Abstract
The establishment of synaptic plasticity and long-term memory requires lasting cellular and molecular modifications that, as a whole, must endure despite the rapid turnover of their constituent parts. Such a molecular feat must be mediated by a stable, self-perpetuating, cellular information storage mechanism. DNA methylation, being the archetypal cellular information storage mechanism, has been heavily implicated as being necessary for stable activity-dependent transcriptional alterations within the central nervous system (CNS). This review details the foundational discoveries from both gene-targeted, as well as whole-genome sequencing, studies that have successfully brought DNA methylation to our attention as a chief regulator of activity- and experience-dependent transcriptional alterations within the CNS. We present a hypothetical framework with which the disparate experimental findings dealing with distinct manipulations of the DNA methylation, and their effect on memory, might be resolved while taking into account the unique impact activity-dependent alterations in DNA methylation potentially have on both memory promoting and memory-suppressing gene expression. And last, we discuss potential avenues for future inquiry into the role of DNA methylation during remote memory formation.
Keywords: DNA methylation, Memory, synaptic plasticity, DNA demethylation, Epigenetics
Introduction
Within the past 20 years, arguably one of the most pivotal discoveries within the field of neurobiology is that long-term memory formation, and it’s cellular correlate synaptic plasticity, are reliant upon persistent alterations in gene transcription within the central nervous system (CNS) (Kandel 2001; Sweatt 2013). Experience-driven alterations in DNA methylation have emerged as a potential governor of the relatively long-lived alterations in gene expression that underlie memory formation. This review will introduce DNA methylation as a mediator of cellular information storage, while detailing the evolution of recent thought and empirical evidence, regarding its role in memory formation. Our ultimate objective is to leave readers across the wide spectrum of the neuroscience community with a sense of the current “state of the field” regarding the role conferred by DNA methylation in the neuroepigenetic regulation of memory formation.
Long-term memory formation requires the establishment of persistent neuronal synaptic connections, which, in turn, requires the induction of complex synapse-to-nuclear signal transduction cascades that lead to stable changes in gene expression (Adams and Sweatt 2002). Paradoxically, the longevity of the cellular and molecular alterations that subserve the memory-promoting enhancement in synaptic plasticity greatly outlives the maximum lifespan of the proteins that collectively constitute the molecular basis of synaptic plasticity (Crick 1984; Lisman 1985; Holliday 1999; Day and Sweatt 2010). Put another way, it is interesting that memories can last a lifetime, yet the proteins that enable synaptic plasticity, and allow for the establishment and maintenance of the memory trace, are subject to perpetual turnover. This paradoxical phenomenon is in many respect analogous to the Greek legend of the ship of Theseus which sailed for 102 years, during which time it’s entire crew, sail, and mast were replaced, thereby raising the question as to how the ship could still be that of Theseus as nothing of the original boat remained.
In order to account for the existence of long-term memory, and thus the prerequisite resolution of the memory paradox, one would expect there to exist some mechanism whereby the information pertaining to the cellular and molecular components of the memory trace could be stably stored. Moreover, such a stable cellular information storage mechanism would necessitate the existence of self-perpetuating biochemical reactions (Day and Sweatt 2010). Theoretically speaking, such a reaction would involve the perpetual self-duplication and, or, transference of cellular information from one information storage medium to another. A biochemical process that achieves cellular information storage in this manner would be referred to as an “mnemogenic”, or memory-forming, reaction (Roberson and Sweatt 2001; Day and Sweatt 2010). This mnemogenic reaction would be achieved by some molecule, after it is modified or activated by an experience, in turn being capable of directly or indirectly catalyzing the conversion of another molecule of itself (autoconvert) from inert into an active form (Day and Sweatt 2010). Such an mnemogenic reaction is therefore the sine qua non of long-term memory, as has been discussed extensively (Crick 1984; Lisman 1985; Holliday 1999; Roberson and Sweatt 2001; Day and Sweatt 2010). As will be discussed at length below, DNA methylation is exquisitely well suited to serve as a self-perpetuating information storage device, and is therefore believed to be the archetypal mnemogenic process capable of underlying long-term memory.
DNA methylation as a putative mnemogenic mechanism
For starters, DNA methylation involves the covalent attachment of a methyl moiety to the fifth position (5′) carbon within the cytosine pyrimidine ring (5-methylcytosine, 5mC) (Bird 2002). The resulting carbon-carbon bond between the 5′ carbon on the cytosine ring and the carbon on the methyl moiety is extremely stable, with demethylation requiring a prohibitively high degree of energy (Day and Sweatt 2010). In order to appreciate the nuanced, context-dependent, function of DNA methylation one must first appreciate its distribution throughout the genome and the manner by which it is regulated.
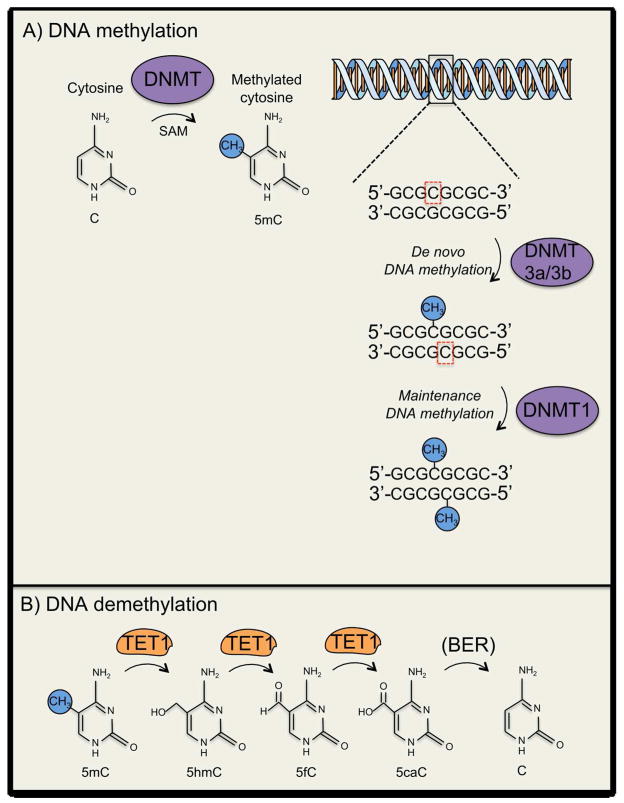
In mammals DNA methylation has been canonically thought to be restricted to cytosines that constitute palindromic cytosine-phosphate-guanine (CpG) dinucleotides that tend to be symmetrically methylated (i.e., mCpG:GpCm) (See Fig. 1A; (Miranda and Jones 2007). There are approximately 28 million CpGs in the human genome and approximately 10% of these CpGs occur in CpG-rich regions referred to as CpG-Islands (Smith and Meissner 2013). DNA methyltransferases, as the name implies, mediate this reaction, while using S-Adenosyl methionine (SAM) as the primary methyl donor (Miranda and Jones 2007). There are three conserved DNA methyltransferases (DNMTs) in mammals: the de novo DNMT3A and DNMT3B, which are canonically thought to methylate CpG pairs for which neither CpG is methylated (e.g., CpG:GpC → DNMT3A/B → mCpG/GpC) (Okano and others 1998; Okano and others 1999). Alternatively, the de novo DNA methyltransferase 1 (DNMT1) maintains the existence of the symmetrically methylated CpGs by recognizing hemimethylated DNA and methylating the currently unmethylated cytosine (i.e., CpG:GpCm → DNMT1 → mCpG/GpCm) (Hermann and others 2004).
Figure 1.
General schematic of DNA methylation and its mechanisms of regulation. (A) Methylation of DNA involves covalent addition of a methyl group to the 5′ position of the cytosine pyrimidine ring by DNMTs. DNA methylation commonly occurs at genes enriched with cytosine-guanine nucleotides (CpG islands). De novo methyltransferases (e.g., DNMT3a) methylate CpG pairs for which neither CpG is methylated (e.g., CpG:GpC → DNMT3A/B → mCpG/GpC), where as the maintenance methyltransferase (i.e., DNMT1) methylates hemimethylated DNA strands (B) General mechanisms of DNA demethylation within the mammalian central nervous system. TET1 participates in sequential 5mC oxidation prior to 5caC being subject to base-excision-repair that results in the regeneration of C. Abbreviations: 5-hydroxymethylcytosine (5hmC); 5-formylcytosine (5fC); 5-carboxylcytosine (5caC).
DNA methylation at gene promoters has been canonically associated with transcriptional suppression (Ng and Bird 1999). DNA methylation-induced transcriptional repression can generally be achieved by either direct interference with transcription factor binding, or through the recruitment of transcriptional repression complexes involving Methyl-CpG-binding protein 2 and histone deacetylases (HDACs) (Nan and others 1998; Ng and Bird 1999; Suzuki and Bird 2008; Deaton and Bird 2011). Yet over the past decade increasing evidence suggests that DNA methylation’s function as a transcriptional regulator may be more nuanced than had been previously suspected, with its influence on transcription being developmental time-point, cell-type, and genomic region-specific (Suzuki and Bird 2008; Deaton and Bird 2011; Baubec and others 2015).
With DNA methylation exhibiting the capacity for self-perpetuation, it has been theorized that it may mediate the perpetual maintenance of cellular phenotype throughout the lifetime of an organism (Crick 1984; Holliday 1999; Day and Sweatt 2010). That is to say, DNA methylation may likely serve as the principle cellular information storage device that is capable of stably and perpetually regulating cellular phenotype. Greatly intrigued that DNA methylation may be the long sought-after mnemogenic mechanism capable of stable cellular phenotype regulation within the context of synaptic plasticity, experimenters set out to ascertain its possible involvement in the persistent experience-dependent transcriptional regulation that underlies memory formation.
Early insights into the role of DNA methylation in memory formation
It was first revealed that in vitro neuronal depolarization resulted in hypomethylation within the transcriptional regulatory region of the brain-derived neurotrophic factor (BDNF) gene, along with a corresponding increase in BDNF mRNA expression (Martinowich and others 2003). This seminal finding pointed towards the dynamic regulation of DNA methylation as a potential mediator of activity-dependent transcriptional regulation within the CNS. The evidence of reduced methylation, and increased BDNF gene expression, in response to neuronal stimulation was consistent with a working model implicating transcriptional regulation as being permissive for synaptic plasticity and memory (Abel and Kandel 1998; Pittenger and Kandel 1998; Kandel 2001; Day and Sweatt 2010). In accordance with this model, HDAC-inhibitors, which were generally thought to promote gene transcription, were found to enhance synaptic plasticity (Levenson and others 2004). Therefore, it was reasoned that an intervention believed to have a positive effect on gene transcription, such as blocking DNA methylation with DNMT-inhibitors, would ultimately lead to an enhancement in synaptic plasticity. A study involving hippocampus slices bath-treated with the non-specific DNMT-inhibitor zebularine (Zeb) detected an acute (40 min) decrease in DNA methylation at the promoters of two genes whose expression is positively correlated with memory formation: Reelin (Rln) and Brain-derived neurotrophic factor exon 1 (Bdnfex1) (Weeber and others 2002; Levenson 2006; Levenson and others 2008). Yet, a perplexing result revealed that, pre-treatment with two structurally distinct DNMT-inhibitions, Zeb and 5-aza-2′-deoxycytidine (5-Aza), led to a diminution in long-term potentiation (LTP), the cellular correlate of memory (Bliss and Collingridge 1993; Levenson 2006). Although this study revealed for the first time evidence of dynamic regulation of DNA methylation within the hippocampus, the results were seemingly antithetical to the working model in which the suppression of gene expression was disruptive towards memory formation.
It was later determined that fear conditioning leads to an increase in the mRNA expression of de novo DNMTs (i.e., DNMT3a and DNMT3b), as well as a decreased in the transcript of protein phosphotase 1, catalytic subunit, beta (Ppp1cb), a gene believed to be suppressive towards synaptic plasticity and memory (Genoux and others 2002; Lee and others 2003; Miller and Sweatt 2007). Along with decreasing the expression of Ppp1cb mRNA expression, fear conditioning resulted in acute methylation of the Ppp1cb promoter (Miller and Sweatt 2007). Importantly, intra-CA1 administration of the DNMT-inhibitor 5-Aza not only impaired hippocampus-dependent contextual fear memory, but also led to an abolishment of the experience-dependent methylation of the Ppp1cb gene promoter. As expected, fear conditioning resulted in an experience-dependent demethylation of Rln and an increase in Rln gene expression. Interestingly, both of the dynamic alterations in DNA methylation at Ppp1cb and Rln were short-lived and dissipated within 24 hours (Miller and Sweatt 2007). This seminal paper served to illustrate the degree to which experience-dependent alterations can evoke gene-specific, bidirectional, and seemingly targeted, alterations in DNA methylation, thereby implicating dynamic DNA methylation in hippocampus-dependent memory consolidation.
The observation of the gene-specific bidirectionality of DNA methylation raises the question of exactly how targeted and precise such methylation changes can be. A study addressing this inquiry demonstrated that DNA methylation can occur in a highly precise, and targeted fashion, as exon-specific hypomethylation of specific exons within the BDNF gene occurred after hippocampus-dependent fear learning (Lubin and others 2008). This finding supports the existence of an experience-dependent epigenetic program that manages to orchestrate DNA methylation in a highly targeted, gene locus-specific, fashion.
One drawback of using non-selective DNMT-inhibitors has to do with the inability to make inferences as to the involvement of specific DNMTs in activity-dependent methylation and memory formation. In an attempt to address this deficiency, post-natal, forebrain excitatory neuron-specific, knockouts of either DNMT1 or DNMT3a, as well as a DNMT1 and DNMT3a double knockout (DKO) mouse were examined (Feng and others 2010). Whereas neither single DNMT knockout evoked an abnormal phenotype of any kind, the double knockouts exhibited deficits in synaptic plasticity as well as hippocampus-dependent memory formation. Interestingly, although the DKO mice exhibited aberrant basal gene expression and DNA methylation alterations within the hippocampus, the observed changes were minimal and did not include any genes commonly linked to learning and memory (Feng and others 2010). Although not tested, it is conceivable that a more dramatic evidence of widespread gene-expression dysregulation would have been observed in an experience-evoked context. Furthermore, there remains the possibility that double deletion of DNMT1 and DNMT3a resulted in the induction of a compensatory gene expression program suited to account for the dramatic loss of methylation enzymes, as a means of preventing a cataclysmic mass-demethylation event. Such a hypothetical compensatory program might involve an up-regulation of a DNA demethylase. Until now I have intentionally left unaddressed a finding, which is as highly repeatable as it is provocative, that being the evidence of activity dependent demethylation of memory-enhancing genes within the CNS. This topic will be explored in detail in the section below.
Demystifying DNA demethylation
A memory suppressor-and-promoter model implies, and the findings presented above evoke, the existence of a DNA demethylation process that until recently was only discussed in a speculative manner to account for the evidence of neuronal activity-induced reductions in DNA methylation. Within the past decade, determining the molecular basis of DNA demethylation has become an endeavor shared by investigators across the wide spectrum of the biological sciences. For a comprehensive review on mammalian DNA demethylation we suggest the following reviews (S.C. Wu and Zhang 2010; H. Wu and Zhang 2014). In keeping with the memory-focused theme of this review we will restrict our commentary to only those studies that best aided the understanding of the role of DNA demethylation in memory formation. Of the various putative enzymatic mediators of activity-dependent DNA demethylation within the brain, the first to be interrogated was Growth arrest and DNA-damage-inducible, beta (GADD45b).
The role of GADD45b in activity-dependent DNA demethylation
GADD45b was demonstrated to be a regulator of activity-induced neurogenesis, dendritic growth, activity-induced demethylation, and gene expression of the genes BDNFexIX and fibroblast growth factor-1B (FGF-1B) (Ma and others 2009). Moreover, GADD45b was shown to be upregulated within the hippocampus after contextual fear conditioning, suggestive of its memory-permissive properties (Leach and others 2012; Sultan and others 2012). Yet the results of GADD45b deletion were difficult to interpret, with one study detecting a subtle, and selective, memory deficit while another group detected a selective memory impairment in the GADD45b KO mice (Guo, Su, and others 2011; Leach and others 2012; Sultan and others 2012). Moreover, the role of GADD45a, the original GADD45 to be implicated in DNA demethylation, as a demethylase in non-neuronal tissues is under debate (Barreto and others 2007; Jin and others 2008). Future studies need to be conducted to further determine the role of GADD45b in activity-dependent DNA demethylation and learning and memory within the mammalian nervous system.
The role of TET1 in activity-dependent DNA demethylation
It should be noted again that the removal of the methyl moiety from the DNA base cytosine is a thermodynamically unfavorable process (Suzuki and Bird 2008; Rudenko and others 2013). With this being the case, despite GADD45b’s having been implicated as being permissive for DNA demethylation, there was still an empirical and conceptual void in terms of a concrete mechanistic solution towards the problem of DNA demethylation. Leading up to this junction, there had been relatively disparate findings regarding the processes that were, in retrospect, harbingers of more complete understanding of activity-dependent DNA demethylation in the CNS. Evidence of hydroxylated methyl cytosine (5hmC) within the brain piqued the interest of the field, as it was hypothesized that a modification of 5mC might be a precursor to demethylation (Kriaucionis and Heintz 2009). The initial observations of base-hydroxylation in protozoa lead to the identification of Ten-eleven translocation methylcytosine dioxygenase (TET) group of enzymes, of which there are three (i.e., TET1, TET2, TET3) that are capable hydroxylating 5mC, and in turn, creating the 5-hydroxy-methylcytosine (5hmC) base in mammalian DNA, as well further oxidation to 5-formylcytosine (5fC) and-carboxylcytosine (5caC) (Tahiliani and others 2009; Ito and others 2010; He and others 2011). There was also evidence that demethylation was mediated by activation-induced deaminases (AID) (Bhutani and others 2010; Popp and others 2010). Lastly, there was the aforementioned evidence that active DNA demethylation occurs within the hippocampus within the context of learning and memory (Miller and Sweatt 2007; Lubin and others 2008; Miller and others 2010). All of these disparate findings were synthesized into one coherent unifying model when it was empirically determined that TET1 mediated the conversion of 5mC to 5hmC which then undergoes APOBEC1-mediated Base Excision Repair (BER), leading to demethylation, and that this process mediates activity-dependent DNA demethylation of memory permissive genes within the dentate gyrus (See Fig 1B; Guo, Su, and others 2011). Thus, this unifying model for the first time elucidated the mechanisms that lead to activity-dependent DNA demethylation within mammals, in general, and within the mammalian CNS, in particular.
Since a coherent mechanism for DNA demethylation had been discovered, it then became necessary to determine the nature of the involvement of TET1, and thus TET1-mediated DNA demethylation, in long-term memory formation. Using mice with a global deletion of TET1 (TET1KO) allowed for, along with the reconfirmation that TET1 was necessary for activity-dependent demethylation of memory related genes, the realization that loss of TET1 led to perturbations in hippocampal long-term depression and impaired memory extinction (Rudenko and others 2013). Additionally, independent studies revealed that hippocampus-specific over-expression of TET1 lead to an increase in 5hmC and a decrease in 5mC, an increase in the expression of many memory-permissive genes, and an impairment in hippocampus-dependent memory (Kaas and others 2013). The finding that TET1OE enhanced the basal expression of memory-permissive genes, but not memory-suppressive genes suggests that either TET1 does not demethylate memory-suppressive genes, or alludes to the possibility that memory-suppressive genes are minimally suppressed under basal conditions and thus making rendering their transcriptional status negligible to the effects of TET1 over-expression.
Hypothetical framework for the dynamic regulation of DNA methylation during memory formation
At this point, we would like to offer a hypothetical framework that resolves many of the findings presented herein while proposing a novel way of thinking about the regulation of memory-dependent DNA methylation and transcription. Some, but not all, of the foundational premises used in this model have been in circulation for some time and we will attempt to give the appropriate credit. This hypothetical framework relies heavily on comparisons between basal conditions vs. neuronal activity-induced conditions, which required we make educated guesses regarding the trends in transcription in a given context.
Purveyors of the hypothesis that methylation-mediated alterations in gene expression drive memory formation have adopted a longstanding conceptual framework in which genes are either classified as being permissive for memory (i.e., memory promoters) or disruptive towards memory formation (i.e., memory suppressor) (Abel and Kandel 1998; Pittenger and Kandel 1998; West and others 2001; Genoux and others 2002; Levenson 2006; Miller and Sweatt 2007; Sultan and Day 2011). With this previously established conceptual framework a model can be devised whereby neuronal activity leads to the induction of a presently unknown molecular signaling cascade that impinges on, and engages, the DNA methylation regulatory machinery, which ultimately gives way to both the transcriptional repression of memory suppressors and the transcriptional activation of memory enhancers (Fig. 2). Ultimately, the combined transcription of memory-promoters and repression of memory-suppressors tips the scale in favor of the cellular and molecular events that promote synaptic plasticity and memory formation (Fig. 2). Importantly, in this hypothetical framework it is posited that the neuronal activity-dependent, relatively prolonged, transcriptional repression of memory suppressors is critical for the establishment of synaptic plasticity and memory formation, an assumption for which there is considerable empirical support (Sultan and Day 2011).
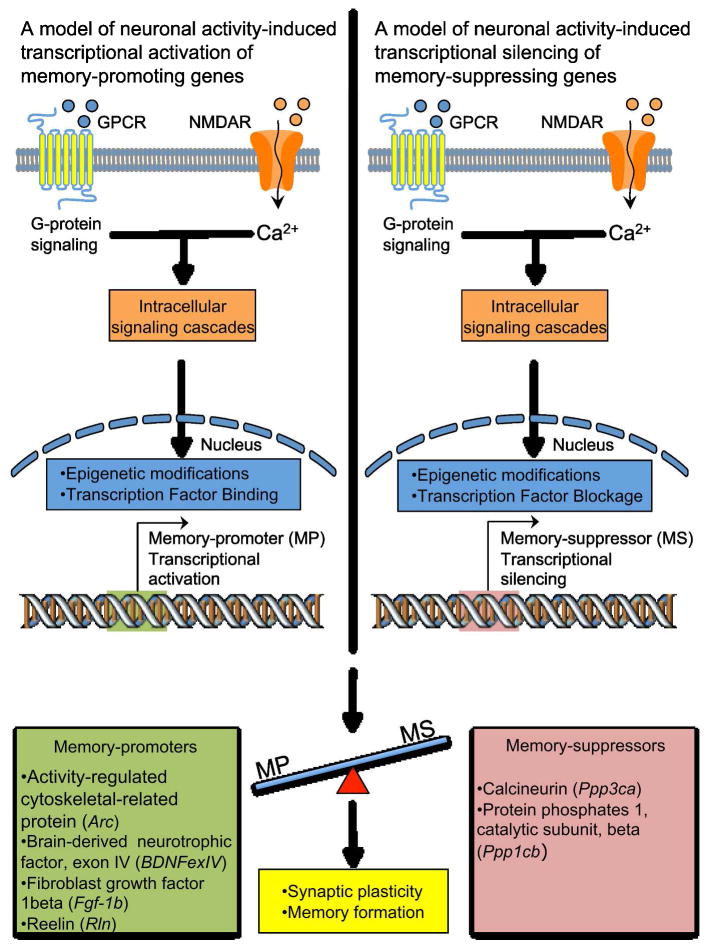
Figure 2.
A model depicting the manner by which experience-dependent stimuli have been proposed to differentially regulate the expression of memory-promoter genes and memory-suppressor genes. Environmental stimuli, which consist primarily of associative learning tasks in animal models, evoke neurotransmitter-induced activation of specific post-synaptic receptors. Receptor activation stimulates specific intracellular signaling cascades that lead to distinct epigenetic patterns and transcriptional regulation at the gene regulatory domain of memory promoters and suppressors. The net increase in memory-promoter gene expression facilitates the establishment of synaptic plasticity and memory formation. List of memory-promoters: Activity-regulated cytoskeletal-related protein (Arc), Brain-derived neurotrophic factor, exon IV (BDNFexIV), Reelin (Rln), Fibroblast growth factor, 1beta (Fgf-1b). List of memory-suppressors: Calcineurin (Ppp3ca), Protein phophatase 1, catalytic subunit, beta (Ppp1cb).
This hypothetical framework builds on the aforementioned model of activity-dependent regulation of memory-related genes. In this hypothetical framework it is posited that the basal degree of expression for both of memory-promoters and memory-suppressors are inversely related, with memory-suppressors being more highly expressed during basal conditions, whereas memory-promoters would be minimally expressed, a phenomenon possibly owning to the CNS drive not to engage in metabolically costly memory-permissive molecular programs until they are necessary (i.e., exposure to a memory-engaging stimulus is encountered) (Fig. 3A). With this being the case, it follows that during basal conditions the relative DNA methylation levels of the memory-suppressors would be relatively lower than that of the memory-promoters. When the appropriate stimulus evokes neuronal activity it results in activity-dependent, DNMT-mediated, methylation of the gene’s transcription regulatory elements, and a subsequent diminution of the expression of the memory-suppressor (Fig. 3A). Finally, we posit that after some time following the stimulus has elapsed, both DNMT1-mediated remethylation and transcriptional suppression, along with TET1-mediated demethylation (i.e., derepression) and transcriptional activation, of memory-promoting and memory-suppressing genes, respectively, occurs (Fig. 3A). Ultimately, as mentioned above, the combined transcriptional regulation of memory promoter and suppressors tips the scale in favor of memory-promoting cellular and molecular events. Vis-à-vis this basic model many of the results highlighted in this review are resolved.
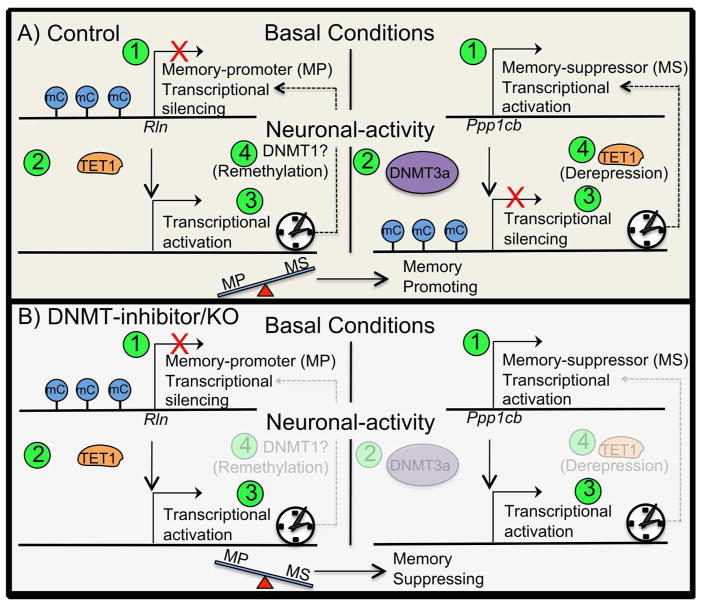
Figure 3.
Hypothetical framework of basal versus activity-dependent gene expression for memory-promoters (screen left) and memory-suppressors (screen right). Numbers in green circles depict the order of events. A) Control mouse. 1) During basal conditions the memory-promoter (MP) gene Rln is transcriptionally silenced, whereas the memory-suppressor (MS) gene Ppp1cb is transcriptionally activated. 2) After neuronal activation and the promoter region of Rln is demethylated by TET1, whereas the promoter of Ppp1cb is methylated by DNMT3a. 3) The MP is now transcriptionally activated, whereas the MS is transcriptionally silenced. The net-memory promoter expression-load does is not outweighed by that of memory suppressor, therefore memory-promoting cellular processes are induced. 4) After sufficient time after the neuronal activating event has passed the MP’s promoter is re-methylated and gene expression is silenced, thus returned in the basal gene expression state, whereas the MS’s promoter is demethylated and gene expression is de-repressed, and thus returned to the basal gene expression state of transcriptional activation. B) DNMT1-inhibition/KO mouse. 1) During basal conditions the gene expression of memory-promoter (MP) gene Rln is silenced, whereas gene expression of the memory-suppressor (MS) gene Ppp1cb is activated. 2) After neuronal activation the promoter region of the MP’s gene is demethylated by TET1, whereas the MS’s gene promoter is not methylated due to the inhibition, or deletion, of DNMT3a. 3) The MP is now transcriptionally activated, whereas the MS is also transcriptional active. The net-memory promoter expression-load does not outweigh that of memory suppressor, therefore memory-suppressing cellular processes are maintained.
Upon utilizing this hypothetical framework, the confounding memory-impairing effects of DNMT-inhibitors can be resolved (Levenson 2006; Miller and Sweatt 2007; Lubin and others 2008). DNMT-inhibitors when administered within the time window of memory consolidation, would serve to block the activity-dependent methylation of memory suppressors, whose transcription typically proceeds in a relatively unencumbered fashion during basal conditions (Abel and Kandel 1998; Genoux and others 2002; Miller and Sweatt 2007; Sultan and Day 2011). Alternatively, DNMT-inhibition would likely have a negligible effect on the expression of memory-promoters, as their transcriptional regulator elements are already relatively highly methylated during basal conditions. Thus DNMT-inhibition would, by default, selectively impede the activity-dependent methylation of memory-suppressors, and would result in the pathologically high expression of memory-suppressors, all during the critical periods of memory consolidation that require the minimal expression of memory-suppressors (Fig. 3B). Moreover, according to this model, DNMT1 and DNMT3a double knockouts (DKO) would be expected to exhibit synaptic plasticity and learning deficits, due to the absence of an activity-induced, DNMT-mediated, diminution in the expression of memory-suppressors (as occurred in the DNMT-inhibition example above). Moreover, it would be reasoned that DKO mice would not exhibit altered basal gene expression, as the memory-suppressor genes, due to their unique intrinsic transcriptional regulatory properties, would exhibit the same high (relative to memory promoter genes) basal expression that is presumed to be present in wild-type mice. Likewise, the basal expression of memory-promoting genes would remain lowly expressed (relative to that of memory suppressors).
Another facet of this model involves DNA demethylation, and speculation regarding its kinetics. To our knowledge, a comprehensive, CNS-specific, rendering of the relative time courses for both DNA methylation and demethylation has yet to be generated either experimentally or computationally. Yet, it would seem that DNA methylation of unmethylated cytosines within the transcriptional regulatory elements of memory suppressor genes would occur more rapidly than DNA demethylation, as DNA methylation involves a mechanism with relatively high processivity, involving TET1-mediated hydroxylation of cytosine, and further cytosine oxidation, followed by AID and APOBEC-mediated BER (H. Wu and Zhang 2014). With both active DNA methylation and demethylation being temporally staggered, the gene-selectivity could possibly be conferred by default based off of the initial methylation status of the gene in question. With this being the case, it would reason that TET1KO-induced a disruption of DNA demethylation, within the context of neuronal activity, would ultimately lead to a net hypermethylation of both memory-promoting genes (as demonstrated), and memory suppressor genes (theoretically, after activity-dependent methylation of memory-suppressors had run its course) (Fig. 4B). In this hypothetical scenario, although memory permissive genes are transcriptionally repressed, there would also be a critical prolonged repression of memory-suppression genes, due to the prolonged memory-suppressors DNA methylation owed to TET1KO. The prolonged repression of memory-suppressor genes may result in a subsequent disinhibition of memory-permissive molecular events. For instance, there might be less PP1-mediated dephosphorylation of the GluR1 subunit of the AMPA (a-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid) receptor, thereby resulting in a decoupling of the effects of net-transcriptional repression of memory-promoting genes from their downstream memory-promoting molecular processes (e.g., GluR1 phosphorylation) (Genoux and others 2002; Lee and others 2003). Put another way, in this TET1KO milieu the relinquishment of the repressive forces on plasticity-promoting molecular changes, due to the hypermethylation and repression of memory-suppressor genes, results in a net shift towards memory-permissive molecular events that favor synaptic plasticity (e.g., increased GluR1 phosphorylation), which would ultimately benefit synaptic plasticity and long-term memory formation (Lee and others 2003).
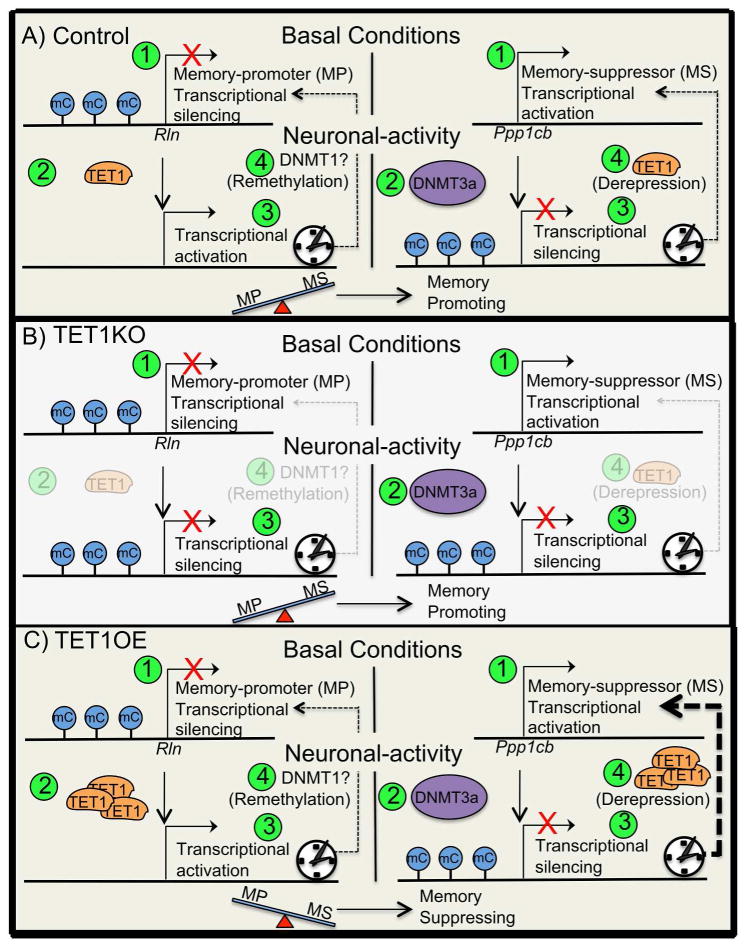
Figure 4.
Hypothetical framework of basal versus activity-dependent gene expression for memory-promoters (screen left) and memory-suppressors (screen right). Numbers in green circles depict the order of events. A) Control mouse. 1) During basal conditions the memory-promoter (MP) gene Rln is transcriptionally silenced, whereas the memory-suppressor (MS) gene is transcriptionally activated. 2) After neuronal activation the MP’s gene promoter is demethylated by TET1, whereas the MS’s gene promoter is methylated by DNMT3a. 3) The MP is now transcriptionally activated, whereas the MS is transcriptionally silenced. The net-memory promoter expression-load is not outweighed by that of memory suppressor, therefore memory-promoting cellular processes are induced. 4) After sufficient time after the neuronal activating event has passed the MP promoter is remethylated and silenced, thus returning to the basal gene expression state, whereas the MS promoter is demethylated and gene expression is derepressed, and thus returned to the basal gene expression state of transcriptional activation. B) TET1KO mouse. 1) During basal conditions the memory-promoter (MP) gene Rln is transcriptionally silenced, whereas the memory-suppressor (MS) gene is transcriptionally activated. 2) After neuronal activation MP’s gene promoter remains methylated due to the lack of TET1 owing to TET1 deletion, whereas the MS’s gene promoter is methylated by DNMT3a. 3) The MP remains transcriptionally silenced, whereas the MS is also transcriptionally silenced. The net-memory promoter expression-load is not outweighed by that of memory suppressor, therefore memory-promoting cellular processes are induced. 4) After sufficient time after the neuronal activating event has passed the MP promoter remains hypermethylated and the gene expression of the MP remains silenced, yet the MS promoter also remains hypermethylated and the gene expression of the MP remains silenced, thus setting the stage for future memory promoting conditions. C) TET1OE mouse. 1) During basal conditions the memory-promoter (MP) gene Rln is transcriptionally silenced, whereas the memory-suppressor (MS) gene is transcriptionally activated. 2) After neuronal activation and the promoter region of the MP’s gene promoter is demethylated by an abundance of TET1 owing to TET1 overexpression, whereas the MS’s gene promoter is methylated by DNMT3a. 3) The MP is now transcriptionally activated, whereas the MS is transcriptionally silenced. 4) Due to the abundance of over-expressed TET1 the MS is rapidly demethylated thus reestablishing its transcriptional activativation. Even before the MP’s gene promoter is remethylated the net-memory promoter expression-load is outweighed by that of memory suppressor, therefore memory-suppressing cellular processes are maintained.
Alternatively, according this hypothetical framework, TET1 over-expression (TET1OE) would result in a net demethylation. Thus, although the activity-dependent methylation, and transcriptional silencing, of memory suppressor genes would occur, it would be greatly outpaced, and thereby nullified, by the TET1-mediated demethylation of memory-suppressors, therefore ultimately leading to a shift away from a memory-permissive state via maintaining the molecular constraints on synaptic plasticity (Fig. 4C). The unabated molecular constrains on synaptic plasticity would offset the benefit of having increased demethylation, and increased expression, of memory-promoting gene products, and ultimately lead to a shift in the molecular milieu that is geared towards the inhibition of plasticity-promoting biochemical processes, thus culminating in impaired memory formation. Finally, TET1OE mice, during basal conditions, would be expected to have elevated memory-promoter gene expression, due to the demethylation of genes whose expression is typically suppressed under basal conditions. Furthermore, an increase in the likelihood of demethylation would not be expected to effect the expression of memory suppressor genes, as they are already constitutively expressed during basal conditions. These expected findings are virtually identical to, and can account for, the published findings by Kaas et al., (2013). As stated above, the basis for the experimental accounts produced with this hypothetical framework are founded in supposition, but hopefully the process of thinking through this hypothetical framework will offer fodder for future experimental inquiry.
Insights from next-generation sequencing studies
During the past decade, much of the progress related towards better understanding the involvement of DNA methylation in regulating the stable transcriptional alterations involved in synaptic plasticity have been achieved via targeted, gene-specific, transcript and DNA methylation analyses. Though informative, targeted gene-specific analyses are limited in that they omit from their analysis potentially salient transcripts, and DNA methylation events, that are presently unknown and have yet to be implicated as being involved in memory formation. Fortunately, the relatively recent development of whole-genome sequencing technologies has enabled researchers to directly address the limitations of targeted, gene-specific, analysis, and in doing so, offer a comprehensive snapshot of the entire transcript and DNA methylation landscape, referred to as the transcriptome and methylome, respectively. In this section I will briefly discuss some of the cutting-edge findings from recent studies that have leveraged whole-genome sequencing technologies and have produced complex data sets that will likely serve as foundational references for future studies investigating the role of DNA methylation in synaptic plasticity and memory formation.
A seminal study characterized activity-induced alterations in DNA CpG methylation at single-nucleotide resolution within the population of dentate granule cells within the hippocampus (Guo, Ma, and others 2011). Neuronal activity induced rapid genome-wide changes (i.e., hypermethylation and hypomethylation) at 1.4% of the 219,991 CpGs that were measured throughout the genome, and did so in a time-dependent and site-specific manner. DNMT3a and GADD45b were found to be required for activity-dependent DNA methylation and demethylation, respectively. CpG islands were rather refractory to activity-induced alterations in DNA methylation, with low-density CpGs found to be the primary targets of activity-induced DNA methylation modifications. CpGs modified by neuronal activity were under-represented in 5′ regions upstream from gene TSS (putative promoters) and exonic regions but were found to be slightly enriched in introns. Intergenic CpGs (>5 kb away from any known genes) were particularly amenable to changes by neuronal activity. CpG methylation near the TSS was inversely correlated with the expression of the corresponding gene. Moreover, the inverse relationship between CpG methylation and gene expression was manifest throughout the entire gene body into the 3′ regions downstream from the translational end sites (TESs) (Guo, Ma, and others 2011).
The canonical substrates for DNA methylation involve cytosines that are found in CpG dinucleotides. Yet, recent evidence has suggested that cytosine methylation in the non-CpG context (mCH, H = A, C, or T) is also detected in the adult mouse and human brain (Xie and others 2012; Varley and others 2013). The idea of DNA methylation not being restricted to CpGs is particularly intriguing as it allows for the possibility that the scope of transcription-regulating DNA methylation with in the brain might be more extensive than previously suspected. One study determined that the degree of mCpH at a gene was inversely related to the expression of the corresponding transcript, consistent with mCpH being transcriptionally repressive (Lister and others 2013). Of the total methylated fraction of adult human neuronal genomes, mCH accounts for ~53%, whereas mCG constitutes ~47% (Lister and others 2013). Another study by Guo et al., (2014) accessed genome-wide DNA methylation from DNA isolated from granule neurons in the adult mouse dentate gyrus. Moreover, transcriptional activity is associated with intragenic hmCG enrichment and the overwhelming majority of hmC was found within the CpG context (99.98%) in mouse adult and fetal frontal cortex, with negligible evidence of hmCpH (Lister and others 2013).
Dnmt3a-binding regions were greatly enriched for mCpH but not mCpG in neurons (Lister and others 2013). Moreover, DNMT3a, but not DNMT1, knockdown reduces CpH, but not CpG, methylation, while increasing the expression of the CpH-associated genes, but not CpG-associated genes like (Bdnf IX and Fgf1B), or the a unmethylated gene (Bdnf IV) (Guo and others 2014). These findings suggest that there is partial independence between the mCH and mCG marks. With DNMT3a being strongly implicated as a mediator of activity-dependent DNA methylation, and now CpH methylation, and with memory suppressors appearing to be the targets of activity-dependent DNA methylation, it would reason that memory suppressors would be the targets of DNMT3a-mediated DNA methylation at CpH residues. Future studies that compare activity-induced alterations in CpH methylation across the gene body of memory-suppressors versus memory-promoters will be needed in order to test this hypothesis.
Moreover, exciting new evidence suggests that DNMT3b1’s recruitment to mCpGs within genic DNA regions is associated with transcriptional activation in mouse stem cells (Baubec and others 2015). Within the field of epigenetics there has been a precedent for intragenic, within the gene body, DNA methylation to be associated with transcriptional activation in non-neuronal cell types (Smith and Meissner 2013). Future studies should determine the extent to which activity-induced alterations in intragenic DNA methylation and DNMT3b1 binding are associated with the activity-induced transcriptional regulation of memory-associated genes.
In all, it is exciting to consider the possibility that mCpG, hmCpG, mCpH, and perhaps even hmCpH, each represent distinct nodes in the epigenetic regulatory system that confer a degree of specificity and directionality, with respect to the gene target and degree of transcriptional regulation. Future studies should continue to harness the power of whole-genome next-generation sequencing (NGS) as a means of further characterizing the spatial and temporal properties of activity-dependent DNA methylation alterations while elucidating their individual and combined potential as a mnemogenic cellular information storage medium involved in memory formation.
Realizing DNA methylation’s mnemogenic potential during cortical consolidation
By now one might have begun to appreciate the conundrum presented by evidence of transient activity-dependent DNA methylation, and demethylation, within the hippocampus, as this seemingly refutes to fundamental premise that stable, long-lived, alterations in DNA methylation underlie the phenomenon of long-term memory. Yet, the evidence of short-lived DNA methylation within the hippocampus is consistent with the established model whereby hippocampus-mediated memory consolidation serves as a temporary precursor to subsequent cortex-mediated long-lasting memory storage (Dash and others 2004; Frankland and others 2004; Wiltgen and others 2004). This compelling, empirically based, model for systems-wide remote memory consolidation has been in circulation for some time. In this model, experience-related information is processed and encoded by discrete neocortical neuronal populations and then rapidly linked to the hippocampus (Wiltgen and others 2004). During periods of inactivity and sleep, unique bursts of activity, called sharp-waves (SPWs), occur within the hippocampus and are thought to drive the playback of the experience-related neocortical activity that was involved in the learning event. Recurring activation of these neocortical areas is suspected to promote intercortical plasticity. Once the cortical connections are sufficiently strengthened the memory is consolidated and independent of the hippocampus (Wiltgen and others 2004).
Exciting findings suggests that gradual DNA methylation alterations within the cortex may promote this process of cortical-consolidation, with DNA methylation of the memory-suppressor gene calcineurin (Ppp3ca) within the prefrontal cortex (PFC) corresponding with the time-dependent establishment of remote memory formation, and DNMT-inhibition within the prefrontal cortex being associated with impaired remote memory formation (Miller and others 2010). On a speculative note, each round of hippocampus-mediated playback of the neocortical regions may lead to the induction of presently unknown nuclear signaling events that gradually build up until some stimulation threshold is reached and the stable change in DNA methylation is produced. One possible, and highly speculative, mechanism would involve SPW-induced activation of neocortical neurons, the resulting activation of DNMT3A/3B, and the subsequent promotion of hemimethylation at memory suppressor genes. This SPW-induced DNA methylation would likely be transient and subject to demethylation, thereby leading to recurring episodes of SPW-induced hemimethylation followed by demethylation. Yet, after numerous rounds playback occur, during multiple days and weeks of sleep-events, some epigenetic switch may occur that allows for SPW-induced, DNMT3a-driven, hemimethylation, followed by maintenance DNMT1-driven methylation of the other DNA strand. This epigenetic switch may be triggered by the strong stimulation converging on neurons form both hippocampal inputs and, finally, the newly formed intercortical inputs. Importantly, in this scenario, the establishment of double-stranded DNA methylation would mark the completion of cortical consolidation. Once double-stranded DNA methylation has been achieved, as discussed above, double-stranded demethylation would be highly resistant to erasure, which might account for the long-lived changes in neocortical plasticity and the establishment of life-long remote memories. Thus, the gradual establishment of double-stranded DNA methylation of memory-suppressor within the cortex may underlie the cortical consolidation of remote memory. With that said, future studies should attempt to investigate the role of gradual double-stranded DNA methylation formation within PFC during the cortical consolidation of remote memories.
DNA methylation has been, and continues to be, a highly promising putative mnemogenic cellular information storage mechanism thought to mediate the stable transcriptional alterations that underlie synaptic plasticity and memory formation. Future studies will continue to further characterize the manner by which neuronal activation modulates the context specific DNA methylation landscape and leads to the complex transcriptional alterations in a panoply memory-related gene targets. Ultimately, future insights into the regulatory relationship between DNA methylation and memory formation may be leveraged to develop neuropsychiatric therapeutic interventions that, through their targeted manipulation of the DNA methylation machinery, ameliorate memory disorders that might owe to perturbed transcriptional regulation within the CNS, such as age-related cognitive decline (Penner and others 2011; Day and others 2015).
Table 1.
High-throughput methods use to characterize DNA methylation in the CNS
| Method | Description | Advantages | Disadvantages |
|---|---|---|---|
| Whole-genome bisulfite sequencing (WGBS) | Denatured DNA is treated with sodium bisulfite which converts unmethylated cytosine to uracil, whereas methylated cytosines are protected from conversion. During subsequent PCR the uracils are amplified as thymines, and the 5mCs are amplified as cytosine. Sequecing the amplified DNA allows reveals single-nucleotide resolution methylation status. information. | Able to query CpG and CpH methylation status. | Does not distinguish 5mC from 5hmC. Mapping the sequence reads is challenging, as a T from a sequencing read can come from either a T or a bisulfite-converted C. 300 million sequencing reads are needed to achieve a 10 fold coverage. |
| Tet-assisted bisulfite sequencing (TAB-seq) | 5hmC is protected by glycosylation, then Tet enzymes oxidize 5mC to 5fC and 5caC, while glycosylated 5-hmC is unaffected. After bisulfite conversion, only 5-hmC is read as C by DNA polymerase, whereas C, 5mC, 5fC and 5caC are all read as T in the sequencing reaction. | Able to query CpG and CpH methylation status. Distinguishes between 5mC from 5hmC. | Mapping sequence reads is challenging, as a T from a sequencing read can come from either a T or from a bisulfite-converted C. 300 million sequencing reads are needed to achieve a 10 fold coverage. |
| Reduced representation bisulfite sequencing (RRBS) | DNA digested with the restriction enzyme Mspl, which cuts CCGG sequences regardless of the methylation status of the central CpG. The library is generated using fully methylated adapters and then bisulfite converted. Short inserts are eliminated and long inserts is not as efficiently amplified. RRBS preferentially queries the methylation status of genomic regions where the density of CCGG sites is relatively high. | Single-nucleotide resolution. Only requires 10 million reads to profile 60% of promoters with 100-fold coverage. | Only queries the methylation status of promoters. Does not discern between the context of DNA methylation (e.g., 5mCpG vs. 5hmCpG). |
| Affinity enrichment-based (e.g., MeDIP-Seq) | DNA is fragmented, the size of which determines the resolution of the assay. Specific modifications of interests are pulled down with antibodies to specific DNA modifications (e.g., 5mC, 5hmC), or high-affinity binding proteins (e.g., MBD1). Immunoprecipitated fragments, which contain a higher amount of the DNA modification of interest, are then made into libraries and sequenced. Regions of the genome with higher enrichment of sequencing reads are considered regions with increased modification. | Precipitates methylated cytosines in different contexts (e.g., 5mC vs. 5hmC). | Sonication is required to produce 200–1000 bp DNA fragments. High density of the DNA modification is required. It can be difficult to appreciate small differences between samples, making affinity enrichment inherently qualitative. |
Acknowledgments
We would like to thank Laura McMeekin, Garrett Kaas, and Andrew Kennedy for valuable discussions related to the preparation of this review. This work was supported by grants awarded by National Institute of Mental Health (MH57014 and MH091122 to JDS). FDH is supported by the UNCF/Merck Graduate Science Research Dissertation Fellowship and a grant award from the National Heart, Lung, and Blood Institute (T32HL105349).
References
- Abel T, Kandel E. Positive and negative regulatory mechanisms that mediate long-term memory storage. Brain Res Brain Res Rev. 1998;26:360–378. doi: 10.1016/s0165-0173(97)00050-7. [DOI] [PubMed] [Google Scholar]
- Adams JP, Sweatt JD. Molecular psychology: roles for the ERK MAP kinase cascade in memory. Annu Rev Pharmacol Toxicol. 2002;42:135–163. doi: 10.1146/annurev.pharmtox.42.082701.145401. [DOI] [PubMed] [Google Scholar]
- Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015 doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Crick F. Memory and molecular turnover. Nature. 1984;312:101. doi: 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- Dash PK, Hebert AE, Runyan JD. A unified theory for systems and cellular memory consolidation. Brain Res Brain Res Rev. 2004;45:30–37. doi: 10.1016/j.brainresrev.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Day JJ, Kennedy AJ, Sweatt JD. DNA methylation and its implications and accessibility for neuropsychiatric therapeutics. Annu Rev Pharmacol Toxicol. 2015;55:591–611. doi: 10.1146/annurev-pharmtox-010814-124527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankland PW, Bontempi B, Talton LE, Kaczmarek L, Silva AJ. The involvement of the anterior cingulate cortex in remote contextual fear memory. Science. 2004;304:881–883. doi: 10.1126/science.1094804. [DOI] [PubMed] [Google Scholar]
- Genoux D, Haditsch U, Knobloch M, Michalon A, Storm D, Mansuy IM. Protein phosphatase 1 is a molecular constraint on learning and memory. Nature. 2002;418:970–975. doi: 10.1038/nature00928. [DOI] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang M-H, Bonaguidi MA, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming G-L, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y-F, Li B-Z, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. Journal of Biological Chemistry. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- Holliday R. Is there an epigenetic component in long-term memory? J Theor Biol. 1999;200:339–341. doi: 10.1006/jtbi.1999.0995. [DOI] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S-G, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming G-L, et al. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach PT, Poplawski SG, Kenney JW, Hoffman B, Liebermann DA, Abel T, et al. Gadd45b knockout mice exhibit selective deficits in hippocampus-dependent long-term memory. Learn Mem. 2012;19:319–324. doi: 10.1101/lm.024984.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-K, Takamiya K, Han J-S, Man H, Kim C-H, Rumbaugh G, et al. Phosphorylation of the AMPA receptor GluR1 subunit is required for synaptic plasticity and retention of spatial memory. Cell. 2003;112:631–643. doi: 10.1016/s0092-8674(03)00122-3. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. Journal of Biological Chemistry. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Qiu S, Weeber EJ. The role of reelin in adult synaptic function and the genetic and epigenetic regulation of the reelin gene. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2008;1779:422–431. doi: 10.1016/j.bbagrm.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Levenson JM. Evidence That DNA (Cytosine-5) Methyltransferase Regulates Synaptic Plasticity in the Hippocampus. Journal of Biological Chemistry. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Lisman JE. A mechanism for memory storage insensitive to molecular turnover: a bistable autophosphorylating kinase. Proc Natl Acad Sci USA. 1985;82:3055–3057. doi: 10.1073/pnas.82.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic Regulation of bdnf Gene Transcription in the Consolidation of Fear Memory. Journal of Neuroscience. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-anpongkul N, et al. Neuronal Activity-Induced Gadd45b Promotes Epigenetic DNA Demethylation and Adult Neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, et al. DNA Methylation–Related Chromatin Remodeling in Activity-Dependent Bdnf Gene Regulation. Science [Internet] 2003;302:890–893. doi: 10.1126/science.1090842. Available from: http://www.sciencemag.org/content/302/5646/890. [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, et al. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent Modification of DNA Regulates Memory Formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Ng HH, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, et al. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiology of Aging. 2011;32:2198–2210. doi: 10.1016/j.neurobiolaging.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger C, Kandel E. A genetic switch for long-term memory. C R Acad Sci III, Sci Vie. 1998;321:91–96. doi: 10.1016/s0764-4469(97)89807-1. [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Sweatt JD. Memory-forming chemical reactions. Rev Neurosci. 2001;12:41–50. doi: 10.1515/revneuro.2001.12.1.41. [DOI] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Sultan FA, Day JJ. Epigenetic mechanisms in memory and synaptic function. Epigenomics. 2011 doi: 10.2217/epi.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD. Genetic deletion of Gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. Journal of Neuroscience. 2012;32:17059–17066. doi: 10.1523/JNEUROSCI.1747-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80:624–632. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber EJ, Beffert U, Jones C, Christian JM, Forster E, Sweatt JD, et al. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. Journal of Biological Chemistry. 2002;277:39944–39952. doi: 10.1074/jbc.M205147200. [DOI] [PubMed] [Google Scholar]
- West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, et al. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci USA. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltgen BJ, Brown RAM, Talton LE, Silva AJ. New circuits for old memories: the role of the neocortex in consolidation. Neuron. 2004;44:101–108. doi: 10.1016/j.neuron.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]