Abstract
Crossover recombination reshuffles genes and prevents errors in segregation that lead to extra or missing chromosomes (aneuploidy) in human eggs, a major cause of pregnancy failure and congenital disorders. Here, we generate genome-wide maps of crossovers and chromosome segregation patterns by recovering all three products of single female meioses. Genotyping > 4 million informative single-nucleotide polymorphisms (SNPs) from 23 complete meioses allowed us to map 2,032 maternal and 1,342 paternal crossovers and to infer the segregation patterns of 529 chromosome pairs. We uncover a novel reverse chromosome segregation pattern in which both homologs separate their sister chromatids at meiosis I; detect selection for higher recombination rates in the female germline by the elimination of aneuploid embryos; and report chromosomal drive against non-recombinant chromatids at meiosis II. Collectively, our findings reveal that recombination not only affects homolog segregation at meiosis I but also the fate of sister chromatids at meiosis II.
INTRODUCTION
Errors in chromosome segregation during the meiotic divisions in human female meiosis are a major cause of aneuploid conceptions, leading to implantation failure, pregnancy loss, and congenital disorders1. The incidence of human trisomies increases exponentially in women from ~ 35 years of age, but despite conservative estimates that 10-30% of natural conceptions are aneuploid2, the underlying causes and their relative contributions are still unclear. In addition to maternal age, one important factor that predisposes to missegregation in both sexes is altered recombination. Recombinant chromosomes in the offspring are the result of crossovers, the reciprocal exchange of DNA between homologous chromosomes (homologs). Together with sister chromatid cohesion, crossovers physically link the homolog pair together during the prophase stage of meiosis (Fig. 1a), which takes place during foetal development in females. The linkages have to be maintained for decades, because the two rounds of chromosome segregation only occur in the adult woman. By following the pattern of genetic markers such as single nucleotide polymorphisms (SNPs) on the two chromosomes inherited from the mother in trisomic conceptions, it has been inferred that some crossovers occur too close to centromeres1, 3-6, where they may disrupt the cohesion between the two sister chromatids7, 8. Other crossovers have been suggested to be too far from the centromeres to mediate correct attachment, or to be lacking altogether (non-exchange, E0)1, 3-6. If these inferences are correct, it follows that events that shape the recombination landscape in oocytes during foetal development affect the risk of women having an aneuploid conception decades later in adult life.
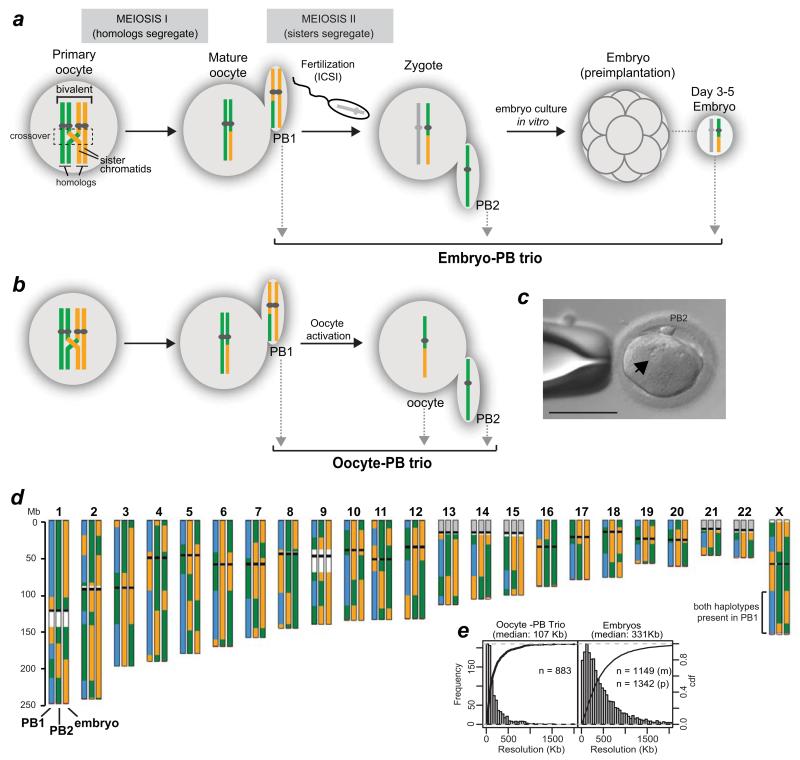
Figure 1.
Human MeioMaps from embryos and oocytes together with their corresponding polar bodies.
(a,b) The genotypes of the two maternal chromosomes are shown as green and yellow. Crossovers, shown in the dashed box, occurs during foetal development. The two polar bodies were sequentially biopsied (grey arrows) to avoid misidentification. Maternal MeioMaps were deduced from the embryo following intracytoplasmic sperm injection (ICSI) or directly assessed in the haploid oocyte, after artificial activation (b).
(c) An activated oocyte with a single pronucleus (arrow) and PB2. Scale bar: 110 μm.
(d) An example of a MeioMap after genome-wide SNP detection and phasing (see Methods). Each chromosome is represented by three vertical columns representing the three cells of the trio (PB1, PB2, and embryo or oocyte). The two phased maternal haplotypes are represented by green and yellow. Blue represents the detection of both haplotypes. Regions where SNPs are not available on the array are shown in white (repetitive sequences on chr. 1 and 9) or gray (rDNA). Black bars illustrate the position of the centromere. Red bars shows the last informative SNPs to call. Crossovers are manifested as reciprocal breakpoints in haplotypes (green to yellow, blue to green, etc.) in two of the three cells. Note that the colours of the haplotype blocks between different chromosomes are not necessarily derived from the same grandparent. Histograms of the resolution of the crossovers are shown in (e). The resolution was 352 Kb and 311 Kb for maternal (m) and paternal (p) crossovers in the embryos, respectively.
A limitation of these extensive population-based studies, however, is that only one of the products of meiosis is analysed (the oocyte). This prevents direct identification of the origin of chromosome segregation errors and provides only partial information on the crossovers during prophase of meiosis I. The ‘missing data’ problem is so significant that even the meiotic origin of age-related trisomies has been challenged recently9. Another confounding factor is that spontaneous miscarriages, still and live births on which our current knowledge is based represent only a minor fraction of the aneuploid embryos at conception. The majority of affected embryos are lost throughout pregnancy resulting in major preclinical and clinical losses2. Thus, to understand the origin of human aneuploidies, we need to assess all three meiotic products in unselected oocytes and embryos.
RESULTS
MeioMaps of single meioses in oocytes and embryos
To follow genome-wide recombination and chromosome segregation simultaneously, we recovered all three products of female meiosis, which include the first and second polar bodies (PB1 and PB2) and the corresponding activated oocytes or fertilised embryos. We refer to these as oocyte-PB or embryo-PB trios (Fig 1a-c). 10 embryo-PB trios were obtained after fertilisation of the oocyte following intracytoplasmic sperm injection (ICSI). The embryos reached various stages of preimplantation development and originated from a single donor having preimplantation genetic screening for recurrent miscarriage and who consented to follow up genetic analysis of her embryos (Fig. 1a, Supplementary Table 1). A further 13 trios were generated without fertilisation by activating mature MII-arrested oocytes with a calcium ionophore, which induced completion of MII and extrusion of the PB2 (Fig. 1b,c). This method was highly successful (85%, n = 40, Supplementary Table 2) and did not alter the rate of meiosis II errors in the activated oocytes compared to embryos generated by ICSI (6 of 299 versus 4 of 230; Table 1). The oocyte-PB trios were obtained from five healthy female donors, who had cryopreserved unfertilised eggs in the course of fertility treatment. Four of the five donors had achieved a pregnancy and live birth following IVF and all five consented to their remaining eggs being activated and undergoing genome analyses. The principle of isolating all three meiotic products is similar to the approach of using the polar bodies and recovering the female pronucleus from zygotes10.
Table 1. Origin and incidence of maternal aneuploidies.
| Chromosome missegregation eventsd | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dataset: | Mean maternal agea | nb | % aneuploid oocytes | All events | Aneuploid outcome in oocyte | Gain in oocyte | Loss in oocyte | Total chr | ||
| MI | MII | MI | MII | |||||||
| Oocyte-PB trios | 37.3 (33-41) | 13 | 62% | 26 | 12 | 2 | 4 | 4 | 2 | 299 |
| Embryo-PB trios | 38.3 | 10 | 70% | 19 | 8 | 4 | 1 | 0 | 3 | 230 |
| Embryo only | 37.1 (34-42) | 29c | 54% | n.d. | 19 | 5 | 4 | n.d | n.d. | 667 |
Mean age and range.
Number of trios or embryos analysed
28 embryos and 1 chorionic villus sample.
Statistical test for significance of MII nondisjunction rates in oocyte-PB and embryo-PB trios: 6 out of 299 compared to 4 out of 230, respectively, G-test with Williams’ correction, p = 0.82.
n.d., not determined since no information from polar bodies.
The trio datasets were complemented with data on recombination and aneuploidy rates from 29 embryos (without polar bodies) in which SNP genotyping and Karyomapping11 had previously been used for preimplantation genetic diagnosis. Because informative SNPs were available from both the mother and father, we were able to compare recombination in paternal and maternal chromosomes and their association with aneuploidy in embryos (Supplementary Table 3).
All samples were amplified by whole genome amplification and genotyped at ~300,000 SNP loci genome-wide11. Across the 23 complete trios (meioses), we detected > 4 million informative SNPs at high stringency with an average resolution of 30 kb. The SNPs spanned > 92% of the genome. For the oocyte-PB trios, genomic DNA from each donor was also genotyped to identify informative heterozygous SNP loci (hetSNPs). For the oocyte-PB trios, all heterozygous SNPs in the mother’s genomic DNA are informative, whereas in embryos, maternal and paternal hetSNPs may be shared. Hence, the pattern of recombination in the paternal chromosomes was analysed by Karyomapping11, 12 and only the two subsets of SNP loci which were heterozygous in the father and homozygous in the mother (or vice versa) were identified and used to phase the two haplotypes from the given parent in the embryo11, 12. The informative SNPs were phased using ‘siblings’10 that contain only a single chromatid from their mother (PB2, oocytes or maternal chromatid in embryo) or father (embryos). The informative SNPs were phased by selecting a PB2 or oocyte/embryo as a reference (also known as ‘assumed ancestor’)10 and inferring the crossover positions in the assumed offsprings (i.e. trios from the same parent; Supplementary Figure 1). Crossovers in the same position in the assumed offspring are highly unlikely to occur and these common crossovers can therefore be used to re-form the reference genome from which the two haplotypes can be deduced (Supplementary Figure 1). Since many of our samples were single cells, we validated our workflow by comparing recombination maps in 15 individual cells from a donor to the genomic DNA of the child, and by assessing 10 individual blastomeres from the same embryo for direct comparison. In all cases, concordance of recombination frequencies and their positions was > 99%.
A typical MeioMap from a normal embryo-PB trio is shown in Fig 1d. MeioMaps reveal Mendelian segregation of sequence polymorphisms (green and yellow segregate 2:2 across haplotype regions) and independent assortment of different chromosomes in meiosis I (pericentromeric SNPs are used as a chromosome’s fingerprint). Crossovers, which result in recombinant chromosomes, are evident by transitions between the two maternal haplotypes (green or yellow) in the PB2 and oocyte, or between a single maternal haplotype and heterozygous regions (PB1). 39 cases of aneuploidy were detected by the absence or presence of SNPs from an entire chromosome (Table 1). The inferred chromosomal aneuploidies can be observed by array CGH (Supplementary Figure 2). We also detected three gross structural rearrangements to chromosomes. Since two of the three meiotic products were affected (reciprocal gain and loss), it rules out that these rearrangements occurred during germline development and demonstrates that such rearrangements can occur during meiosis (Supplementary Table 4). Aneuploidy rates and the contribution of MI and MII errors were similar to those expected for this age range (33-41 years; Table 1)13-17.
All gains and losses were reciprocal and involved two meiotic products, such that a gain in the oocyte was matched by the loss of the chromosome in the PB1 or PB2. Of the 529 chromosome pairs assessed in the trios, we did not detect any deviation from the four chromatids expected to participate in meiosis. These observations firmly establish meiotic errors as the main contributor of aneuploid conceptions and do not support germline mosaicism in chromosome number prior to meiosis9 as a significant factor in the maternal age-related increase in human trisomies.
A novel, reverse segregation pattern in human meiosis
To understand the nature of missegregation, we inferred chromosome segregation from the trios by following the informative SNPs at the pericentromere. Trisomies that occur at a high rate in the natural population of women of advanced maternal age4 were originally hypothesized to arise by MI nondisjunction (MI NDJ) where both homologs segregate to the oocyte at meiosis I, followed by a normal second division18 (Fig. 2a, Supplementary Figure 3a). However, cytological examination of human oocytes that failed to fertilise in IVF clinics suggested that precocious separation of sister chromatids (PSSC) was the major cause of human age-related trisomies19, at least in a clinical setting (Fig. 2a, Supplementary Figure 3b). Having the genetic identity of the chromatids not only from the embryos or oocytes but also their matched polar bodies allows the two segregation patterns to be distinguished, because the chromosome signatures in the two PBs will differ (Fig. 2a). Confirming previous studies using array CGH for copy number analysis in trios20, classical meiosis I nondisjunction was relatively rare and precocious separation of sister chromatids was more frequent, at least in stimulated IVF-treated patients (Fig. 2a-c). The preponderance of PSSC compared to meiosis I nondisjunction is consistent with findings in oocytes from younger Chinese donors, although aneuploidy rates are much lower in this age group (Fig. 2b)10.
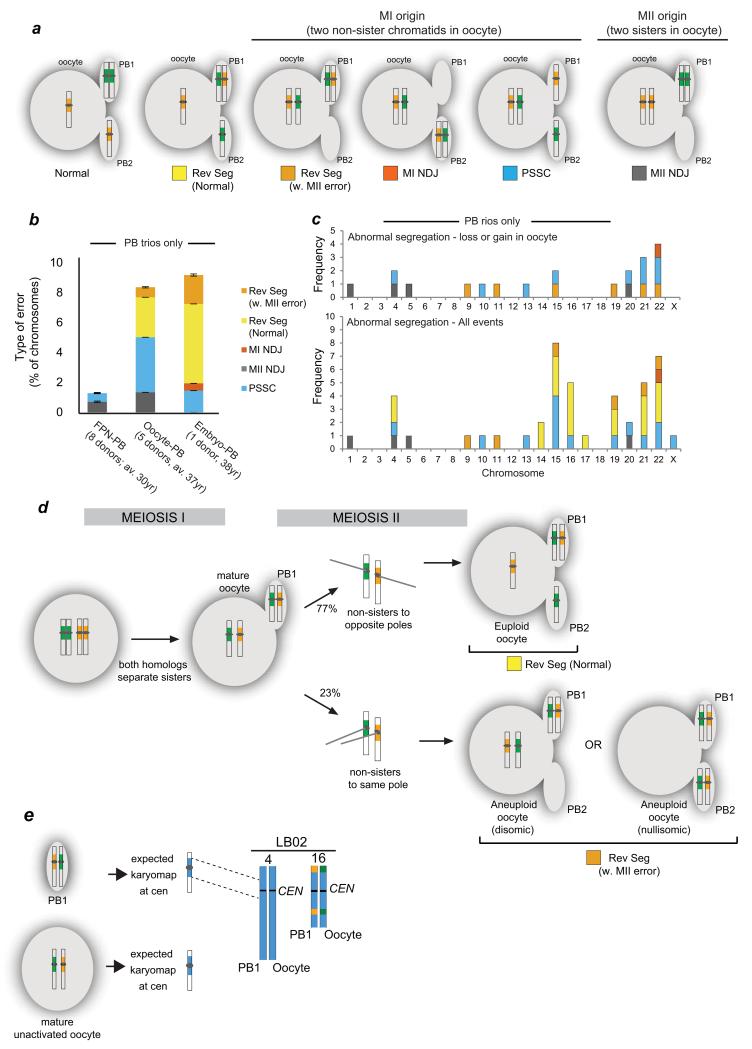
Figure 2.
MeioMaps reveal origin of aneuploidies and a novel chromosome segregation pattern.
(a) Segregation patterns revealed from following the pericentromeric haplotypes (yellow and green around centromere) in all three products of female meiosis. Only examples leading to trisomic conceptions are shown. For all possible segregation patterns detected by Meiomapping see Supplementary Figure 3. MI NDJ: meiosis I nondisjunction; Rev Seg reverse segregation; PSSC: precocious separation of sister chromatids; MII NDJ: meiosis II nondisjunction.
(b) Incidence and type of segregation errors in oocyte-PB and embryo-PB trios. Errors detected in MeioMaps generated from the female pronucleus (FPN-PB) from a younger donor population10 are shown for comparison. The number of donors and average (av.) age are shown. Age ranges were 25-35 for FPN-PB10 and 33-41 for oocyte-PB trios. The embryo donor was 38 years (Supplementary Table 1 and 2). Bars: standard error of a proportion.
(c) PB trios.
(d) Inferred mode of reverse segregation (Rev Seg). Frequencies are shown in Table 2. Alternative segregation outcomes at meiosis II (euploid and aneuploid, n = 26; p < 0.025; binomial exact test with correction for continuity).
(e) Detection of the inferred intermediate of reverse segregation, a mature oocyte and PB1 containing two non-sister chromatids each. Two mature oocytes that contained a PB1 but were unactivated were biopsied and the SNPs detected genome-wide. The expected chromosome fingerprints that contained heterozygous SNPs around the centromeres are shown in blue. Two examples were found in this egg (chromosomes 4 and 16; Table 2).
Unexpectedly, the most frequent non-canonical segregation pattern gave rise to a PB1 that contained two non-sister chromatids (green and yellow fingerprints around the centromere, n = 26). In 20 of the 26 instances, both the oocyte and the PB2 contained a normal chromosome content, but with non-sisters instead of sister chromatids (Fig. 2a, Rev Seg). This pattern cannot be detected by copy number analysis used previously20, since the complement of chromosomes in the three cells is normal. We refer to this novel pattern as reverse segregation, since we infer that sister chromatids of both homologs separated first in meiosis I, followed by non-sister chromatids in meiosis II (Fig. 2d). The equational division at MI is unlikely to be the result of two independent PSSC events, because the observed frequency of both homologs separating their sister chromatids is more than 100× greater than the predicted frequency based on two independent PSSC events (p < 0.001). Consistent with equational divisions of both homologs at meiosis I, we observed the predicted intermediate of reverse segregation, a mature oocyte and PB1 that contain two non-sister chromatids (Fig. 2e, Table 2). Both acrocentric and larger metacentric chromosomes displayed this reverse segregation pattern (Fig. 2c, Table 2), which was observed in all donors, ruling out that it was specific to certain women (Supplementary Table 5). In the remaining six cases, the two non-sister chromatids missegregated into the egg or the PB2, resulting in an aneuploid oocyte (Fig. 2a, Rev Seg w. MII, Supplementary Figure 3d). In summary, we have observed a novel segregation pattern where both homologs undergo an equational division at meiosis I, followed by a weak preference for accurate disjunction of the two non-sister chromatids at meiosis II. This is reminiscent of ‘inverted meiosis’ in organisms with holocentric chromosomes21-23.
Table 2. Incidence of reverse segregation.
| Sample type | Incidence | Chromosomes involved |
|---|---|---|
| Oocyte-PB1 duos (unactivated)a | 8.7 ± 4.2% (n = 46) | 4, 13, 14, 16 |
| Oocyte-PB1-PB2 trios | 3.7 ± 1.1% (n = 299) | 4, 11, 14, 15, 16, 19, 22 |
| Embryo-PB1-PB2 trios | 7.2 ± 1.8% (n = 207) | 4, 9, 16, 17, 19, 21, 22 |
See Fig. 2e. Reverse segregation was observed in all donors (SupplementalryTable 5).
Variation in global recombination rates in adult oocytes
Variation in recombination in foetal oocytes has been hypothesized to give rise to vulnerable crossover configurations that predispose chromosome pairs to missegregation decades later in the adult woman. To assess recombination in adult oocytes and embryos, we mapped 883 maternal crossovers in the oocyte-PB trios and 1149 and 1342 maternal and paternal crossovers, respectively, in the embryos (Fig. 3a; Supplementary Tables 5-9). 12% of the reciprocal crossover events mapped to non-sister chromatids in the PB2 and oocyte, whilst the PB1 was heterozygous for the SNPs. A similar proportion would be expected to be present in the PB1 and are undetectable, since the two DNA strands cannot currently be separated and phased individually10. Using the 300K SNP arrays gave median resolutions of 107 Kb and 331 Kb for crossovers in the oocyte-PB and embryos, respectively (Fig. 1e). This is similar to high-resolution population-based studies employing SNP arrays24-26.
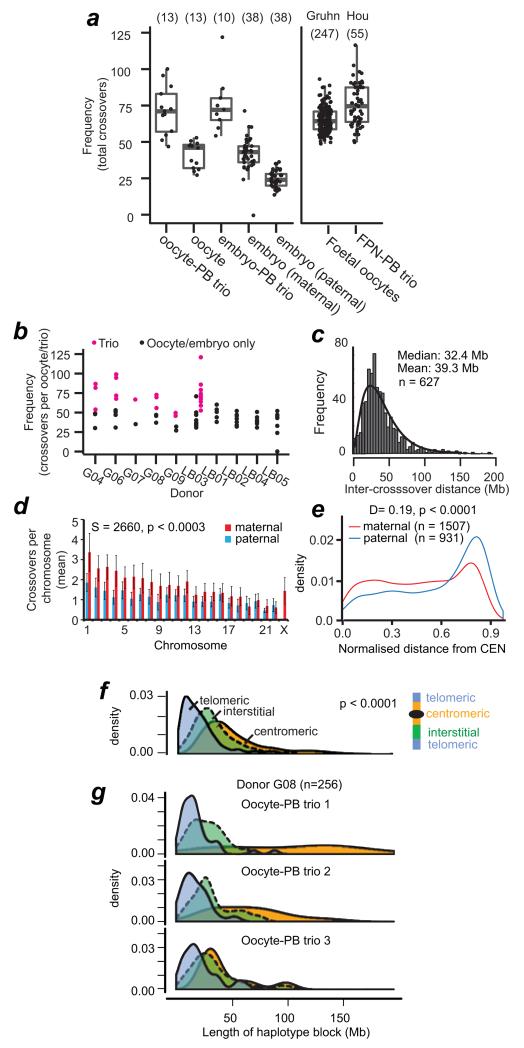
Figure 3.
Variation in genome-wide recombination rates between and within individuals.
(a) Boxplot of global recombination rates showing the interquartile range (box), median (horizontal bar), and whiskers (1.5× IQR). Numbers analysed are in parentheses. Rates from foetal oocytes (‘Gruhn’)29 and female pronucleus-PB trios (‘Hou’)10.
(b) Recombination rates for the 10 donors. Black: rates using information from oocyte or embryo only. Magenta: rates using the information from the complete oocyte-PB trios.
(c) Inter-crossover distances, excluding centromeric distances. The fitted curve is based on maximum likelihood estimation of the gamma distribution, shape: 2.6141 ± 0.14 (S.E.), rate 0.066 ± 0.0039 (S.E.). Estimated fitted mean: 39.3 Mb, log-likehood of fitting: −2802.738 ; AIC: 5609.476.
(d) Average and standard deviation of chromosome-specific recombination (Supplementary Table 3). GLM analysis revealed that chromosome size had a significant effect on sex-specific recombination frequencies. Spearman correlation test is shown for the p-value for individual, pair-wise comparisons between maternal and paternal recombination frequencies per chromosome. As chromosome size decreases, the contribution of sex to crossover frequencies decreases (see Source Data for Fig. 3d).
(e) Crossover position relative to centromeres (CEN), normalized to chromosome length. Statistics: Two-sided Kolmogorov-Smirnov test of normalized and absolute lengths; p < 0.0005; X chr. excluded; Supplementary Figure 5).
(f) Length of haplotype blocks (not inter-crossover distances), according to position relative to telomeres (blue), centromeres (yellow), or interstitial (green). Statistics: non-parametric ANOVA (p < 0.0001). Centromeric blocks excluded the ~ 3 × 106 base pairs of alpha-satellite DNA.
(g) Variation in centromere repression of crossovers in oocyte-PB trios from the same donor.
Several observations support the conclusion that recombination rates in the adult oocytes and embryos are highly variable, like those seen in unselected, foetal oocytes27-29. At the same time, the average recombination frequencies are reminiscent of those reported for human populations. The average number of maternal crossovers in the oocyte or embryos was 41.6 ± 11.3 S.D. (n = 51 ; Supplementary Tables 5-9). This rate is consistent with estimates from foetal oocytes and population-based assessments10, 24, 26, 30-36 and those detected in the female pronucleus (42.5 ± 9.0 S.D., n = 52)10. The frequencies of crossovers detected in the egg correlated well with those in the PB1 or PB2 (Supplementary Figure 4). The maternal recombination rates and the lengths of haplotype blocks were highly variable between donors as well as within donors27, 28, 35, 37, varying by as much as two-fold (Fig. 3b, f and g; Supplementary Figure 4). Using the oocyte-PB trios, maternal crossovers displayed a median distance of 32.4 Mb, which was in excess of the 18.3 Mb predicted by random distribution of crossovers along chromosomes (Online Methods). This is consistent with crossover interference along homolog pairs10.
Embryos contain informative markers of both maternal and paternal origin. This allows us to assess recombination of both sexes in unselected embryos for the first time. Maternal recombination rates were 1.63-fold higher than paternal rates in the embryos, consistent with population-based studies and molecular approaches on single sperm and foetal oocytes26-32. The additional maternal recombination events are in part from female-only recombination along the X chromosome and in part from higher crossover frequencies on larger autosomes (Fig. 3d). Maternal recombination was more centromeric compared to paternal events (Fig. 3e, Supplementary Figure 5), although centromeres tended to suppress nearby recombination10, 26-32 (Fig. 3f). However, the suppression of centromeric crossovers varied amongst oocyte-PB trios, even within the same woman (Fig. 3g). This variation may predispose some oocytes to crossovers positioned too close to centromeres that may interfere with segregation. Collectively, these observations reveal that the variation in total crossover numbers detected in adult oocytes is analogous to the variation in Mlh1 counts observed in foetal oocytes27-29, suggesting that Mlh1 foci serve as a good proxy for crossover recombination events in human oocytes. Simultaneously, the average recombination rates are reminiscent of those in the human population. This validates our approach and lends support to the hypothesis that the variability in the rates and distribution of recombination events between and within individuals give rise to vulnerable crossover configurations in foetal oocytes that are propagated to adult oocytes and, ultimately, embryos.
Global recombination rates as a risk factor for aneuploidy
To understand how the variability of maternal recombination rates affects human aneuploidy, we addressed whether the global, genome-wide recombination rates were correlated with the incidence of aneuploidy in individual oocytes and embryos. Indeed, the global recombination rate was a strong predictor of aneuploidy (Fig. 4a), even when we excluded an outlier embryo, which contained 12 aneuploidies and no detectable crossovers amongst any of the chromosome pairs. The recombination rate is an important factor, accounting for 18% of the variation in the incidence of aneuploidy (outlier excluded; permutation test).
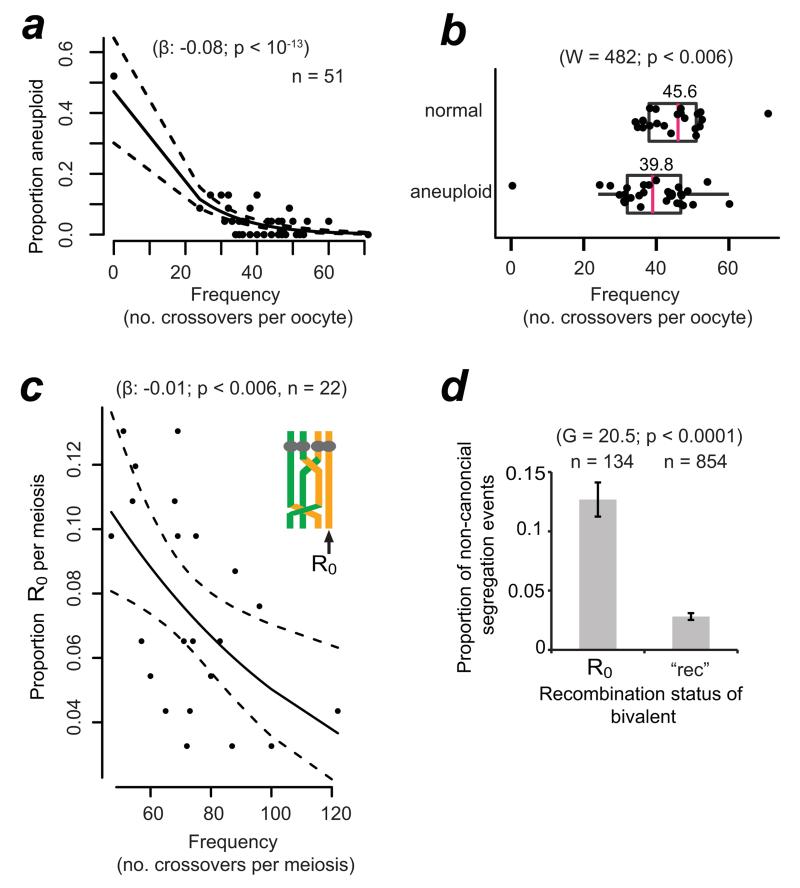
Figure 4.
Higher global recombination rates protect against aneuploidy and are selected for in the human female germline.
(a) Logistic regression of the frequency of aneuploid chromosomes as a function of global recombination rate in the embryo or oocyte. Black lines shows logistic regression model and 95% confidence interval (dashed line; binomial family). When the outlier with 0 recombination events was omitted, the regression coefficient β was −0.06 and still highly significant (p < 0.003). The outlier was omitted from all subsequent statistical analyses.
(b) Recombination rates in normal versus aneuploid oocytes and embryos. The arithmetic mean is shown above of the median (magenta, vertical bar). Statistics: Mann-Whitney-Wilcoxon test; one-sided.
(c) Incidence of bivalents containing at least one non-recombinant chromatid (R0) as a function of global recombination rates in oocyte-PB and embryo-PB trios. Statistics as in (a).
(d) Segregation errors amongst chromosomes that contained one or more R0 or where all four chromatids had recombined (‘rec’). p-values from G-test of heterogeneity (two-sided) are shown. Bars represent standard errors of a proportion (√[p ×(1−p)/n]).
If lower global recombination rates predispose oocytes to meiotic chromosome segregation errors, then normal euploid embryos should contain chromosomes that underwent higher maternal genome-wide recombination frequencies than those of aneuploid embryos. To examine whether this was the case, we divided the embryos and oocytes into two groups (euploid or aneuploid) and determined their respective recombination rates (Fig. 4b). Normal, euploid oocytes and embryos had on average 5.8 recombination events more than aneuploid ones. This difference was significant even when we accounted for crossovers that may not be detected due to the presence of two chromosomes in the aneuploid oocyte10. Notably, the overlap in the distribution of recombination rates between the euploid and aneuploid groups is consistent with the presence of other factors that influence the fidelity of chromosome segregation1. Our findings suggest that higher global recombination frequencies, which are determined during foetal development, protect against errors in chromosome segregation decades later in the adult woman. When errors do occur, they give rise to aneuploidy, many of which are selected against prior to the implantation of the embryo38. One implication of this is that recombination rates may be under selection in women as they enter their 30s, increasing rates by as much as 14% in women of advanced maternal age (5.8/41.5, the overall average).
Non-recombinant chromatids are at risk of PSSC
How do global recombination rates affect the segregation outcomes of individual homolog pairs? We hypothesized that lower global recombination rates might increase the risk of generating vulnerable crossover configurations. We first considered non-exchange E0 homolog pairs, which would give rise to trios, where the PB1 contains one homolog (green or yellow) and the oocyte and PB2 one sister each from the other homolog (Supplementary Figure 6a). Of 529 chromosome pairs, no such example was observed in our data, although one case was observed by Hou et al. (Hou, pers. comm.). E0 may be extremely rare, or another possibility is that they missegregate. Indeed, we observed 13 putative E0 from the 529 chromosome pairs across the 23 trios (Supplementary Figure 6d-g). The overall incidence (2.6%, n = 506) and the overrepresentation of the two smallest chromosomes (21 and 22) are reminiscent of observations of cytological markers for crossovers on foetal chromosomes in meiotic prophase27-29. The observed incidence of presumed E0 was much lower than expected if crossovers were randomly distributed amongst chromosomes (Supplementary Figure 6h), suggestive of crossover assurance mechanism(s) in human oocytes. None of the presumed E0 chromosomes followed a classical meiotic segregation pattern. Instead they all underwent PSSC or reverse segregation (with or without MII missegregation; Supplementary Figure 6). This is consistent with the bi-orientation of sister chromatids of univalent chromosomes at meiosis I in model organisms39, 40.
Informative SNPs on missegregated chromosomes cannot be phased, making crossovers undetectable (Supplementary Figure 6). However, most of the presumed E0 contained non-recombinant chromatids (R0). Fig. 4c shows that global recombination rates are important for determining the generation of R0, which in turn are at increased risk of missegregation compared to fully recombinant bivalents (all four chromatids engaged in recombination; “rec”, Fig. 4d). Bivalents that contained a R0 were preferentially involved in PSSC, suggesting that non-recombinant chromatids are at risk of precociously separating from their sister at meiosis I. It is possible that non-recombinant chromatids are at elevated risk of becoming dissociated from the rest of the bivalent during the decades-long dictyate arrest41, 42. We conclude that recombination affects not only the generation and segregation of putative non-exchange homolog pairs, but also influences the dynamics of sister chromatid segregation.
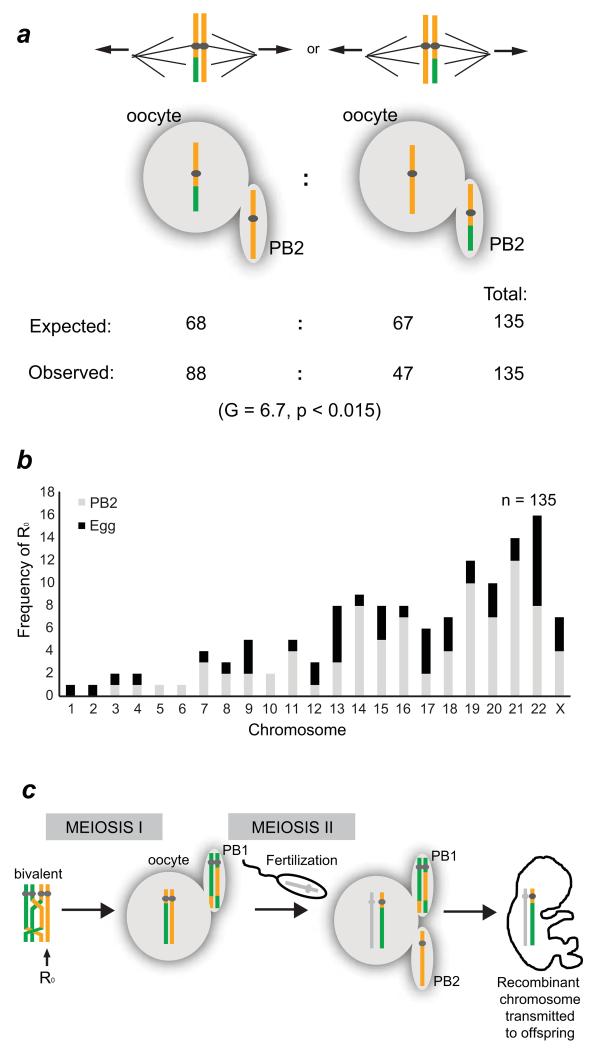
Meiotic drive for recombinant chromatids at meiosis II
Nonrecombinant chromatids are not only at risk of PSSC, but their segregation at meiosis II is also affected by the lack of recombination. The MeioMaps revealed 135 chromatids in the oocyte or PB2 that were non-recombinant and had segregated normally (Fig. 5a). These R0 are expected to be randomly distributed amongst the oocyte and the PB2. Contrary to this expectation, R0 were nearly twice as likely to be found in the PB2 than the oocyte. The selection appears to be against non-recombinant chromatids, because when both sisters recombined, their segregation was random and the recombination rates were similar in the oocyte and PB2 (Supplementary Table 5). We infer that when the two sister chromatids segregated at meiosis II, non-recombinant chromatids were preferentially driven into the PB2 and thus eliminated from the human germline (Fig. 5b,c). The use of the asymmetric cell divisions during oogenesis for the preferential inclusion of an allele43 or even whole chromosomes44-46 is referred to as meiotic or chromosomal drive. The meiotic drive against non-recombinant chromatids resulted in a 6.6% elevation in the recombination rates in oocytes compared to the PB2s (Supplementary Table 5). These findings imply that recombination is not only important for the accurate segregation of homologs at meiosis I, but also acts as a driving force during sister chromatid segregation at meiosis II. Selection against non-recombinant chromatids may prevent entire chromosomes from being inherited as a single haplotype block, thereby reducing the probability of inbreeding or propagation of segregation distorters47-49. This may be significant in terms of population structure and the genomic health of children. The difference in genome structure between the PB2 and oocyte is particularly relevant, because the PB2 has been proposed for use in treatment of mitochondrial disease50.
Figure 5.
Meiotic drive for recombinant chromatids at meiosis II increases recombination rates in the human female germline.
(a) Sister chromatids are expected to segregate randomly at meiosis II. However, when chromosomes that contained one non-recombinant chromatid and one recombinant one segregated, the recombinant chromatid was twice as likely to segregate to the oocyte. G-test for proportions (two-sided).
(b) Chromosome-specific frequencies of R0 chromatids segregating to the PB2 or oocyte.
(c) Diagrammatic representation of meiotic drive against non-recombinant chromatids at meiosis II in the human female germline. Paternal chromosome is shown in gray.
DISCUSSION
Until recently, recombination and chromosome segregation were studied in populations, where missing polar body information was not available; or in foetal oocytes, which arise decades prior to the segregation events being studied. MeioMaps from unselected adult oocytes, the female pronucleus in zygotes10, and embryos, now provide a ‘missing link’ between events that occur during foetal development and their influence on chromosome segregation outcomes decades later in the adult oocyte.
Recombination rates in the unselected oocytes were 1.6-fold higher than in males and showed a broad distribution, similar to the high degree of variation in foetal oocytes27-29. Sex-specific differences in chromosome structure during meiotic prophase have been suggested to explain this difference, with female chromosomes having a longer axis and shorter chromatin loops51. The increased loop number correlates with the increased recombination rate in female meiosis29, 51. Although the mean female recombination rates were similar to those seen in populations, the range was substantially broader. We found that lower genome-wide recombination rates were selected against because they were less likely give rise to an euploid oocyte. This is consistent with findings that Down Syndrome individuals have lower genome-wide recombination rates compared to their euploid siblings25. The degree of selection is not observed in younger women10 and could contribute to the higher recombination rates in children as mothers age24, 32, 52. This model predicts that children born to younger mothers should display a broader range in recombination frequencies compared to those born to women of advanced maternal age.
Lower genome-wide recombination rates increase the risk of at least two types of vulnerable crossover configurations: non-recombinant chromatids (R0) and putative non-exchange homologs (E0). Non-recombinant chromatids (R0) are a risk factor for PSSC and their preferential segregation to the second polar body at meiosis II (meiotic drive). The putative E0 either underwent PSSC or a novel reverse segregation pattern, where sister chromatids of both homologos separated at meiosis I, followed by a weak preference of accurate division of the two non-sister chromatids at meiosis II. The reverse segregation pattern is not limited to E0 and could be the result of centromeric crossovers that fall at or within 1-2 Mb of the centromeres, the positions of the last informative SNPs (Supplementary Table 10). Centromeric crossovers interfere with segregation of sister chromatids in Drosophila7 and budding yeast8, and are associated with an increased risk of aneuploidy in humans1. The relatively high incidence of MII nondisjunction (23%, n = 26) associated with reverse segregation could be explained by crossover in the extreme vicinity of centromeres. Another possible mechanism that seems particularly plausible for the larger metacentric chromosomes where two crossovers would have to occur within 1 Mb on both sides of the centromere, is that homologs segregated their sister chromatids in an equational fashion in MI, followed by a weak preference for accurate non-sister chromatid segregation at MII (77% compared to 50% expected from random; n = 26; p < 0.05). It is possible that failure to establish crossovers (E0) or maintain the bivalent structure during the extended dictyate arrest may predispose to the equational division at meiosis I. This could occur by deterioration of cohesion between sister chromatids, sister kinetochores, or bivalents falling apart into univalents. There is evidence for the latter in human MI oocytes53, 54, but it is unknown whether their frequencies and chromosome-specific effects match the maternally-derived, age-related component underlying human aneuploidies. In mouse oocytes, univalents preferentially segregate sister chromatids at meiosis I39, 40 and this may also be case in humans. At meiosis II, non-sister chromatids could be physically attached by unresolved recombination intermediates (joint molecules)55, other threads21, 22, 56, or the oocyte may use segregation mechanisms that do not rely on physical attachment between chromosomes57. The relative contributions of reverse segregation mechanisms and centromeric crossovers remain to be determined, but in either case demonstrate that events attributed to mistakes in chromosome segregation in meiosis II can have their origin at meiosis I in human female meiosis.
ONLINE METHODS
Ethics statement
All material for the study was ethically sourced with fully informed patient consent. All oocytes for the study were obtained from donors after completion of their IVF treatment and were destroyed for analysis. The oocytes used were vitrified in accordance with Italian law in place at the time of oocyte retrieval for IVF treatment. Use of the oocytes for the study was approved by the Institutional Review Board of the Valle Giulia Clinic where the oocytes were stored and did not influence patient treatment. All embryo samples for the study were either obtained by tubing embryos in their entirety (destroyed) for analysis following a previous abnormal outcome in clinical tests or reanalysis of a clinical biopsy samples after embryos were transferred, stored or discarded depending on the clinical result. SNP genotyping was performed as clinical follow up/validation of clinical genetic analysis and covered by the HFEA code of practice. All primary data were encoded such that informative SNPs were represented as A and B. Only secondary data with informative SNPs encoded A and B were used for data analysis.
Oocyte-PB trios
Patient participation and consent
All MII oocytes for the study were obtained from patients undergoing ICSI treatment in the Centre for Reproductive Medicine GENERA in Rome between 2 September 2008 and 15 May 2009 following controlled ovarian hyperstimulation performed using two different protocols: GnRH-agonist long protocol and GnRH-antagonist protocol. According to the Italian law in force when these oocytes were collected, a maximum of three oocytes could be inseminated per patient. The remaining MII oocytes were vitrified and later recruited for the study after informed consent was obtained from the patients. The study and the informed consent were approved by the Institutional Review Board of the Valle Giulia Clinic and did not influence patient treatment.
Oocyte collection
Oocyte collection was performed at 35 h post-hCG administration. Removal of the cumulus mass was performed by brief exposure to 40 IU/ml hyaluronidase solution in Sage fertilization medium + 10% human serum albumin (HSA) (Cooper Surgical, USA), followed by mechanical removal of the corona radiata with the use of plastic “denuding” pipettes of defined diameters (COOK Medical, Ireland) in a controlled 6% CO2 and 37°C environment. This procedure was performed between 37 and 40 h post-hCG administration. MII oocytes were then identified for vitrification.
Oocyte vitrification and warming
The vitrification and warming procedures were performed according to Kuwayama et al. 58, 59. Commercially available vitrification and warming kits were used (Kitazato BioPharma Co., Japan). The vitrification procedure was performed a maximum of 40 hours post hCG administration. The oocytes were stored on a cryotop vitrification tool (Kitazato BioPharma Co., Japan) with a plastic cap for protection during storage in liquid nitrogen. All oocytes were stored submerged in liquid nitrogen until warming was performed. Following oocyte warming degenerated oocytes were discarded and the surviving oocytes were cultured before biopsy of the first polar body (PB1) and activation.
Oocyte culture and activation
All oocyte culture was performed at 37°C in 6% CO2 and 5% O2. To enable tracking of the oocytes and PBs, individual culture was performed and culture drops and wells were numbered to allow traceability throughout the experiment.
Immediately after warming, the surviving oocytes were allocated to individually numbered 35 μl microdrops of Sage cleavage medium + 10% HSA under mineral oil (Cooper Surgical, USA) and cultured for 2 hours prior to PB1 biopsy and activation.
Oocytes were activated by exposure to activation medium: 100 μM calcium ionophore (A23187, C7522 Sigma-Aldrich) in Sage cleavage + 10% HSA (Cooper Surgical, USA) from a stock solution in DMSO (Sigma-Aldrich) diluted 1:40. Oocytes were transferred to 35 μl drops of the activation medium under Sage oil, numbered appropriately. Activation culture was performed for 40-120 mins. The oocytes were then moved to post activation culture.
Post activation culture was performed in separate wells of EmbryoScope slides (Unisence Fertilitech, Denmark) in cleavage medium - medium as used in post warm culture under Sage oil. The slides were placed in the EmbryoScope time lapse incubator (Unisence Fertilitech, Denmark) for assessment of second polar body (PB2) extrusion and appearance of pronuclei prior to PB2 biopsy.
Polar body biopsy
Polar bodies were biopsied sequentially in order to discriminate between the 3 products of meiosis using micromanipulators (Narishige, Japan) on an inverted microscope (Nikon Ltd, Japan) equipped with Hoffman Modulation contrast and a 37°C heating stage (Linkam Scientific Instruments, UK). The first polar body (PB1) was biopsied prior to oocyte activation and the second polar body (PB2) was biopsied following its extrusion, post activation as previously described by Capalbo et al.16. All biopsies were performed in individually numbered 10 μl microdrops of HEPES medium + 10% HSA under Sage oil (Cooper Surgical, USA) for traceability. For both PB1 and PB2 biopsies, oocytes were positioned on the microscope to give a clear view of the PB and secured by suction with the holding pipette (TPC, Australia). An aperture was made in the zona pelucida with a series of laser pulses (Saturn laser; Research Instruments, UK) working inwards from the outer surface of the zona. The aspiration pipette (zona drilling pipette; TPC, Australia) was then inserted through the opening and the PB removed with gentle suction. PB1 biopsy: Once biopsied the oocytes were moved to activation culture leaving the biopsied PB1 in the microdrop for immediate transfer to a 0.2 ml, RNase and DNase free thin walled, flat cap PCR tube (Corning, Sigma-Aldrich) for DNA amplification. PB2 biopsy: once biopsied the PB2 was immediately transferred to a PCR tube for DNA amplification with the oocyte still in the microdrop. The oocyte was then returned to the micromanipulator for full zona removal. The zonae were removed from the oocytes using the same setup for the biopsy procedure. The oocyte was anchored to the holding pipet and a larger aperture was made in the zona using laser pulses. The oocyte was removed from the zona using both displacement and zona manipulation techniques with the aspiration pipette. Once free from the zonae, the oocytes were transferred to PCR tubes for DNA amplification.
Transfer of the samples to PCR tubes was performed using a plastic denuding pipette (COOK Medical, Ireland) with a 130 μm lumen. Individually labelled PCR tubes were primed with 2 μl Dulbecco’s phosphate buffered saline (DPBS) (Gibco, Life technologies) with 0.1% polyvinyl alcohol (Sigma-Aldrich). Individual samples were expelled into the DPBS in around 1 μl of the medium containing the samples, leaving a final volume of no more than 4 μl of medium with the sample in the PCR tubes. The PCR tubes were then briefly centrifuged, snap frozen in liquid nitrogen and stored at −20 °C prior to whole genome amplification.
DNA extraction and Whole Genome Amplification (WGA)
Genomic DNA (gDNA) from all oocyte donors was obtained using buccal cell swabs (Isohelix, Cell Projects Ltd). Extraction of the gDNA from the swabs was performed using a proteinase K extraction kit to a final volume of 30 μl, following the manufacturer’s instructions. DNA from all three products of meiosis was obtained by lysis of the cells and whole-genome amplication (WGA). The PCR tubes containing the samples were brought to an end volume of 4 μl with PBS and REPLI-g Single Cell Kit multiple displacement amplification (SureMDA, Illumina) or PCR library based SurePlex amplification (Illumina) was performed according to the manufacturer’s instructions. MDA was performed with a short 2h incubation.
Embryos and embryo-PB trios
Embryo samples
Thirty five embryos diagnosed as affected and/or aneuploid were analysed from four clinical cases for either preimplantation genetic diagnosis (PGD) of single gene defects or preimplantation genetic screening (PGS) for aneuploidy following standard IVF protocols at The Bridge Centre, London with patients informed consent. SNP genotyping was performed for quality control purposes following clinical biopsy and genetic testing of the embryos under the HFEA clinic licence L0070-14-a using similar methods to those described for the processing of the oocyte-PB trios.
In one of the PGD cases, two surplus denuded MI oocytes were allowed to mature in vitro by overnight culture in Sage fertilisation medium +10% HSA under mineral oil (Cooper Surgical). Biopsy of PB1, tubing and WGA of the oocyte and PB1 were then performed as described for the oocyte-PB trios.
Embryo-PB trios
In another PGS case, in which array CGH had been used to detect aneuploidy by copy number analysis of both polar bodies, the WGA products (Sureplex; Illumina, San Diego, CA, USA) from both polar bodies were SNP genotyped along with parental genomic DNAs and, with patients informed consent, WGA products (SureMDA; Illumina, San Diego, CA, USA) of nine corresponding fertilised embryos which had all been diagnosed as aneuploid.
Array CGH, SNP bead array and data analysis
For array CGH analysis, 4 μl aliquots of Sureplex single cell amplified DNA Products (PB1, PB2, oocyte or blastomere) were processed on microarray slides (24Sure; Illumina, USA). The data was imported and analysed using dedicated software (BlueFuse Multi v 4.0; Illumina, USA).
For SNP genotyping, 400 ng of genomic DNA or 8 μl of WGA products from the single cell and embryo samples (PB1, PB2, oocyte, single blastomere or whole embryo) were processed on a SNP genotyping beadarray (Human CytoSNP-12 or Human Karyomapping beadarray; Illumina, San Diego, CA, USA) for ~300K SNPs, using a shortened protocol and the genotype data analysed using a dedicated software programme for karyomapping (Bluefuse Multi v 4.0; Illumina, San Diego, CA, USA) or exported as a text file for analysis in Micrsoft Excel12.
MeioMap analysis
Following SNP genotyping, MeioMaps were constructed and displayed by importing the data into Microscoft Excel and processing using custom macros written in Visual Basic for Applications. For the oocyte-PB trios, a simple algorithm was used to phase all heterozygous maternal SNP loci using a haploid PB2 or oocyte sample as a reference. This defined a reference set of homozygous SNP loci (haplotype) genome wide (AA or BB), across each chromosome. The genotype of each of the samples including the reference were then interrogated at each of these informative SNP loci and displayed as either the same as the reference (yellow) or opposite to the reference (green) or heterozygous (blue) indicating the presence of both maternal haplotypes. Phase transitions at crossovers were then manually tagged in Excel by copying the closest SNP calls bracketing the crossover and the type and position of these SNPs imported into a second spreadsheet for further processing. Because phasing is achieved using a reference sample, any phase transitions caused by crossovers in that particular sample appear in identical positions in all other samples analysed (with the exception of any crossover between the reference and the PB2 or oocyte in that trio). Macros in the second spreadsheet therefore identified these common crossovers, restored them to the reference sample and removed them from all of the other samples. The meiomaps were then displayed, checked and further edited manually as necessary. All oocyte-PB trios were run with at least two references to MeioMap any aneuploid chromosomes in the reference trio and to double-check all crossovers.
For embryo-PB trios, two methods were used. Where the SNP genotype of a close relative or, in some cases, a sibling embryo was available, the samples were karyomapped using the standard algorithm which identifies informative SNP loci for all four parental haplotypes in either Excel or using dedicated software (Bluefuse Multi v 4.0; Illumina, San Diego, CA) 11, 12. Alternatively to improve resolution, a modified karyomapping algorithm, with a PB2 or oocyte as reference, was used. This algorithm identified all combinations of parental genotypes that were informative for the maternal haplotype only. In either case, the phase transitions were manually tagged and imported into the second spreadsheet for further processing, display and final editing as above.
We validated our workflow on single cells by comparing recombination maps in 15 individual cells from a donor to the genomic DNA of the child, and by assessing 10 individual blastomeres from the same embryo for direct comparison. In all cases, concordance of recombination rates and positions was >99% (data not shown).
Simulation: crossover distribution amongst chromosomes and distances between crossovers along chromosomes
12% of crossovers were mapped in the PB2 and oocyte. A similar proportion would be expected to be present in the PB1, but are undetectable since the heterozygous SNPs (blue regions) cannot be phased.
Maternal recombination rates in the oocyte-PB and embryo-PB trios were similar to those reported from foetal oocytes27-29 as well as female pronucleus-PB trios from young women10. When only the embryo or oocyte was used, >50% of crossovers went undetected in the two polar bodies (Fig.3a-b). The recombination rates in the oocyte or embryo correlated well with those discerned from polar bodies (Supplementary Figure 3). This supports the notion that recombination occurs randomly amongst non-sister chromatids. Indeed, when homolog pairs had engaged in crossing over twice, no evidence of increased or decreased probability of the same two chromatids engaging in the second crossover was detected (Methods). This is consistent with reports that the preference for two sister chromatids to re-engage in a second crossover given their involvement in the first (negative chromatid interference) is very weak10.
Simulations were performed to allocate a specified number of crossover events to set of chromosomes. Chromosomes were allocated a specified length using the minimum and maximum crossover locations mapped within the experimental dataset. Crossovers were allocated randomly to chromosomes with weighted probability using the chromosome length, thus longer chromosomes receive more crossovers. The allocation was either totally random (non-obligate) or random following allocation of one crossover per chromosome (obligate). For each chromosome the positions of the allocated crossovers was determined iteratively by randomly selecting an available location. The available locations were all possible positions not within a minimum distance (107 kb) from the existing crossover positions. The simulation reported the total number of crossovers per chromosome and the inter-crossover distances. The distance from the outermost crossover to the chromosome termini was not included. 10,000 simulations were performed to create the distributions. The scripts (Ottolini_Scripts_CrossoverData.pl) are freely available under copyright and GNU public licence.
To estimate the fraction of missed crossovers, we randomly distributed 125 crossovers amongst chromosomes with a minimum distance of 0 kb between them (Ottolini_Scripts_CrossoverData.pl). A cumulative distribution of inter-crossover distances was constructed, ignoring crossover distances that were adjacent to telomeres. The cumulative frequencies was 0.04% at 10 kb, 0.15% at 30 kb, 0.52% at 107 kb, 0.75% at 150 kb, and 1% at 200 kb.
Chromatid interference
To detect chromatid interference, we identified 134 chromosome pairs with two crossovers and we asked whether the same two chromatids were less or more likely to be involved in both crossover events compared to random participation. We were unable to reject the null hypothesis of no chromatid interference (p > 0.5; t-test for proportions), consistent with reports that negative chromatid interference is weak 10.
Statistics, modelling, and graphics
Statistical tests and modelling were carried out in Perl or R. All tests were permutation and non-parametric tests, or logistic regression analysis as indicated throughout the manuscript. For logistic regression, we used the AIC to choose the appropriate link function. Binomial distribution of error variances were assessed using the plot(model) function of R. Residual variance and degrees of freedom was tested using chi-square and rejected if below 5%. Two-sided tests were employed, unless otherwise indicated. We used the lme4, lmPerm, psperman libraries in R. Graphics were rendered using the basic functions in R or the ggplot2 library60.
Supplementary Material
Supplementary Table 1. Donor information of embryos.
Supplementary Table 2. Donor information of oocyte-PB trios.
Supplementary Table 3. Recombination frequencies in embryos.
Supplementary Table 4. Structural rearrangements to chromosomes in meiosis.
Supplementary Table 5. Summary of recombination and chromosome segregation in all trios.
Supplementary Table 6. Map distances in embryos.
Supplementary Table 7. Crossovers in oocytes.
Supplementary Table 8. Recombination frequencies in oocytes.
Supplementary Table 9. Map distances in oocytes.
Supplementary Table 19. hetSNP or informative SNP resolution near centromeres of chromosomes undergoing reverse segregation.
ACKNOWLEDGEMENTS
We gratefully acknowledge Celia May, Adam Eyre-Walker, Jennifer Gruhn, Ross Rowsey, James Turner, Francesca Cole, and members of the Hoffmann lab for discussion and critical reading of the manuscript. We thank Yu Hou, Wei Fan, and Sunney Xie for freely sharing data and discussion on their study of female pronucleus-PB trios. Financial support for this research was provided by the MRC (Senior Research Fellowship to ERH; G0902043) and an EMBO Young Investigator award to ERH.
Footnotes
Competing financial interests: The authors declare no competing financial interests.
REFERENCES
- 1.Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet. 2012;13:493–504. doi: 10.1038/nrg3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 3.Zaragoza MV, et al. Nondisjunction of human acrocentric chromosomes: studies of 432 trisomic fetuses and liveborns. Hum Genet. 1994;94:411–417. doi: 10.1007/BF00201603. [DOI] [PubMed] [Google Scholar]
- 4.Hassold TJ, Jacobs PA. Trisomy in man. Annu Rev Genet. 1984;18:69–97. doi: 10.1146/annurev.ge.18.120184.000441. [DOI] [PubMed] [Google Scholar]
- 5.Hassold T, et al. A cytogenetic study of 1000 spontaneous abortions. Ann Hum Genet. 1980;44:151–178. doi: 10.1111/j.1469-1809.1980.tb00955.x. [DOI] [PubMed] [Google Scholar]
- 6.Freeman SB, et al. The National Down Syndrome Project: design and implementation. Public Health Rep. 2007;122:62–72. doi: 10.1177/003335490712200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koehler KE, et al. Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nat Genet. 1996;14:406–414. doi: 10.1038/ng1296-406. [DOI] [PubMed] [Google Scholar]
- 8.Rockmill B, Voelkel-Meiman K, Roeder GS. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics. 2006;174:1745–1754. doi: 10.1534/genetics.106.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulten MA, Patel S, Jonasson J, Iwarsson E. On the origin of the maternal age effect in trisomy 21 Down syndrome: the oocyte mosaicism selection model. Reproduction. 2010;139:1–9. doi: 10.1530/REP-09-0088. [DOI] [PubMed] [Google Scholar]
- 10.Hou Y, et al. Genome analyses of single human oocytes. Cell. 2013;155:1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 11.Handyside AH, et al. Karyomapping: a universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J Med Genet. 2010;47:651–658. doi: 10.1136/jmg.2009.069971. [DOI] [PubMed] [Google Scholar]
- 12.Natesan SA, et al. Genome-wide karyomapping accurately identifies the inheritance of single-gene defects in human preimplantation embryos in vitro. Genet Med. 2014 doi: 10.1038/gim.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alfarawati S, et al. The relationship between blastocyst morphology, chromosomal abnormality, and embryo gender. Fertil Steril. 2011;95:520–524. doi: 10.1016/j.fertnstert.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Kuliev A, Zlatopolsky Z, Kirillova I, Spivakova J, Cieslak Janzen J. Meiosis errors in over 20,000 oocytes studied in the practice of preimplantation aneuploidy testing. Reprod Biomed Online. 2011;22:2–8. doi: 10.1016/j.rbmo.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Gutierrez-Mateo C, et al. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril. 2011;95:953–958. doi: 10.1016/j.fertnstert.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Capalbo A, et al. Sequential comprehensive chromosome analysis on polar bodies, blastomeres and trophoblast: insights into female meiotic errors and chromosomal segregation in the preimplantation window of embryo development. Hum Reprod. 2013;28:509–518. doi: 10.1093/humrep/des394. [DOI] [PubMed] [Google Scholar]
- 17.Franasiak JM, et al. The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril. 2014;101:656–663. e651. doi: 10.1016/j.fertnstert.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Reiger R, Michaelis A, M.M. G. Glossary of genetics and cytogenetics. Springer Verlag; Berlin, Heidelberg, New York: 1968. [Google Scholar]
- 19.Angell RR. Predivision in human oocytes at meiosis I: a mechanism for trisomy formation in man. Hum Genet. 1991;86:383–387. doi: 10.1007/BF00201839. [DOI] [PubMed] [Google Scholar]
- 20.Handyside AH, et al. Multiple meiotic errors caused by predivision of chromatids in women of advanced maternal age undergoing in vitro fertilisation. Eur J Hum Genet. 2012;20:742–747. doi: 10.1038/ejhg.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heckmann S, et al. Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nat Commun. 2014;5:4979. doi: 10.1038/ncomms5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabral G, Marques A, Schubert V, Pedrosa-Harand A, Schlogelhofer P. Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat Commun. 2014;5:5070. doi: 10.1038/ncomms6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viera A, Page J, Rufas JS. Inverted meiosis: the true bugs as a model to study. Genome Dyn. 2009;5:137–156. doi: 10.1159/000166639. [DOI] [PubMed] [Google Scholar]
- 24.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 25.Middlebrooks CD, et al. Evidence for dysregulation of genome-wide recombination in oocytes with nondisjoined chromosomes 21. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong A, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 27.Lenzi ML, et al. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis I in human oocytes. Am J Hum Genet. 2005;76:112–127. doi: 10.1086/427268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng EY, et al. Meiotic recombination in human oocytes. PLoS Genet. 2009;5:e1000661. doi: 10.1371/journal.pgen.1000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruhn JR, Rubio C, Broman KW, Hunt PA, Hassold T. Cytological studies of human meiosis: sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PLoS One. 2013;8:e85075. doi: 10.1371/journal.pone.0085075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tease C, Hartshorne GM, Hulten MA. Patterns of meiotic recombination in human fetal oocytes. Am J Hum Genet. 2002;70:1469–1479. doi: 10.1086/340734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong A, et al. Recombination rate and reproductive success in humans. Nat Genet. 2004;36:1203–1206. doi: 10.1038/ng1445. [DOI] [PubMed] [Google Scholar]
- 33.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 34.Kong A, et al. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science. 2008;319:1398–1401. doi: 10.1126/science.1152422. [DOI] [PubMed] [Google Scholar]
- 35.Kong A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 36.Kong A, et al. Common and low-frequency variants associated with genome-wide recombination rate. Nat Genet. 2014;46:11–16. doi: 10.1038/ng.2833. [DOI] [PubMed] [Google Scholar]
- 37.Chowdhury R, Bois PR, Feingold E, Sherman SL, Cheung VG. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 2009;5:e1000648. doi: 10.1371/journal.pgen.1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capalbo A, et al. Correlation between standard blastocyst morphology, euploidy and implantation: an observational study in two centers involving 956 screened blastocysts. Hum Reprod. 2014 doi: 10.1093/humrep/deu033. [DOI] [PubMed] [Google Scholar]
- 39.LeMaire-Adkins R, Hunt PA. Nonrandom segregation of the mouse univalent X chromosome: evidence of spindle-mediated meiotic drive. Genetics. 2000;156:775–783. doi: 10.1093/genetics/156.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouznetsova A, Lister L, Nordenskjold M, Herbert M, Hoog C. Bi-orientation of achiasmatic chromosomes in meiosis I oocytes contributes to aneuploidy in mice. Nat Genet. 2007;39:966–968. doi: 10.1038/ng2065. [DOI] [PubMed] [Google Scholar]
- 41.Wolstenholme J, Angell RR. Maternal age and trisomy--a unifying mechanism of formation. Chromosoma. 2000;109:435–438. doi: 10.1007/s004120000088. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Cruz R, et al. Dynamics of cohesin proteins REC8, STAG3, SMC1 beta and SMC3 are consistent with a role in sister chromatid cohesion during meiosis in human oocytes. Hum Reprod. 2010;25:2316–2327. doi: 10.1093/humrep/deq180. [DOI] [PubMed] [Google Scholar]
- 43.Sandler L, Novitski E. Meiotic drive as an evolutionary force. Am Nat. 1957;91:105–110. [Google Scholar]
- 44.Bongiorni S, Fiorenzo P, Pippoletti D, Prantera G. Inverted meiosis and meiotic drive in mealybugs. Chromosoma. 2004;112:331–341. doi: 10.1007/s00412-004-0278-4. [DOI] [PubMed] [Google Scholar]
- 45.Pardo-Manuel de Villena F, Sapienza C. Nonrandom segregation during meiosis: the unfairness of females. Mamm Genome. 2001;12:331–339. doi: 10.1007/s003350040003. [DOI] [PubMed] [Google Scholar]
- 46.Dawe RK, Cande WZ. Induction of centromeric activity in maize by suppressor of meiotic drive 1. Proc Natl Acad Sci U S A. 1996;93:8512–8517. doi: 10.1073/pnas.93.16.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haig D, Grafen A. Genetic scrambling as a defence against meiotic drive. J Theor Biol. 1991;153:531–558. doi: 10.1016/s0022-5193(05)80155-9. [DOI] [PubMed] [Google Scholar]
- 48.Chmatal L, et al. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 2014;24:2295–2300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brandvain Y, Coop G. Scrambling eggs: meiotic drive and the evolution of female recombination rates. Genetics. 2012;190:709–723. doi: 10.1534/genetics.111.136721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang T, et al. Polar body genome transfer for preventing the transmission of inherited mitochondrial diseases. Cell. 2014;157:1591–1604. doi: 10.1016/j.cell.2014.04.042. [DOI] [PubMed] [Google Scholar]
- 51.Lynn A, et al. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science. 2002;296:2222–2225. doi: 10.1126/science.1071220. [DOI] [PubMed] [Google Scholar]
- 52.Campbell CL, Furlotte NA, Eriksson N, Hinds D, Auton A. Escape from crossover interference increases with maternal age. Nat Commun. 2015;6:6260. doi: 10.1038/ncomms7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Angell R. First-meiotic-division nondisjunction in human oocytes. Am J Hum Genet. 1997;61:23–32. doi: 10.1086/513890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia-Cruz R, et al. Cytogenetic analyses of human oocytes provide new data on non-disjunction mechanisms and the origin of trisomy 16. Hum Reprod. 2010;25:179–191. doi: 10.1093/humrep/dep347. [DOI] [PubMed] [Google Scholar]
- 55.Copsey A, et al. Smc5/6 coordinates formation and resolution of joint molecules with chromosome morphology to ensure meiotic divisions. PLoS Genet. 2013;9:e1004071. doi: 10.1371/journal.pgen.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hughes SE, et al. Heterochromatic threads connect oscillating chromosomes during prometaphase I in Drosophila oocytes. PLoS Genet. 2009;5:e1000348. doi: 10.1371/journal.pgen.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hughes-Schrader S. Distance segregation and compound sex chromosomes in mantispids (Neuroptera:Mantispidae) Chromosoma. 1969;27:109–129. doi: 10.1007/BF00326139. [DOI] [PubMed] [Google Scholar]
Methods-only references
- 58.Kuwayama M. Highly efficient vitrification for cryopreservation of human oocytes and embryos: the Cryotop method. Theriogenology. 2007;67:73–80. doi: 10.1016/j.theriogenology.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 59.Kuwayama M, Vajta G, Kato O, Leibo SP. Highly efficient vitrification method for cryopreservation of human oocytes. Reprod Biomed Online. 2005;11:300–308. doi: 10.1016/s1472-6483(10)60837-1. [DOI] [PubMed] [Google Scholar]
- 60.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Use R. 2009:1–212. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Donor information of embryos.
Supplementary Table 2. Donor information of oocyte-PB trios.
Supplementary Table 3. Recombination frequencies in embryos.
Supplementary Table 4. Structural rearrangements to chromosomes in meiosis.
Supplementary Table 5. Summary of recombination and chromosome segregation in all trios.
Supplementary Table 6. Map distances in embryos.
Supplementary Table 7. Crossovers in oocytes.
Supplementary Table 8. Recombination frequencies in oocytes.
Supplementary Table 9. Map distances in oocytes.
Supplementary Table 19. hetSNP or informative SNP resolution near centromeres of chromosomes undergoing reverse segregation.