tuberculosis expressed during preclinical tuberculosis: serological immunodominance of proteins with repetitive amino acid sequences. Infect Immun. 2001;69:4185–4191. doi: 10.1128/IAI.69.6.4185-4191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galagan JE. Genomic insights into tuberculosis. Nat Rev Genet. 2014;15:307–320. doi: 10.1038/nrg3664. [DOI] [PubMed] [Google Scholar]
- 19.Gutacker MM, Mathema B, Soini H, Shashkina E, Kreiswirth BN, Graviss EA, Musser JM. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J Infect Dis. 2006;193:121–8. [DOI] [PubMed]
- 20.Musser JM, Amin A, Ramaswamy S. Negligible Genetic Diversity of Mycobacterium tuberculosis Host Immune System Protein Targets: Evidence of Limited Selective Pressure. Genetics. 2000;155:7–16. doi: 10.1093/genetics/155.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Copin R, Coscollá M, Seiffert SN, Bothamley G, Sutherland J, Mbayo G, Gagneux S, Ernst JD. Sequence diversity in the pe_pgrs genes of Mycobacterium tuberculosis is independent of human T cell recognition. MBio. 2014;5:e00960–13. [DOI] [PMC free article] [PubMed]
- 22.Mycobacterium tuberculosis 18b genome. [http://www.ncbi.nlm.nih.gov/nuccore/CP007299.1]
- 23.Rodríguez JG, Pino C, Tauch A, Murcia MI. Complete Genome Sequence of the Clinical Beijing-Like Strain Mycobacterium tuberculosis 323 Using the PacBio Real-Time Sequencing Platform. Genome Announc. 2015;3. [DOI] [PMC free article] [PubMed]
- 24.Pepperell CS, Casto AM, Kitchen A, Granka JM, Cornejo OE, Holmes EC, Birren B, Galagan J, Feldman MW. The role of selection in shaping diversity of natural M. tuberculosis populations. PLoS Pathog. 2013;9:e1003543. [DOI] [PMC free article] [PubMed]
- 25.McEvoy CRE, Cloete R, Müller B, Schürch AC, van Helden PD, Gagneux S, Warren RM, Gey van Pittius NC. Comparative analysis of Mycobacterium tuberculosis pe and ppe genes reveals high sequence variation and an apparent absence of selective constraints. PLoS One. 2012;7:e30593. [DOI] [PMC free article] [PubMed]
- 26.Liu X, Gutacker MM, Musser JM, Fu Y-X. Evidence for recombination in Mycobacterium tuberculosis. J Bacteriol. 2006;188:8169–8177. doi: 10.1128/JB.01062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, Wong WSW, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 30.Graña M, Bellinzoni M, Bellalou J, Haouz A, Miras I, Buschiazzo A, Winter N, Alzari PM. Crystal structure of Mycobacterium tuberculosis LppA, a lipoprotein confined to pathogenic mycobacteria. Proteins. 2010;78:769–72. [DOI] [PubMed]
- 31.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, Nicol M, Niemann S, Kremer K, Gutierrez MC, Hilty M, Hopewell PC, Small PM. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–73. [DOI] [PMC free article] [PubMed]
- 32.Tsolaki AG, Hirsh AE, DeRiemer K, Enciso JA, Wong MZ, Hannan M, Goguet de la Salmoniere Y-OL, Aman K, Kato-Maeda M, Small PM. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci U S A. 2004;101:4865–70. [DOI] [PMC free article] [PubMed]
- 33.Målen H, Berven FS, Fladmark KE, Wiker HG. Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics. 2007;7:1702–1718. doi: 10.1002/pmic.200600853. [DOI] [PubMed] [Google Scholar]
- 34.Sampson SL. Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin Dev Immunol. 2011;2011:497203. doi: 10.1155/2011/497203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karboul A, van Pittius NC G, Namouchi A, Vincent V, Sola C, Rastogi N, Suffys P, Fabre M, Cataldi A, Huard RC, Kurepina N, Kreiswirth B, Ho JL, Gutierrez MC, Mardassi H. Insights into the evolutionary history of tubercle bacilli as disclosed by genetic rearrangements within a PE_PGRS duplicated gene pair. BMC Evol Biol. 2006;6:107. [DOI] [PMC free article] [PubMed]
- 36.Rocha EPC, Smith JM, Hurst LD, Holden MTG, Cooper JE, Smith NH, Feil EJ. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239:226–35. [DOI] [PubMed]
- 37.Chaitra MG, Shaila MS, Nayak R. Evaluation of T-cell responses to peptides with MHC class I-binding motifs derived from PE_PGRS 33 protein of Mycobacterium tuberculosis. J Med Microbiol. 2007;56(Pt 4):466–474. doi: 10.1099/jmm.0.46928-0. [DOI] [PubMed] [Google Scholar]
- 38.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, Ernst JD, Gagneux S. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. [DOI] [PMC free article] [PubMed]
- 39.Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, Warren RM, Streicher EM, Calver A, Sloutsky A, Kaur D, Posey JE, Plikaytis B, Oggioni MR, Gardy JL, Johnston JC, Rodrigues M, Tang PKC, Kato-Maeda M, Borowsky ML, Muddukrishna B, Kreiswirth BN, Kurepina N, Galagan J, Gagneux S, Birren B, Rubin EJ, Lander ES, Sabeti PC, Murray M. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet. 2013;45:1183–9. [DOI] [PMC free article] [PubMed]
- 40.Mehaffy C, Guthrie JL, Alexander DC, Stuart R, Rea E, Jamieson FB. Marked microevolution of a unique Mycobacterium tuberculosis strain in 17 years of ongoing transmission in a high risk population. PLoS One. 2014;9:e112928. doi: 10.1371/journal.pone.0112928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coll F, Preston M, Guerra-Assunção JA, Hill-Cawthorn G, Harris D, Perdigão J, Viveiros M, Portugal I, Drobniewski F, Gagneux S, Glynn JR, Pain A, Parkhill J, McNerney R, Martin N, Clark TG. PolyTB: a genomic variation map for Mycobacterium tuberculosis. Tuberculosis (Edinb). 2014;94:346–54. [DOI] [PMC free article] [PubMed]
- 42.Coll F, McNerney R, Preston M, Guerra-Assunção JA, Warry A, Hill-Cawthorn G, Mallard K, Nair M, Miranda A, Alves A, Perdigão J, Viveiros M, Portugal I, Hasan Z, Hasan R, Glynn JR, Martin N, Pain A, Clark TG. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med. 2015, In Press. [DOI] [PMC free article] [PubMed]
- 43.Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. The MaSuRCA genome assembler. Bioinformatics. 2013;29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012;22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin A V, Sirotkin A V, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. [DOI] [PMC free article] [PubMed]
- 47.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed]
- 49.Tsai IJ, Otto TD, Berriman M. Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol. 2010;11:R41. doi: 10.1186/gb-2010-11-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. [DOI] [PMC free article] [PubMed]
- 51.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vilella AJ, Blanco-Garcia A, Hutter S, Rozas J. VariScan: Analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics. 2005;21:2791–2793. doi: 10.1093/bioinformatics/bti403. [DOI] [PubMed] [Google Scholar]
- 53.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 54.Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S, Røder G, Peters B, Sette A, Lund O, Buus S. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2:e796. [DOI] [PMC free article] [PubMed]
- 55.Guerra-Assunção JA, Houben RMGJ, Crampin AC, Mzembe T, Mallard K, Coll F, et al. Recurrence due to Relapse or Reinfection With Mycobacterium tuberculosis: A Whole-Genome Sequencing Approach in a Large, Population-Based Cohort With a High HIV Infection Prevalence and Active Follow-up. J Infect Dis. 2014 doi: 10.1093/infdis/jiu574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guerra-Assunção J, Crampin A, Houben R, Mzembe T, Mallard K, Coll F, et al. Large-scale whole genome sequencing of M. tuberculosis provides insights into transmission in a high prevalence area. Elife. 2015;4. doi:10.7554/eLife.05166. [DOI] [PMC free article] [PubMed]
- 57.Bergval I, Coll F, Schuitema A, de Ronde H, Mallard K, Pain A, et al. A proportion of mutations fixed in the genomes of in vitro selected isogenic drug-resistant Mycobacterium tuberculosis mutants can be detected as minority variants in the parent culture. FEMS Microbiol Lett. 2015;362:1–7. doi: 10.1093/femsle/fnu037. [DOI] [PubMed] [Google Scholar]
- 58.Hasan Z, Ali A, McNerney R, Mallard K, Hill-Cawthorne G, Coll F, et al. Whole genome sequencing-based characterization of extensively drug resistant (XDR) strains of Mycobacterium tuberculosis from Pakistan. Int J Mycobacteriology Elsevier. 2015;4:11–12. doi: 10.1016/j.ijmyco.2014.10.050. [DOI] [Google Scholar]
- 59.Perdigão J, Silva H, Machado D, Macedo R, Maltez F, Silva C, et al. Unraveling Mycobacterium tuberculosis genomic diversity and evolution in Lisbon, Portugal, a highly drug resistant setting. BMC Genomics. 2014;15:991. doi: 10.1186/1471-2164-15-991. [DOI] [PMC free article] [PubMed] [Google Scholar]


