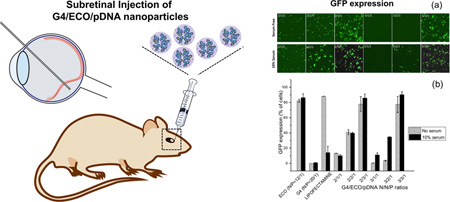
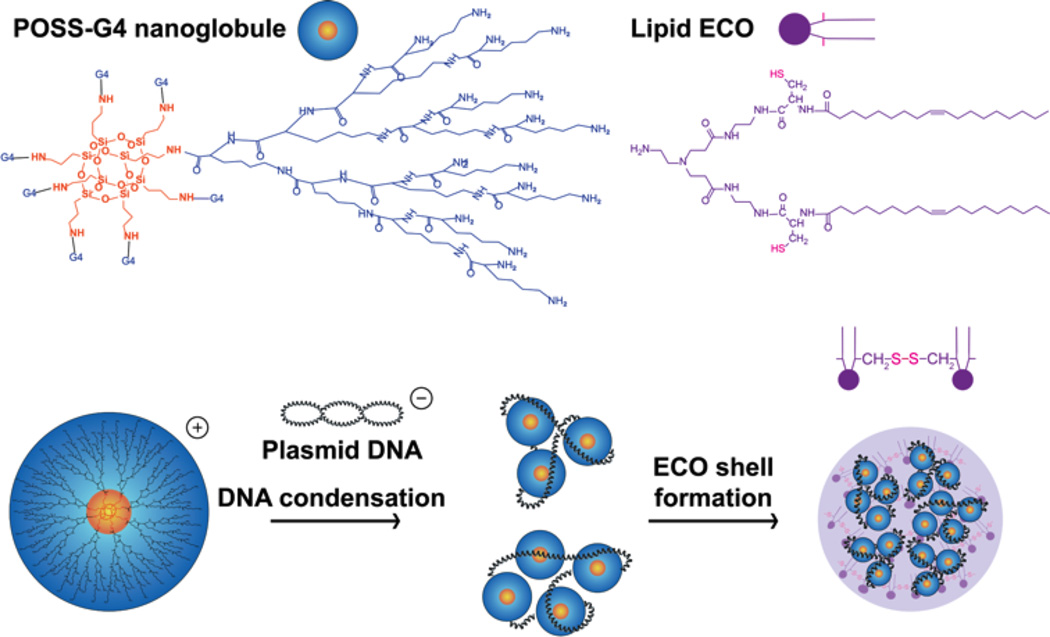
Abstract
Gene therapy is a promising approach to treat genetic diseases. Development of gene delivery systems with high efficiency and low toxicity is a high priority in clinical application of gene therapy. In this work, we designed and evaluated a hybrid nonviral gene delivery system composed of a multifunctional lipid ECO and a core-shell dendrimer G4 nanoglobule for efficient intracellular gene delivery at low charge ratios (N/P). The dendrimeric nanoglobule was designed to effectively condense the gene cargo and the cationic lipid was used to enhance cellular uptake. As expected, this hybrid system formed stable nanoparticles at low N/P ratios (overall N/P = 6) and had low cytotoxicity. The hybrid nanoparticles induced higher GFP (green fluorescent protein) expression in ARPE-19 cells in the culture medium containing 10% serum than in serum-free medium at low N/P ratios. The nanoparticles also mediated significant reporter gene GFP expression in the retina ex-vivo from wild type C57 mice and in vivo with BALB/c mice. Together, these results demonstrate that the rationally designed G4 nanoglobule/pDNA/ECO nanoparticles are a promising delivery system for in vitro and in vivo gene delivery at low charge ratios.
Keywords: gene delivery, nanoglobule, ECO lipid, DNA plasmid, subretinal gene delivery
Graphical abstract
INTRODUCTION
Gene therapy has garnered much attention in treating ocular diseases due to the well-defined anatomy of the eye and well-characterized genetic defects of visual disorders[1]. Currently, therapies for genetic abnormalities in photoreceptor cells and retinal pigmented epithelial (RPE) cells have been developed and tested[2–4]. For example, viral-based gene therapies are tested clinically in individuals with RPE65-LCA (Leber's congenital amaurosis caused by mutations in retinal pigment epithelium-specific 65 kDa protein)[5, 6], CHM (the gene encoding the Rab escort protein 1)[7], and ABCA4 (an ATP-binding cassette protein)[8] mutations. Despite promising results[9–11], some viral-based delivery systems suffer from limitations due to their toxic side effects, including unwanted immunogenic and inflammatory responses[12, 13]. Therefore, non-viral delivery systems with improved safety and efficiency have been designed to overcome the limitations of viral-based systems.
Gene therapy requires safe and efficient delivery of therapeutic nucleic acids into target cells. Non-viral gene delivery systems hold great promise for safe treatment of human genetic diseases. Nucleic acid-based therapeutics are negatively charged, making it difficult for them to enter cells through negatively charged cell membranes. Thus, an efficient non-viral delivery system should possess at least three key features: i) a robust transfection efficiency of the genetic cargo into target cells; ii) an ability to penetrate or bypass biological barriers during the in vivo delivery process; and iii) minimal adverse side effects in healthy cells and tissues[14]. Cationic polymers[15, 16] and lipids[17] have been extensively used to condense or encapsulate nucleic acids to form nanoparticles through electrostatic interactions. A large excess of cationic materials is often used to enhance the delivery of non-viral systems[18, 19]. Despite promising results demonstrated by cationic lipid and cationic polymer based systems, excessive positive charges of these cationic non-viral systems could be cytotoxic[20] and unsafe for in vivo applications due to the high positive to negative charge ratio involved in nanoparticle formulation. Therefore, a non-viral delivery system that has low charge ratio but good transfection efficiency is desirable.
We intended to design an efficient non-viral gene delivery system with a low positive to negative charge ratio (N/P ratio) suitable for safe gene therapy of retinal diseases. Previously, we designed core/shell polylysine dendrimers with a cubic octa(3-aminopropyl)silsesquioxane (POSS) core or nanoglobules with a relatively rigid spheric structure to mimic histones, a class of natural proteins involved in DNA packing. These core-shell dendrimers have well-defined nanostructures and are highly efficient to condense plasmid DNA at low N/P ratios[21–23]. Recently, we also have developed multifunctional pH-sensitive lipids that form stable nanoparticles with nucleic acids and possess the capability to facilitate cellular uptake, pH-responsive endosomal escape, and cytosolic delivery of nucleic acids[24–27]. We hypothesized that the combination of a core/shell nanoglobule and a multifunctional lipid could result in a safe and highly efficient hybrid delivery system with a low N/P ratio by utilizing the distinct and advantageous features of both the nanoglobules and lipids for in vivo gene therapy of retinal diseases.
In this study, we designed a hybrid delivery system featuring the combination of the generation 4 (G4) nanoglobule with a multifunctional lipid ECO for gene delivery into RPE cells (Figure 1). The G4 nanoglobule has a relatively rigid globular structure and a molecular weight of 16,283 Da, similar to that of histones[28, 29]. ECO is a highly efficient multifunctional lipid carrier for cytosolic delivery of nucleic acids31. We investigated the formulation of hybrid G4/ECO/DNA nanoparticles over a range of N/P ratios and their physicochemical properties, including morphology and stability. The effect of the composition of G4/ECO/DNA nanoparticles on transfection efficiency in ARPE-19 cells was determined in vitro. The in vivo gene expression of a lead nanoparticle formulation in retina and RPE cells was assessed in mice via subretinal injection.
Figure 1.
Formation of G4/ECO/pDNA nanoparticles. Hybrid G4/ECO/pDNA nanoparticles are formed following two stepwise electrostatic complexations: plasmid DNA first is condensed by G4 nanoglobules and lipid ECO then is incorporated into the delivery system through electrostatic interactions between the cationic head group of ECO and the negatively charged surface of the G4/pDNA complexes.
RESULTS AND DISCUSSION
The design of hybrid G4/ECO/DNA delivery system is depicted in Figure 1. The fourth generation nanoglobule (G4) has 128 surface primary amine groups, allowing effective electrostatic complexation with the DNA cargo. Due to the multifunctionality of the octa(3-aminopropyl)silsesquioxane core, the G4 nanoglobule has eight branches, and a size and globular shape reminiscent of histones. Histones are a class of natural proteins with molecular weights in the range of 10,000 – 22,000 Da and condensate DNA molecules via electrostatic interactions of their positively charged Lys residue rich regions and negatively charged genetic materials[28]. This process helps to pack DNA in high efficiency, regulates DNA transcription and replication, and protects genetic materials from degradation. G4 nanoglobule can condense DNA cargo in the similar manner. The other component in our delivery system is a multifunctional cationic lipid ECO. Our previous studies have shown that ECO can complex with nucleic acids forming nanoparticles through charge complexation and hydrophobic condensation of the lipid tails. The thiol groups in ECO can be oxidized for disulfide bond crosslinks to further stabilize the nanoparticle formulations, which are cleaved through a glutathione-dependent reduction inside cytosol to release nucleic acids after escaping from endosomal-lysosomal pathway[30, 31]. These features of ECO facilitate cellular uptake and endosomal escape for efficient cytosolic delivery of nucleic acids in response to the pH change during subcellular trafficking[32–34]. Therefore, the incorporation of ECO into the G4/DNA condensates would result in a hybrid plasmid DNA delivery system to overcome the barriers for efficient intracellular gene at low N/P ratios.
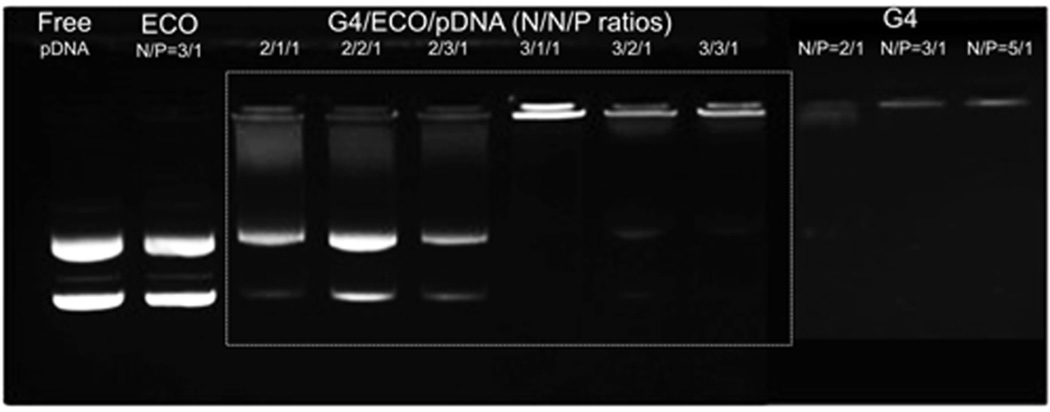
A two-step self-assembly process was used to form the hybrid G4/ECO/DNA nanoparticles. Plasmid DNA (pDNA) was first condensed by the G4 nanoglobule. Lipid ECO was then added to form a lipid shell surrounding the G4/DNA plasmid complexes (Figure 1). Six hybrid formulations were prepared with different N/P (positive/negative) charge ratios. Stabilities of all formulations were evaluated by agarose gel electrophoresis (Figure 2). When only ECO and pDNA were in the formulation, they could not form stable nanoparticle at N/P ratios of 3 and lower. For formulations that had only G4 and pDNA, DNA was efficiently condensed at the N/P ratio of 3. For the G4/ECO/pDNA formulations, when N/N/P ratios for G4/ECO/pDNA were 2/1/1, 2/2/1 and 2/3/1, the hybrid formulations were unable to effectively condense pDNA and form stable nanoparticles. When N/N/P ratios were 3/1/1, 3/2/1 and 3/3/1, the formulations were able to effectively condensed pDNA and form stable hybrid nanoparticles. These results indicate that the hybrid systems were able to form stable nanoparticles at low overall N/P ratios with the N/P ratio of G4/pDNA as 3.
Figure 2.
Agarose gel electrophoresis showing retardation of G4/ECO/pDNA nanoparticles over a range of N/N/P ratios as compared with free pDNA, G4/pDNA nanoparticles and ECO/pDNA nanoparticles.
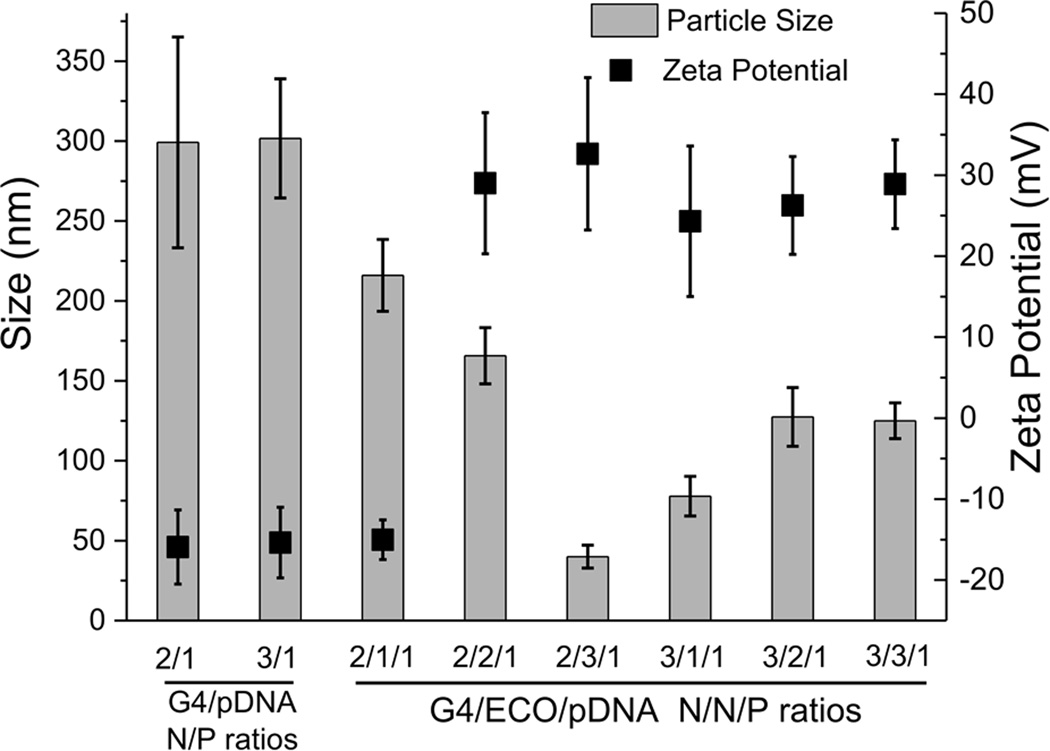
The sizes and surface charges of G4/pDNA and G4/ECO/pDNA nanoparticles were determined by dynamic light scattering measurements (Figure 3). The size of G4/pDNA nanoparticles was around 300 nm at N/P ratio 2 and 3. The complexation with ECO resulted in a significant reduction of the size of the nanoparticles. The sizes for G4/ECO/pDNA were distributed from 50 nm to 200 nm dependent of the N/N/P ratios. All of our stable G4/ECO/pDNA nanoparticles (3/1/1, 3/2/1 and 3/3/1) had sizes smaller than 200 nm, which were suitable sizes for efficient cellular uptake[14]. All formulations displayed positive surface charges except for that with N/N/P ratio 2/1/1 and also G4/pDNA particles (Figure 3). The negative surface charge of those particles might be caused by incomplete condensation, which resulted in more coiled DNA plasmid molecules than positively charged dendrimer or lipid. The net positive charge on the particle surface is an important factor affecting cellular uptake. Because cell membranes carry net negative charges, positive surface charges of the nanoparticles mediate their interactions with cell membrane, facilitating efficient endocytosis of the nanoparticles. Our stable nanoparticles possessed a moderate amount of positive charges with zeta potentials in the range of 25 –30 mV. Taken together, the hybrid nanoparticles with N/N/P of 3/1/1, 3/2/1, and 3/3/1 had the size and zeta potential needed for efficient cellular gene transfection.
Figure 3.
Size and zeta potential of G4/pDNA and G4/ECO/pDNA nanoparticles over a range of N/P or N/N/P ratios.
The morphology of the nanoparticles with the N/N/P ratio of 3/3/1 was imaged with AFM and TEM (Figure 4). The size of the particles were around 200 nm as shown in the AFM image, comparable to what was measured by dynamic light scattering. It appears that some of the nanoparticles formed loose aggregates in the dry state. It is known that aggregation of plasmid DNA/lipid complexes is a function of the plasmid DNA/cationic lipid ratio and the DNA concentrations[35–37]. Also, when these ratios and concentrations are low, there is non-uniformity in particle morphology[35–37]. The amount of cationic lipid and nanoglobule was greatly reduced in our particle formulation. Thus, we expected a non-uniform morphology for some of G4/ECO/pDNA nanoparticles. The morphological characteristic was also confirmed by TEM (Figure 4c). Single particles and particle aggregations were both observed in the TEM image. The sizes turned out to be smaller because of the shrinkage caused by air-drying process.
Figure 4.
Morphological characterization of a G4/ECO/pDNA N/N/P ratio 3/3/1 formulation. (a) 2D image from a tapping mode AFM scan. (b) Corresponding 3D image from the same tapping mode AFM scan. (c) TEM image of dry form G4/ECO/pDNA N/N/P ratio 3/3/1 particles.
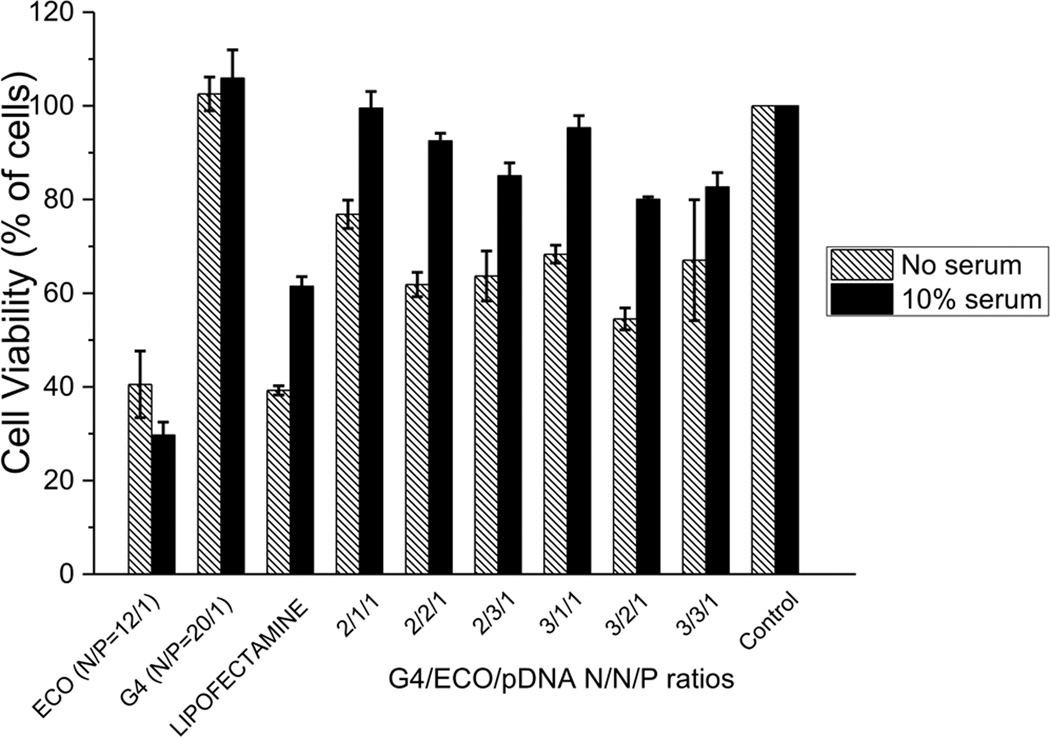
The toxicity of the G4/ECO/pDNA nanoparticles, along with the controls of lipofectamine 2000/pDNA, ECO/pDNA particles, G4/pDNA nanoparticles and PBS, were evaluated in ARPE-19 cell line (human retinal pigment epithelium) (Figure 5). Our hybrid nanoparticles exhibited significantly less cytotoxic effects on ARPE-19 cells in 10 % serum media than lipofectamine 2000. With serum free media, ECO/pDNA (N/P=20/1) and lipofectamine 2000/pDNA nanoparticles exhibited low cell viability (ca. 40 %), while the G4/ECO/pDNA nanoparticles had at least 60 % viability. Cell viability was greatly improved for the G4/ECO/pDNA nanoparticles in serum media.
Figure 5.
Viability of ARPE-19 cells incubated without or with 10 % serum transfection media. G4/ECO/pDNA nanoparticles were tested over a range of N/N/P ratios with lipofectamine 2000, ECO/pDNA nanoparticle and G4/pDNA nanoparticle serving as controls.
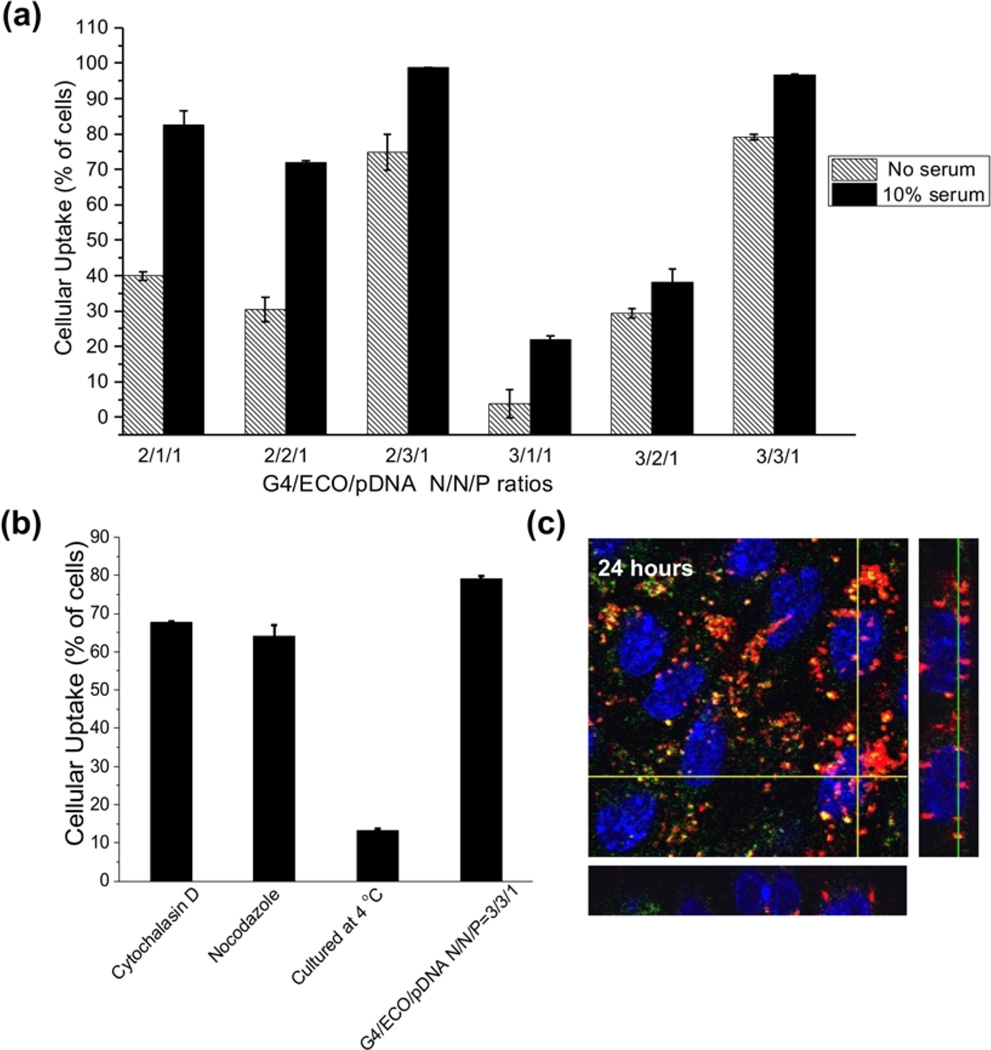
Cellular uptake of G4/ECO/pDNA nanoparticles was evaluated in the ARPE-19 cells. Cellular uptake studies were carried out with a Cy3-labeled plasmid DNA. The percentage of cells transfected with G4/ECO/pDNA nanoparticles after incubation for 4 h was quantified by flow cytometry (Figure 6a). Generally, an increase in pDNA uptake associated with the increase in lipid ECO amount. pDNA uptake was higher in 10 % serum transfection media than in serum-free media as reflected by the greater cell viability in the former media. The nanoparticles with N/N/P of 3/3/1 exhibited transfection efficiency over 95 % in 10 % serum media.
Figure 6.
(a) Cellular uptake of G4/ECO/Cy3 pDNA nanoparticles cultured with ARPE-19 cells for 4 h. A range of N/P ratios were tested under serum-free and 10 % serum culture media. (b) Cellular uptake of G4/ECO/pDNA N/P ratio 3/3/1 particles cultured with ARPE-19 cells for 4 h in serum-free media under different inhibitory conditions (cytochalasin D (5 µg/mL), nocodazole (20 µM) and 4 C). (c) Comfocal fluorescence image of cytosolic delivery of G4/ECO/pDNA N/P ratio 3/3/1 particles. Late endosomes were stained with LysoTracker Green (green), nuclei were stained with Hoechst 33342 (blue) and DNA plasmid was labeled with Cy3 (red). At 24 h, nanoparticles mostly escape endosomal entrapment, shown by the red signals dispersed throughout the cytoplasm. Green signals represent late endosomes/lysosomes. Co-localization with late endosomes (yellow) appears minor.
Cellular uptake was further investigated using G4/ECO/pDNA nanoparticles with N/N/P ratio of 3/3/1 in the presence of different inhibitors to understand the pathway through which the formulations are internalized. The nanoparticles were cultured with either cytochalasin D a phagocytosis inhibitor[38] or nocodazole, an inhibitor of actin and microtubules polymerization[39]. Another group of cells was cultured under 4 °C, which inhibits all energy dependent trafficking pathways. The results showed that the groups cultured with inhibitors had a slightly lower uptake than the control group at 37 °C (Figure 6b). However, a much larger difference was observed between the group cultured under 4 °C and the other groups, suggesting that G4/ECO/pDNA nanoparticles were mainly internalized through an energy dependent pathway. The positively charged nanoparticles might electrostatically interact with the negatively charged cellular membrane, followed by their non-specific absorptive endocytosis[40].
The cytosolic delivery of G4/ECO/pDNA nanoparticles was studied using the nanoparticle with an N/N/P ratio of 3/3/1, which was captured by 3D confocal microscopy at 24 h (Figure 6c). An excellent endosomal escape property was observed which was shown by a predominant red fluorescence signals over yellow signals were observed in the cytoplasm. The yellow color indicated the co-localization of the nanoparticles of pDNA labeled with red fluorescence with late endosomes/lysosomes labeled with green fluorescence, while red signals suggested the endosomal escape of the nanoparticles in the cytoplasm.
Intracellular transfection efficiency of the G4/ECO/pDNA nanoparticles, along with some controls, such as lipofectamine 2000, ECO/pDNA particles and G4/pDNA particles, was tested with GFP plasmid DNA. GFP expression was tested in transfection media with no serum and with 10 % serum using confocal microscopy and flow cytometry (Figure 7). GFP expression was increased in 10 % serum transfection media samples relative to those with serum-free media for the hybrid nanoparticles. For hybrid nanoparticles, an increase of the ECO ratio in the formulation resulted in higher GFP expression in 10 % serum media. The nanoparticles with N/N/P ratios of 2/3/1 and 3/3/1 showed the higher GFP expression in 10 % serum transfection media than in the other formulations. ECO/pDNA (N/P=12/1) nanoparticles exhibited high GFP expression in all transfection media. Whereas the G4/pDNA (N/P=20/1) control group failed to evoke GFP expression in the ARPE-19 cells, although G4/pDNA nanoparticles are reportedly effective in transfecting the MDA-MB-231 cells[21]. The presence of ECO in the hybrid nanoparticles clearly enhanced transfection of the hybrid nanoparticles. Unfortunately, ECO/pDNA nanoparticles resulted in relatively low cell viability at a high N/P ratio (Figure 5). The combination of the G4 nanoglobule and ECO resulted in the hybrid nanoparticle formulation with high transfection efficiency in low serum concentration at low N/P ratios, which was critical for high cell viability. The commercial control lipofectamine 2000 group evidenced high GFP expression in the absence of serum but this system was not as effective in 10 % serum transfection media. Moreover, cell viability was also low with lipofectamine 2000 in 10 % serum media as compared with G4/ECO/pDNA formulations (Figure 5). Notably, GFP expression for G4/ECO/pDNA N/N/P ratio 2/3/1 and 3/3/1 formulations was much higher than lipofectamine 2000 in 10 % serum media, and less toxic. Overall, the G4/ECO/pDNA nanoparticles showed high gene expression with less cytotoxicity as compared with the commercialized delivery system.
Figure 7.
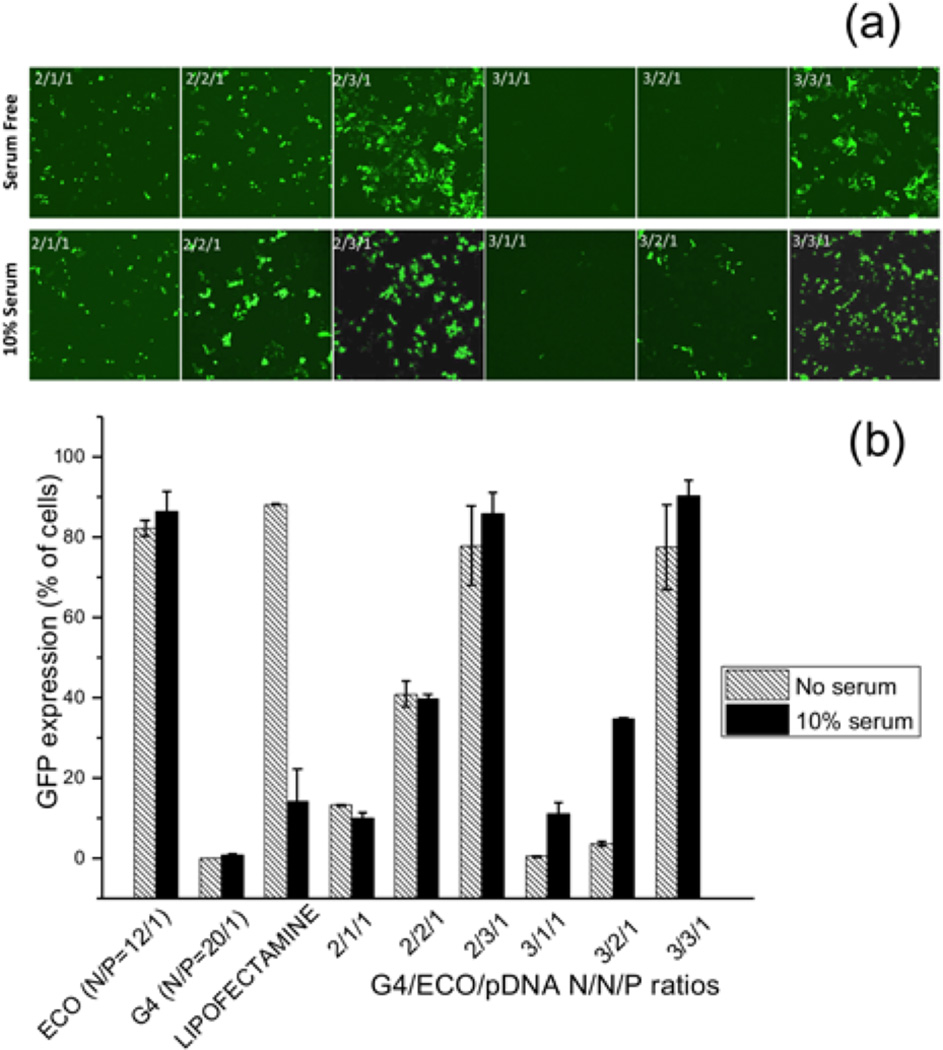
(a) Confocal microscopic image of GFP expression 48 h post transfection in ARPE-19 cell line under serum-free and 10 % serum transfection media. Transfection of G4/ECO/pDNA N/N/P ratios 2/1/1, 2/2/1, 2/3/1, 3/1/1, 3/2/1 and 3/3/1 nanoparticles were evaluated. (b) Flow cytometry of GFP expression 48 h post transfection of ARPE-19 cell line under serum-free and 10 % serum transfection media. G4/ECO/pDNA nanoparticles were tested over a range of N/N/P ratios with lipofectamine 2000, ECO/pDNA particles and G4/pDNA particles used as controls.
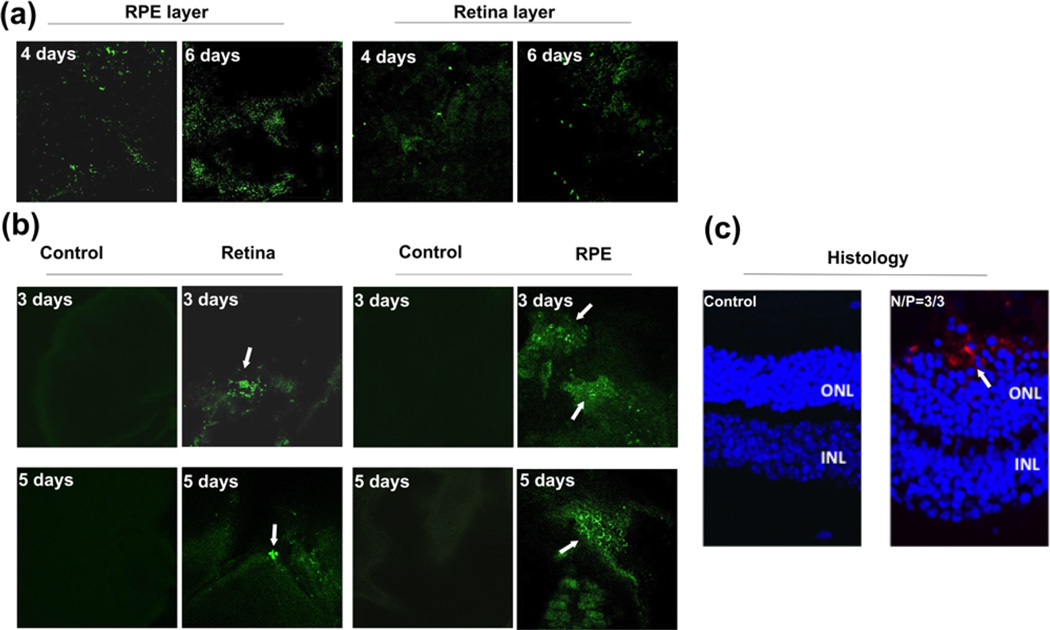
Some of the ocular genetic disorders involve monogenetic mutations in the photoreceptor cells or retinal pigment epithelium (RPE) cells. Introduction of a therapeutic gene to the photoreceptor cells or RPE cells has the potential to correct the monogenetic disorders. The hybrid G4/ECO/pDNA nanoparticles of N/N/P=3/3/1 showed a good combination of stability, safety and gene transfection, which were selected for gene delivery with ex-vivo tissue culture and an animal model. An organotypic culture experiment was carried out with retina and RPE tissue from C57B6 wild type mice. The RPE layer and retinal layer were cultured with the nanoparticles expressing GFP for 8 h and the culture media was replaced with fresh media. GFP expression then was assessed by confocal microscopy after 4 and 6 days later, Figure 8a. GFP expression was noted from both RPE and retina layers 4 and 6 days post transfection indicating good transfection properties for live tissue.
Figure 8.
(a) Ex-vivo transfection of RPE and retinal tissues accomplished with an organotypic culture method. G4/ECO/pDNA N/N/P ratio 3/3/1 nanoparticles were cultured with RPE and retinal tissue from C57/BL6J wild type mice for 8 h. Confocal microscopic images of GFP expression (green) after either 4 days or 6 days are shown. (b) In vivo transfection. G4/ECO/pDNA N/N/P ratio 3/3/1 particles were subretinally injected into the eyes of wild type BALB/c mice. Confocal microscopic images of GFP expression (green) in flat mounted retina and the RPE layer are shown 3 days and 5 days after the injection. Control groups consisted of mice treated with only DNA plasmid at the same dose. (c) Histology of eye cups from BALB/c mice 7 days post-transfection. GFP antibody and a secondary fluorescent labeling antibody (red) were applied to identify GFP expression.
The nanoparticles were also subretinally injected to the eyes of BALB/c mice. After 3, 5 and 7 days, eyes were collected and eyecups prepared. GFP expression of the flat mounted RPE layer and retina layer was visualized with a confocal microscope after 3 and 5 days. GFP expression was found in both the RPE and retina layers, Figure 8b. It appears that high GFP signal in the retina layer was mainly observed at the site of injection and the nanoparticles mediated gene expression in broader area of the RPE layer. GFP expression was further validated by immunohistochemistry with an anti-GFP antibody a secondary red fluorescence labeled antibody at 7 days after injection. Figure 8c shows immunofluorescence staining of GFP in histological slides from eye cups. Green fluorescent protein was shown in the RPE layer. The results suggest that the hybrid G4/ECO/pDNA nanoparticles are able to mediate in vivo gene expression in retina and RPE cells after subretinal injection.
The hybrid G4/ECO/pDNA with N/N/P of 3/3/1 showed high stability and in vitro and in vivo gene transfection efficiency. The overall N/P ratio of the nanoparticles was only 6/1, which was much lower than some reported non-viral gene delivery systems. Consequently, the nanoparticles demonstrated lower cytotoxicity to ARPE-19 cells in serum media than a commercial transfection agent. These findings provide a paradigm to enhance transfection efficiency through the design of an improved delivery system. Subretinal injection is a commonly used approach of gene therapy with both viral and non-viral gene delivery systems. It avoids the complications associated with systemic gene therapy approaches. We have shown in this study that the hybrid nanoglobule/ECO/pDNA nanoparticles are promising for subretinal delivery of genes to retina or RPE cells. Our hybrid nanoparticles can be further optimized by surface modification and incorporation of targeting moieties specific to retina or RPE cells to further improve gene transfection and expression. Plasmid DNA also can be constructed with a specific promoter to control gene expression specifically in retina or RPE cells.
CONCLUSION
Hybrid nanoglobule/ECO/pDNA nanoparticles at low charge ratios were designed and assessed for efficient intracellular gene delivery and for subretinal gene delivery to treat retinal disorders. The compact G4 nanoglobule effectively condensed DNA at low N/P ratios while multifunctional cationic lipid ECO mediated efficient cellular uptake and gene expression. The G4/ECO/pDNA nanoparticles with an N/N/P ratio of 3/3/1 or overall N/P of 6/1 showed high stability, low cytotoxicity and efficient intracellular gene transfection and expression in ARPE-19 cells in 10 % serum media. The nanoparticles also mediated significant gene transfection in both mouse retina and RPE layers ex-vivo. Subretinal injection of the nanoparticles also resulted in significant gene expression in both retina and RPE cells in mice for at least 7 days. The hybrid G4/ECO/pDNA nanoparticles provide a promising platform for safe and efficient delivery of gene therapeutics to treat genetic eye diseases.
EXPERIMENTAL SECTION
Cell Culture
ARPE-19 cells were cultured in Dulbecco’s modified Eagle’s medium and supplemented with 10 % fetal bovine serum, 100 µg/mL streptomycin, and 100 unites/mL penicillin (all reagents were from Invitrogen, Waltham, MA). Cells were maintained in a humidified incubator at 37 °C and 5 % CO2.
Animal
BALB/c and C57BL/6J wild type mice were obtained from the Jackson Laboratory (Bar Harbor, ME). All mice were housed and cared for in the animal facility at the School of Medicine, Case Western Reserve University. All animal procedures and experiments were approved by CWRU Institutional Animal Care and Use Committee.
Preparation of G4/ECO/DNA Nanoparticles
G4 nanoglobule and cationic lipid ECO were synthesized as previously reported[21, 32]. G4/ECO/pDNA nanoparticles were prepared by a stepwise self-assembly of the G4 nanoglobule and ECO with plasmid DNA at N/N/P ratios of 2/1/1, 2/2/1, 2/3/1, 3/1/1, 3/2/1, and 3/3/1 for G4/ECO/DNA. The G4 nanoglobule stock solution (1 mg/mL) and plasmid DNA stock solution (0.5 mg/mL) at predetermined amounts based on desired N/N/P ratios were diluted into equal volumes with nuclease-free water, mixed and shaken for 30 min at room temperature. Then ECO stock solution (2.5 mM in ethanol) was added to the G4/DNA mixture and the resulting mixture was shaken for another 30 min prior to each experiment. The amount of G4 and ECO was determined by N/P ratios in the formulations. G4/pDNA and ECO/pDNA nanoparticles used as controls were prepared by mixing plasmid DNA with G4 or ECO solutions for 1 h on a shaker. Lipofectamine 2000 (Invitrogen, Waltham, MA)/DNA nanoparticles were prepared according to the manufacturer’s recommendation.
Nanoparticle Characterization
The sizes and zeta potentials of G4/pDNA and G4/ECO/pDNA nanoparticles at different N/P or N/N/P ratios were determined by dynamic light scattering with a Brookhaven ZetaPALS Particle Size and Zeta Potential Analyzer (Brookhaven Instruments, Holtsville, NY).
Atomic Force Microscope
Atomic force microscopy (AFM) images were obtained on a Veeco DI Atomic Force Microscope (Veeco, Plainview, NY) at room temperature under ambient conditions, by employing a tapping mode oscillating imaging technique. G4/ECO/pDNA nanoparticles at N/N/P ratio of 3/3/1 were evaluated in this experiment. The nanoparticle solution (10 µL) was deposited onto a glass slide and left to dry for 24 h in a lyophilizer. AFM images then were obtained by scanning the slide surface.
Transmission Electron Microscope
The morphology of G4/ECO/pDNA N/N/P ratios 3/3/1 nanoparticles were also checked with Transmission Electron Microscope (Zeiss Libra 200EF). The sample for TEM observation was prepared by depositing 20 µL of the particle solution onto a 300-mesh copper grid for electron microscopy covered by thin amorphous carbon film (20 nm). Immediately after deposition, the excess of liquid was removed by touching the grid to filter paper. The sample was dried and images were taken.
Gel Electrophoresis for Particle Stability
Solutions (15 µL) of G4/ECO/pDNA nanoparticles with the N/N/P ratios of either 2/1/1, 2/2/1, 2/3/1, 3/1/1, 3/2/1 or 3/3/1 were mixed with 3 µL of loading dye (Promega) and loaded onto a 1% agarose gel containing ethidium bromide. The gel was submerged in 0.5× Tris/Borate/EDTA (TBE) buffer at room temperature and run at 100 V for 25 min. Free pDNA was used as a control. DNA plasmid bands were visualized with an AlphaImager ultraviolet imaging system (Biosciences, USA).
Cell Viability
ARPE-19 cells were incubated with G4/ECO/pDNA nanoparticles at a pDNA concentration of 1 µg/mL in a 12-well plate with a seeding density of 4 × 104 cells/well. After 48 hours, MTT reagent (Invitrogen) was added to the cells for 4 hours followed by the addition of SDS-HCl and incubation at 37 °C for another 4 h. The absorbance was measured at 570 nm with a SpectraMax spectrophotometer (Molecular Devices, Sunnyvale, CA). Cellular viability was calculated as the average of triplicates for each N/N/P ratio normalized to the untreated control.
Cellular Uptake
Cellular uptake of G4/ECO/pDNA nanoparticles was quantitatively evaluated by flow cytometry. G4/ECO/pDNA nanoparticles of Cy3-labeled plasmid DNA (Mirus Bio, Madison, WI) were similarly prepared at different N/N/P ratios as described above. ARPE-19 cells (4 × 104 /well) were seeded onto 12-well plates and grown until they reached 90 % confluence. These cells were transfected with the G4/ECO/Cy3-pDNA nanoparticles at a pDNA concentration of 0.125 µg/mL in serum-free or 10% serum media for 4 h. The transfection media then was removed and each well was washed twice with PBS (1.06 mM KH2PO4, 155 mM NaCl and 2.96 mM Na2HPO4-7H2O). Cells were harvested after treatment with 0.25% trypsin containing 0.26 mM EDTA (Invitrogen, Waltham, MA) for 5 min at 37 °C by centrifugation at 1000 rpm for 5 min, and fixed in 750 µL PBS containing 4% paraformaldehyde, and finally passed through a 35µm cell strainer (BD Biosciences, San Jose, CA). Cellular internalization of G4/ECO/Cy3-pDNA nanoparticles was quantified by the fluorescence intensity measurement for 10,000 cells per each sample by using a BD FACSCalibur flow cytometer (BD FACSCalibur flow cytometer). Nanoparticles at each N/P ratio were assayed in triplicates.
Endosomal Escape
ARPE-19 cells (4 × 104/well) were seeded onto glass-bottom micro-well dishes and allowed to grow to 90 % confluence. The cells were stained with 4 µg/mL Hoechst 33342 (Invitrogen) and 100 mM LysoTracker Green (Life Technologies, Carlsbad, CA). Cells were then treated with G4/ECO/Cy3-pDNA (N/P ratio 3/3/1) nanoparticles in serum-free medium. After culturing for 8 h in serum-free medium before the transfection medium was removed and replaced with fresh serum containing medium (10 % FBS). Cells were cultured for 24 h when the medium was removed and they were washed with PBS for three times before fixation with PBS containing 4 % paraformaldehyde. Fluorescence images were taken with an Olympus FV1000 confocal microscope.
In Vitro Transfection
ARPE-19 cells were seeded in 12-well plates at a density of 4 × 104 cells per well and allowed to grow for 24 h at 37 °C. Transfections were carried out in serum-free or 10 % serum media with the nanoparticles of GFP plasmid DNA (Altogen Biosystems, Las Vegas, NV) at a DNA concentration of 1 µg/mL. G4/ECO/pGFP nanoparticles were incubated with ARPE-19 cells for 8 hours at 37 °C. The media then was replaced with fresh serum-containing media (10 % serum) and the cells were then cultured for an additional 48 h. GFP expression was monitored with an Olympus FV1000 confocal microscope (Olympus, Center Valley, PA). After the culture media was removed, each well was washed twice with PBS. Cells were harvested after treatment with 0.25% trypsin containing 0.26 mM EDTA, (Invitrogen) by centrifugation at 1000 rpm for 5 min, fixed in 750 µL PBS containing 4 % paraformaldehyde, and finally passed through a 35 µm cell strainer (BD Biosciences, San Jose, CA). A BD FACSCalibur flow cytometer (BD Biosciences) was used to determine GFP expression based on quantified by the fluorescence intensity in a total of 10,000 cells for each sample.
Ex Vivo Retinal Transfection
Mouse eyes were enucleated, washed with penicillin-streptomycin solution (Sigma, St. Louis, MO), and rinsed with Hanks' balanced solution (Hyclone, Waltham, MA). Eye cups were prepared and removed retinas were flattened by making retinal flaps. Flattened retinas were transferred onto filter paper, where the retinas were gently peeled away from the RPE layer. All these procedures were performed under a surgical microscope. Each retina and RPE layer on a filter paper was placed in a separate well of a 12-well plate filled with 0.5 mL DMEM containing 10 % serum and incubated for 16 h at 37 °C. After the retinas and RPE layers were washed twice with 0.5 mL of fresh DMEM with 10% serum, they were incubated with G4/ECO/pDNA nanoparticles (N/N/P ratio 3/3/1) of 4.5 g pDNA expressing GFP for 8 h at 37 °C. The culture media then was replaced by fresh culture media and the retinas and RPE layers were further cultured for either 4 days or 6 days when GFP expression was determined with an Olympus FV1000 confocal microscope.
In Vivo Subretinal Transfection with G4/ECO/pDNA Nanoparticles
All surgical manipulations were carried out under a surgical microscope (Leica M651 MSD). Mice were anesthetized by intraperitoneal injection of 20 µL/g of body weight of 6 mg/mL ketamine and 0.44 mg/mL xylazine in 10 mM sodium phosphate and 100 mM NaCl buffer solution (pH=7.2). Pupils were dilated with 1.0 % tropicamide ophthalmic solution (Bausch & Lomb, Rochester, NY). A 33-gauge beveled needle (World Precision Instruments, Sarasota, FL) was used as a lance to make a full thickness cut through sclera at 1.0 mm posterior to the limbus. This needle was replaced with a 36-gauge beveled needle attached to an injection system (UMP-II microsyringe pump and Micro4 controller with a footswitch, World Precision Instruments). This needle was aimed toward the inferior nasal area of the retina, and a G4/ECO/pDNA (3/3/1) nanoparticles solution was injected at a pDNA (GFP plasmid) dose of 18 ng into the subretinal space. Successful administration was confirmed by observing bleb formation. The tip of the needle remained in the bleb for 10 s after bleb formation, when the needle was gently withdrawn. A solution with pDNA alone (18 ng) was also injected into the subretinal space of the contra eye served as a control (mock group). After 3 days or 5 days, eyes were collected, washed with penicillin-streptomycin solution (Sigma), and rinsed with Hanks' balanced solution (Hyclone, Waltham, MA). Eye cups were prepared just as previously described. The retina and RPE layers were placed in glass bottom confocal plate and fixed with 1 mL of PBS with 4 % paraformaldehyde. An Olympus FV1000 confocal microscope was used to assess GFP expression.
Histology
The eye cups for histology were fixed in 2% glutaraldehyde, 4% paraformaldehyde and processed for visualization by OCT (optimum cutting temperature formulation). Sections were cut at 1 µm. Slides samples were permeabilized and fixed sequentially with 4% PFA and 0.25% Triton X-100 followed by blocking with 0.5% BSA blocking solution for 1 h at room temperature. Antibodies were applied at with proper concentrations for 1h at room temperature and washed 3 times with a 0.1% TBST, 5 min each wash. Slides were counter-stained with DAPI and mounted with coverslip using the Prolong Gold regent (Invitrogen) before imaging. Stained tissue was imaged with an Olympus FV1000 confocal microscope.
Statistical Analysis
Experiments were performed in triplicate and presented as the means and standard deviations. Statistical analysis was conducted with ANOVA and two-tailed Student’s t-tests using a 95% confidence interval. Statistical significance was accepted when p ≤ 0.05.
AKNOWLEGEMENTS
This work was supported by grant R24EY021126 from the National Eye Institute of the National Institutes of Health (NIH). ZRL is M. Frank Rudy and Margaret Domiter Rudy Professor of Biomedical Engineering and KP is John H. Hord Professor of Pharmacology.
References
- 1.Bainbridge JW, Tan MH, Ali RR. Gene Ther. 2006;13:1191. doi: 10.1038/sj.gt.3302812. [DOI] [PubMed] [Google Scholar]
- 2.Liu X, Brandt CR, Rasmussen CA, Kaufman PL. Can. J. Ophthalmol. 2007;42:447. [PubMed] [Google Scholar]
- 3.Boye SE, Boye SL, Lewin AS, Hauswirth WW. Mol. Ther. 2013;21:509. doi: 10.1038/mt.2012.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koirala A, Conley SM, Naash MI. Biomaterials. 2013;34:7158. doi: 10.1016/j.biomaterials.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maguire AM, Simonelli F, Pierce EA, Pugh EN, Mingozzi F, Bennicelli J, Banfi S, Marshall KA, Testa F, Surace EM, Rossi S, Lyubarsky A, Arruda VR, Konkle B, Stone E, Sun J, Jacobs J, Dell'Osso L, Hertle R, Ma J-x, Redmond TM, Zhu X, Hauck B, Zelenaia O, Shindler KS, Maguire MG, Wright JF, Volpe NJ, McDonnell JW, Auricchio A, High KA, Bennett J. N. Engl. J. Med. 2008;358:2240. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bainbridge JWB, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, Viswanathan A, Holder GE, Stockman A, Tyler N, Petersen-Jones S, Bhattacharya SS, Thrasher AJ, Fitzke FW, Carter BJ, Rubin GS, Moore AT, Ali RR. N. Engl. J. Med. 2008;358:2231. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 7.MacLaren RE, Groppe M, Barnard AR, Cottriall CL, Tolmachova T, Seymour L, Clark KR, During MJ, Cremers FP, Black GC, Lotery AJ, Downes SM, Webster AR, Seabra MC. Lancet. 2014;383:1129. doi: 10.1016/S0140-6736(13)62117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsybovsky Y, Molday RS, Palczewski K. Adv. Exp. Med. Biol. 2010;703:105. doi: 10.1007/978-1-4419-5635-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Annear MJ, Mowat FM, Bartoe JT, Querubin J, Azam SA, Basche M, Curran PG, Smith AJ, Bainbridge JW, Ali RR, Petersen-Jones SM. Hum. Gene Ther. 2013;24:883. doi: 10.1089/hum.2013.146. [DOI] [PubMed] [Google Scholar]
- 10.Cideciyan AV, Jacobson SG, Beltran WA, Sumaroka A, Swider M, Iwabe S, Roman AJ, Olivares MB, Schwartz SB, Komaromy AM, Hauswirth WW, Aguirre GD. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E517. doi: 10.1073/pnas.1218933110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen-Jones SM. Vet. Ophthalmol. 2012;15(Suppl 2):29. doi: 10.1111/j.1463-5224.2012.01054.x. [DOI] [PubMed] [Google Scholar]
- 12.Jooss K, Chirmule N. Gene Ther. 2003;10:955. doi: 10.1038/sj.gt.3302037. [DOI] [PubMed] [Google Scholar]
- 13.Provost N, Le Meur G, Weber M, Mendes-Madeira A, Podevin G, Cherel Y, Colle MA, Deschamps JY, Moullier P, Rolling F. Mol. Ther. 2005;11:275. doi: 10.1016/j.ymthe.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 14.Neu M, Fischer D, Kissel T. J. Gene Med. 2005;7:992. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- 15.Shcharbin DG, Klajnert B, Bryszewska M. Biochemistry (Mosc.) 2009;74:1070. doi: 10.1134/s0006297909100022. [DOI] [PubMed] [Google Scholar]
- 16.Thomas M, Klibanov AM. Appl. Microbiol. Biotechnol. 2003;62:27. doi: 10.1007/s00253-003-1321-8. [DOI] [PubMed] [Google Scholar]
- 17.Junquera E, Aicart E. Curr. Top. Med. Chem. 2014;14:649. doi: 10.2174/1568026614666140118203128. [DOI] [PubMed] [Google Scholar]
- 18.Kolli S, Wong SP, Harbottle R, Johnston B, Thanou M, Miller AD. Bioconjug. Chem. 2013;24:314. doi: 10.1021/bc3004099. [DOI] [PubMed] [Google Scholar]
- 19.Demeneix B, Behr J, Boussif O, Zanta MA, Abdallah B, Remy J. Adv Drug Deliv Rev. 1998;30:85. doi: 10.1016/s0169-409x(97)00109-9. [DOI] [PubMed] [Google Scholar]
- 20.Kim ST, Saha K, Kim C, Rotello VM. Acc. Chem. Res. 2013;46:681. doi: 10.1021/ar3000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneshiro TL, Wang X, Lu ZR. Mol. Pharm. 2007;4:759. doi: 10.1021/mp070036z. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, Yu G, Luo C, Maeda A, Zhang N, Sun D, Zhou Z, Puntel A, Palczewski K, Lu ZR. ACS Nano. 2014;8:153. doi: 10.1021/nn4054107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Z, Lu ZR. Nanomedicine (Lond) 2014;9:2387. doi: 10.2217/nnm.14.133. [DOI] [PubMed] [Google Scholar]
- 24.Collins DS, Unanue ER, Harding CV. J. Immunol. 1991;147:4054. [PubMed] [Google Scholar]
- 25.Xu R, Wang X, Lu Z-R. Chin. Sci. Bull. 2012;57:3979. [Google Scholar]
- 26.Xu R, Lu Z-R. Science China Chemistry. 2011;54:359. [Google Scholar]
- 27.Xu R, Wang XL, Lu ZR. Langmuir. 2010;26:13874. doi: 10.1021/la1024185. [DOI] [PubMed] [Google Scholar]
- 28.DeLange RJ, Smith EL. Annu. Rev. Biochem. 1971;40:279. doi: 10.1146/annurev.bi.40.070171.001431. [DOI] [PubMed] [Google Scholar]
- 29.Canine BF, Wang Y, Hatefi A. J. Control. Release. 2008;129:117. doi: 10.1016/j.jconrel.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Won YW, Yoon SM, Lee KM, Kim YH. Mol. Ther. 2011;19:372. doi: 10.1038/mt.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manickam DS, Li J, Putt DA, Zhou QH, Wu C, Lash LH, Oupicky D. J. Control. Release. 2010;141:77. doi: 10.1016/j.jconrel.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malamas AS, Gujrati M, Kummitha CM, Xu R, Lu ZR. J. Control. Release. 2013;171:296. doi: 10.1016/j.jconrel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gujrati M, Malamas A, Shin T, Jin E, Sun Y, Lu ZR. Mol. Pharm. 2014;11:2734. doi: 10.1021/mp400787s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parvani JG, Gujrati MD, Mack MA, Schiemann WP, Lu ZR. Cancer Res. 2015;75:2316. doi: 10.1158/0008-5472.CAN-14-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer D, von Harpe A, Kunath K, Petersen H, Li Y, Kissel T. Bioconjug. Chem. 2002;13:1124. doi: 10.1021/bc025550w. [DOI] [PubMed] [Google Scholar]
- 36.Wheeler CJ, Felgner PL, Tsai YJ, Marshall J, Sukhu L, Doh SG, Hartikka J, Nietupski J, Manthorpe M, Nichols M, Plewe M, Liang X, Norman J, Smith A, Cheng SH. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11454. doi: 10.1073/pnas.93.21.11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett MJ, Aberle AM, Balasubramaniam RP, Malone JG, Nantz MH, Malone RW. Journal of Liposome Research. 1996;6:545. [Google Scholar]
- 38.Ribes S, Ebert S, Czesnik D, Regen T, Zeug A, Bukowski S, Mildner A, Eiffert H, Hanisch UK, Hammerschmidt S, Nau R. Infect. Immun. 2009;77:557. doi: 10.1128/IAI.00903-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kastl L, Sasse D, Wulf V, Hartmann R, Mircheski J, Ranke C, Carregal-Romero S, Martinez-Lopez JA, Fernandez-Chacon R, Parak WJ, Elsasser HP, Rivera Gil P. ACS Nano. 2013;7:6605. doi: 10.1021/nn306032k. [DOI] [PubMed] [Google Scholar]
- 40.Erbacher P, Remy JS, Behr JP. Gene Ther. 1999;6:138. doi: 10.1038/sj.gt.3300783. [DOI] [PubMed] [Google Scholar]