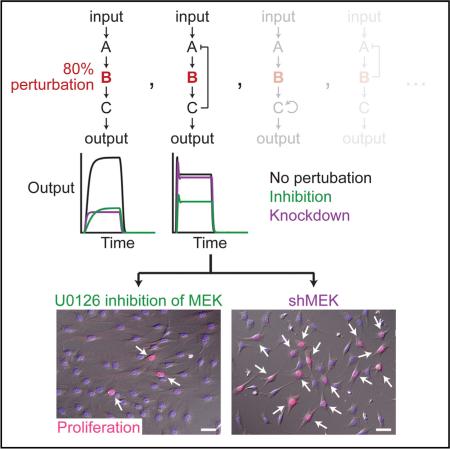
SUMMARY
Chemical inhibition and genetic knockdown of enzymes are not equivalent in cells, but network-level mechanisms that cause discrepancies between knockdown and inhibitor perturbations are not understood. Here we report that enzymes regulated by negative feedback are robust to knockdown but susceptible to inhibition. Using the Raf–MEK–ERK kinase cascade as a model system, we find that ERK activation is resistant to genetic knockdown of MEK but susceptible to a comparable degree of chemical MEK inhibition. We demonstrate that negative feedback from ERK to Raf causes this knockdown-versus-inhibitor discrepancy in vivo. Exhaustive mathematical modeling of three-tiered enzyme cascades suggests that this result is general: negative autoregulation or feedback favors inhibitor potency, whereas positive autoregulation or feedback favors knockdown potency. Our findings provide a rationale for selecting pharmacologic versus genetic perturbations in vivo and point out the dangers of using knockdown approaches in search of drug targets.
Graphical Abstract
INTRODUCTION
Cascades of kinases, proteases, and other enzymes are an important component of cellular regulation. Quantitative analyses have shown that enzymatic cascades can be configured to provide ultrasensitivity (Bagowski et al., 2003; Huang and Ferrell, 1996; Romano et al., 2014), adaptation (Ma et al., 2009), bistability (Bagowski and Ferrell, 2001; Ferrell and Machleder, 1998; Shah and Sarkar, 2011), thresholding (Bentele et al., 2004), feedback amplification (Sturm et al., 2010), oscillations (Liu et al., 2011; Shankaran et al., 2009), and ligand discrimination (Marshall, 1995; Murphy et al., 2002; Nakakuki et al., 2010). By contrast, the response of such cascades to different targeted perturbations has not been examined systematically despite reports of counterintuitive findings in specific settings (Albeck et al., 2008; Fritsche-Guenther et al., 2011).
From a biochemical perspective, enzymatic cascades are unique because pathway activity can be perturbed in two fundamentally different ways. Using molecular-genetic approaches such as stable RNA interference (Brummelkamp et al., 2002) or dCas9-mediated clustered regularly interspaced short palindromic repeats (CRISPR) interference (Qi et al., 2013), one can reduce the abundance of enzymes within a cascade to decrease overall catalysis. Alternatively, one can use small molecules to pharmacologically inhibit the rate of product formation by competing with the native substrate(s) of an enzyme in the cascade. If knockdown of an enzyme and pharmacologic competition for substrate are both ~100% effective, then these two perturbations should yield identical results, provided that the enzyme does not have a catalysis-independent function (Knight and Shokat, 2007). However, because knockdowns are often partial and small molecule doses are limited by pharmacokinetics and off-target toxicities, molecular genetics and pharmacology typically yield only a fractional inhibition in vivo (Bollag et al., 2010; Knott et al., 2014). Given a fractional perturbation, it is unclear whether knockdown and small molecule approaches are truly equal, especially in cascades that contain feedback, feedforward, and autoregulatory mechanisms.
Here we compared the equivalence of enzyme knockdown and inhibition in a three-tiered cascade (A → B → C) that is elaborated with various combinations of internal regulation. Using simple and more detailed chemical-kinetic models of enzymatic cascades, we report that addition of a negative feedback (C –| A) around a targeted enzyme (B) is sufficient to cause the overall efficacy of target knockdown and inhibition to diverge. This prediction is tested experimentally in the canonical Raf → MEK → ERK cascade of protein kinases, where Raf is regulated negatively through its hyperphosphorylation catalyzed by ERK (Dougherty et al., 2005). ERK –| Raf feedback causes an inhibitor of MEK phosphorylation to block pathway activity far more potently than a short hairpin RNA (shRNA) that knocks down MEK abundance by 80%. To examine the role of network wiring more broadly, we comprehensively simulate the knockdown and inhibitor response of three-tiered enzymatic cascades with all possible combinations of single and double feedback. Network topologies with negative feedback generally render pharmacologic inhibition more potent than knockdown, whereas the opposite is predicted for cascades containing positive feedback. The wiring of enzymatic networks provides a new explanation for why genetics and pharmacology could disagree in specific biological settings.
RESULTS
Targeting a Michaelian Enzyme in a Cascade Flanked by Negative Feedback
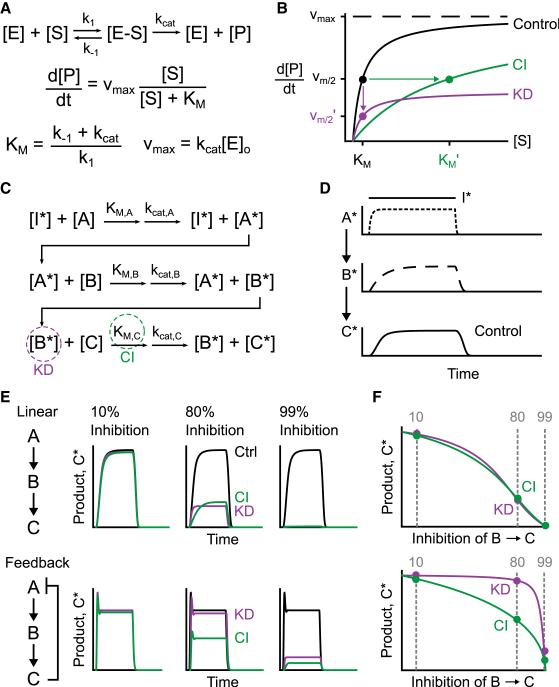
The classic model of enzyme-mediated catalysis involves an enzyme (E), its substrate (S), a reversible enzyme-substrate complex (E-S), and the product formed (P). Assuming that E-S is at pseudo-steady state and that the concentration of S is much greater than P yields the familiar Michaelis-Menten equation. This equation relates the rate of P formation to the concentration of S, the maximum velocity (Vmax), and the Michaelis constant (KM) (Figure 1A). More complicated reaction schemes can be modeled that avoid simplifying assumptions or incorporate multi-substrate reactions. However, a Michaelian system is an adequate starting point for illustrating how the efficacy of pharmacologic and genetic perturbations can diverge.
Figure 1. Negative Feedback Diminishes the Effective Potency of Enzyme Knockdown in a Three-Tiered Michaelian Cascade.
(A) Standard reaction scheme for a Michaelian enzyme, yielding the Michaelis-Menten equation that describes the rate of P formation as a function of Vmax, KM, and the concentration of S.
(B) Knockdown (KD, purple) and competitive inhibition (CI, green) affect the rate of product formation differently as a function of S. For the representative perturbations, Vmax’ = 0.4 Vmax and KM’ = 10 KM.
(C) Abbreviated reaction scheme for a three-tiered cascade of Michaelian enzymes. The parameters are as in (A).
(D) Signal amplification along the three-tiered cascade. The A* trace is shown at one-fifth the arbitrary scale of B* and C*.
(E and F) Fractional perturbation of (E) a linear three-tiered cascade and (F) a negative feedback cascade by KD (purple) or CI (green). The left three plots show C* traces as a function of time. The right plot shows the time-integrated C* traces as a function of the extent of perturbation.
The reaction parameters for the models were KM = 0.04, kcat = 1.0, E0 = 1.0, and Vmax = 1.0. See also Table S1 and Data S1.
In the Michaelian framework, a purely competitive pharmacologic inhibitor acts by precluding the binding of S to E, which increases KM but leaves Vmax unaltered. Conversely, knockdown of a Michaelian enzyme is mathematically equivalent to a perfect non-competitive inhibitor that reduces Vmax without any effect on KM. The distinct mechanisms of competitive inhibition and enzyme knockdown are sufficient to complicate comparisons of relative efficacy. Namely, the potency of the two perturbations can change relative to each other depending on the concentration of S in the system. At low concentrations of S, competitive inhibitors should be more potent than knockdown, whereas the opposite is predicted at high concentrations of S (Figure 1B). Therefore, reaction networks that push an enzyme toward or away from saturation can theoretically give rise to discrepancies in potency.
We examined this principle computationally by modeling a linear cascade of three Michaelian enzymes. In the model, an upstream input (I*) catalytically activates the proximal enzyme of the cascade (A → A*), which then activates the middle enzyme (B → B*) that activates the distal enzyme (C → C*) as the output (Figure 1C). A*, B*, and C* reversibly deactivate according to Michaelian rate processes that return baseline activities to zero in the absence of I* (Experimental Procedures). As expected, we found that a transient step increase in I* led to progressive amplification of the signal from A* to B* to C* (Figure 1D). Amplification is consistent with past studies of signal propagation within three-tiered cascades (Alessi et al., 1995; Schoeberl et al., 2002), and the amount of C* was used to gauge the efficacy of targeted perturbations within the cascade.
Model perturbations focused on the middle enzyme B, with a competitive inhibitor that increases KM,C and knockdown that decreases total B. We optimized rate parameters in the model so that fractional perturbation of B by knockdown or inhibition yielded virtually identical C* profiles for the entire perturbation range (Figure 1E). Using the optimized model, we then supplemented the cascade with a negative feedback from C* to A and compared the efficacy of knockdown with competitive inhibition. When perturbations were negligible (10%) or nearly complete (99%), adding C –| A feedback did not affect the relative impact of knockdown or competitive inhibition on C* (Figure 1F). However, for a realistic intermediate perturbation of 80%, we found that competitive inhibition of B was much more potent than knockdown when negative feedback was present. In the model, the divergent efficacies arise from the kinetics of C*, which accumulate more slowly with a competitive inhibitor (Figure 1E). The delayed accumulation provides time for the C –| A feedback to restrain the steady-state level of C* and, therefore, the overall pathway output upon competitive inhibition. For 80% knockdown of B, the early spike of C* is unaffected, and C –| A feedback from C* causes its trajectory to be almost indistinguishable from the unperturbed network (Figure 1F). These models indicate that simple variations in network topology can alter the apparent potency of genetic and pharmacologic perturbations within Michaelian cascades.
Feedback Causes Knockdown/Inhibitor Discrepancies in a Mass Action Model of the Raf-MEK-ERK Kinase Cascade
We next sought to evaluate perturbations in a more authentic model of an enzyme cascade with or without feedback. One example of a highly modular cascade is the three-tiered phosphorelay system of the mitogen-activated protein kinases (MAPKs) (Johnson and Lapadat, 2002; Schulze et al., 2004). In the prototypical cascade, active Raf (a MAPK kinase kinase) doubly phosphorylates and activates MEK (a MAPK kinase), which then doubly phosphorylates and activates ERK (a MAPK; Figure 2A; Seger and Krebs, 1995). Prior MAPK experiments have estimated rate parameters for association, dissociation, and catalysis (Aoki et al., 2011; Fujioka et al., 2006), enabling detailed mass action models of the cascade that do not require Michaelis-Menten assumptions (Aldridge et al., 2006). Accordingly, the Raf → MEK → ERK signaling pathway has been modeled with various mathematical formalisms in different biological settings (Ahmed et al., 2014; Birtwistle et al., 2007; Chen et al., 2009; Schoeberl et al., 2006; Sturm et al., 2010; Wang et al., 2009).
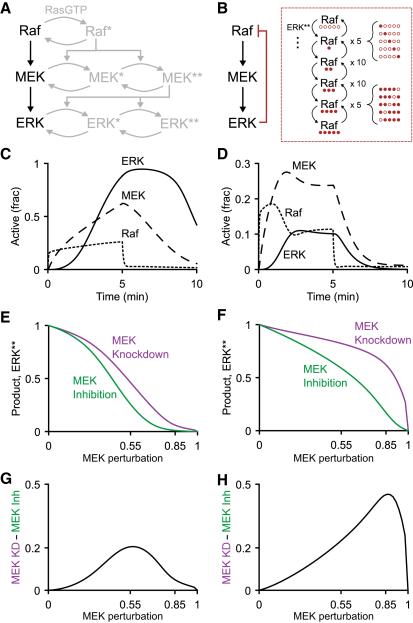
Figure 2. ERK –| Raf Feedback Hyperphosphorylation Dampens the Efficacy of MEK Knockdown in a Mass Action Model.
(A) Standard reaction scheme for the Raf-MEK-ERK MAPK signaling cascade. RasGTP is the upstream activator of Raf. Both MEK and ERK are doubly phosphorylated by a distributive mechanism.
(B) Elaboration of Raf inhibitory hyperphosphorylation by ERK. Raf is directly phosphorylated by ERK on five sites (Dougherty et al., 2005), yielding combinatorial proteoforms of Raf in the model.
(C and D) Active Raf, MEK, and ERK traces in response to a 5-min step increase in RasGTP for the mass action model (C) without feedback or (D) with feedback.
(E and F) Time-integrated trace of active ERK as a function of MEK perturbation by inhibition (green) or knockdown (purple) for the mass action model (E) without feedback or (F) with feedback.
(G and H) Knockdown inhibitor discrepancy as a function of MEK perturbation for the mass action model (G) without feedback or (H) with feedback. See also Figure S1 and Data S1.
Despite its modularity, the architecture of the MAPK cascade is not always linear. In some cell types, active ERK feeds back negatively on the cascade by inhibitory multisite phosphorylation of Raf (Dougherty et al., 2005). ERK –| Raf feedback adds robustness to the steady-state levels of ERK phosphorylation and affects the efficacy of pathway inhibitors (Fritsche-Guenther et al., 2011; Sturm et al., 2010). In the generic Michaelian cascade described above, negative feedback from C to A suppressed the apparent potency of B knockdown compared with pharmacologic inhibition (Figures 1E and 1F). The MAPK pathway and its elaboration with ERK –| Raf feedback provided a three-tiered module to extend this prediction by using more detailed models that were parameterized empirically.
We began by extracting the Raf-MEK-ERK cascade from a mass action model of receptor tyrosine kinase signaling that did not include ERK –| Raf feedback (Chen et al., 2009). A variation of the extracted cascade was built by using a reaction scheme for multisite phosphorylation (Thomson and Gunawardena, 2009) to capture the various stoichiometries of inhibited phospho-Raf (Figure 2B; Experimental Procedures). In both models, deactivation processes were increased slightly (1.5- to 2.5-fold higher compared with Chen et al. [2009]) so that the isolated cascades yielded smooth trajectories in response to a 5-min step increase in Raf activator (Figures 2C and 2D). These two models served as the baseline architectures for investigating knockdown or inhibitor potency when targeting the middle enzyme MEK.
Knockdown of specific proteins is straightforward in mass action models, but encoding small-molecule inhibitors is nontrivial and depends on the mechanism of action (Kleiman et al., 2011). The most specific MEK inhibitors bind to MEK in a way that prevents its activation by Raf (Ballif and Blenis, 2001; Davies et al., 2000). Therefore, a prototypical MEK inhibitor (MEKi) was appended to both models as a species that forms a reversible complex with inactive MEK (Ballif and Blenis, 2001). MEKi-MEK complexes were prohibited from binding Raf but could interact with ERK at the same rate as inactive MEK without MEKi (Supplemental Experimental Procedures). In each model, we incrementally perturbed MEK by increasing MEKi (inhibition) or decreasing total MEK (knockdown). Downstream ERK activation was integrated over time as a final measure of signaling output from the perturbed MAPK cascade.
For the no feedback model, we found that MEK inhibition and knockdown were similar in the context of strong (>80%) MEK perturbations that block most activation of ERK (Figure 2E). By contrast, MEK inhibition and knockdown diverged substantially in the model that included negative feedback (Figure 2F). Earlier work showed that ERK –| Raf feedback confers robustness to the levels of active ERK and resistance to MEK inhibition (Fritsche-Guenther et al., 2011; Sturm et al., 2010). We likewise found that negative feedback within the MAPK cascade dampened the efficacy of MEKi, requiring a 1.5-fold greater perturbation of MEK to achieve 50% inhibition of ERK (Figures 2E and 2F; Figure S1A). However, response damping was even more pronounced for MEK knockdown, with a 91% reduction needed to inhibit ERK activation by 50% (Figure 2F; Figure S1B). Without feedback, the discrepancy between MEK knockdown and inhibition was low except for marginal (~50%) perturbations that would not likely disrupt MAPK function (Figure 2G). The peak discrepancy more than doubled with the addition of ERK –| Raf feedback and, importantly, shifted to ~85% perturbation of MEK (Figure 2H). Rightward shifts in knockdown/inhibitor discrepancy, which peaked at ~85% perturbation, were also observed with various other measures of MAPK signaling output (Figures S1C–S1E). 80+% knockdown efficiency is often assumed to be acceptable for functional studies (Knott et al., 2014). However, our results here suggested that this extent of targeting would be insufficient for disrupting a MAPK cascade with negative feedback, especially when compared with a MEK inhibitor.
ERK –| Raf Feedback Causes MEK Knockdown/Inhibitor Discrepancies in Cells
ERK –| Raf feedback was first described in NIH 3T3 mouse fibroblasts stimulated with platelet-derived growth factor (PDGF), and the kinetics of PDGF-induced MAPK activation have been studied intensely in these cells (Cirit and Haugh, 2012; Dougherty et al., 2005; Murphy et al., 2002; Wang et al., 2009). To test the predictions of the mass action model (Figures 2E–2H), we required a genetic perturbation of NIH 3T3 cells that knocked down MEK strongly but incompletely. Near the 5′ end of the sequence encoding the MEK kinase domain, we identified a sequence that was 100% identical in the two isoforms of MEK and engineered two lentiviral shRNA hairpins (Supplemental Experimental Procedures; Figure S2A). Transduction and selection of NIH 3T3 cells expressing the first shMEK hairpin yielded knockdown of 80% compared with control cells (Figure 3A). Given this level of MEK perturbation, our goal was to determine whether ERK –| Raf feedback influenced the potency of shMEK relative to the selective and established MEK inhibitor U0126 (Davies et al., 2000; Favata et al., 1998), whose mechanism of action is similar to MEK-targeted drugs used clinically (Yoshida et al., 2012).
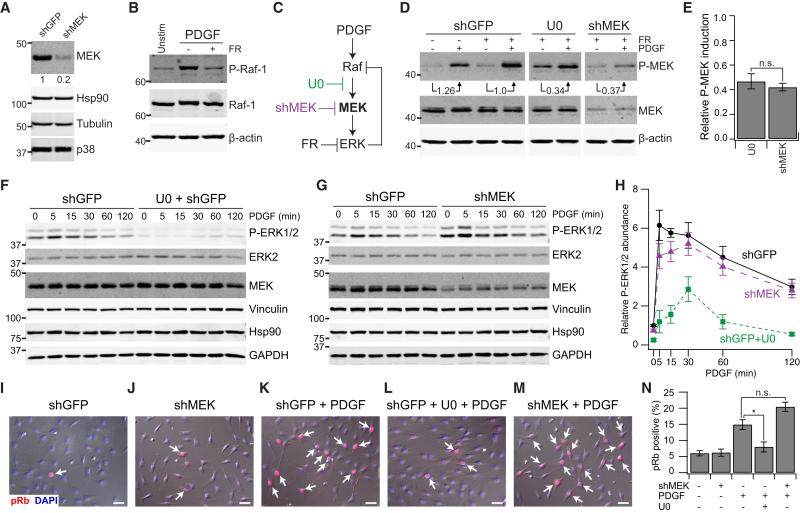
Figure 3. ERK –| Raf Feedback Hyperphosphorylation Dampens the Efficacy of MEK Knockdown in NIH 3T3 Cells Stimulated with PDGF.
(A) Knockdown of total MEK with shRNA. NIH 3T3 cells were transduced with the indicated lentiviruses, lysed, and immunoblotted for endogenous MEK, with Hsp90, tubulin, and p38 used as loading controls.
(B) Cells were pretreated with 20 μM FR180204 (FR) and stimulated with 100 ng/ml PDGF for 30 min, lysed, and immunoblotted for hyperphosphorylated Raf (P-Raf), with total Raf and β-actin used as loading controls.
(C) PDGF-stimulated MAPK cascade and its perturbation by U0126 (U0) and shMEK, with ERK –| Raf feedback disabled by FR.
(D) Quantification of PDGF-induced MEK phosphorylation with ERK –| Raf feedback disabled by FR. Cells were pretreated with 20 μM FR + 2 μM U0, stimulated with 100 ng/ml PDGF for 5 min, lysed, and immunoblotted for phosphorylated MEK (P-MEK) and total MEK, with β-actin used as a loading control. The fold induction of P-MEK is indicated relative to unperturbed cells expressing a control shRNA.
(E) Relative PDGF-induced P-MEK in shMEK cells is equivalent to 2 μM U0126. Data are shown as the mean ± SEM of six independent experiments.
(F and G) PDGF-stimulated ERK phosphorylation is perturbed by U0 but not by shMEK. Cells were pretreated with 2 μM U0 or 0.1% DMSO, stimulated with 1 ng/ml PDGF for the indicated time points, lysed, and immunoblotted for phosphorylated ERK1/2 (P-ERK1/2) and MEK, with total ERK2, vinculin, Hsp90, and tubulin used as loading controls.
(H) Replicated densitometry of the experiment in (F) and (G). Data are shown as the mean ± SEM of four additional independent experiments.
(I–M) PDGF-stimulated proliferation is perturbed by U0 but not by shMEK. Cells were serum-starved, pretreated with 2 μM U0 (L) or 0.1% DMSO (I, K, and M), stimulated with 1 ng/ml PDGF for 24 hr (K–M), and immunostained for hyperphosphorylated Rb (pRb, red), with DAPI (blue) used as a nuclear counterstain. Fluorescence images are shown overlaid on the accompanying differential interference contrast (DIC) image. Scale bar, 20 μm.
(N) Frequency of pRb-positive cells scored for the indicated conditions. Data are shown as the mean ± SEM of three independent experiments. *p < 0.05 by Welch's two-sided t test. n.s., not significant.
See also Figure S2.
To compare inhibition and knockdown fairly, it was important to identify the dose of U0126 that matched the perturbation of shMEK when ERK –| Raf feedback was absent. Feedback is easily added and subtracted in a mass action model (Figure 2), but such reconfigurations are more difficult to achieve cleanly by experiment. Because cells adapt over time to changes in regulatory circuitry (Chandarlapaty et al., 2011; Pratilas et al., 2009), we preferred a small-molecule approach that could acutely disable ERK –| Raf feedback. We found that a 30-min pretreatment with the ERK inhibitor FR180204 (Ohori et al., 2005) completely blocked the PDGF-stimulated hyperphosphorylation of Raf in NIH 3T3 cells (Figure 3B). Therefore, by adding or withholding FR180204, we could examine MAPK signal transduction in the absence or presence of ERK –| Raf feedback and compare shMEK with U0126 (Figure 3C).
Using FR180204 to block feedback, we quantified the PDGF-induced MEK phosphorylation in shMEK-expressing cells as well as in control cells with or without U0126 pretreatment (Figure 3D). Compared with control cells, shMEK reduced stimulated MEK phosphorylation by ~60%, which was matched by pretreating control cells with 2 μM U0126 (Figure 3E). To complement the U0126-shMEK results based on pharmacologic blockade of ERK –| Raf feedback, we engineered a truncated, hormone-responsive Raf allele lacking all ERK phosphorylation sites reported by Dougherty et al. (2005) (Supplemental Experimental Procedures; Figure S2B). Rapid hormone-induced activation of MEK phosphorylation with this feedback-resistant Raf was blocked by 2 μM U0126 as with shMEK (Figures S2C and S2D), corroborating the equivalency found with FR180204. Remarkably, when FR180204 was withheld to allow ERK –| Raf feedback, we found that the matched U0126-shMEK perturbations diverged qualitatively (Figures 3F–3H). U0126 inhibited PDGF-stimulated ERK phosphorylation with surprising potency considering the partial reduction of induced MEK phosphorylation (Figures 3E and 3F). Conversely, an 80% reduction in enzyme abundance with shMEK was largely ineffective as a perturbation in the presence of ERK –| Raf feedback (Figures 3G and 3H). shMEK reduced ERK phosphorylation by ~25% at 5 min after PDGF stimulation, roughly the inhibition caused by 0.01 μM U0126 with ERK –| Raf feedback intact (Figure 3H; Figures S2E and S2F). Therefore, negative feedback within the MAPK cascade can give the appearance of a 2 / 0.01 = ~200-fold difference in potency for perturbations that are grossly equivalent.
We next repeated experiments with a second shMEK hairpin that yielded 97% knockdown, a level of perturbation that should cause knockdown and inhibition to converge (Figure 2H; Figure S2G). Nearly complete knockdown of MEK reduced PDGF-stimulated ERK phosphorylation strongly, comparable with the efficacy of U0126 (Figures S2H–S2J). These results corroborate key predictions of the earlier models (Figures 1 and 2) and demonstrate experimentally that network architecture controls the dynamic signaling response to perturbations.
PDGF stimulation of ERK causes quiescent NIH 3T3 cells to enter into the cell cycle (Murphy et al., 2002). To examine whether the observed differences in ERK phosphorylation were propagated phenotypically, proliferating cells were scored by immunostaining for hyperphosphorylated Rb (Wang et al., 2011). We found that 97% knockdown of MEK significantly decreased proliferation like U0126, which blocked PDGF-induced cell cycle entry (Figures S2K–S2P). In shMEK cells with 80% knockdown, however, basal and PDGF-induced proliferation was comparable with unperturbed cells, showing no resemblance to the U0126-induced phenotype (Figures 3I–3N). In knockdown screens based on cell phenotype, enzyme networks with negative feedback may be prone to false negatives compared with what could be targeted pharmacologically.
Exhaustive Rewiring of Three-Tiered Enzymatic Cascades Reveals Network Motifs for Enhanced Knockdown or Inhibitor Efficacy
The specific importance of ERK –| Raf feedback in the MAPK pathway (Figures 2 and 3) prompted us to examine network topology more generally by returning to the basic Michaelian cascade (Figure 1). Starting with the linear A → B → C pathway, we enumerated all possible additions of one or two regulatory edges within the cascade (14 single-edge topologies and 84 double-edge topologies; Figure S3A). These 98 networks were individually encoded and simulated for their responsiveness to perturbation by knockdown or inhibition as before with the linear cascade (Supplemental Experimental Procedures; Figure 1E). Differences in responsiveness (knockdown – inhibition) were integrated from 0%–100% perturbation of B and sorted to identify motifs that repeatedly created discrepancies in knockdown-inhibitor potency (Milo et al., 2002). If such motifs corresponded to actual enzymatic cascades, one could reevaluate perturbation responses from the perspective of network topology.
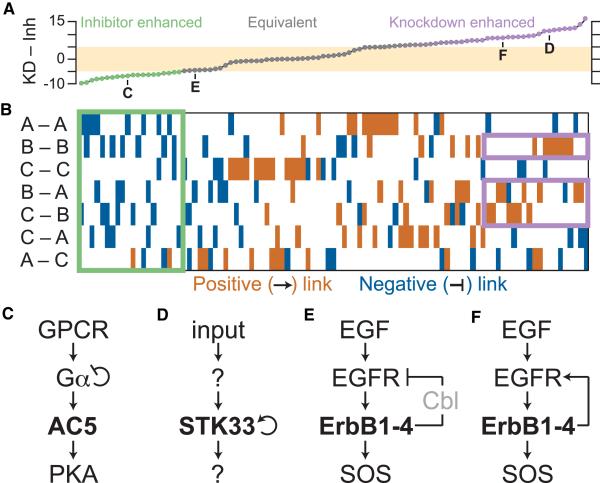
Overall, we found that the network configurations were incredibly diverse in their response to genetic versus pharmacologic perturbation (Figure 4A). Many topologies were negligibly different from the linear case, with 39% showing integrated differences between −5 and 5. At the extremes, however, we identified configurations with discrepancies 2–3 times larger in magnitude than described previously (minimum = −9.8 and maximum = 17 compared with −5.2 for C –| A). Therefore, some enzymatic networks could respond to perturbations in an even more biased manner than what we observed for the MAPK cascade (Figures 2 and 3).
Figure 4. Network Motifs Give Rise to Knockdown/Inhibitor Discrepancies in Three-Tiered Enzyme Cascades.
(A) Exhaustive reconfiguration of a three-tiered Michaelian cascade (Figures 1C and 1D) with all combinations of one to two regulatory edges (Figure S3A). The integrated differences between KD and inhibition (Inh) were calculated across the full range of perturbation as in Figures 1E and 1F. Networks were considered approximately equivalent when the magnitude of KD – Inh discrepancy was less than five (tan). Bold letters refer to the network motifs in Figures 4C–4F.
(B) Color map illustrating the network topologies giving rise to the knockdown/inhibitor discrepancies in (A). Activating edges between enzymes are indicated in brown, and inhibitory edges are indicated in blue. Motifs for enhanced inhibitor and knockdown potency are shown in green and purple, respectively.
(C) G protein-coupled receptor (GPCR) signaling from Gα to AC5 to protein kinase A (PKA).
(D) Autoactivation of STK33 within an unknown enzymatic cascade.
(E and F) EGF signaling from EGFR to ErbB family receptors to SOS, including (E) negative feedback from Cbl or (F) positive feedback from transphosphorylation.
See also Figure S3, Table S2, and Data S1.
When the sorted networks were annotated according to their connectivity, we identified various motifs for strong knockdown/inhibitor discrepancy (Figure 4B). Networks with negative feedback (B –| A, C –| B, C –| A) or negative autoregulation (A –| A, B –| B, C –| C) were associated broadly with inhibitor potency (Figure 4B, green). Although the precise ordering of such networks depended upon initial conditions and rate constants, the set of inhibitor-enhanced motifs was robust to model parameters (Figures S3B–S3E). Among the top 20 networks with the greatest inhibitor potency, 34 of 36 added edges encoded for negative regulation (p < 10−9, binomial test). The most striking example was the A –| A autoregulatory motif, which was contained in eight of 20 networks with the greatest inhibitor potency, including the four most potent (Figure 4B, green). Therefore, networks with A –| A autoregulation would be predicted to be very good candidates for pharmacologic targeting of the effector downstream of A.
A –| A autoregulation occurs in heterotrimeric G proteins, which transmit signals from G protein-coupled receptors (Figure 4C). The Gα subunit of the α-βγ heterotrimer becomes activated by GTP loading but also harbors a GTPase domain that hydrolyzes guanosine triphosphate (GTP) to guanosine diphosphate (GDP) (Neer, 1995). GTP-loaded Gα activates adenylyl cyclase, an enzyme that synthesizes cyclic AMP (cAMP) as a second messenger for protein kinase A (Gilman, 1987). In the heart, one dominant isoform of adenylyl cyclase is adenylyl cyclase 5 (AC5) (Ishikawa et al., 1992), and AC5-selective inhibitors of enzyme activity have been sought as treatments for heart failure (Pierre et al., 2009). A selective AC5 inhibitor has been reported to block cardiomyocyte apoptosis induced by high-dose isoproterenol (Iwatsubo et al., 2004). Despite reducing total adenylyl cyclase activity by 55% and cAMP accumulation by only ~20%, the inhibitor elicited phenotypes as potent as those observed in knockout cardiomyocytes lacking AC5 entirely. Adenylyl cyclases have been underemphasized as drug targets (Pierre et al., 2009), but our results suggest that they are positioned for enhanced pharmacologic efficacy within G protein-coupled receptor signaling cascades.
A reciprocal motif that favored knockdown potency over inhibition of B was positive feedback or autoregulation of B itself (Figure 4B, purple). This result was largely insensitive to the specific parameters used in the models (Figures S3B–S3E). Seventeen of 37 edges in the top 20 networks with the greatest knockdown potency were B → A, B → B, or C → B (p < 10−3, binomial test). Among these, positive B → B autoregulation was the most notable in that it was contained in six of ten networks with the most enhanced knockdown efficacy. Therefore, pharmacology should be less effective than genetics when perturbing proteases, kinases, and E3 ligases that are activated by self-cleavage, autophosphorylation, and autoubiquitination, respectively.
We found evidence for such knockdown/inhibitor discrepancy in RNA interference screens for “druggable” targets. An oncology-focused screen of kinase-phosphatase enzymes discovered that knockdown of the serine-threonine kinase STK33 was lethal to cancer cells harboring mutant KRAS (Scholl et al., 2009). This study prompted the development of potent small-molecule inhibitors of STK33 kinase activity, which, regrettably, had no effect on KRAS-mutant cancers (Luo et al., 2012; Weïwer et al., 2012). Although some have questioned whether STK33 truly interacts genetically with mutant KRAS (Babij et al., 2011), we asked whether network context could provide an alternative explanation for drug failure. The signaling network surrounding STK33 is unknown, but there is evidence that STK33 autophosphorylates in vitro (Brauksiepe et al., 2008) and in cells (Scholl et al., 2009). Moreover, for other kinases in the calcium/calmodulin-dependent protein kinase family to which STK33 belongs, autophosphorylation is known to increase kinase catalytic activity (Colbran et al., 1989). Postulating that STK33 autoactivates by phosphorylation (Figure 4D), we asked whether any surrounding network configuration could influence the superior potency of knockdown. For networks with B → B autoregulation, we found that the median discrepancy was 8.6 (90% nonparametric confidence interval, [6.9–11.6]). Our models raise the possibility that STK33 arose in the original knockdown screen and failed in the pharmacologic follow-up because of its mechanism of autoregulation (Luo et al., 2012; Scholl et al., 2009; Weïwer et al., 2012). Indeed, STK33 continues to autophosphorylate in vitro at inhibitor concentrations >50-fold higher than the half-maximal inhibitory concentration (IC50) for trans-substrate phosphorylation (Luo et al., 2012). An STK33-like knockdown/inhibitor discrepancy has also been described recently for ATM (Lee et al., 2015), a protein kinase whose activity is governed by autophosphorylation (Bakkenist and Kastan, 2003).
Enzymatic cascades are not static in their regulatory edges and can be reconfigured in response to selective pressures from drugs and mutations (Lito et al., 2013). Dynamic rewiring could alter perturbation responses even when the target itself has not been affected directly. We found an example of perturbation potency switching in the epidermal growth factor receptor (EGFR), a receptor tyrosine kinase of the ErbB family. EGFR dimerizes with and transphosphorylates receptors of the ErbB family (including EGFR/ErbB1) to create binding sites for Grb2 and its associated guanine exchange factor, SOS (Yarden and Sliwkowski, 2001). ErbB receptor activation feeds back positively to transphosphorylate EGFR, but strong ErbB family signaling also promotes receptor internalization and degradation through activation of the ubiquitin ligase Cbl (Levkowitz et al., 1998). These competing feedbacks create different wirings for EGFR/ErbB perturbation (Figures 4E and 4F).
In the models, B –| A feedback (Figures 4A and 4E) gave rise to a modest preference for inhibition of B over knockdown (knockdown – inhibitor discrepancy = −4.5). Consistent with this prediction, non-small-cell lung cancer lines showing the fastest inducible downregulation of EGFR have the greatest sensitivity to an EGFR/ErbB1 kinase inhibitor (Ono et al., 2004). Interestingly, in lung cancer lines with evolved or intrinsic resistance to EGFR/ErbB1 inhibition, receptor internalization is disrupted (Kwak et al., 2005; Shtiegman et al., 2007), creating an opportunity for transphosphorylation to dominate (Figure 4F). With B → A positive feedback encoded, the model predicted that knockdown would be much more potent than inhibition of B (knockdown – inhibitor discrepancy = 8.7). Accordingly, inhibitor-resistant lung cancer lines remain susceptible to EGFR/ErbB1 knockdown as well as irreversible EGFR/ErbB1 inhibitors that effectively reduce EGFR/ErbB1 abundance in cells (Kwak et al., 2005). Together, our results indicate that the druggability of an enzyme target is critically dependent on the regulatory edges that surround it.
DISCUSSION
Functional studies using reverse genetics or pharmacology are overwhelmingly focused on the target and downstream consequences of its perturbation. There are known mechanisms by which target knockdown and inhibition can yield discordant results, especially for proteins that signal and scaffold or promote compensatory changes in the cell (Knight and Shokat, 2007). Our work here with enzymes illustrates that the mechanism of perturbation and the associated response are intertwined with the location of that enzyme in the broader signaling network. Perturbation responses can diverge with the addition of one regulatory edge, which we demonstrate by using simple and detailed kinetic models complemented with experiments. In the MAPK literature, there are reports of surprising discrepancies between inhibitor and knockdown responses (Beliveau et al., 2010; Woodson and Kedes, 2012). As with the other wiring motifs, we believe that these discrepancies could be reconciled with the strength of ERK –| Raf feedback, which varies depending on cellular context (Pratilas et al., 2009).
For simplicity, our analysis focused purely on enzymatic cascades, ignoring accessory proteins such as scaffolds and additional kinetic steps such as protein translocation. Likewise, we did not consider more sophisticated mechanisms of small-molecule inhibition other than competitive inhibitors (Figures 1 and 4) and the allosteric, noncompetitive inhibitors in the U0126 class (Figures 2 and 3). Such details will undoubtedly affect the kinetics of signal activation and propagation, but they are unlikely to equalize genetic and pharmacologic perturbations for all network configurations. As long as enzyme perturbations are partial rather than complete (Bollag et al., 2010; Knott et al., 2014), there is the potential for knockdowns and inhibitors to diverge in a topology-dependent manner.
Why do knockdown/inhibitor discrepancies segregate according to the positive and negative regulation in the network? The answer relates back to the fact that genetics and pharmacology make fundamentally different perturbations to enzyme kinetics (Figure 1A). Networks with positive feedback or autoregulation are poised to exhibit switch-like responses that drive an enzymatic reaction to saturation (Tyson et al., 2003). If S is also E (autoregulation), or P promotes the turnover of S (feedback), then it will be difficult for all but the most potent inhibitors to slow a reaction appreciably. Knockdown, by contrast, puts an upper limit on an enzyme-catalyzed reaction even at saturation, thereby favoring increased potency. A reciprocal logic holds for networks with negative feedback or autoregulation, which attenuate enzyme kinetics (Amit et al., 2007). When the effective concentration of S is low, inhibitors provide a superior perturbation compared with partial knockdown of E (Figure 1A). The systems-level rules provided here can help to guide perturbation strategies that seek to tie enzymatic cascades to the biological functions they regulate.
EXPERIMENTAL PROCEDURES
Modeling
For the generic three-tiered enzymatic cascade (Figures 1 and 4), Michaelis-Menten kinetics for activation and deactivation reactions were coded in MATLAB and solved with ode15s. Initial conditions were set to arbitrary values (range, 0–1), and rate parameters were optimized to produce nearly identical perturbations of signaling output upon knockdown or inhibition of B in the linear cascade. Model parameters are included in Tables S1 and S2, a sensitivity analysis of the network topology models is included in Figures S3B–S3E, and the models are archived in Data S1.
For the MAPK model, a mass action model of the Raf-MEK-ERK cascade was extracted from the larger model of Chen et al. (2009) and elaborated with ERK –| Raf feedback inhibition based on five phosphorylation sites that can be attributed directly to ERK (Dougherty et al., 2005). Rate parameters for ERK-mediated feedback phosphorylation of Grb2 (Chen et al., 2009) were used to encode ERK –| Raf feedback hyperphosphorylation. U0126 inhibition was modeled as a reversible binding reaction with inactive MEK that prevented its binding to active Raf. The model is archived in Data S1. Additional modeling details are included in the Supplemental Experimental Procedures.
Experiments
Cell culture, stimulation and lysis, cloning, viral transduction, immunoblotting, and immunofluorescence were performed by standard procedures as described previously (Janes, 2015; Wang et al., 2011). Additional experimental details are included in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
The network around an enzyme affects its susceptibility to knockdown or inhibition
Negative feedback enhances pharmacologic inhibitor potency compared with knockdown
Positive feedback enhances knockdown potency compared with pharmacologic inhibition
ACKNOWLEDGMENTS
We thank Mike Weber, Dan Gioeli, Stuart Licht, and Cheryl Borgman for comments regarding the manuscript, Deborah Morrison and Simon Cook for plasmids, Jun Lin for assistance with cloning, and Chun-Chao Wang for assistance with cell culture. This work was supported by the NIH (Grants 1-R21-AI105970 and 1-DP2-OD006464), the American Cancer Society (Grant 120668-RSG-11-047-01-DMC), the Pew Charitable Trusts (Grant 2008-000410-006), and The David and Lucile Packard Foundation (Grant 2009-34710).
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, K.J.J., C.B.M., and K.A.J.; Investigation, K.J.J. and K.A.J.; Software, C.B.M.; Formal Analysis, K.J.J., C.B.M., and K.A.J.; Writing – Original Draft, K.J.J., C.B.M., and K.A.J.; Writing – Review & Editing, K.J.J., C.B.M., and K.A.J.; Visualization, K.J.J., C.B.M., and K.A.J.; Supervision, K.A.J.; Project Administration, K.A.J.; Funding Acquisition, K.A.J.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, three figures, two tables, and one data file and can be found with this article online at http://dx.doi.org/10.1016/j.cels.2016.01.012.
REFERENCES
- Ahmed S, Grant KG, Edwards LE, Rahman A, Cirit M, Goshe MB, Haugh JM. Data-driven modeling reconciles kinetics of ERK phosphorylation, localization, and activity states. Mol. Syst. Biol. 2014;10:718. doi: 10.1002/msb.134708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albeck JG, Burke JM, Spencer SL, Lauffenburger DA, Sorger PK. Modeling a snap-action, variable-delay switch controlling extrinsic cell death. PLoS Biol. 2008;6:2831–2852. doi: 10.1371/journal.pbio.0060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat. Cell Biol. 2006;8:1195–1203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, et al. A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- Aoki K, Yamada M, Kunida K, Yasuda S, Matsuda M. Processive phosphorylation of ERK MAP kinase in mammalian cells. Proc. Natl. Acad. Sci. USA. 2011;108:12675–12680. doi: 10.1073/pnas.1104030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babij C, Zhang Y, Kurzeja RJ, Munzli A, Shehabeldin A, Fernando M, Quon K, Kassner PD, Ruefli-Brasse AA, Watson VJ, et al. STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res. 2011;71:5818–5826. doi: 10.1158/0008-5472.CAN-11-0778. [DOI] [PubMed] [Google Scholar]
- Bagowski CP, Ferrell JE., Jr. Bistability in the JNK cascade. Curr. Biol. 2001;11:1176–1182. doi: 10.1016/s0960-9822(01)00330-x. [DOI] [PubMed] [Google Scholar]
- Bagowski CP, Besser J, Frey CR, Ferrell JE., Jr. The JNK cascade as a biochemical switch in mammalian cells: ultrasensitive and all-or-none responses. Curr. Biol. 2003;13:315–320. doi: 10.1016/s0960-9822(03)00083-6. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Ballif BA, Blenis J. Molecular mechanisms mediating mammalian mitogen-activated protein kinase (MAPK) kinase (MEK)-MAPK cell survival signals. Cell Growth Differ. 2001;12:397–408. [PubMed] [Google Scholar]
- Beliveau A, Mott JD, Lo A, Chen EI, Koller AA, Yaswen P, Muschler J, Bissell MJ. Raf-induced MMP9 disrupts tissue architecture of human breast cells in three-dimensional culture and is necessary for tumor growth in vivo. Genes Dev. 2010;24:2800–2811. doi: 10.1101/gad.1990410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentele M, Lavrik I, Ulrich M, Stösser S, Heermann DW, Kalthoff H, Krammer PH, Eils R. Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J. Cell Biol. 2004;166:839–851. doi: 10.1083/jcb.200404158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtwistle MR, Hatakeyama M, Yumoto N, Ogunnaike BA, Hoek JB, Kholodenko BN. Ligand-dependent responses of the ErbB signaling network: experimental and modeling analyses. Mol. Syst. Biol. 2007;3:144. doi: 10.1038/msb4100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauksiepe B, Mujica AO, Herrmann H, Schmidt ER. The Serine/threonine kinase Stk33 exhibits autophosphorylation and phosphorylates the intermediate filament protein Vimentin. BMC Biochem. 2008;9:25. doi: 10.1186/1471-2091-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Schoeberl B, Jasper PJ, Niepel M, Nielsen UB, Lauffenburger DA, Sorger PK. Input-output behavior of ErbB signaling pathways as revealed by a mass action model trained against dynamic data. Mol. Syst. Biol. 2009;5:239. doi: 10.1038/msb.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirit M, Haugh JM. Data-driven modelling of receptor tyrosine kinase signalling networks quantifies receptor-specific potencies of PI3K- and Ras-dependent ERK activation. Biochem. J. 2012;441:77–85. doi: 10.1042/BJ20110833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbran RJ, Smith MK, Schworer CM, Fong YL, Soderling TR. Regulatory domain of calcium/calmodulin-dependent protein kinase II. Mechanism of inhibition and regulation by phosphorylation. J. Biol. Chem. 1989;264:4800–4804. [PubMed] [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty MK, Müller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, Conrads TP, Veenstra TD, Lu KP, Morrison DK. Regulation of Raf-1 by direct feedback phosphorylation. Mol. Cell. 2005;17:215–224. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- Ferrell JE, Jr., Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- Fritsche-Guenther R, Witzel F, Sieber A, Herr R, Schmidt N, Braun S, Brummer T, Sers C, Blüthgen N. Strong negative feedback from Erk to Raf confers robustness to MAPK signalling. Mol. Syst. Biol. 2011;7:489. doi: 10.1038/msb.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka A, Terai K, Itoh RE, Aoki K, Nakamura T, Kuroda S, Nishida E, Matsuda M. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J. Biol. Chem. 2006;281:8917–8926. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- Gilman AG. G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Huang CY, Ferrell JE., Jr. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. USA. 1996;93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y, Katsushika S, Chen L, Halnon NJ, Kawabe J, Homcy CJ. Isolation and characterization of a novel cardiac adenylylcyclase cDNA. J. Biol. Chem. 1992;267:13553–13557. [PubMed] [Google Scholar]
- Iwatsubo K, Minamisawa S, Tsunematsu T, Nakagome M, Toya Y, Tomlinson JE, Umemura S, Scarborough RM, Levy DE, Ishikawa Y. Direct inhibition of type 5 adenylyl cyclase prevents myocardial apoptosis without functional deterioration. J. Biol. Chem. 2004;279:40938–40945. doi: 10.1074/jbc.M314238200. [DOI] [PubMed] [Google Scholar]
- Janes KA. An analysis of critical factors for quantitative immunoblotting. Sci. Signal. 2015;8:rs2. doi: 10.1126/scisignal.2005966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- Kleiman LB, Maiwald T, Conzelmann H, Lauffenburger DA, Sorger PK. Rapid phospho-turnover by receptor tyrosine kinases impacts downstream signaling and drug binding. Mol. Cell. 2011;43:723–737. doi: 10.1016/j.molcel.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Knott SR, Maceli AR, Erard N, Chang K, Marran K, Zhou X, Gordon A, El Demerdash O, Wagenblast E, Kim S, et al. A computational algorithm to predict shRNA potency. Mol. Cell. 2014;56:796–807. doi: 10.1016/j.molcel.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak EL, Sordella R, Bell DW, Godin-Heymann N, Okimoto RA, Brannigan BW, Harris PL, Driscoll DR, Fidias P, Lynch TJ, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc. Natl. Acad. Sci. USA. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Bohrson C, Pike AM, Wheelan SJ, Greider CW. ATM Kinase Is Required for Telomere Elongation in Mouse and Human Cells. Cell Rep. 2015;13:1623–1632. doi: 10.1016/j.celrep.2015.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat. Med. 2013;19:1401–1409. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- Liu P, Kevrekidis IG, Shvartsman SY. Substrate-dependent control of ERK phosphorylation can lead to oscillations. Biophys. J. 2011;101:2572–2581. doi: 10.1016/j.bpj.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Masson K, Jaffe JD, Silkworth W, Ross NT, Scherer CA, Scholl C, Fröhling S, Carr SA, Stern AM, et al. STK33 kinase inhibitor BRD-8899 has no effect on KRAS-dependent cancer cell viability. Proc. Natl. Acad. Sci. USA. 2012;109:2860–2865. doi: 10.1073/pnas.1120589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138:760–773. doi: 10.1016/j.cell.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 2002;4:556–564. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- Nakakuki T, Birtwistle MR, Saeki Y, Yumoto N, Ide K, Nagashima T, Brusch L, Ogunnaike BA, Okada-Hatakeyama M, Kholodenko BN. Ligand-specific c-Fos expression emerges from the spatiotemporal control of ErbB network dynamics. Cell. 2010;141:884–896. doi: 10.1016/j.cell.2010.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer EJ. Heterotrimeric G proteins: organizers of transmembrane signals. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- Ohori M, Kinoshita T, Okubo M, Sato K, Yamazaki A, Arakawa H, Nishimura S, Inamura N, Nakajima H, Neya M, et al. Identification of a selective ERK inhibitor and structural determination of the inhibitor-ERK2 complex. Biochem. Biophys. Res. Commun. 2005;336:357–363. doi: 10.1016/j.bbrc.2005.08.082. [DOI] [PubMed] [Google Scholar]
- Ono M, Hirata A, Kometani T, Miyagawa M, Ueda S, Kinoshita H, Fujii T, Kuwano M. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signal-regulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol. Cancer Ther. 2004;3:465–472. [PubMed] [Google Scholar]
- Pierre S, Eschenhagen T, Geisslinger G, Scholich K. Capturing adenylyl cyclases as potential drug targets. Nat. Rev. Drug Discov. 2009;8:321–335. doi: 10.1038/nrd2827. [DOI] [PubMed] [Google Scholar]
- Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, Rosen N. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc. Natl. Acad. Sci. USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano D, Nguyen LK, Matallanas D, Halasz M, Doherty C, Kholodenko BN, Kolch W. Protein interaction switches coordinate Raf-1 and MST2/Hippo signalling. Nat. Cell Biol. 2014;16:673–684. doi: 10.1038/ncb2986. [DOI] [PubMed] [Google Scholar]
- Schoeberl B, Eichler-Jonsson C, Gilles ED, Müller G. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat. Biotechnol. 2002;20:370–375. doi: 10.1038/nbt0402-370. [DOI] [PubMed] [Google Scholar]
- Schoeberl B, Pace E, Howard S, Garantcharova V, Kudla A, Sorger PK, Nielsen UB. A data-driven computational model of the ErbB receptor signaling network. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006;1:53–54. doi: 10.1109/IEMBS.2006.259754. [DOI] [PubMed] [Google Scholar]
- Scholl C, Fröhling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, Silver SJ, Tamayo P, Wadlow RC, Ramaswamy S, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Schulze A, Nicke B, Warne PH, Tomlinson S, Downward J. The transcriptional response to Raf activation is almost completely dependent on Mitogen-activated Protein Kinase Kinase activity and shows a major auto-crine component. Mol. Biol. Cell. 2004;15:3450–3463. doi: 10.1091/mbc.E03-11-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger R, Krebs EG. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- Shah NA, Sarkar CA. Robust network topologies for generating switch-like cellular responses. PLoS Comput. Biol. 2011;7:e1002085. doi: 10.1371/journal.pcbi.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran H, Ippolito DL, Chrisler WB, Resat H, Bollinger N, Opresko LK, Wiley HS. Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Mol. Syst. Biol. 2009;5:332. doi: 10.1038/msb.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtiegman K, Kochupurakkal BS, Zwang Y, Pines G, Starr A, Vexler A, Citri A, Katz M, Lavi S, Ben-Basat Y, et al. Defective ubiquitinylation of EGFR mutants of lung cancer confers prolonged signaling. Oncogene. 2007;26:6968–6978. doi: 10.1038/sj.onc.1210503. [DOI] [PubMed] [Google Scholar]
- Sturm OE, Orton R, Grindlay J, Birtwistle M, Vyshemirsky V, Gilbert D, Calder M, Pitt A, Kholodenko B, Kolch W. The mammalian MAPK/ERK pathway exhibits properties of a negative feedback amplifier. Sci. Signal. 2010;3:ra90. doi: 10.1126/scisignal.2001212. [DOI] [PubMed] [Google Scholar]
- Thomson M, Gunawardena J. Unlimited multistability in multisite phosphorylation systems. Nature. 2009;460:274–277. doi: 10.1038/nature08102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- Wang CC, Cirit M, Haugh JM. PI3K-dependent cross-talk interactions converge with Ras as quantifiable inputs integrated by Erk. Mol. Syst. Biol. 2009;5:246. doi: 10.1038/msb.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brugge JS, Janes KA. Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc. Natl. Acad. Sci. USA. 2011;108:E803–E812. doi: 10.1073/pnas.1103423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weïwer M, Spoonamore J, Wei J, Guichard B, Ross NT, Masson K, Silkworth W, Dandapani S, Palmer M, Scherer CA, et al. A Potent and Selective Quinoxalinone-Based STK33 Inhibitor Does Not Show Synthetic Lethality in KRAS-Dependent Cells. ACS Med. Chem. Lett. 2012;3:1034–1038. doi: 10.1021/ml300246r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson EN, Kedes DH. Distinct roles for extracellular signal-regulated kinase 1 (ERK1) and ERK2 in the structure and production of a primate gammaherpesvirus. J. Virol. 2012;86:9721–9736. doi: 10.1128/JVI.00695-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Kakegawa J, Yamaguchi T, Hantani Y, Okajima N, Sakai T, Watanabe Y, Nakamura M. Identification and characterization of a novel chemotype MEK inhibitor able to alter the phosphorylation state of MEK1/2. Oncotarget. 2012;3:1533–1545. doi: 10.18632/oncotarget.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.