Abstract
Cancer spread (metastasis) is responsible for 90% of cancer-related fatalities. Informing patient treatment to prevent metastasis, or kill all cancer cells in a patient’s body before it becomes metastatic is extremely powerful. However, aggressive treatment for all non-metastatic patients is detrimental, both for quality of life concerns, and the risk of kidney or liver-related toxicity. Knowing when and where a patient has metastatic risk could revolutionize patient treatment and care. In this review, we attempt to summarize the key work of engineers and quantitative biologists in developing strategies and model systems to predict metastasis, with a particular focus on cell interactions with the extracellular matrix (ECM), as a tool to predict metastatic risk and tropism.
Graphical abstract
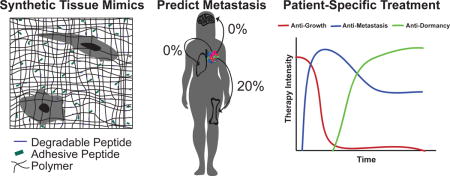
Introduction
Metastasis, the spread of cancer cells from an initial tumor site to other areas of the tissue, or to other tissues entirely, is the cause of 90% of cancer-related deaths. Cancer does not metastasize randomly, rather each type of cancer exhibits a tissue-specific pattern of spread (called tropism) [3]. Some types, like colon [6] and ovarian cancer [7], are dictated by circulation patterns and anatomical proximity. But other types of cancer metastasize to distant organs independently of circulation. Prostate cancer metastasizes nearly exclusively to the bone [8], pancreatic cancer to the lung and liver, and other types, like breast and non-small cell lung cancer, spread to many tissues ([3], Figure 1).
Figure 1.
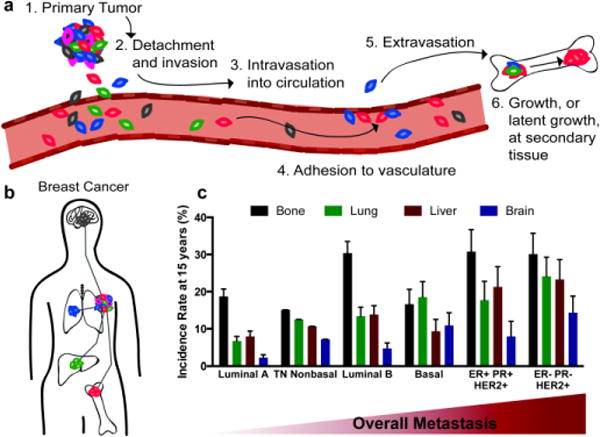
Breast cancer metastatic cascade. a) Overview of metastatic process. We focus on steps 2 and 6 in this review. b) Common sites of breast cancer metastasis, adapted from Barney et al [1]. c) Through a large clinical study, Kennecke et al. found correlations with breast cancer subtype and frequency of metastases found at specific tissue sites, data reproduced from [5]. (ER=estrogen receptor, PR=progesterone receptor, and HER2=human epidermal growth factor receptor 2, and TN=triple negative)
By and far, evidence for what might control metastasis to certain tissues, but not others, has focused on the genetic determinants of the seed (the cancer cell) that dictate tissue selectivity (tropism) [9–13]. Here, we summarize engineered model systems to study the role of the soil (the extracellular matrix, ECM) in mediating metastasis. We discuss cell-ECM interactions and physical forces in guiding metastasis, with a focus on breast cancer. We discuss the community’s ability to predict when and where cancer cells will metastasize, and speculate what these predictions mean for patient prognosis, surveillance, and drug treatment.
Predicting Invasion
Preceding eventual metastatic colonization at a secondary tissue, primary tumor cells must first invade through the basement membrane into the surrounding interstitial tissue. Biopsies are the gold standard for determining the local invasiveness of a patient’s tumor upon diagnosis. There are many biomarkers, such as proteins, miRNAs, and copy number alterations that can identify a patient’s tumor as invasive [14–17]. However, biopsies only provide one snapshot of a tumor, with limited ability to predict any stochastic changes in phenotype or mutations that may occur, thus limiting the future efficacy of a patient’s drug treatment regimen. Recently, biologists and engineers have sought to understand how the ECM impacts the invasiveness of cancer, and if the ECM itself could be used as a predictive biomarker.
Matrix and/or cell stiffness is one such potential predictive marker. Recently a study found that mice with more mechanically compliant primary tumors had more aggressive relapsed tumors at the time of euthanasia [18]. Shear stress due to fluid flow also induces an epithelial to mesenchymal transition (EMT) in ovarian cancer, suggesting that more porous tumors are more metastatic [19]. On in vitro 2D surfaces, Leight et al. found that TGF-β1 induced invasion occurs only on rigid surfaces, thereby linking chemical and physical factors on the initiation of EMT [20]. A large study spearheaded by the National Cancer Institute (NCI) recently showed that physical differences (such as cell compliance, traction forces) are enough to distinguish between a malignant (MDA-MB-231) and normal (MCF-10A) cell line [21]. This study required massive efforts across many laboratories, so more sophisticated tools, perhaps those that rely on electrical impedance [22] instead of individual cell analysis, are required for high-throughput application.
In addition to stiffness, many ECM proteins have documented roles in augmenting or abrogating metastasis (for review, see [23]). In fact, Weinberg’s Hallmarks of Cancer was recently updated to include the role of the ECM in each step of metastasis [24]. The potential for metastasis and the particular distant tissue site may be associated with an identifying “ECM signature” [25]. For example, using ECM-binding as a predictive marker showed that cancer cell binding to combinations of ECM proteins was sufficient to separate metastatic from non-metastatic cell lines [26]. While these are seminal studies, a consistent predictive biomarker of disease prognosis, such as the structure of the ECM, or perhaps an individual or set of proteins within the ECM, has yet to be developed. This is imperative to translate understanding of the physical relationship between a cell and its ECM to clinical treatment.
Where will cancer metastasize?
Given what we know about the heterogeneous nature of tropism, how do we predict where a cell will metastasize? Ideally clinicians would be able to predict metastatic location with biomarkers in a tumor biopsy or in a blood draw, or with quick in vitro assays. An ideal in vitro platform would have controlled, decoupled properties, be highly reproducible, and be easily transferrable to different laboratories and clinics. In in vitro models, high invasion, motility, and proliferation are generally correlated with metastatic potential. One example recently shown by Sheetz and colleagues used simple control of mechanics to report that MDA-MB-231 variant cells proliferate in vitro on materials mechanically similar to the tissue to which they metastasize to in vivo, (Figure 2, [2]). This work was reproduced in a comparative study between ovarian and breast cancer cells (Figure 2, [4]). These examples suggest that physical features may filter for populations of breast cancer cells that exhibit enhanced proliferative ability at specific secondary tissue sites, and that biomaterial platforms with controlled mechanics could be used to identify this proliferative capacity in vitro.
Figure 2.
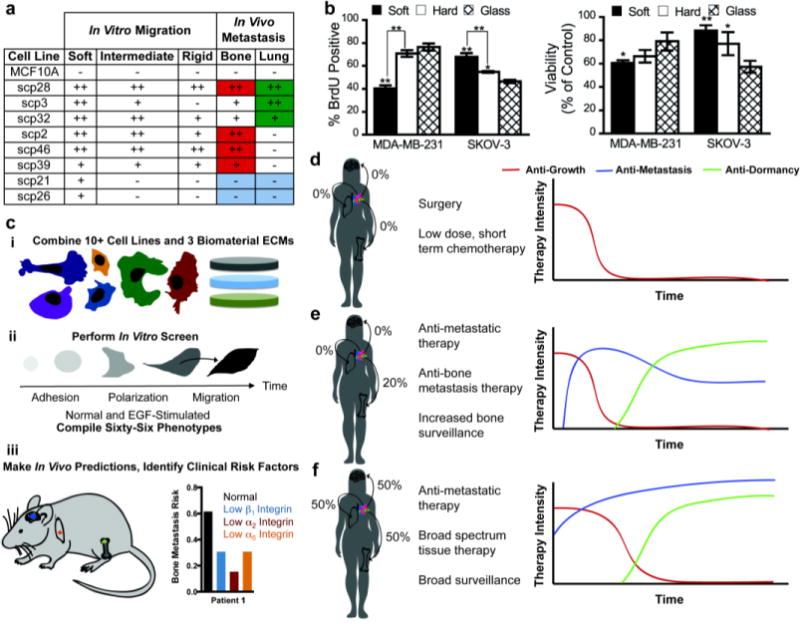
Predicting metastasis and potential impact on therapy. a) Cells that metastasized to bone and lung in vivo shared similar in vitro motility characteristics, adapted from Kostic et al. [2]. b) Mechanosensitivity can be used to distinguish between breast cancer metastatic cells (MDA-MB-231) and ovarian cancer metastatic cells (SKOV-3), from McGrail et al. [4]. c) Barney et al. developed an in vitro screen that used integrin-binding to predict in vivo metastasis to the bone, brain, and lung, as well as identify integrin subunits as tissue-specific risk factors [1] ADD REFERENCE. d–f) One can envision that these types of predictive screens could impact therapy for patients with different risk factors for developing metastatic disease. Patients with low risk of metastasis could be shielded from harsh treatment modalities (d), whereas patients at risk for metastasis (e, f) could be treated aggressively to help prevent metastasis or relapse.
Because the tissues recipient of breast cancer metastasis have striking differences in microenvironment composition, our lab and others have hypothesized that cell-matrix interactions may mediate tropism. We recently showed that integrin-mediated adhesion and motility phenotypes of breast cancer cells, compiled into a phenotypic fingerprint using a systems biology-like approach, can predict bone, brain, or lung metastasis in breast cancer (Figure 2, [1]). In contrast to tools that rely on genetics or fixed tumor tissue (Oncotype DX, MammaPrint, MetaSite Breast, and Prosigna), these engineering approaches use live cell interactions with the microenvironment and may reveal prognostic results missed by traditional approaches.
Tissue-Specific Drivers of Metastasis
Within breast cancer, the pathological subtype of the disease is correlated with different rates of metastasis to different tissues (Figure 1c, [5]). Bone metastases are by far the most common, occurring in 75% of all metastatic breast cancer patients. Brain metastases, on the other hand, are rare, occurring in roughly 15% of all patients, with ER− negative and HER2+ subtypes most commonly presenting. Metastasis to the lungs occurs in approximately 25% of metastatic patients, with the highest presentations in HER2+, ER−/PR− breast cancer.
Pioneering genetic research has identified a subset of genetic markers associated with tissue-specific metastasis. These include COX2, ST6GALNAC5, EGFR, HBEGF [12] L1CAM and SERPINS [27], and GABA for brain metastasis [28]. Bone tropic populations overexpress genes that facilitate metastasis: CXCR4 promotes homing and extravasation, MMP1 facilitates invasion, CTGF and FGF5 aid angiogenesis, and IL11 and OPN are involved with osteolysis and remodeling the bone matrix [10]. Similar studies using lung-tropic human cells have identified up to 54 potential genes in mice, most prominently IL13Rα2, SPARC, ID1, and VCAM1, to be involved in breast-to-lung metastasis [13]. These genetic markers have thus far provided the only possible predictive biomarkers in the primary tumor, and could serve as future therapeutic targets in the clinic.
To augment these genetic efforts, engineers are creating model systems, from simple-to-complex, to represent these tissue sites. These representations typically focus on healthy cells commonly found in these tissues, ECM protein composition, ECM stiffness, and tissue dimensionality. Hydrogels are popular tissue mimics because they recapitulate 3D tissue structure and physiological water content (Figure 3b) [29]. Natural hydrogels, such as collagen-based systems, elicit biochemical cues well, but over-sequester media proteins and cannot simultaneously recapitulate tissue stiffness and biochemical properties. While synthetic polymers can be independently tuned mechanically and chemically, they can be over-simplified, and more fundamental research on tissue properties is needed to make these models more tissue-specific. To combine the advantages of both, Beck et al. combined tunable, synthetic PEG gels with the common cancer model system Matrigel to investigate cell invasion in response to ECM stiffness [30]. Interestingly, they found very similar responses by both cancerous and normal cells, as both had reduced proliferation/morphogenesis in stiff environments. An analogous study used interpenetrating networks of Matrigel and alginate (both natural materials with very different properties), and somewhat in contradiction to the previous study, could force a malignant transformation in healthy breast cells via ECM stiffening [31]. Here, we briefly review approaches used to make these platforms more tissue-specific, and research that can be used to improve future tissue design parameters, with the hopes of uncovering a consensus of how these matrix features drive metastasis.
Figure 3.
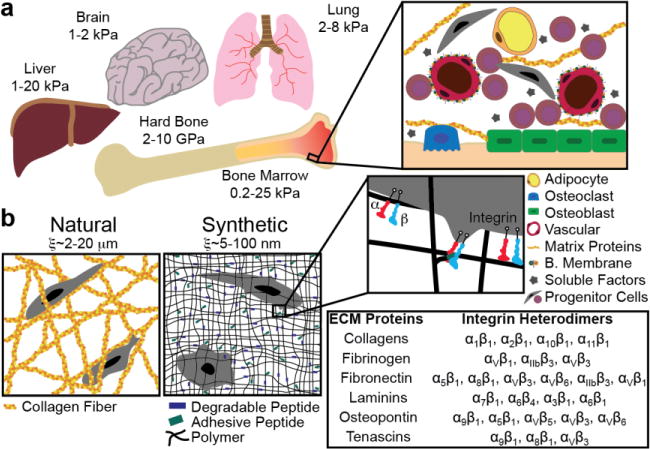
In vitro models of the metastatic niche. a) Common sites of breast cancer metastasis and their published stiffnesses (Young’s modulus). These sites consist of tissue-specific cell types that are part of the metastatic niche. b) These tissues can be represented by natural protein fiber gels or synthetic polymer materials. Polymer matrices can be modified to allow for tissue-specific cell-ECM interactions (via integrin binding) by including oligopeptides responsible for the integrin-binding domains of full-length ECM proteins typically found in these tissues.
The most comprehensive study on tissue proteomics is available through the Proteinatlas (proteinatlas.org), which has annotated ~17,000 protein-coding genes across 32 tissues using both antibodies and RNAseq [32]. The brain ECM consists mainly of hyaluronan (HA) bound by lecticans and tenascin-R with some laminin, fibronectin, collagen IV, and heparin sulfate proteoglycans [33, 34]. Cancer cell binding to the brain ECM through integrins αvβ3 and αvβ6 [35] is also a possible biomarker of brain metastasis risk. The composition of the lung ECM is primarily elastin, collagen I, laminins, and glycosaminoglycans [36]. Upregulation and production of tenascin C, a transient glycoprotein, has been found along with lung metastases and is a sign of poor prognosis [13, 37]. Bone is a composite tissue, dominated by collagen I on the hard trabecular/cortical bone, and fibrillar collagen I, fibronectin, fibrinogen, and proteoglycans throughout the marrow [32].
Researchers are using a variety of techniques to quantify the rigidity of these tissue sites (Figure 3). Reports on the mechanical properties (stiffness) of brain tissue are primarily limited to elastography [38, 39], a technique not commonly performed on in vitro model systems. Obtaining human brain tissue for ex vivo mechanical testing is challenging, therefore indentation studies are limited to animal models, and report Young’s moduli ranging from 1–2 kPa in Young’s modulus [40]. The stiffness of the lung parenchyma has been measured using a variety of techniques in multiple animal models with a Young’s modulus ranging from 2 to 8 kPa [41–43]. Trabecular and cortical bone is markedly stiff (~10GPa). Our lab recently reported the first physiological values for intact bone marrow tissue stiffness (0.3–24.7 kPa), and we anticipate that improving the mechanical relevance of in vitro systems will elucidate mechanisms involved in breast-to-bone cancer progression [44].
A key defining characteristic of the skeleton in addition to its high stiffness is its cyclic mechanical nature - the primary regulator of bone cell function and remodeling [45], alongside increased pressure and fluid shear forces on cells within. When cancer cells arrive in the skeleton, they too are exposed to these cyclic signals, such as compression, hydrostatic pressure, and fluid shear stress. Tumor formation in the bone is inhibited in both breast and ovarian cancer during mechanical load [46, 47]. However, the magnitude of shear stresses and strains in the bone ECM are sufficient for driving malignancy in primary cancer cells in vitro [48], so understanding these relationships may help predict breast-to-bone metastasis.
Engineers have used these analyses as inspiration to create in vitro model systems of brain, bone, and lung tissue, to quantify interactions cancer cells with tissue-specific ECMs. In vitro models of brain include those based on HA [49], HA crosslinked with PEG [50], silk [51, 52], brain tissue slices, microfluidic devices, and some have begun using 3-D bioprinting [53, 54]. In vitro, cells that have been selected for lung metastasis have been found to grow optimally on intermediately stiff 3D hydrogel environments [2]. Decellularized tissues have been used to mimic bone ECM [55], as well as synthetic and silk-based scaffolds with either inverted colloidal geometry or electrospinning, functionalized with proteins found in bone [56, 57]. When implanted, they can capture circulating tumor cells [58].
Microfluidic systems can also capture features of cancer cell extravasation into bone-like matrices in the presence of local mesenchymal stem cells (MSCs) and endothelial cells [59]. One such lab developed a 3D metastasis model, called rMet, which is composed of ECM proteins characterizing of primary and secondary (bone) tissues sites of cancer metastasis [60]. Cancer cells that were highly invasive into their bone ECM were also highly metastatic to the bone in mice, demonstrating that an in vitro system can be predictive of in vivo behavior.
Combined with experimental results, predictive algorithms can be derived and incorporated into mechanics simulations. For example, Wang et al. incorporated osteocyte viability algorithms into their models simulating disuse and recovery to predict when bone tissue would undergo resorption or formation [61]. These types of systems could be used for metastasis studies, to predict how cancer cells will behave under a particular mechanical environment (e.g. will they be dormant, will they migrate elsewhere, fracture risk after a potential drug, etc).
The next step for model systems is to incorporate these matrices with vasculature [62, 63] for a more complete assessment of the metastatic cascade, and to translate these phenomenological behaviors into biomarkers useful for clinicians and patients at risk of metastasis, such as those identified in vivo [64].
Dormancy
Breakthrough research has revealed the existence of dormant tumor cells: metastasized cells quiescent at the metastatic site that may not proliferate, or “awaken” for several years, even decades, and are extremely difficult to detect and treat. Local stromal cells in the metastatic niche likely play a dominant role in paracrine activation of dormant cells, such as glial cells in the brain [65], and hepatocytes and non-parenchymal cells in the liver [66]. Vasculature plays a dual role, in some cases maintaining quiescence [67], and releasing the factors critical for re-activation in others [68]. Both Ghajar [69] and Aguirre-Guiso et al. [70] have proposed a therapeutic regimen to systemically treat possible dormant cells both during and after adjuvant therapy.
In vitro, cell adhesion to specific stromal-derived matrices [71], or substrates of particular stiffnesses [72] can permit dormancy. Specific combinations of bone marrow or hepatic stromal cells can induce breast cancer cell dormancy via cytokine secretion [66, 73]. For example, the Griffith lab used an engineered liver tissue to study stromal cell and ECM re-activation of dormant breast cancer cells in the liver [66]. They found that roughly half of infiltrated breast cancer cells were dormant, and not surprisingly, the more aggressive MDA-MB-231 cells were more sensitive to re-activation by non-parenchymal cells than the luminal MCF7s.
In combination with these systems, traditional culture platforms allow for selection of quiescent subpopulations, via isolating dormant cells after therapeutic administration [74] or from biomaterial systems that induce dormancy. By coupling these subpopulations with engineered microenvironments, it will be possible to accurately determine how therapy may be directed to induce dormancy and eradicate quiescent cells, preventing metastatic outgrowth altogether [69].
Predicting drug response for metastatic patients
Ideally, once metastasis risk is assessed, developing patient-specific drug regimes will follow. Anti-metastasis drugs include those targeted at vascularization, growth in certain microenvironments, cell-ECM binding, receptor kinases, and cancer stem cells (Figure 2d–f) [75]. In vitro model systems are being developed to test for drug response in more physiologically relevant microenvironments that could be tailored to specific tissue sites. A recent example is a 4D lung model, in which circulating tumor cells were as resistant to cisplatin in the model as they were in vivo, results not captured on 2D surfaces [76]. Similarly, combining ovarian cancer cells with fibroblasts in a representative ECM system recapitulated in vivo drug response [77]. These types of organotypic representations of tissue for drug screening are likely to become increasingly popular with the advent of patient-derived cultures [78]. These systems can be used with a more reductionist approach as well, e.g., ECM stiffness has been reported to be a powerful mediator of drug response for paclitaxel [79], ibuprofen [80], and sorafenib [81]. Ideally, the findings demonstrated here can be related to quantifiable, predictive biomarkers, informing drug treatment and monitoring.
Conclusions
Our discussion here was limited to experimental model systems to understand, predict, and treat metastatic spread. There is a keen need for computational methods to better inform and guide these experiments, such as how cells can resist chemotherapy treatment on the basis of likely heterogeneous drug distribution [82]. To our knowledge, no in silico approaches have been employed at point of care to predict metastatic spread. In addition, we suggest a need for computational experts to apply population-level survival models alongside kinetics to create a global map of cell trafficking from the primary tumor to eventual metastasis and growth in a secondary tissue site. This could include a probabilistic model of mutations inducing invasion, trafficking and dispersion throughout the body via circulation, and eventual rates of dormancy or rapid growth at a distant site. Experimental model systems are growing, and an increasing number of engineers are applying their expertise to cancer metastasis. Model system improvements include increasing their throughput, including patient-derived cells, and translating their predictive findings into clinically useful biomarkers.
Highlights.
In vitro systems are needed to predict if, when, and where a tumor will metastasize.
The ECM is a major driver of metastasis to specific tissue destinations.
Mechanical forces play a major role in driving invasion and metastatic outgrowth.
Predictive tools need to be moved toward clinical use, e.g. biomarkers.
Acknowledgments
We focused on publications within the last two years, and we acknowledge the substantial foundational work published before this time that we regret we cannot include here. SRP is a Pew Biomedical Scholar supported by the Pew Charitable Trusts. This work was funded by an NIH New Innovator award (1DP2CA186573-01), a grant from the NSF and NCI (DMR-1234852), and start-up funds from the University of Massachusetts, Amherst. LEB was partially supported by National Research Service Award T32 GM008515 from the National Institutes of Health. SRP was supported by a Barry and Afsaneh Siadat faculty development award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1**.Barney LE, Dandley EC, Jansen LE, Reich NG, Mercurio AM, Peyton SR. A cell-ECM screening method to predict breast cancer metastasis. Integr Biol (Camb) 2015;7(2):198–212. doi: 10.1039/c4ib00218k. The authors used a simple, quick 2D adhesion and motility phenotyping platform to predict metastatic outcomes using living cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2*.Kostic A, Lynch CD, Sheetz MP. Differential matrix rigidity response in breast cancer cell lines correlates with the tissue tropism. PLoS One. 2009;4(7):e6361. doi: 10.1371/journal.pone.0006361. Breast cancer cells preferentially proliferate and migrate on biomaterials with stiffness similar to the stiffness of the tissue they preferentially metastasize to. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hess KR, Varadhachary GR, Taylor SH, Wei W, Raber MN, Lenzi R, Abbruzzese JL. Metastatic patterns in adenocarcinoma. Cancer. 2006;106(7):1624–33. doi: 10.1002/cncr.21778. [DOI] [PubMed] [Google Scholar]
- 4.McGrail DJ, Kieu QM, Iandoli JA, Dawson MR. Actomyosin tension as a determinant of metastatic cancer mechanical tropism. Phys Biol. 2015;12(2):026001. doi: 10.1088/1478-3975/12/2/026001. [DOI] [PubMed] [Google Scholar]
- 5**.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO, Gelmon K. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28(20):3271–7. doi: 10.1200/JCO.2009.25.9820. A large, comprehensive clinical study of metastatic relapse at specific tissue sites across the heterogeneous breast cancer clinical subtypes. [DOI] [PubMed] [Google Scholar]
- 6.Weiss L, G E, Torhorst J, Hartveit F, Moberg I, Eder M, Fenoglio-Preiser CM, Napier J, Horne CH, Lopez MJ, et al. Haematogenous metastatic patterns in colonic carcinoma: an analysis of 1541 necropsies. J Pathol. 1986;150(3):195–203. doi: 10.1002/path.1711500308. [DOI] [PubMed] [Google Scholar]
- 7.Rose PG, Piver M Steven, Tsukada Yoshiaki, Lau Taishing. Metastatic Patterns in Histologic Variants of Ovarian Cancer. Cancer. 1989;64(7):1580–13. doi: 10.1002/1097-0142(19891001)64:7<1508::aid-cncr2820640725>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 8.Bubendorf L, Schöpfer A, Wagner U, Sauter G, Moch H, Willi N, Gasser TC, Mihatsch MJ. Metastatic patterns of prostate cancer: An autopsy study of 1,589 patients. Human Pathology. 2000;31(5):578–583. doi: 10.1053/hp.2000.6698. [DOI] [PubMed] [Google Scholar]
- 9.Lorusso G, Ruegg C. New insights into the mechanisms of organ-specific breast cancer metastasis. Semin Cancer Biol. 2012;22(3):226–33. doi: 10.1016/j.semcancer.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–49. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 11.Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, Foekens JA, Massague J. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16(1):67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bos PD, Zhang XH-F, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens Ja, Massagué J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aleskandarany MA, Negm OH, Green AR, Ahmed MA, Nolan CC, Tighe PJ, Ellis IO, Rakha EA. Epithelial mesenchymal transition in early invasive breast cancer: an immunohistochemical and reverse phase protein array study. Breast Cancer Res Treat. 2014;145(2):339–48. doi: 10.1007/s10549-014-2927-5. [DOI] [PubMed] [Google Scholar]
- 15.Afghahi A, Forgo E, Mitani AA, Desai M, Varma S, Seto T, Rigdon J, Jensen KC, Troxell ML, Gomez SL, Das AK, Beck AH, Kurian AW, West RB. Chromosomal copy number alterations for associations of ductal carcinoma in situ with invasive breast cancer. Breast Cancer Res. 2015;17:108. doi: 10.1186/s13058-015-0623-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Volinia S, Nuovo G, Drusco A, Costinean S, Abujarour R, Desponts C, Garofalo M, Baffa R, Aeqilan R, Maharry K, Sana ME, Di Leva G, Gasparini P, Dama P, Marchesini J, Galasso M, Manfrini M, Zerbinati C, Corra F, Wise T, Wojcik SE, Previati M, Pichiorri F, Zanesi N, Alder H, Palatini J, Huebner KF, Shapiro CL, Negrini M, Vecchione A, Rosenberg AL, Croce CM, Garzon R. Pluripotent stem cell miRNAs and metastasis in invasive breast cancer. J Natl Cancer Inst. 2014;106(12) doi: 10.1093/jnci/dju324. The authors demonstrated that there is a differential rigidity response to TGF-beta1, where on soft substrates, cells undergo apoptosis, while on stiff substrates, cells undergo an epithelial to mesenchymal transition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Harbeck N, Sotlar K, Wuerstlein R, Doisneau-Sixou S. Molecular and protein markers for clinical decision making in breast cancer: today and tomorrow. Cancer Treat Rev. 2014;40(3):434–44. doi: 10.1016/j.ctrv.2013.09.014. This large, collaborative effort identified many biophysical differences between non-metastatic and metastatic cells, providing insight into the physical evolution of cancer. [DOI] [PubMed] [Google Scholar]
- 18.Fenner J, Stacer AC, Winterroth F, Johnson TD, Luker KE, Luker GD. Macroscopic stiffness of breast tumors predicts metastasis. Sci Rep. 2014;4:5512. doi: 10.1038/srep05512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizvi I, Gurkan UA, Tasoglu S, Alagic N, Celli JP, Mensah LB, Mai Z, Demirci U, Hasan T. Flow induces epithelial-mesenchymal transition, cellular heterogeneity and biomarker modulation in 3D ovarian cancer nodules. Proc Natl Acad Sci U S A. 2013;110(22):E1974–83. doi: 10.1073/pnas.1216989110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leight JL, Wozniak MA, Chen S, Lynch ML, Chen CS. Matrix rigidity regulates a switch between TGF-beta1-induced apoptosis and epithelial-mesenchymal transition. Mol Biol Cell. 2012;23(5):781–91. doi: 10.1091/mbc.E11-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Physical Sciences - Oncology Centers N. Agus DB, Alexander JF, Arap W, Ashili S, Aslan JE, Austin RH, Backman V, Bethel KJ, Bonneau R, Chen WC, Chen-Tanyolac C, Choi NC, Curley SA, Dallas M, Damania D, Davies PC, Decuzzi P, Dickinson L, Estevez-Salmeron L, Estrella V, Ferrari M, Fischbach C, Foo J, Fraley SI, Frantz C, Fuhrmann A, Gascard P, Gatenby RA, Geng Y, Gerecht S, Gillies RJ, Godin B, Grady WM, Greenfield A, Hemphill C, Hempstead BL, Hielscher A, Hillis WD, Holland EC, Ibrahim-Hashim A, Jacks T, Johnson RH, Joo A, Katz JE, Kelbauskas L, Kesselman C, King MR, Konstantopoulos K, Kraning-Rush CM, Kuhn P, Kung K, Kwee B, Lakins JN, Lambert G, Liao D, Licht JD, Liphardt JT, Liu L, Lloyd MC, Lyubimova A, Mallick P, Marko J, McCarty OJ, Meldrum DR, Michor F, Mumenthaler SM, Nandakumar V, O’Halloran TV, Oh S, Pasqualini R, Paszek MJ, Philips KG, Poultney CS, Rana K, Reinhart-King CA, Ros R, Semenza GL, Senechal P, Shuler ML, Srinivasan S, Staunton JR, Stypula Y, Subramanian H, Tlsty TD, Tormoen GW, Tseng Y, van Oudenaarden A, Verbridge SS, Wan JC, Weaver VM, Widom J, Will C, Wirtz D, Wojtkowiak J, Wu PH. A physical sciences network characterization of non-tumorigenic and metastatic cells. Sci Rep. 2013;3:1449. doi: 10.1038/srep01449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen TA, Yin TI, Reyes D, Urban GA. Microfluidic chip with integrated electrical cell-impedance sensing for monitoring single cancer cell migration in three-dimensional matrixes. Anal Chem. 2013;85(22):11068–76. doi: 10.1021/ac402761s. [DOI] [PubMed] [Google Scholar]
- 23.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196(4):395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15(12):1243–53. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naba A, Clauser KR, Whittaker CA, Carr SA, Tanabe KK, Hynes RO. Extracellular matrix signatures of human primary metastatic colon cancers and their metastases to liver. BMC Cancer. 2014;14:518. doi: 10.1186/1471-2407-14-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reticker-Flynn NE, Malta DF, Winslow MM, Lamar JM, Xu MJ, Underhill GH, Hynes RO, Jacks TE, Bhatia SN. A combinatorial extracellular matrix platform identifies cell-extracellular matrix interactions that correlate with metastasis. Nat Commun. 2012;3:1122. doi: 10.1038/ncomms2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XHF, Lee DJ, Chaft JE, Kris MG, Huse JT, Brogi E, Massagué J. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell. 2014;156:1002–16. doi: 10.1016/j.cell.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neman J, Termini J, Wilczynski S, Vaidehi N, Choy C, Kowolik CM, Li H, Hambrecht AC, Roberts E, Jandial R. Human breast cancer metastases to the brain display GABAergic properties in the neural niche. Proc Natl Acad Sci U S A. 2014;111(3):984–9. doi: 10.1073/pnas.1322098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kharkar PM, Kiick KL, Kloxin AM. Designing degradable hydrogels for orthogonal control of cell microenvironments. Chem Soc Rev. 2013;42(17):7335–72. doi: 10.1039/c3cs60040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck JN, Singh A, Rothenberg AR, Elisseeff JH, Ewald AJ. The independent roles of mechanical, structural and adhesion characteristics of 3D hydrogels on the regulation of cancer invasion and dissemination. Biomaterials. 2013;34(37):9486–95. doi: 10.1016/j.biomaterials.2013.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhuri O, Koshy ST, Branco da Cunha C, Shin JW, Verbeke CS, Allison KH, Mooney DJ. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat Mater. 2014;13(10):970–8. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 32.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, Feilitzen KV, Forsberg M, Persson L, Johansson F, Zwahlen M, Heijne GV, Nielsen J, Pontén F. Tissue-based map of the human proteome. Science. 2015;347(6220) doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 33.Lau LW, Cua R, Keough MB, Haylock-Jacobs S, Yong VW. Pathophysiology of the brain extracellular matrix: a new target for remyelination. Nature reviews Neuroscience. 2013;14:722–9. doi: 10.1038/nrn3550. [DOI] [PubMed] [Google Scholar]
- 34.Suttkus A, Rohn S, Weigel S, Glöckner P, Arendt T, Morawski M. Aggrecan, link protein and tenascin-R are essential components of the perineuronal net to protect neurons against iron-induced oxidative stress. Cell death & disease. 2014;5:e1119. doi: 10.1038/cddis.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schittenhelm J, Klein A, Tatagiba MS, Meyermann R, Fend F, Goodman SL, Sipos B. Comparing the expression of integrins αvβ3, αvβ5, αvβ6, αvβ8, fibronectin and fibrinogen in human brain metastases and their corresponding primary tumors. International Journal of Clinical and Experimental Pathology. 2013;6:2719–2732. [PMC free article] [PubMed] [Google Scholar]
- 36.O’Neill JD, Anfang R, Anandappa A, Costa J, Javidfar J, Wobma HM, Singh G, Freytes DO, Bacchetta MD, Sonett JR, Vunjak-Novakovic G. Decellularization of human and porcine lung tissues for pulmonary tissue engineering. Annals of Thoracic Surgery. 2013:1046–1055. doi: 10.1016/j.athoracsur.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oskarsson T, Acharyya S, Zhang XHF, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massagué J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nature medicine. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy MC, Huston J, 3rd, Jack CR, Jr, Glaser KJ, Senjem ML, Chen J, Manduca A, Felmlee JP, Ehman RL. Measuring the characteristic topography of brain stiffness with magnetic resonance elastography. PLoS One. 2013;8(12):e81668. doi: 10.1371/journal.pone.0081668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo J, Hirsch S, Fehlner A, Papazoglou S, Scheel M, Braun J, Sack I. Towards an elastographic atlas of brain anatomy. PLoS One. 2013;8(8):e71807. doi: 10.1371/journal.pone.0071807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Budday S, Nay R, de Rooij R, Steinmann P, Wyrobek T, Ovaert TC, Kuhl E. Mechanical properties of gray and white matter brain tissue by indentation. Journal of the mechanical behavior of biomedical materials. 2015;46:318–30. doi: 10.1016/j.jmbbm.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown AC, Fiore VF, Sulchek Ta, Barker TH. Physical and chemical microenvironmental cues orthogonally control the degree and duration of fibrosis-associated epithelial-to-mesenchymal transitions. Journal of Pathology. 2013;229:25–35. doi: 10.1002/path.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Booth AJ, Hadley R, Cornett AM, Dreffs AA, Matthes SA, Tsui JL, Weiss K, Horowitz JC, Fiore VF, Barker TH, Moore BB, Martinez FJ, Niklason LE, White ES. Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. American Journal of Respiratory and Critical Care Medicine. 2012;186:866–876. doi: 10.1164/rccm.201204-0754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luque T, Melo E, Garreta E, Cortiella J, Nichols J, Farré R, Navajas D. Local micromechanical properties of decellularized lung scaffolds measured with atomic force microscopy. Acta Biomaterialia. 2013;9:6852–6859. doi: 10.1016/j.actbio.2013.02.044. [DOI] [PubMed] [Google Scholar]
- 44.Jansen LE, Birch NP, Schiffman JD, Crosby AJ, Peyton SR. Mechanics of intact bone marrow. Journal of the Mechanical Behavior of Biomedical Materials. 2015 doi: 10.1016/j.jmbbm.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lanyon LE, Sugiyama T, Price JS. Regulation of Bone Mass: Local Control or Systemic Influenceor Both? IBMS Bonekey. 2009;6(6):218–226. [Google Scholar]
- 46*.Lynch ME, Brooks D, Mohanan S, Lee MJ, Polamraju P, Dent K, Bonassar LJ, van der Meulen MC, Fischbach C. In vivo tibial compression decreases osteolysis and tumor formation in a human metastatic breast cancer model. J Bone Miner Res. 2013;28(11):2357–67. doi: 10.1002/jbmr.1966. These authors showed that mechanical loading decreases bone metastasis of a highly aggressive breast cancer cell line by altering growth, rather than inducing apoptosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pagnotti GM, Adler BJ, Green DE, Chan ME, Frechette DM, Shroyer KR, Beamer WG, Rubin J, Rubin CT. Low magnitude mechanical signals mitigate osteopenia without compromising longevity in an aged murine model of spontaneous granulosa cell ovarian cancer. Bone. 2012;51(3):570–7. doi: 10.1016/j.bone.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munson JM, Shieh AC. Interstitial fluid flow in cancer: implications for disease progression and treatment. Cancer Manag Res. 2014;6:317–28. doi: 10.2147/CMAR.S65444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedron S, Becka E, Harley BAC. Regulation of glioma cell phenotype in 3D matrices by hyaluronic acid. Biomaterials. 2013;34:7408–17. doi: 10.1016/j.biomaterials.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 50.Wang C, Tong X, Yang F. Bioengineered 3D brain tumor model to elucidate the effects of matrix stiffness on glioblastoma cell behavior using PEG-based hydrogels. Mol Pharm. 2014;11(7):2115–25. doi: 10.1021/mp5000828. [DOI] [PubMed] [Google Scholar]
- 51.Hopkins AM, De Laporte L, Tortelli F, Spedden E, Staii C, Atherton TJ, Hubbell JA, Kaplan DL. Silk Hydrogels as Soft Substrates for Neural Tissue Engineering. Advanced Functional Materials. 2013;23:5140–5149. [Google Scholar]
- 52.Zhang Q, Liebeck BM, Yan K, Demco DE, Körner A, Popescu C. Alpha-Helix Self-Assembly of Oligopeptides Originated From Beta-Sheet Keratin. Macromolecular Chemistry and Physics. 2012;213:2628–2638. [Google Scholar]
- 53.Hopkins AM, DeSimone E, Chwalek K, Kaplan DL. 3D in vitro modeling of the central nervous system. Progress in Neurobiology. 2014;125:1–25. doi: 10.1016/j.pneurobio.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naik P, Cucullo L. In vitro blood-brain barrier models: current and perspective technologies. Journal of pharmaceutical sciences. 2012;101:1337–54. doi: 10.1002/jps.23022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Villasante A, Marturano-Kruik A, Vunjak-Novakovic G. Bioengineered human tumor within a bone niche. Biomaterials. 2014;35(22):5785–94. doi: 10.1016/j.biomaterials.2014.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seib FP, Berry JE, Shiozawa Y, Taichman RS, Kaplan DL. Tissue engineering a surrogate niche for metastatic cancer cells. Biomaterials. 2015;51:313–9. doi: 10.1016/j.biomaterials.2015.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thibaudeau L, Taubenberger AV, Holzapfel BM, Quent VM, Fuehrmann T, Hesami P, Brown TD, Dalton PD, Power CA, Hollier BG, Hutmacher DW. A tissue-engineered humanized xenograft model of human breast cancer metastasis to bone. Dis Model Mech. 2014;7(2):299–309. doi: 10.1242/dmm.014076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee J, Li M, Milwid J, Dunham J, Vinegoni C, Gorbatov R, Iwamoto Y, Wang F, Shen K, Hatfield K, Enger M, Shafiee S, McCormack E, Ebert BL, Weissleder R, Yarmush ML, Parekkadan B. Implantable microenvironments to attract hematopoietic stem/cancer cells. PNAS. 2012;109(48):19638–19643. doi: 10.1073/pnas.1208384109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bersini S, Jeon JS, Dubini G, Arrigoni C, Chung S, Charest JL, Moretti M, Kamm RD. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials. 2014;35(8):2454–61. doi: 10.1016/j.biomaterials.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parikh MR, Minser KE, Rank LM, Glackin CA, Kirshner J. A reconstructed metastasis model to recapitulate the metastatic spread in vitro. Biotechnol J. 2014;9(9):1129–39. doi: 10.1002/biot.201400121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H, Ji B, Liu XS, van Oers RF, Guo XE, Huang Y, Hwang KC. Osteocyte-viability-based simulations of trabecular bone loss and recovery in disuse and reloading. Biomech Model Mechanobiol. 2014;13(1):153–66. doi: 10.1007/s10237-013-0492-1. [DOI] [PubMed] [Google Scholar]
- 62.Chen MB, Whisler JA, Jeon JS, Kamm RD. Mechanisms of tumor cell extravasation in an in vitro microvascular network platform. Integr Biol (Camb) 2013;5(10):1262–71. doi: 10.1039/c3ib40149a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ehsan SM, Welch-Reardon KM, Waterman ML, Hughes CC, George SC. A three-dimensional in vitro model of tumor cell intravasation. Integr Biol (Camb) 2014;6(6):603–10. doi: 10.1039/c3ib40170g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ell B, Mercatali L, Ibrahim T, Campbell N, Schwarzenbach H, Pantel K, Amadori D, Kang Y. Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell. 2013;24(4):542–56. doi: 10.1016/j.ccr.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kodack DP, Askoxylakis V, Ferraro GB, Fukumura D, Jain RK. Emerging strategies for treating brain metastases from breast cancer. Cancer Cell. 2015;27(2):163–75. doi: 10.1016/j.ccell.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wheeler SE, Clark AM, Taylor DP, Young CL, Pillai VC, Stolz DB, Venkataramanan R, Lauffenburger D, Griffith L, Wells A. Spontaneous dormancy of metastatic breast cancer cells in an all human liver microphysiologic system. Br J Cancer. 2014;111(12):2342–50. doi: 10.1038/bjc.2014.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67**.Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, Almeida D, Koller A, Hajjar KA, Stainier DY, Chen EI, Lyden D, Bissell MJ. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol. 2013;15(7):807–17. doi: 10.1038/ncb2767. This work combined in vitro and two in vivo models to demonstrate that dormant breast cancer cells can evade detection and potentially treatment via binding to thrombospondin-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Magnus N, Garnier D, Meehan B, McGraw S, Lee TH, Caron M, Bourque G, Milsom C, Jabado N, Trasler J, Pawlinski R, Mackman N, Rak J. Tissue factor expression provokes escape from tumor dormancy and leads to genomic alterations. Proc Natl Acad Sci U S A. 2014;111(9):3544–9. doi: 10.1073/pnas.1314118111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghajar CM. Metastasis prevention by targeting the dormant niche. Nat Rev Cancer. 2015;15(4):238–47. doi: 10.1038/nrc3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aguirre-Ghiso JA, Bragado P, Sosa MS. Metastasis awakening: targeting dormant cancer. Nat Med. 2013;19(3):276–7. doi: 10.1038/nm.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hurst RE, Hauser PJ, You Y, Bailey-Downs LC, Bastian A, Matthews SM, Thorpe J, Earle C, Bourguignon LY, Ihnat MA. Identification of novel drugs to target dormant micrometastases. BMC Cancer. 2015;15:404. doi: 10.1186/s12885-015-1409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schrader J, Gordon-Walker TT, Aucott RL, van Deemter M, Quaas A, Walsh S, Benten D, Forbes SJ, Wells RG, Iredale JP. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology. 2011;53(4):1192–205. doi: 10.1002/hep.24108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marlow R, Honeth G, Lombardi S, Cariati M, Hessey S, Pipili A, Mariotti V, Buchupalli B, Foster K, Bonnet D, Grigoriadis A, Rameshwar P, Purushotham A, Tutt A, Dontu G. A novel model of dormancy for bone metastatic breast cancer cells. Cancer Res. 2013;73(23):6886–99. doi: 10.1158/0008-5472.CAN-13-0991. [DOI] [PubMed] [Google Scholar]
- 74.Li S, Kennedy M, Payne S, Kennedy K, Seewaldt VL, Pizzo SV, Bachelder RE. Model of tumor dormancy/recurrence after short-term chemotherapy. PLoS One. 2014;9(5):e98021. doi: 10.1371/journal.pone.0098021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stock AM, Troost G, Niggemann B, Zanker KS, Entschladen F. Targets for anti-metastatic drug development. Curr Pharm Des. 2013;19(28):5127–34. doi: 10.2174/1381612811319280011. [DOI] [PubMed] [Google Scholar]
- 76.Vishnoi M, Mishra DK, Thrall MJ, Kurie JM, Kim MP. Circulating tumor cells from a 4-dimensional lung cancer model are resistant to cisplatin. J Thorac Cardiovasc Surg. 2014;148(3):1056–63. doi: 10.1016/j.jtcvs.2014.05.059. discussion 1063–4. [DOI] [PubMed] [Google Scholar]
- 77.Kenny HA, Lal-Nag M, White EA, Shen M, Chiang CY, Mitra AK, Zhang Y, Curtis M, Schryver EM, Bettis S, Jadhav A, Boxer MB, Li Z, Ferrer M, Lengyel E. Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nat Commun. 2015;6:6220. doi: 10.1038/ncomms7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VS, Seepo S, Sekhar Pedamallu C, Cibulskis K, Carter SL, McKenna A, Lawrence MS, Lichtenstein L, Stewart C, Koster J, Versteeg R, van Oudenaarden A, Saez-Rodriguez J, Vries RG, Getz G, Wessels L, Stratton MR, McDermott U, Meyerson M, Garnett MJ, Clevers H. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–45. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79*.Zustiak S, Nossal R, Sackett DL. Multiwell stiffness assay for the study of cell responsiveness to cytotoxic drugss. Biotechnol Bioeng. 2013 doi: 10.1002/bit.25097. Using a highthroughput platform, the authors found that paclitaxel resistance is dependent upon substrate stiffness. This is one of the first efforts to make soft gels in a high-throughput method, allowing for rapid screening of cell reponse to drugs in tunable environments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Timm DM, Chen J, Sing D, Gage JA, Haisler WL, Neeley SK, Raphael RM, Dehghani M, Rosenblatt KP, Killian TC, Tseng H, Souza GR. A high-throughput three-dimensional cell migration assay for toxicity screening with mobile device-based macroscopic image analysis. Sci Rep. 2013;3:3000. doi: 10.1038/srep03000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81*.Nguyen TV, Sleiman M, Moriarty T, Herrick WG, Peyton SR. Sorafenib resistance and JNK signaling in carcinoma during extracellular matrix stiffening. Biomaterials. 2014;35(22):5749–59. doi: 10.1016/j.biomaterials.2014.03.058. This is the first work identifying signaling mechanisms possibly responsible for stiffness-mediated drug resistance. The authors improved on existing high-throughput gel platforms by allowing in situ polymerization. [DOI] [PubMed] [Google Scholar]
- 82.Fu F, Nowak MA, Bonhoeffer S. Spatial heterogeneity in drug concentrations can facilitate the emergence of resistance to cancer therapy. PLoS Comput Biol. 2015;11(3):e1004142. doi: 10.1371/journal.pcbi.1004142. [DOI] [PMC free article] [PubMed] [Google Scholar]


