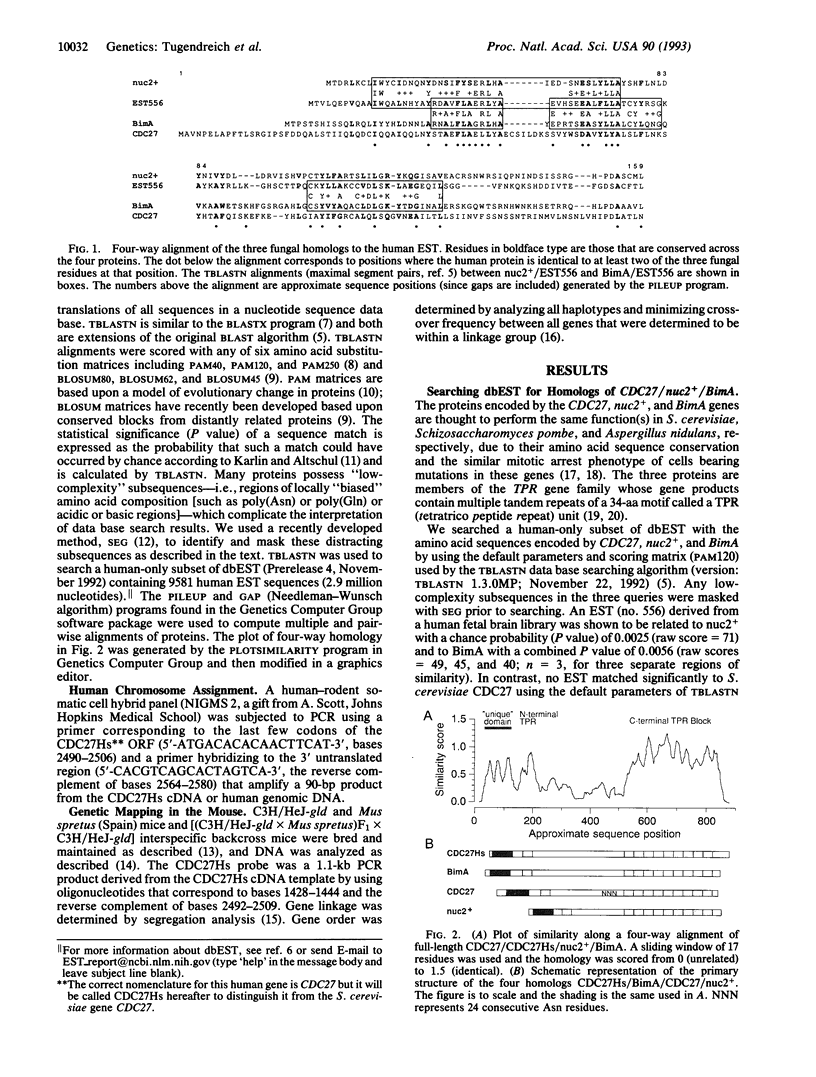
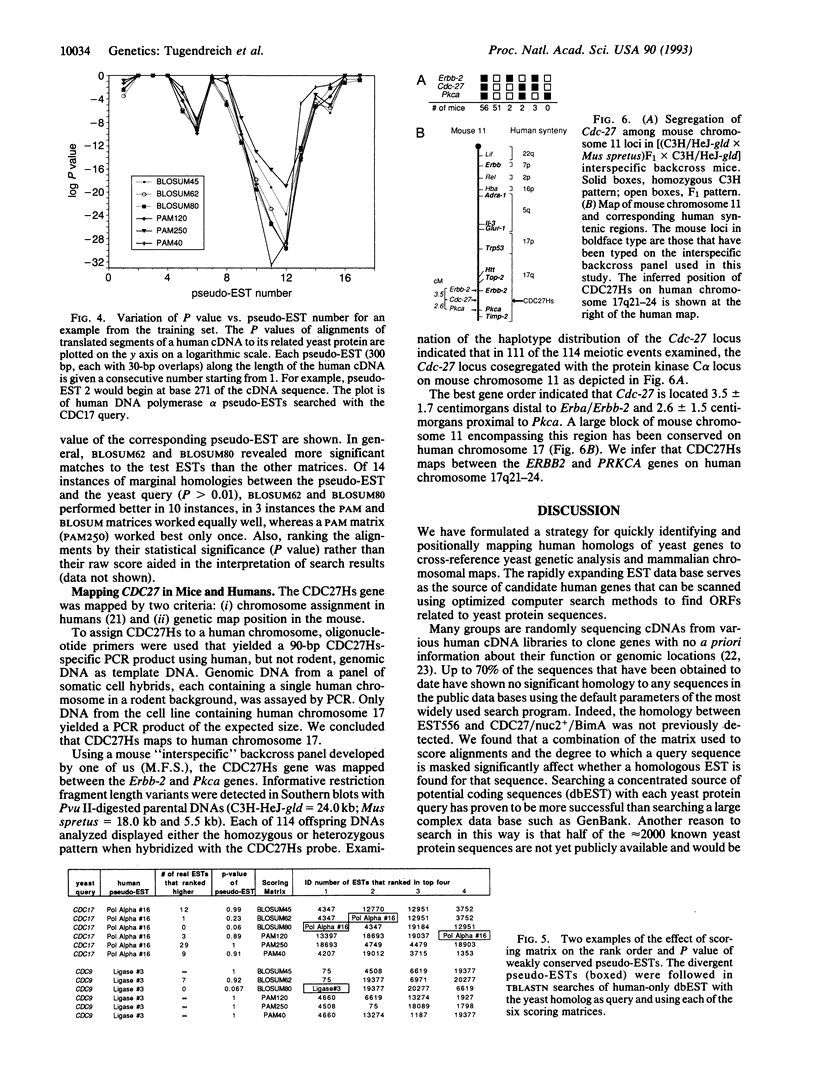
Abstract
We describe a strategy for quickly identifying and positionally mapping human homologs of yeast genes to cross-reference the biological and genetic information known about yeast genes to mammalian chromosomal maps. Optimized computer search methods have been developed to scan the rapidly expanding expressed sequence tag (EST) data base to find human open reading frames related to yeast protein sequence queries. These methods take advantage of the newly developed BLOSUM scoring matrices and the query masking function SEG. The corresponding human cDNA is then used to obtain a high-resolution map position on human and mouse chromosomes, providing the links between yeast genetic analysis and mapped mammalian loci. By using these methods, a human homolog of Saccharomyces cerevisiae CDC27 has been identified and mapped to human chromosome 17 and mouse chromosome 11 between the Pkca and Erbb-2 genes. Human CDC27 encodes an 823-aa protein with global similarity to its fungal homologs CDC27, nuc2+, and BimA. Comprehensive cross-referencing of genes and mutant phenotypes described in humans, mice, and yeast should accelerate the study of normal eukaryotic biology and human disease states.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F. Amino acid substitution matrices from an information theoretic perspective. J Mol Biol. 1991 Jun 5;219(3):555–565. doi: 10.1016/0022-2836(91)90193-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ballabio A. The rise and fall of positional cloning? Nat Genet. 1993 Apr;3(4):277–279. doi: 10.1038/ng0493-277. [DOI] [PubMed] [Google Scholar]
- Ballester R., Marchuk D., Boguski M., Saulino A., Letcher R., Wigler M., Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990 Nov 16;63(4):851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- Bishop D. T. The information content of phase-known matings for ordering genetic loci. Genet Epidemiol. 1985;2(4):349–361. doi: 10.1002/gepi.1370020404. [DOI] [PubMed] [Google Scholar]
- Boguski M. S., Lowe T. M., Tolstoshev C. M. dbEST--database for "expressed sequence tags". Nat Genet. 1993 Aug;4(4):332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Gish W., States D. J. Identification of protein coding regions by database similarity search. Nat Genet. 1993 Mar;3(3):266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Goebl M., Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991 May;16(5):173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Hiraoka Y., Yanagida M. A temperature-sensitive mutation of the Schizosaccharomyces pombe gene nuc2+ that encodes a nuclear scaffold-like protein blocks spindle elongation in mitotic anaphase. J Cell Biol. 1988 Apr;106(4):1171–1183. doi: 10.1083/jcb.106.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde S. C., Emsley P., Hartshorn M. J., Mimmack M. M., Gileadi U., Pearce S. R., Gallagher M. P., Gill D. R., Hubbard R. E., Higgins C. F. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature. 1990 Jul 26;346(6282):362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- Karlin S., Altschul S. F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K., Sterne R. E., Thorner J. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 1989 Dec 20;8(13):3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K. L., Osmani A. H., Osmani S. A., Morris N. R. bimA encodes a member of the tetratricopeptide repeat family of proteins and is required for the completion of mitosis in Aspergillus nidulans. J Cell Sci. 1991 Aug;99(Pt 4):711–719. doi: 10.1242/jcs.99.4.711. [DOI] [PubMed] [Google Scholar]
- Saunders A. M., Seldin M. F. A molecular genetic linkage map of mouse chromosome 7. Genomics. 1990 Nov;8(3):525–535. doi: 10.1016/0888-7543(90)90040-2. [DOI] [PubMed] [Google Scholar]
- Seldin M. F., Morse H. C., 3rd, Reeves J. P., Scribner C. L., LeBoeuf R. C., Steinberg A. D. Genetic analysis of autoimmune gld mice. I. Identification of a restriction fragment length polymorphism closely linked to the gld mutation within a conserved linkage group. J Exp Med. 1988 Feb 1;167(2):688–693. doi: 10.1084/jem.167.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikela J. M., Auffray C. Finding new genes faster than ever. Nat Genet. 1993 Mar;3(3):189–191. doi: 10.1038/ng0393-189. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Boguski M. S., Goebl M., Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990 Jan 26;60(2):307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- Teem J. L., Berger H. A., Ostedgaard L. S., Rich D. P., Tsui L. C., Welsh M. J. Identification of revertants for the cystic fibrosis delta F508 mutation using STE6-CFTR chimeras in yeast. Cell. 1993 Apr 23;73(2):335–346. doi: 10.1016/0092-8674(93)90233-g. [DOI] [PubMed] [Google Scholar]
- Watson M. L., D'Eustachio P., Mock B. A., Steinberg A. D., Morse H. C., 3rd, Oakey R. J., Howard T. A., Rochelle J. M., Seldin M. F. A linkage map of mouse chromosome 1 using an interspecific cross segregating for the gld autoimmunity mutation. Mamm Genome. 1992;2(3):158–171. doi: 10.1007/BF00302874. [DOI] [PubMed] [Google Scholar]
- Wilcox A. S., Khan A. S., Hopkins J. A., Sikela J. M. Use of 3' untranslated sequences of human cDNAs for rapid chromosome assignment and conversion to STSs: implications for an expression map of the genome. Nucleic Acids Res. 1991 Apr 25;19(8):1837–1843. doi: 10.1093/nar/19.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. F., Lin B., Tanaka K., Dunn D., Wood D., Gesteland R., White R., Weiss R., Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990 Nov 16;63(4):835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]




