Abstract
Purpose of review
The use of biomarkers in rheumatology can help identify disease risk, improve diagnosis and prognosis, target therapy, assess response to treatment, and further our understanding of the underlying pathogenesis of disease. Here, we discuss the recent advances in biomarkers for rheumatic disorders, existing impediments to progress in this field, and the potential of biomarkers to enable precision medicine and thereby transform rheumatology.
Recent findings
Although significant challenges remain, progress continues to be made in biomarker discovery and development for rheumatic diseases. The use of next-generation technologies, including large-scale sequencing, proteomic technologies, metabolomic technologies, mass cytometry, and other single-cell analysis and multianalyte analysis technologies, has yielded a slew of new candidate biomarkers. Nevertheless, these biomarkers still require rigorous validation and have yet to make their way into clinical practice and therapeutic development. This review focuses on advances in the biomarker field in the last 12 months as well as the challenges that remain.
Summary
Better biomarkers, ideally mechanistic ones, are needed to guide clinical decision making in rheumatology. Although the use of next-generation techniques for biomarker discovery is making headway, it is imperative that the roadblocks in our search for new biomarkers are overcome to enable identification of biomarkers with greater diagnostic and predictive utility. Identification of biomarkers with robust diagnostic and predictive utility would enable precision medicine in rheumatology.
Keywords: biomarker development, biomarkers, mechanistic biomarkers, precision medicine, rheumatic diseases
INTRODUCTION
The development of more robust biomarkers for rheumatic diseases will lay the foundation for precision medicine in rheumatology. Even though some biomarkers are already routinely used to diagnose and treat rheumatic diseases, there is an unmet need for novel biomarkers both in clinical practice and in drug development. For instance, biomarkers that can facilitate early clinical diagnosis offer the potential to enable therapeutic intervention to prevent development of disease. Predictive biomarkers are needed to guide clinical decision making and to reduce the costs of drug development by enabling identification of individuals likely to respond to a specific therapy. Pharmacodynamic biomarkers are needed to monitor response to therapy, both in clinical practice and clinical trials. In this review, we provide examples of different types of biomarkers for rheumatic diseases and discuss the recent progress and remaining challenges in biomarker discovery and validation, as well as the potential of novel biomarkers to transform clinical practice and therapeutic development.
WHAT ARE BIOMARKERS?
A biomarker is defined as a characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention [1]. Some common types of biomarkers in rheumatology are described below.
Molecular biomarkers
Molecular biomarkers are biochemical variables such as measurements of nucleic acids, proteins, lipids, carbohydrates, metabolites and other biomolecules in the blood, synovial fluid and other bodily fluids, and tissues. Objective, quantitative measurements of molecular biomarkers through a variety of techniques serve as indicators of normal or pathologic processes, or indicators of response to therapy. The advent of new technologies such as large-scale nucleic acid sequencing, proteomics, lipidomics, glycomics, metabolomics, and mass cytometry have enabled the identification of the next-generation molecular biomarkers [2■,3,4].
Imaging biomarkers
Imaging technologies such as MRI [5], PET–computed tomography [6■] and ultrasound [7–9] provide biomarkers that enable assessment of disease activity and response to treatments by visualizing anatomical and structural changes. Imaging methods are generally noninvasive, obviating the need for collecting samples from patients. Compared with molecular biomarkers, to date imaging biomarkers have frequently been more closely associated with the phenotypic manifestations of established diseases. Moreover, imaging allows structural and functional assessments of disease activity and therapy.
Clinical biomarkers
Clinical biomarkers are typically physical variables or symptoms, such as joint counts (i.e. the number of swollen and tender joints) [10], pain scores [11], level of proteinuria, and other clinical findings. Although contributing to the diagnosis and assessment of established disease, clinical biomarkers have generally not provided utility in guiding selection of therapies for the treatment of patients with rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), or vasculitis.
DESCRIPTIVE VERSUS MECHANISTIC BIOMARKERS
Descriptive biomarkers reflect the state of a disease but are not directly involved in disease pathogenesis. For example, erythrocyte sedimentation rate and C-reactive protein (CRP) are components of the RA Disease Activity Score [12], but these clinical laboratory biomarkers are not specific to RA and are also elevated in many infectious and inflammatory diseases. Because descriptive biomarkers do not directly mediate disease pathogenesis, the value of the diagnostic and prognostic information that they provide is limited [13].
In contrast, mechanistic biomarkers, which are rooted in the biologic mechanisms of disease, have the greatest potential for guiding clinical decision making [13]. Because they are directly involved in disease pathogenesis, mechanistic biomarkers make for more useful predictive and pharmacodynamic biomarkers as they reflect the dysregulation of molecular pathways directly involved in pathogenesis.
APPLICATION OF BIOMARKERS IN RHEUMATOLOGY
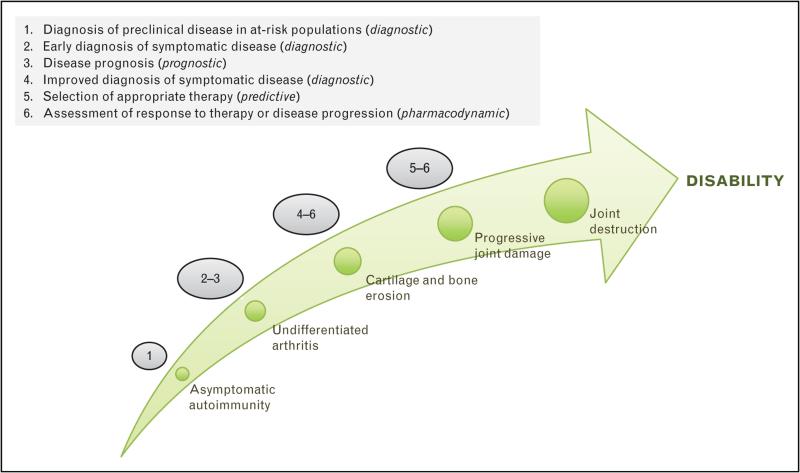
Biomarkers are important tools that have significant untapped potential for guiding both the clinical management of rheumatic diseases and therapeutic development. Their versatility is illustrated in Fig. 1.
FIGURE 1.
Applications of biomarkers at different stages in the development of rheumatic diseases.
Diagnosis of disease
In the clinic, a number of actionable biomarkers are already used to diagnose established disease, with the specific diagnosis then directing therapeutic intervention. For example, detection of autoantibodies including rheumatoid factor [14] and anticitrullinated protein antibodies [15,16] are important components of the diagnostic criterion for RA. The presence of antinuclear antibodies including anti-Sm and anti-DNA are routinely tested for in cases of suspected SLE [17,18]. And detection of antineutrophil cytoplasmic antibodies forms part of the diagnostic criteria for antineutrophil cytoplasmic antibodies-associated vasculitis [19,20]. Advances have recently been made in potential diagnostic biomarkers for RA and other rheumatic diseases [21,22] and are further discussed below.
Another important application of biomarkers is in identifying individuals with disease before the onset of clinical symptoms, as well as individuals with susceptibility to disease, thus offering an opportunity for therapeutic intervention aimed at preventing or slowing the development of symptomatic disease. In RA, for instance, the presence of autoantibodies, elevations in cytokine/chemokine levels, and carriage of certain predisposing genetic factors identify individuals at increased risk of developing the clinically apparent disease [23–25], enabling the institution of therapeutic intervention that can reduce the incidence or severity of RA [26]. In SLE, the presence of specific subsets of antinuclear antibodies in the blood of asymptomatic individuals predicts the development of clinical symptoms [27], potentially enabling treatment to prevent development of this disorder.
Assessment of disease activity and prognosis
Biomarkers that allow monitoring of disease activity, such as erythrocyte sedimentation rate, CRP, and complement proteins C3 and C4, provide information about disease activity but have not exhibited sufficient predictive utility to date for their results in isolation to be actionable. In contrast, a 12-plex inflammatory marker panel offers increased predictive value in assessing disease activity in RA [28]. There is potential for next-generation biomarkers to be even more robust in assessing disease activity and to afford even greater predictive utility in guiding therapeutic decision making. Such biomarkers are particularly valuable in facilitating the selection of an appropriate therapeutic regimen in cases where there is discordance between symptoms and disease activity [29].
Another important potential use of biomarkers is the assessment of clinical disease remission to identify individuals in which disease-modifying therapies should be tapered or discontinued. Despite clinical remission, a subset of patients exhibit structural and functional deterioration in multiple studies [30]. Current disease activity scores and remission criteria are largely based on clinical findings, and do not integrate subclinical molecular inflammation. Potential biomarkers for assessing remission include imaging and immunological bio-markers.
Prediction of response to therapy
Predictive biomarkers allow clinicians to assess the likelihood that a patient will respond to a particular therapy before therapy is started. A major challenge in both clinical practice and drug development is patient selection, because many drugs target discrete molecular aberrations and are usually effective in only a subset of the patient population [31]. The ability to identify the responsive subpopulation of patients prior to treatment could allow treatments to be personalized, reduce healthcare costs, and accelerate the development of new therapeutics. Such predictive biomarkers have the potential to be developed as companion diagnostics, for use in clinical trials to enrich enrollment of responsive study participants and in clinical practice to guide selection of individuals to receive a particular therapy.
Assessment of response to therapy and drug toxicity
Pharmacodynamic biomarkers measure the effect of therapy on the disease, and can be mechanistic or descriptive. These biomarkers can facilitate and reduce the costs of therapeutic development, particularly by enabling more rapid assessment of response to treatment compared with clinical assessment. Pharmacodynamic biomarkers are particularly valuable for rapidly assessing the activity of therapeutics in early-stage clinical proof of concept trials. This information is critical for deciding whether or not to move the drug development process forward to the next stage. Recently, development of pharmacodynamic biomarkers has been prioritized by the US Food and Drug Administration's Critical Path Initiative, with the aim of substituting pharmacodynamic biomarkers for clinical endpoints in early-stage clinical trials [32].
Pharmacodynamic biomarkers of response to rituximab therapy in RA include flow cytometry analysis of peripheral blood B cells and peripheral blood plasmablasts, or immunoglobin J transcripts, a marker for antibody-secreting plasmablasts [33,34■,35]. A biomarker of disease activity and/or of response to therapy in RA is a Multibiomarker Disease Activity score based on 12 serum analytes [28].
CHALLENGES IN BIOMARKER DEVELOPMENT IN RHEUMATOLOGY
For rheumatic diseases, concerted efforts in the hunt for biomarkers have yet to deliver on their promises. The advent of large-scale genomic, transcriptomic, proteomic, and other omic technologies and their continued refinement have provided a rich source for biomarker discovery and validation, but few biomarkers have been adopted into the clinical practice of rheumatology. Current patient care in rheumatology still relies primarily on a combination of traditional evaluations based on clinical assessments and standard laboratory tests. Therapeutic development for rheumatic diseases, which measures its success largely with clinical endpoints, requires clinical trials on large numbers of patients and exceedingly long timelines, and the cost of developing a new drug has skyrocketed in the past decades [36,37]. There remains a pressing need for new, robust biomarkers that can improve patient care and reduce medical costs.
Progress in biomarker development has been hampered by a number of challenges inherent in rheumatology. First, most rheumatic diseases are highly heterogeneous in their pathophysiology, disease course, and therapeutic response. For example, the synovium, the affected site in rheumatic diseases involving the joints, is a complex tissue with a large number of cell lineages and, when inflamed, can be highly variable in the location of the pathobiology within a joint or the pathobiology findings between different joints [38]. This makes identification and validation of biomarkers in these diseases a monumental task, but also highlights the need for next-generation biomarkers to aid patient stratification and targeting therapies.
Second, although peripheral blood, saliva, and urine are the most accessible materials for biomarker studies, they are often not the sites of rheumatic diseases – unique, mechanistic biomarkers are more likely to be found in the diseased tissues but certain mechanistic biomarkers can be found in the peripheral blood. Biomarker discovery in diseased tissues, such as joint tissues in arthritis studies, requires biopsies, remnant tissue from surgical procedures, or autopsy tissue. Biomarkers that are detectable only through tissue are limited in their usefulness compared with those obtained via noninvasive means. An important objective in ongoing efforts is to identify molecular biomarkers that can be detected in peripheral blood, urine, or other readily accessible biologic samples. The development of such biomarkers can help to avoid invasive procedures, such as synovial biopsies in RA and osteoarthritis, renal biopsies in SLE, and muscle biopsies in juvenile dermatomyositis [39].
Third, rheumatic diseases are frequently multi-factorial. The success of predictive biomarkers in oncology stems in part from the fact that many cancers are caused by a single-gene mutation or a discrete chromosomal event. Thus, a handful of predictive biomarkers are routinely used in the management of cancers [40] and others are gaining clinical acceptance as objective measurements that inform on the patients’ response to a particular treatment [31]. For example, overexpression of human epidermal growth factor receptor 2 predicts responsiveness of breast cancer to antibody therapies such as trastuzumab, and breakpoint cluster region-Abelson 1 translocation is indicative of responsiveness of chronic myelocytic leukemia to imatinib. In contrast, RA and other rheumatic diseases are not driven by single-gene mutations, and single or multiple-gene biomarkers have not proven useful as predictive biomarkers [41].
Finally, the identification of new biomarkers from omic studies faces daunting statistical hurdles owing to the need to correct for multiple comparisons as well as nonbiological signals. These methods generate large multivariate data that need to be mined by robust bioinformatics analyses, and the candidate biomarkers selected from these data require rigorous validation [42].
RECENT PROGRESS
Considerable progress has been made in addressing the heterogeneity in rheumatic diseases by evaluating synovial tissue through blind needle biopsy, visually guided arthroscopic biopsy or ultrasound-guided biopsy [43]. By targeting tissue collection to areas of inflammation and allowing imaging assessments of synovial lining thickness and vascularity, these techniques mitigate sampling errors that frequently complicate analysis of diseased tissues that are difficult to obtain. Recently, synovial biopsies from individuals with RA have yielded information on possible patient stratification based on histologic patterns that might have utility as predictive biomarkers [44–46]. Specifically, synovial pathotypes have emerged as potential biomarkers for patient stratification and individualized therapy [43,47,48], with histopathological (e.g. follicular, diffuse or pauci-immune) patterns and their distinct cellular and molecular signatures providing the potential to inform disease mechanisms and correlate with response to therapy [43,47]. In parallel to the above approaches, development of new imaging biomarkers [5,6■,7,8,49] could offer a noninvasive means to guide clinical decision making in rheumatology.
Blood-based biomarkers, such as antibodies in the blood or RNA transcripts in peripheral blood cells, are another promising class of biomarkers in rheumatic diseases. In RA, a new set of antibodies, anticarbamylated protein antibodies, have been identified as a potential biomarker for early diagnosis and assessing prognosis [24,25,50■,51,52]. Similarly, an association has been detected between anti-Porphyromonas gingivalis antibody and disease activity in RA, and the titers of this antibody in the serum may correlate with diagnosis and/or disease activity [53]. As an example of RNA transcripts as potential biomarkers for rheumatic diseases, transcript profiles of peripheral blood can predict RA patients’ response to rituximab [54] and antitumour necrosis factor therapies [55■]. In SLE, transcripts of genes associated with the type I interferon pathway in blood samples have the potential as predictive biomarkers [56].
Large-scale sequencing of antibody repertoires in peripheral blood samples provides the potential to uncover biomarkers for the analysis and prediction of disease activity as well as the design of personalized therapies. In acute SLE, deep sequencing of blood antibody repertoires, in combination with proteomic profiling and single-cell analysis, revealed autoantibodies that could become mechanistic biomarkers [57■■]. In RA, barcode-enabled sequencing of antibody repertoires in peripheral blood plasmablasts identified antibodies that are likely integral to the active immune response and either detection of their sequences in blood or could potentially serve as mechanistic biomarkers [58■,59].
Mass cytometry analysis of peripheral blood samples or solid tissues, in combination with advanced data analysis tools and algorithms, also holds promise for the discovery of cellular bio-markers [2■,3]. Using this approach, researchers can tease apart heterogeneous cell populations at single-cell resolution on the basis of their phenotypes and define each cell according to more than 40 parameters. For example, mass cytometry analysis of signaling responses to clinically meaningful physiologic and pharmaceutical stimuli (e.g. toll-like receptor ligands or drug action) in distinct cell populations in the blood has the potential to uncover cellular signatures that could serve as biomarkers for SLE and other rheumatic diseases [3].
Recently, the important contribution of microbiome to the cause of rheumatic diseases such as RA has been recognized, and the microbial signatures, identified through next-generation sequencing, of affected individuals may serve as a new promising class of biomarkers [60–62]. An elegant example of this is the case-control Metagenome-Wide Association Study by Zhang and colleagues [63■■] that found imbalances in the fecal, dental, and salivary microbiome of individuals with RA. Separately, Scher and colleagues [64] discovered that expansion of intestinal Prevotella copri is associated with new-onset untreated RA.
A promising approach to the development of next-generation therapeutics is the investigation of epigenetic biomarkers in rheumatic disorders [65,66]. Epigenetic regulators, including DNA methylation, histone modification, and microRNAs (miRNAs), have been implicated in pathogenic mechanisms underlying autoimmunity [67]. In RA, changes in imprinted DNA methylation in fibroblast-like synoviocytes modulate the cells’ migration, matrix regulation and immune responses [68]. Among miRNAs, miR-146a is expressed by activated T cells, in which it suppresses apoptosis and IL-2 production [69], and its expression in the synovium is associated with increased disease activity in RA. Another candidate miRNA biomarker is miR-155, which induces the development of Th1 cells and Th17 cells [70] and whose expression is increased in peripheral blood mononuclear cells in RA. These observations suggest that miRNAs could be used as mechanistic biomarkers. Further studies are needed to rigorously assess the predictive value of such miRNAs, the reproducibility with which they can be detected, and their usefulness as biomarkers for rheumatic diseases.
An alternative approach to omic assays and the single-analyte assays in biomarker discovery is multiplexed panels of analytes. The strategy of using relatively small panels of candidate biomarkers has intrinsic statistical advantages, allowing the panel itself to be considered a composite biomarker [71]. In a multiplex panel, the performance of individual analytes can be modeled and assigned a statistical weight [72]. The resulting multiplex assay can be performed in automated assays with good technical reproducibility. In such multiplex approaches, the biomarkers in a panel should be independent of each other and not form part of a clinical disease assessment score. For example, many analytes, such as CRP and serum amyloid A protein, are regulated by IL-6. Thus, any process that affects IL-6 would affect CRP and serum amyloid A protein in a dependent fashion. Similarly, if an analytesuch as CRP is included in both the clinical and the biomarker analysis, then interpretation of any correlation between the Disease Activity Score and biomarkers becomes difficult.
CONCLUSION
Moving forward, we anticipate a future where the use of biomarkers is fully integrated with clinical practice and therapeutic development in rheumatology. Biomarker tests will be performed on at-risk individuals to allow detection of disease at its earliest stage, and hence initiation of therapy to prevent the development of disease. Predictive biomarkers will enable therapies that are targeted to the subset of patients most likely to respond, and pharmacodynamic biomarkers used to monitor response to the therapy. When developing a new therapy, predictive biomarkers will facilitate patient selection, and pharmacodynamic biomarkers will help monitor response to treatment. This shift from traditional approaches to patient stratification and targeted therapies will likely dramatically improve patient care and reduce medical costs.
KEY POINTS.
Next-generation biomarkers with robust diagnostic and predictive utility are needed to enable precision medicine in rheumatology.
Compared with descriptive biomarkers, mechanistic biomarkers have greater potential for guiding clinical decision making.
Despite the advent of new technologies and recent concerted efforts on biomarker discovery, few candidate biomarkers have made their way into the clinical practice because of inherent challenges in biomarker development for rheumatic diseases.
The applications of latest technologies, including large-scale sequencing, proteomic technologies, metabolomic technologies, mass cytometry, other single-cell analysis, and multianalyte analysis technologies, has yielded a multitude of new candidate biomarkers, which still requires rigorous validation.
Acknowledgements
We would like to thank members of the Robinson Lab for their scientific discussion on this topic. We thank T. Lindstrom for her editorial input.
Financial support and sponsorship
This work was supported by the Department of Veteran Affairs, NIH NHLBI Proteomics Center N01-HV 00242, NIH NIAMS R01 AR063676, NIH NIAID U01 AI101981, NIH NIAID U01 AI057229, NIH NIAID U19 AI110491, NIH NCI R33 CA183659, NIH NIAMS/NIAID/FNIH AMP Program UH2 AR067681, the Brennan Family, the Northern California Chapter of the Arthritis Foundation (NCCAF) Center of Excellence, and the Bill and Melinda Gates Foundation.
Footnotes
Conflicts of interest
W.H.R. is a consultant to, owns equity in, and serves on the board of directors of Atreca, Inc. R.M. has no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
■ of special interest
■■ of outstanding interest
- 1.Biomarkers Definitions Working Group Biomarkers and surrogate endpoints: preferred definitions and conceptual, framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 2■.Nair N, Mei HE, Chen SY, et al. Mass cytometry as a platform for the discovery of cellular biomarkers to guide effective rheumatic disease therapy. Arthritis Res Ther. 2015;17:127. doi: 10.1186/s13075-015-0644-z. [The article reviewed mass cytometry-based approaches to identify novel cellular biomarkers for rheumatic diseases.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Gorman WE, Hsieh EW, Savig ES, et al. Single-cell systems-level analysis of human toll-like receptor activation defines a chemokine signature in patients with systemic lupus erythematosus. J Allergy Clin Immunol. 2015;136:1326–1336. doi: 10.1016/j.jaci.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castro-Santos P, Laborde CM, Diaz-Pena R. Genomics, proteomics and metabolomics: their emerging roles in the discovery and validation of rheumatoid arthritis biomarkers. Clin Exp Rheumatol. 2015;33:279–286. [PubMed] [Google Scholar]
- 5.Eckstein F, Collins JE, Nevitt MC, et al. Cartilage thickness change as an imaging biomarker of knee osteoarthritis progression: data from the fnih OA biomarkers consortium. Arthritis Rheumatol. 2015 doi: 10.1002/art.39324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6■.Skeoch S, Williams H, Cristinacce P, et al. Evaluation of carotid plaque inflammation in patients with active rheumatoid arthritis using 18f-fluorodeoxyglucose PET-CT and MRI: a pilot study. Lancet. 2015;385(Suppl 1):S91. doi: 10.1016/S0140-6736(15)60406-8. [The study demonstrated the potential of using PET–computed tomography and MRI as biomarkers for predicting cardiovascular risk in individuals with RA.] [DOI] [PubMed] [Google Scholar]
- 7.Iagnocco A, Finucci A, Ceccarelli F, et al. Power Doppler ultrasound monitoring of response to anti-tumour necrosis factor alpha treatment in patients with rheumatoid arthritis. Rheumatology (Oxford) 2015;54:1890–1896. doi: 10.1093/rheumatology/kev211. [DOI] [PubMed] [Google Scholar]
- 8.Peluso G, Bosello SL, Gremese E, et al. Detection of bone erosions in early rheumatoid arthritis: 3D ultrasonography versus computed tomography. Clin Rheumatol. 2015;34:1181–1186. doi: 10.1007/s10067-015-2938-6. [DOI] [PubMed] [Google Scholar]
- 9.Bevers K, Vriezekolk JE, Bijlsma JW, et al. Ultrasonographic predictors for clinical and radiological progression in knee osteoarthritis after 2 years of follow-up. Rheumatology (Oxford) 2015;54:2000–2003. doi: 10.1093/rheumatology/kev224. [DOI] [PubMed] [Google Scholar]
- 10.Scott DL, Antoni C, Choy EH, Van Riel PC. Joint counts in routine practice. Rheumatology (Oxford) 2003;42:919–923. doi: 10.1093/rheumatology/keg235. [DOI] [PubMed] [Google Scholar]
- 11.Sokka T. Assessment of pain in rheumatic diseases. Clin Exp Rheumatol. 2005;23(Suppl 39):S77–S84. [PubMed] [Google Scholar]
- 12.Fransen J, van Riel PL. The Disease Activity Score and the EULAR response criteria. Rheum Dis Clin North Am. 2009;35:745–757. doi: 10.1016/j.rdc.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Robinson WH, Lindstrom TM, Cheung RK, Sokolove J. Mechanistic biomarkers for clinical decision making in rheumatic diseases. Nat Rev Rheumatol. 2013;9:267–276. doi: 10.1038/nrrheum.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waaler E. On the occurrence of a factor in human serum activating the specific agglutintion of sheep blood corpuscles. APMIS 2007. 1939;115:422–438. doi: 10.1111/j.1600-0463.2007.apm_682a.x. [DOI] [PubMed] [Google Scholar]
- 15.Schellekens GA, de Jong BA, van den Hoogen FH, et al. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Immunol 2015. 1998;195:8–16. [PubMed] [Google Scholar]
- 16.Sebbag M, Simon M, Vincent C, et al. The antiperinuclear factor and the so-called antikeratin antibodies are the same rheumatoid arthritis-specific auto-antibodies. J Clin Invest. 1995;95:2672–2679. doi: 10.1172/JCI117969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arana R, Seligmann M. Antibodies to native and denatured deoxyribonucleic acid in systemic lupus erythematosus. J Clin Invest. 1967;46:1867–1882. doi: 10.1172/JCI105677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan EM, Schur PH, Carr RI, Kunkel HG. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J Clin Invest. 1966;45:1732–1740. doi: 10.1172/JCI105479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falk RJ, Jennette JC. Antineutrophil cytoplasmic autoantibodies with specificity for myeloperoxidase in patients with systemic vasculitis and idiopathic necrotizing and crescentic glomerulonephritis. N Engl J Med. 1988;318:1651–1657. doi: 10.1056/NEJM198806233182504. [DOI] [PubMed] [Google Scholar]
- 20.van der Woude FJ, Rasmussen N, Lobatto S, et al. Autoantibodies against neutrophils and monocytes: tool for diagnosis and marker of disease activity in Wegener's granulomatosis. Lancet. 1985;1:425–429. doi: 10.1016/s0140-6736(85)91147-x. [DOI] [PubMed] [Google Scholar]
- 21.Pratesi F, Migliorini P. Something old, something new: biomarkers in rheumatoid arthritis. J Rheumatol. 2014;41:2091–2093. doi: 10.3899/jrheum.141069. [DOI] [PubMed] [Google Scholar]
- 22.Willemze A, Toes RE, Huizinga TW, Trouw LA. New biomarkers in rheumatoid arthritis. Neth J Med. 2012;70:392–399. [PubMed] [Google Scholar]
- 23.Holers VM. Insights from populations at risk for the future development of classified rheumatoid arthritis. Rheum Dis Clin North Am. 2014;40:605–620. doi: 10.1016/j.rdc.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brink M, Verheul MK, Ronnelid J, et al. Anticarbamylated protein antibodies in the presymptomatic phase of rheumatoid arthritis, their relationship with multiple anticitrulline peptide antibodies and association with radiological damage. Arthritis Res Ther. 2015;17:25. doi: 10.1186/s13075-015-0536-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gan RW, Trouw LA, Shi J, et al. Anticarbamylated protein antibodies are present prior to rheumatoid arthritis and are associated with its future diagnosis. J Rheumatol. 2015;42:572–579. doi: 10.3899/jrheum.140767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seegobin SD, Ma MH, Dahanayake C, et al. ACPA-positive and ACPA-negative rheumatoid arthritis differ in their requirements for combination DMARDS and corticosteroids: secondary analysis of a randomized controlled trial. Arthritis Res Ther. 2014;16:R13. doi: 10.1186/ar4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arbuckle MR, McClain MT, Rubertone MV, et al. Development of autoanti-bodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349:1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 28.Hambardzumyan K, Bolce R, Saevarsdottir S, et al. Pretreatment multibio-marker disease activity score and radiographic progression in early RA: Results from the SWEFOT trial. Ann Rheum Dis. 2015;74:1102–1109. doi: 10.1136/annrheumdis-2013-204986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu R, Wang X, Aihara K, Chen L. Early diagnosis of complex diseases by molecular biomarkers, network biomarkers, and dynamical network biomarkers. Med Res Rev. 2014;34:455–478. doi: 10.1002/med.21293. [DOI] [PubMed] [Google Scholar]
- 30.Gul HL, Ferreira JF, Emery P. Remission in rheumatoid arthritis: is it all the same? Expert Rev Clin Pharmacol. 2015;8:575–586. doi: 10.1586/17512433.2015.1061429. [DOI] [PubMed] [Google Scholar]
- 31.La Thangue NB, Kerr DJ. Predictive biomarkers: a paradigm shift towards personalized cancer medicine. Nat Rev Clin Oncol. 2011;8:587–596. doi: 10.1038/nrclinonc.2011.121. [DOI] [PubMed] [Google Scholar]
- 32.Parekh A, Buckman-Garner S, McCune S, et al. Catalyzing the critical path initiative: FDA's progress in drug development activities. Clin Pharmacol Ther. 2015;97:221–233. doi: 10.1002/cpt.42. [DOI] [PubMed] [Google Scholar]
- 33.Edwards JC, Szczepanski L, Szechinski J, et al. Efficacy of B-cell-targeted therapy with rituximab in patients with rheumatoid arthritis. N Engl J Med. 2004;350:2572–2581. doi: 10.1056/NEJMoa032534. [DOI] [PubMed] [Google Scholar]
- 34■.Cambridge G, Perry HC, Nogueira L, et al. The effect of B-cell depletion therapy on serological evidence of B-cell and plasmablast activation in patients with rheumatoid arthritis over multiple cycles of rituximab treatment. J Autoimmun. 2014;50:67–76. doi: 10.1016/j.jaut.2013.12.002. [The study identified new mechanistic biomarkers (soluble CD23, serum free light chains, and immunoglobulin M-rheumatoid factor) for replase during B-cell depletion therapy.] [DOI] [PubMed] [Google Scholar]
- 35.Owczarczyk K, Lal P, Abbas AR, et al. A plasmablast biomarker for non-response to antibody therapy to cd20 in rheumatoid arthritis. Sci Transl Med. 2011;3:101ra192. doi: 10.1126/scitranslmed.3002432. [DOI] [PubMed] [Google Scholar]
- 36.DiMasi JA, Grabowski HG, Hansen RW. The cost of drug development. N Engl J Med. 2015;372:1972. doi: 10.1056/NEJMc1504317. [DOI] [PubMed] [Google Scholar]
- 37.Dickson M, Gagnon JP. Key factors in the rising cost of new drug discovery and development. Nat Rev Drug Discov. 2004;3:417–429. doi: 10.1038/nrd1382. [DOI] [PubMed] [Google Scholar]
- 38.Wechalekar MD, Smith MD. Utility of arthroscopic guided synovial biopsy in understanding synovial tissue pathology in health and disease states. World J Orthop. 2014;5:566–573. doi: 10.5312/wjo.v5.i5.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consolaro A, Varnier GC, Martini A, Ravelli A. Advances in biomarkers for paediatric rheumatic diseases. Nat Rev Rheumatol. 2015;11:265–275. doi: 10.1038/nrrheum.2014.208. [DOI] [PubMed] [Google Scholar]
- 40.August J. Market watch: Emerging companion diagnostics for cancer drugs. Nat Rev Drug Discov. 2010;9:351. doi: 10.1038/nrd3173. [DOI] [PubMed] [Google Scholar]
- 41.Arron JR, Townsend MJ, Keir ME, et al. Stratified medicine in inflammatory disorders: from theory to practice. Clin Immunol. 2015;161:11–22. doi: 10.1016/j.clim.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Gibson DS, Rooney ME, Finnegan S, et al. Biomarkers in rheumatology, now and in the future. Rheumatology (Oxford) 2012;51:423–433. doi: 10.1093/rheumatology/ker358. [DOI] [PubMed] [Google Scholar]
- 43.Pitzalis C, Kelly S, Humby F. New learnings on the pathophysiology of RA from synovial biopsies. Curr Opin Rheumatol. 2013;25:334–344. doi: 10.1097/BOR.0b013e32835fd8eb. [DOI] [PubMed] [Google Scholar]
- 44.Slansky E, Li J, Haüpl T, et al. Quantitative determination of the diagnostic accuracy of the synovitis score and its components. Histopathology. 2010;57:436–443. doi: 10.1111/j.1365-2559.2010.03641.x. [DOI] [PubMed] [Google Scholar]
- 45.Townsend MJ. Molecular and cellular heterogeneity in the rheumatoid arthritis synovium: clinical correlates of synovitis. Best Pract Res Clin Rheumatol. 2014;28:539–549. doi: 10.1016/j.berh.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 46.Dennis G, Jr, Holweg CT, Kummerfeld SK, et al. Synovial phenotypes in rheumatoid arthritis correlate with response to biologic therapeutics. Arthritis Res Ther. 2014;16:R90. doi: 10.1186/ar4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Astorri E, Nerviani A, Bombardieri M, Pitzalis C. Towards a stratified targeted approach with biologic treatments in rheumatoid arthritis: role of synovial pathobiology. Curr Pharm Des. 2015;21:2216–2224. doi: 10.2174/1381612821666150310145758. [DOI] [PubMed] [Google Scholar]
- 48.Romao VC, Pitzalis C. Synovial heterogeneity in rheumatoid arthritis: the key for rational patient stratification? Acta Reumatol Port. 2015;40:6–8. [PubMed] [Google Scholar]
- 49.Glimm AM, Werner SG, Burmester GR, et al. Analysis of distribution and severity of inflammation in patients with osteoarthitis compared to rheumatoid arthritis by ICG-enhanced fluorescence optical imaging and musculoskeletal ultrasound: a pilot study. Ann Rheum Dis. 2015 doi: 10.1136/annrheumdis-2015-207345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50■.Humphreys J, Verheul M, Barton A, et al. Association of anticarbamylated protein antibodies with long-term disability and increased disease activity in patients with early inflammatory arthritis: results from the Norfolk Arthritis Register. Lancet. 2015;(Suppl 1):S44. doi: 10.1016/S0140-6736(15)60359-2. [The study along with others cited here suggest that anticarbamylated protein antibodies are a potential mechanistic biomarker for assessing disease activity in RA.] [DOI] [PubMed] [Google Scholar]
- 51.Verheul MK, Shiozawa K, Levarht EW, et al. Anticarbamylated protein antibodies in rheumatoid arthritis patients of Asian descent. Rheumatology (Oxford) 2015;54:1930–1932. doi: 10.1093/rheumatology/kev250. [DOI] [PubMed] [Google Scholar]
- 52.Yee A, Webb T, Seaman A, et al. Anticarp antibodies as promising marker to measure joint damage and disease activity in patients with rheumatoid arthritis. Immunol Res. 2015;61:24–30. doi: 10.1007/s12026-014-8560-x. [DOI] [PubMed] [Google Scholar]
- 53.Lee JY, Choi IA, Kim JH, et al. Association between antiporphyromonas gingivalis or antialpha-enolase antibody and severity of periodontitis or rheumatoid arthritis (RA) disease activity in RA. BMC Musculoskelet Disord. 2015;16:190. doi: 10.1186/s12891-015-0647-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raterman HG, Vosslamber S, de Ridder S, et al. The interferon type I signature towards prediction of nonresponse to rituximab in rheumatoid arthritis patients. Arthritis Res Ther. 2012;14:R95. doi: 10.1186/ar3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55■.Oswald M, Curran ME, Lamberth SL, et al. Modular analysis of peripheral blood gene expression in rheumatoid arthritis captures reproducible gene expression changes in tumor necrosis factor responders. Arthritis Rheumatol. 2015;67:344–351. doi: 10.1002/art.38947. [The study showed the utility of using transcriptional profiling of peripheral blood samples from individuals with RA to identifying novel biomarkers for rheumatic diseases.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flint SM, McKinney EF, Lyons PA, Smith KG. The contribution of transcriptomics to biomarker development in systemic vasculitis and SLE. Curr Pharm Des. 2015;21:2225–2235. doi: 10.2174/1381612821666150313130256. [DOI] [PubMed] [Google Scholar]
- 57■■.Tipton CM, Fucile CF, Darce J, et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol. 2015;16:755–765. doi: 10.1038/ni.3175. [By deep-sequencing immunoglobulin variable-segment genes from the blood, this study identified a specific population of B cells from which autoreactive antibody-secreting cells are recruited during SLE flares. These results not only provided insight into the B-cell recruitment and differentiation processes that contribute to SLE but also demonstrated the possibility of finding new mechanistic biomarkers by sequencing autoantibodies from patients' blood.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58■.Tan YC, Kongpachith S, Blum LK, et al. Barcode-enabled sequencing of plasmablast antibody repertoires in rheumatoid arthritis. Arthritis Rheumatol. 2014;66:2706–2715. doi: 10.1002/art.38754. [The report describes a novel approach for biomarker discovery in rheumatology using a cell-barcoding technology to perform large-scale sequencing of the paired immunoglobulin heavy and light chains expressed by peripheral blood plasma-blasts in RA. Both antibody sequences and antibody specificities that could serve as mechanistic biomarkers are identified by this approach.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson WH. Sequencing the functional antibody repertoire: diagnostic and therapeutic discovery. Nat Rev Rheumatol. 2015;11:171–182. doi: 10.1038/nrrheum.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benham H, Robinson PC, Baillet AC, et al. Role of genetics in infection-associated arthritis. Best Pract Res Clin Rheumatol. 2015;29:213–225. doi: 10.1016/j.berh.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 61.McLean MH, Dieguez D, Jr, Miller LM, Young HA. Does the microbiota play a role in the pathogenesis of autoimmune diseases? Gut. 2015;64:332–341. doi: 10.1136/gutjnl-2014-308514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sandhya P, Danda D, Sharma D, Scaria V. Does the buck stop with the bugs?: an overview of microbial dysbiosis in rheumatoid arthritis. Int J Rheum Dis. 2015 doi: 10.1111/1756-185X.12728. [DOI] [PubMed] [Google Scholar]
- 63■■.Zhang X, Zhang D, Jia H, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905. doi: 10.1038/nm.3914. [Using next-generation sequencing, this study revealed distinct RA-associated microbiome in fecal, dental, and salivary samples and showed that these microbial imbalances can be partially restored by disease-modifying antirheumatic drugs.] [DOI] [PubMed] [Google Scholar]
- 64.Scher JU, Sczesnak A, Longman RS, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klein K, Gay S. Epigenetics in rheumatoid arthritis. Curr Opin Rheumatol. 2015;27:76–82. doi: 10.1097/BOR.0000000000000128. [DOI] [PubMed] [Google Scholar]
- 66.Loughlin J, Reynard LN. Osteoarthritis: epigenetics of articular cartilage in knee and hip OA. Nat Rev Rheumatol. 2015;11:6–7. doi: 10.1038/nrrheum.2014.189. [DOI] [PubMed] [Google Scholar]
- 67.Bottini N, Firestein GS. Epigenetics in rheumatoid arthritis: a primer for rheumatologists. Curr Rheumatol Rep. 2013;15:372. doi: 10.1007/s11926-013-0372-9. [DOI] [PubMed] [Google Scholar]
- 68.Ai R, Whitaker JW, Boyle DL, et al. DNA methylome signature in early rheumatoid arthritis synoviocytes compared with longstanding rheumatoid arthritis synoviocytes. Arthritis Rheumatol. 2015;67:1978–1980. doi: 10.1002/art.39123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Curtale G, Citarella F, Carissimi C, et al. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood. 2010;115:265–273. doi: 10.1182/blood-2009-06-225987. [DOI] [PubMed] [Google Scholar]
- 70.O'Connell RM, Kahn D, Gibson WS, et al. MicroRNA-155 promotes auto-immune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hirata S, Dirven L, Shen Y, et al. A multi-biomarker score measures rheumatoid arthritis disease activity in the BeSt study. Rheumatology (Oxford) 2013;52:1202–1207. doi: 10.1093/rheumatology/kes362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Centola M, Cavet G, Shen Y, et al. Development of a multibiomarker disease activity test for rheumatoid arthritis. PLoS One. 2013;8:e60635. doi: 10.1371/journal.pone.0060635. [DOI] [PMC free article] [PubMed] [Google Scholar]