Abstract
Neural stem cells (NSCs) are isolated from primary brain tissue and propagated as a heterogeneous mix of cells, including neural progenitors. To date, NSCs have not been purified in vitro to allow study of their biology and utility in regenerative medicine. In this study, we identify C1qR1as a novel marker for NSCs and show that it can be used along with Lewis-X (LeX) to yield a highly purified population of NSCs. Using time-lapse microscopy, we are able to follow NSCs forming neurospheres, allowing their visualization. Finally, using single-cell polymerase chain reaction (PCR), we determine the molecular signature of NSCs. The single-cell PCR data suggest that along with the Notch and Shh pathways, the Hippo pathway plays an important role in NSC activity.
Introduction
Neural stem cells (NSCs) are present throughout the development of the central nervous system (CNS) and persist into adulthood in certain locations. In the adult, endogenous NSCs hold great potential to be harnessed for treatment of neurological diseases [1]. NSCs are multipotent cells that are able to give rise to the three major cell types in the CNS: neurons, astrocytes, and oligodendrocytes [2]. NSCs are also capable of self-renewal over an extended period of time. NSCs can be robustly maintained and expanded in vitro within free-floating aggregates termed neurospheres [3]. However, neural progenitors (NPs) can also give rise to neurospheres. Thus, neurosphere formation is not a direct readout of the presence of NSCs. Furthermore, neurospheres themselves are not a pure population of NSCs; they comprise a heterogeneous mix of cells, which includes lineage-restricted NPs [3,4].
In previous work, we analyzed messenger RNA (mRNA) at the single-cell level and 48 genes per cell with cells derived from neurospheres [5]. Our analysis revealed three populations of cells within cells derived directly from neurospheres; populations that resembled early, intermediate, and late progenitors following a developmental timeline [6]. The early progenitors express high Bmi1 and Hes5 and low Myc and Klf12, while the late progenitors express low Bmi1 and Hes5 and high Myc and Klf12. NSCs are potentially contained within the early progenitors in neurospheres; however, we could not define an NSC population due to its low numbers.
Crucial to the understanding (functional properties and molecular regulation) and clinical use of NSCs is the isolation of a pure population of cells. The development of strategies capable of distinguishing NSCs from other cells contained within neurospheres is of critical importance. The most useful strategy would be the identification of cell surface markers that are unique to or enrich NSCs. At present, various cell surface markers have been associated with NSCs. The marker list includes p75 receptor [7], CD24a, peanut agglutinin ligand [8], syndecan-1, Notch-1, integrin-β1 [9], brain-specific chondroitin sulfate proteoglycans [10], biantennary complex-type N-glycans recognized by Phaseolus vulgaris erythroagglutinating lectin [11], and GD3 ganglioside [12]. However, the most widely used markers for NSCs are Lewis-X (LeX) [13] and CD133/Prominin1 [14,15].
Quantification of NSC frequency is essential to ascertain whether there is an enrichment of NSCs by a surface marker. A single cell forming a neurosphere and potency determination on that single neurosphere are prerequisites for the calculation of NSC frequency. NSC frequency can be defined in vitro as the summation of neurosphere-forming units (NFUs) × neurosphere multipotency under clonal conditions [16,17]. Previously, we have shown that neurospheres generated from culture density of 50 cells/mL are clonal [16]. Hence, in this study, we use this density to generate neurospheres, unless otherwise stated. Louis et al. [18] have recently put forward a colony-forming assay to enumerate NSC frequency. Using selection markers for HSAlo, PNAlo, and cell size, they estimated an NSC frequency of 12%. Similar NSC frequencies have been reported for side population/LeX [19].
In this study, we report that C1qR1 (also known as C1QR1) is a novel marker for NSCs. We found that C1qR1+ cells possess high proliferative potential and the capacity for self-renewal with an NSC frequency of 5.8%. C1qR1 antibodies stained cells in the subventricular zone (SVZ) and ventricular zone (VZ) colocalizing with Nestin-positive cells. Combining the C1qR1 selection with LeX gave a 35-fold enrichment over unsorted populations and an NSC frequency of 46%. Importantly, 80% of neurospheres formed from LeX+/C1qR1+ cells were tripotent and therefore NSCs. We performed time-lapse microscopy on this highly enriched (LeX+/C1qR1+) population over 5 days and were able to visualize NSC-forming neurospheres. Last, using single-cell mRNA profiling of 48 genes of the LeX+/C1qR1+ population, we obtained a molecular signature for NSCs. The mRNA profile of NSCs reaffirms that these cells self-renew, are highly proliferative, and are undifferentiated. In particular, we identify the Hippo pathway to be characteristic of NSCs.
Materials and Methods
Growth of NSCs/NPs in neurosphere culture
The treatment of animals was performed in accordance with the IACUC and NACLAR guidelines and approved by the Biological Resource Centre (Singapore) (www.brc.a-star.edu.sg/index.php?sectionID=11). Neurosphere cultures were prepared from the forebrain of embryonic (E14) C57BL/6 mice as described previously [20]. Briefly, pregnant C57BL/6 mice were sacrificed by cervical dislocation. The cerebral cortices were rapidly excised from the embryos and mechanically dissociated by gentle pipetting. Dissociated cells were cultured at a density of 2 × 104/mL in Dulbecco's modified Eagle's medium/nutrient mixture F-12 (1:1) mixture medium (Invitrogen) containing B27 supplement (Invitrogen), 20 ng/mL fibroblast growth factor 2 (FGF2; PeproTech), 20 ng/mL epidermal growth factor (EGF) (Invitrogen), and 1% penicillin/streptomycin (Invitrogen). The cells were grown as neurospheres at 37°C in 5% CO2 atmosphere in a humidified incubator. Neurospheres were passaged every 5–7 days. In this study, we used NSCs/NPs within five passages.
For secondary neurosphere formation assay (NFA), neurospheres derived from C1qR1+ and C1qR1− cells were collected and dissociated mechanically into single cells. Each pool of the cells was then reseeded under identical neurosphere growth conditions for another 5 days. To assess their differentiation potential, single neurospheres were plated onto glass coverslips coated with laminin (10 μg/mL; Invitrogen) and poly-l-lysine (10 μg/mL; Sigma). The neurospheres were incubated at 37°C in culture medium without FGF2 and EGF for 4 days and processed further for immunocytochemistry.
Time-lapse analysis
Cells were seeded at clonal levels on 3.5-cm Mattek dishes and imaged on a Zeiss Axiovert 200 M motorized microscope. The imaging system utilizes the Pecon XL3 incubator to ensure focus stability. Pecon heating insert P was used to provide the cells at 37°C, 5% CO2, and required level of humidification. Further components of the imaging system included a Marzhauser motorized X–Y stage and a CoolSnap HQ CCD camera used for multipoint visiting and imaging, respectively. The instrument was driven by the multidimensional acquisition (MDA) feature provided by MetaMorph or Micro-Manager [21]. In most cases, imaging was done to maximize signal to noise and to use low level of illumination to prolong cell survivability. To maximize the amount of data collected, multiple cells in the dish were identified, marked, and revisited every 30 min over 5 days for NFA using the MDA feature in MetaMorph or Micro-Manager. The data were analyzed with MetaMorph software.
Fluorescent-activated cell sorting
The primary neurospheres after 5–7 days were dissociated into single cell by gently pipetting. The cells were then resuspended in phosphate-buffered saline (PBS), blocked with 2% bovine serum albumin (Sigma-Aldrich), and stained with selected antibodies for 30 min each at 4°C in the dark. The selected antibodies were against mouse Prom1 (allophycocyanin conjugated; Miltenyi Biotec), mouse LeX (fluorescein isothiocyanate conjugated; R&D System), mouse c-Kit (phycoerythrin [PE] conjugated; BD Pharmingen), mouse End (PE conjugated; eBioscience), and mouse C1qR1 (PE conjugated; R&D System). Isotype-matched mouse immunoglobulin served as controls. After three washes in PBS, the cells were sorted using a Becton Dickinson FACS Aria. Gating parameters were set by forward and side scatter to exclude debris and dead and aggregated cells. The flow rate was ∼1,000 events/s for high purity and recovery.
Neurosphere formation assay
NFAs were adapted from methods previously described [22–26]. Cells were (1) unsorted, (2) sorted with markers, including C1qR1, Prom1, or LeX, (3) double selected with C1qR1+/LeX+ and C1qR1+/Prom1+, or (4) C1qR1+/LeX−, C1qR1+/Prom1−, C1qR1−/LeX+, and C1qR1−/Prom1+, or (5) C1qR1−/LeX− and C1qR1−/Prom1− combinations. Each cell population was sorted into a 96-well ultra-low attachment plate (Corning) at a density of one cell per well. The cells were analyzed after culturing for 14 days. The numbers of wells that contained neurospheres were determined. NFUs are calculated by expressing the neurospheres formed per 100 seeded cells. Relative NFU is NFU normalized to that of control cultures.
Single-cell mRNA profiling and data processing
A TaqMan assay pool was prepared by adding each of the 48 TaqMan assays (20×; Applied Biosystems) to a final concentration of 0.2× for each assay. Neurospheres were dissociated and single cells were sorted by fluorescence-activated cell sorting (FACS) directly into 10 μL of RT-PreAmp Master Mix (5 μL CellsDirect 2× Reaction Mix [Invitrogen], 2.5 μL 0.2× Assay pool, 0.5 μL SuperScript® III RT/Platinum® Taq mix [Invitrogen], and 2 μL TE buffer). Cells were frozen at −80°C and thawed to induce lysis. Sequence-specific reverse transcription (50°C for 20 min) and reverse transcriptase inactivation (95°C for 2 min) were performed to generate complementary DNAs (cDNAs) of the 48 genes, following which sequence-specific preamplification (18 cycles at 95°C for 15 s and 60°C for 4 min) was performed. The preamplified cDNA was diluted 5-fold and used for single-cell mRNA profiling in 48.48 dynamic arrays on a BioMark system (Fluidigm). Single-cell mRNA profiling was run using the BioMark Data Collection software (Fluidigm) and Ct values were calculated using the BioMark Real-time PCR analysis software (Fluidigm). Cells with Ct value for the endogenous control β-actin between 15 and 25 were considered for analysis. Ct values for a specific cell were normalized to the endogenous control by subtracting the Ct value of β-actin for the same cell. The assumed baseline Ct value is 31.
Clustering of cells based on their mRNA profile
Cells were clustered using nonmetric multidimensional scaling (nMDS) and model-based clustering (Mclust). After dimension reduction by nMDS, Mclust was performed, partitioning cells into clusters. The R-packages, heatmap and mclust, were used for performing nMDS and Mclust, respectively (for information on nMDS, see Weiss et al. [24] and Nakamura et al. [25]).
Quantitative RT-PCR gene expression analysis
Quantitative reverse transcription polymerase chain reaction (RT-PCR) was performed using the CellsDirect One-Step RT-PCR kit (Invitrogen) and an Applied Biosystems 7500 Fast Real-Time PCR System. TaqMan probes for each target gene were designed and synthesized by Applied Biosystems. The following gene expression assays were detected: Nestin:Mm00450205_m1, Pax6:Mm00443081_m1, Sox2:Mm03053810_s1, Hes1:Mm01342805_m1, Hes5:Mm00439311_g1, Musashi1:Mm00485224_m1, and FoxG1:Mm02059886_s1. Cells were FACS-sorted into a 96-well polymerase chain reaction (PCR) plate containing lysis buffer. After sorting, each plate was immediately sealed and incubated at 75°C for 10 min. Cell lysate was then subjected to quantitative real-time RT-PCR according to the manufacturer's instructions. The Ct threshold value was determined by using the automatic baseline determination. All expression values were normalized against β-actin and the relative expression of the genes was determined by using the expression 2−ΔCt.
Immunocytochemistry: potency
Cells were fixed in 4% paraformaldehyde for 20 min and immunostained overnight at 4°C with the following primary antibodies: anti-βIII-tubulin (1:500; Covance), anti-O4 (1:500; Chemicon), and anti-glial fibrillary acidic protein (GFAP; 1:1,000; Sigma). Subsequently, after repeated rinses in PBS, the cells were incubated with the appropriate secondary antibody (Alexa Fluor 488, Alex Fluor 594, Alexa Fluor 647, 1:500; Molecular Probes) for 1 h at room temperature and counterstained with 4′,-6-diamidino-2-phenylindole (DAPI; Invitrogen). Immunocytochemistry was also performed by omitting the incubation with primary antibodies as negative control. Stained cultures were examined and photographed by a Zeiss AxioImager Z1 upright microscope with AxioVision 4.4 software.
To evaluate differentiation properties, primary neurospheres were differentiated on a poly-l-lysine/laminin (10 μg/mL; Invitrogen)-coated 50-well coverglass (Sigma) and examined for the neuronal (βIII-tubulin), astrocytic (GFAP), and oligodendrocytic (O4) markers as described above. The number of tripotent, bipotent, and unipotent neurospheres was quantified and expressed as a percentage of all examined neurospheres.
Immunohistochemistry: in situ brain slices
Embryonic brains were dissected from E14.5 litters obtained from pregnant mice. Coronal brain sections, 10–20 μm thick, were cut on a cryostat and mounted directly onto poly-l-lysine-coated slides. The sections were air-dried and subsequently fixed with 50% ice-cold methanol–acetone mixture for 10 min at −20°C. After being blocked with 10% goat serum, sections were incubated with anti-Nestin (1:500; Novus Biologicals) and anti-C1qR1 (1:200; Millipore) overnight at 4°C. Preincubation with a 10-fold excess of specific blocking peptide (1st BASE) against C1qR1 antibody was used as a negative control for this reaction. The following day, the sections were washed several times with PBS and incubated with anti-chicken Alexa Fluor 488 IgG and anti-mouse Alexa Fluor 568 IgG (1:1,000; Molecular Probes) secondary antibodies for 1 h at room temperature. The sections were counterstained with DAPI (Invitrogen). Fluorescent mounting media were applied before placing coverslips onto the slides. The cells immunoreactive for C1qR1 and Nestin were examined under an Olympus FV1000 confocal microscope.
Cell cycle analysis
A single-cell suspension containing approximately 1 million cells was incubated with antibody against mouse C1qR1 (PE conjugated; R&D System) for 30 min at 4°C in the dark. After repeated washing, the cell suspension was fixed in 70% ethanol at −20°C for 30 min. Cells were then centrifuged and incubated in 5 mL PBS at room temperature for 15 min. Finally, the cells were resuspended in PBS containing 10 μg/mL DAPI. Cell cycle analysis was carried out on an LSR II flow cytometer (Becton Dickinson) and using FlowJo software (Tree Star, Inc).
Statistical analysis
All experiments were done in a minimum of three times. Statistical comparison of datasets was performed by two-tailed paired Student's t-test. P values are denoted in the figures with *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Single-cell mRNA analysis of neurosphere cultures
To identify novel NSC markers, we carried out single-cell mRNA analysis of cells grown in neurosphere cultures (Supplementary Fig. S1; Supplementary Data are available online at http://www.liebertpub.com/scd). Genes encoding cell surface markers that were expressed in 20% or less of the cells analyzed were used as selection markers in cell sorting experiments (Supplementary Fig. S1E); these included LeX (as control), endoglin, c-Kit, Prom1, and C1qR1. These results suggested that C1qR1 may be a novel NSC marker.
Identification of C1qR1 as a potential NSC marker
To determine whether C1qR1 selection may be useful for NSC isolation, NFAs were performed. After selection, cells were plated in the culture medium containing EGF and FGF2 at a density of 667 cells/mL. A control corresponding to unsorted live cells was passed through the FACS and isolated. Neurosphere formation for C1qR1-positive cells was significantly higher than that for control cells as well as that for negative cells (Fig. 1A–C). The purified population generated neurospheres in vitro at 2.4-fold enrichment compared with the control cells.
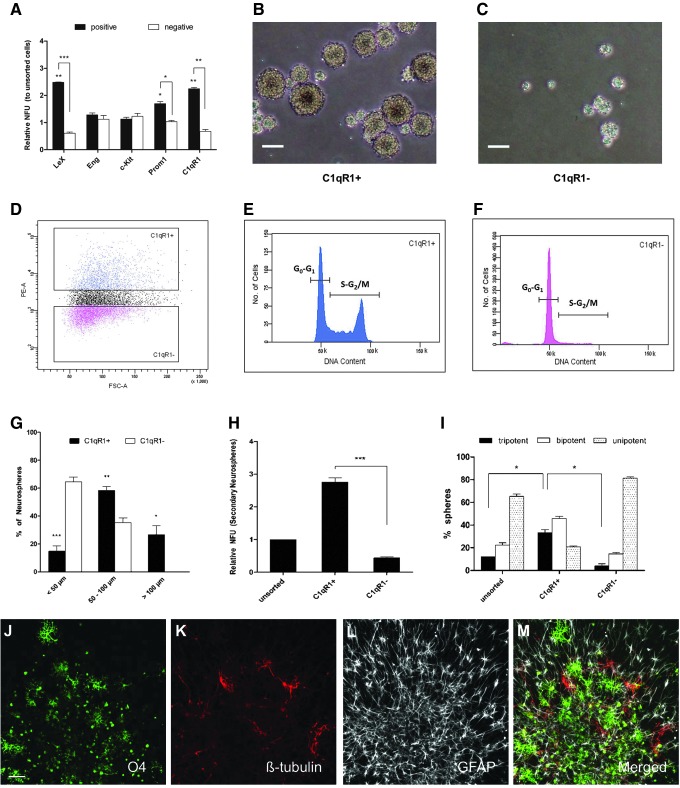
FIG. 1.
In vitro characterization of C1qR1+ cells. (A) Cells obtained from dissociated neurospheres were incubated with anti-Lewis-X (LeX), anti-Eng, anti-cKit, anti-Prom1, and anti-C1qR1 antibodies. Positive and negative cells for each marker were fluorescence-activated cell sorting (FACS)-sorted at 667 cells/mL into the culture medium containing epidermal growth factor (EGF) and fibroblast growth factor 2 (FGF2). The number of neurospheres formed was counted after 5 days. “Unsorted cells” refers to sorting live cells with no gating on any marker. Data are presented as mean ± SEM from three experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (B, C) Bright-field images of neurospheres generated from C1qR1+ and C1qR1− cells. Scale bar = 100 μm. (D–F) Cell cycle analysis of neurosphere-dissociated cells stained with anti-C1qR1-phycoerythrin (PE); 4′-6-diamidino-2-phenylindole (DAPI) was used to separate the cells according to their cell cycle status (G0/G1 phases from sorted [S]-G2/M phases). Analysis gates were set as illustrated in (D). The C1qR1+ and C1qR1− cells are colored blue and red, respectively. Ungated cells are colored black. The cell cycle profile of each population is shown in (E, F). (G) The diameter of neurospheres generated from C1qR1+ and C1qR1− cells was measured and divided into three categories, <50 μm, 50–100 μm, and >100 μm. The bar chart shows the percentage of neurospheres in each size category. (H) Unsorted, C1qR1+, and C1qR1− cells were plated at 667 cells/mL and neurosphere formation followed for 5 days. Neurospheres were then collected and triturated into single cells and reseeded at 667 cells/mL. The number of secondary neurospheres formed was counted 5 days later. (I) Single neurospheres derived from C1qR1+ and C1qR1− cells grown under clonal conditions were differentiated and scored as tripotent, bipotent, or unipotent, according to the cell-type staining observed in each differentiated colony (see the Materials and Methods section). Tripotent neurospheres contain cells that stained positive with glial fibrillary acidic protein (GFAP), ßIII-tubulin, and O4. Bipotent neurospheres contain cells that stained positive with GFAP/ßIII-tubulin or GFAP/O4. Unipotent neurospheres contain cells stained positive with GFAP alone. (J–M) Representative images of differentiated neurospheres stained for the lineage markers, O4 (oligodendrocyte; green), βIII-tubulin (neuron; red), and GFAP (astrocyte; gray). Scale bar = 50 μm.
C1qR1+ neurospheres were larger than C1qR1−, suggesting that they had higher proliferative capacity (Fig. 1B, C). To ascertain whether C1qR1-positive cells exhibit the characteristics of NSCs, their proliferative potential, self-renewal, and multipotency were next examined. Dissociated cells were stained with DAPI and analyzed by flow cytometer to determine their DNA content. Using this approach, cells can be assigned to G0/G1, or G2/M phases of the cell cycle. Flow cytometry analysis indicated that a greater fraction of C1qR1+ population was undergoing DNA synthesis and mitosis compared with C1qR1− population; 47.8% ± 7.2% of C1qR1+ cells were in G1/G0, while 45.6% ± 5.1% were in S-G2/M. Conversely, only 9.4% ± 1.2% of C1qR1− cells were in S-G2/M (Fig. 1D–F). Therefore, C1qR1+ cells appeared to be a rapidly cycling population compared with C1qR1− cells. Next, we categorized the neurospheres generated from both populations into three size groups (<50 μm, 50–100 μm, and >100 μm). Analysis demonstrated that the majority (85%) of neurospheres generated from C1qR1+ cells were larger than 50 μm and 27% larger than 100 μm. In contrast, only 35% of neurospheres generated from C1qR1− cells were larger than 50 μm and none was larger than 100 μm in diameter (Fig. 1G).
Self-renewal capacity was assessed by the ability of cells grown as clonal neurospheres to form secondary neurospheres. Neurospheres generated from C1qR1+ and C1qR1− cells were dissociated into single cells and cultured for another 5 days. C1qR1+ cells gave rise to 6-fold higher number of secondary neurospheres compared with cells derived from C1qR1− neurospheres (Fig. 1H).
To evaluate the multipotency, individual clonal neurospheres derived from C1qR1+ and C1qR1− populations were differentiated and stained for markers of the three lineages (neuron, astrocyte, and oligodendrocyte). An example of a trilineage staining analysis is shown in Fig. 1J–M. About 33% of the differentiated C1qR1+-derived neurospheres contained three cell types. However, only 4% of the neurospheres derived from C1qR1− cells were multipotent (Fig. 1I). Taken together, these results demonstrate that the C1qR1+ cells possess high neurosphere formation, proliferative potential, self-renewal capacity, and multipotency.
C1qR1 expression in brain tissue
To study the localization of C1qR1 in developing embryonic brain cortex, we used immunohistochemistry. During development, NSCs/NPs reside predominantly in the VZ of embryonic cortex from days 10.5 to 14.5. We found that C1qR1 is localized throughout both the VZ and SVZ with stronger expression in the apical VZ. There were occasional intense C1qR1-positive cells in the SVZ or VZ, as shown in Fig. 2C, G. C1qR1 colocalized with Nestin in the embryonic cortex (Fig. 2), suggesting that it is expressed in NSCs in vivo.
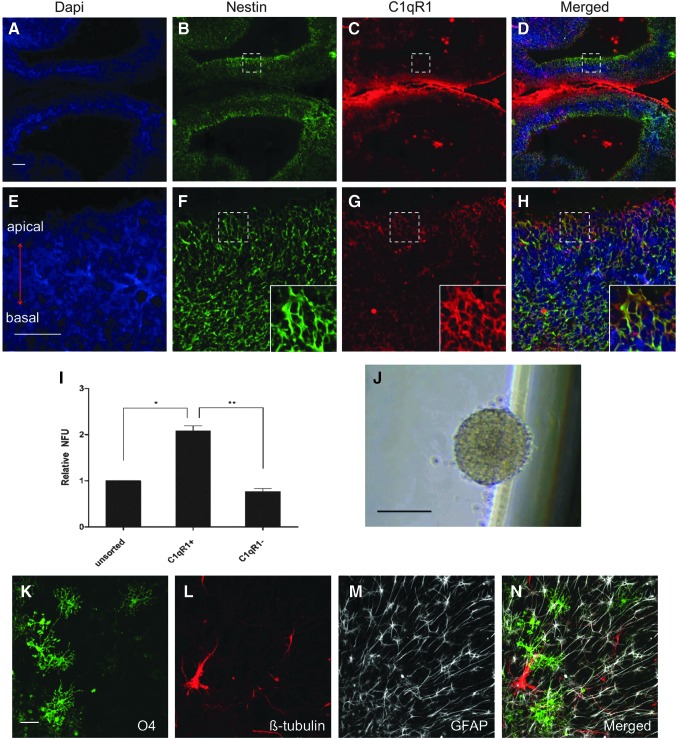
FIG. 2.
Immunohistochemical analysis of C1qR1+ cells. (A–H) Expression and localization of C1qR1 protein in the developing cerebral cortex at E14.5. Immunohistochemistry of cerebral cortex at low magnification (10×) (A–D) and high magnification (40×) (E–H). Labeling for DAPI in (A) and (E), Nestin in (B) and (F), C1qR1 in (C) and (G), and Merged in (D) and (H). The boxes in (B–D) span the ventricular zone (VZ) from the apical to basal region and are shown at higher magnification in (F–H). The inserts in (F–H) show optical zoom of the boxed areas. C1qR1 is localized in the membrane throughout the neural progenitor cells of the cortical ventricular zones (VZ and subventricular zone) with an intense staining in the VZ. The insert in (H) shows the cellular colocalization of C1qR1 and nestin (as seen in yellow). Scale bars (A) = 100 μm and (E) = 40 μm. (I) Cells obtained from freshly dissociated embryonic brain were stained with anti-C1qR1-PE. Unsorted, C1qR1+, and C1qR1− cells were sorted and plated at 667 cells/mL. The number of primary neurospheres formed in the presence of EGF and FGF2 was counted 5 days later. Data are presented as mean ± SEM from three experiments. *P < 0.05, **P < 0.01. (J) Bright-field image of secondary neurosphere generated from C1qR1+ cells. Scale bar = 100 μm, at 4× magnification. (K–N) C1qR1+ population gives rise to neurospheres that differentiate into oligodendrocytes (O4; green), neurons (βIII-tubulin; red), and astrocytes (GFAP; gray). Scale bar = 50 μm.
If C1qR1 is a marker for NSCs, it should enrich for NFUs from dissociated E14.5 brain tissue. To examine this possibility, cells obtained from freshly dissociated E14.5 brain tissue were labeled with PE-C1qR1, and then sorted with a flow cytometer. Sorted cells were cultured at low density (667 cells/mL) in serum-free growth medium containing FGF2 and EGF. C1qR1+ cells formed more neurospheres than the control and C1qR1− cells. The NFUs of C1qR1+ cells were 2.1-fold higher compared with unsorted cells and 2.5-fold higher compared with C1qR1− cells (Fig. 2I).
To verify that C1qR1+ cells from E14.5 brain cells possess the NSC phenotype, primary neurospheres were dissociated into single cells and cultured for 5 days. Formation of secondary neurospheres demonstrated their self-renewal capacity (Fig. 2J). Next, neurospheres were plated onto poly-l-lysine/laminin-coated coverslips and withdrawn from growth factor expansion to confirm the multipotency. Adherent cells displayed distinct morphologies consistent with their expression βIII-tubulin, GFAP, or O4 (Fig. 2K–N). Hence, C1qR1 enriches for self-renewing multipotent neurosphere-forming cells from primary E14.5 brain tissue.
Characteristics of C1qR1/LeX and C1qR1/Prom1 selected cells
Cell surface markers, LeX and Prom1, have been recognized as NSC markers and have been applied to enrich for NSCs cells from various sources [13–15,27,28]. To investigate whether greater enrichment of NSCs may be achieved by combining LeX and Prom1 expression with additional marker, C1qR1, cells were fractionated into different populations by flow cytometry using the gates illustrated in Fig. 3A–D.
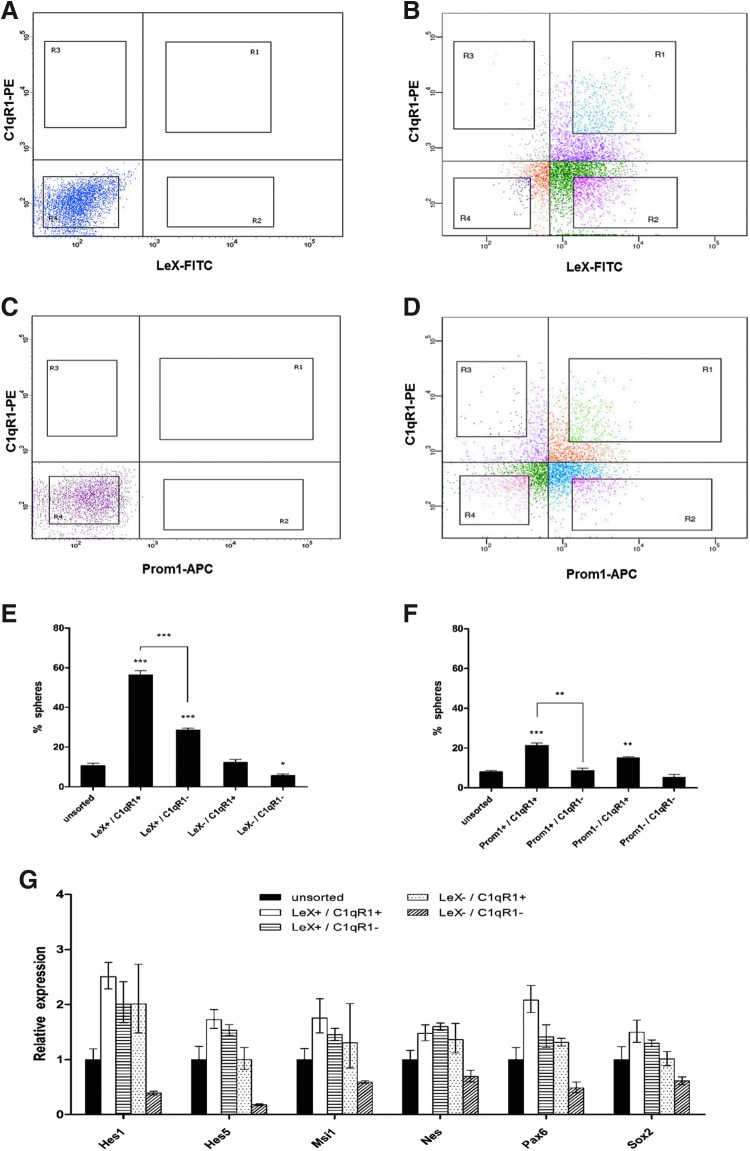
FIG. 3.
Characteristics of LeX/C1qR1-sorted cells. Distribution and sorting gates (R1–R4) of the neurosphere-dissociated cells in combined assays. (A) Negative control for LeX/C1qR1, (B) LeX/C1qR1, (C) negative control for Prom1/C1qR1, (D) Prom1/C1qR1. Four regions (R1: doubly positive; R2 and R3: singly positive; and R4: doubly negative) as well as the unsorted (all live cells) from each combined assay were sorted. (E, F) Bar chart showing the percentage of neurospheres generated per single sorted cell for each subpopulation after 14 days of culture. Data are presented as mean ± SEM from four experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (G) Relative expression of neural stem cell (NSC)-specific genes in each of the FACS-sorted subpopulations from (A) was analyzed by quantitative RT-PCR. The fold change in the gene expression profile was compared with unsorted cells (black bar). RT-PCR, reverse transcription polymerase chain reaction.
Clonal analysis was performed by depositing single NSC/NPs from each purified cell fraction into a 96-well plate. Sorted cells were then cultured to allow neurosphere formation. The NFU of the LeX+/C1qR1+ population was significantly higher compared with the LeX+/C1qR1− population (Fig. 3E). Similarly, the Prom1+/C1qR1+ population generated more neurospheres compared with the Prom1+/C1qR1− population (Fig. 3F). However, the LeX+/C1qR1+ subpopulation gave the highest value for NFU, representing a 2.6-fold enrichment compared with the Prom1+/C1qR1+ population.
We also performed quantitative RT-PCR to test whether any difference in the gene expression pattern could be detected among these FACS-sorted populations. The expression level of markers that have been linked to NSCs was evaluated, including the Sox family member, Sox2 [29], the intermediate filament protein, Nestin [30], the RNA-binding protein, Musashi1 [31], the Notch signaling pathway, Hes1 and Hes5, and the Pax family member, Pax-6. Analysis demonstrated a significantly higher expression of all these genes in LeX+/C1qR1+ cells compared with LeX+/C1qR1−, LeX−/C1qR1+, and unsorted cells (Fig. 3G).
To determine whether LeX and C1qR1 are coexpressed in NSCs/NPs in vivo, we turned to immunohistochemistry analysis of brain slices. E14.5 brain slices were probed with DAPI, LeX, and C1qR1 antibodies. We were able to colocalize LeX with C1qR1 in the VZ and SVZ (Fig. 4A–D). Cellular colocalization of LeX with C1qR1 can be seen in the insert in Fig. 4D. The data clearly show that both LeX and C1qR1 are coexpressed in areas of embryonic brain where NSCs reside.
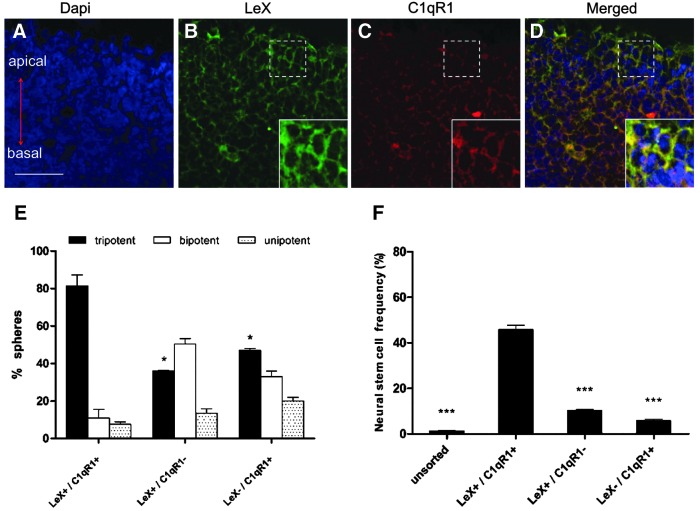
FIG. 4.
NSC frequency of LeX/C1qR1-sorted cells. (A–D) Embryo forebrain (E14.5) coronal sections were counterstained with DAPI (blue) (A), nestin (green) (B), and NogoR1 (red) (C). Merged is shown in (D). The boxes show the location of the inserts in (B–D). The inserts show optical zoom of the boxed areas. The signals of the insets have been adjusted to visualize colocalization of LeX and C1qR1. Scale bar = 40 μm. (E) Individual clonal neurospheres were differentiated on coverslips coated with PLL/laminin and scored as follows: tripotent (astrocytes, neurons, and oligodendrocytes), bipotent (astrocytes/neurons or astrocytes/oligodendrocytes), and unipotent (astrocytes only). Data are presented as mean ± SEM from three experiments. *P < 0.05, ***P < 0.001. Antibodies and methods as described in the Materials and Methods section. (F) NSC frequency was calculated by multiplying the absolute neurosphere-forming units by the percentage of multipotent neurospheres.
Given the higher number of neurospheres formed in LeX+/C1qR1+ combination, multipotency assays were carried out to score for NSC frequency. Individual neurospheres from LeX+/C1qR1+, LeX+/C1qR1−, and LeX−/C1qR1+ populations were differentiated and stained for markers of the three lineages. Upon differentiation, more than 80% of the LeX+/C1qR1+-derived neurospheres were found to be tripotent. Conversely, the tripotent neurospheres in LeX+/C1qR1− and LeX−/C1qR1+ populations were 36% and 47%, respectively. Interestingly, the bipotent neurospheres in the LeX+/C1qR1− population were 40% more compared with LeX+/C1qR1+ population (Fig. 4E). NSC frequency was calculated as the product of NFUs × percentage of tripotent neurospheres; doubly positive selection obtained higher NSC frequency (46%) compared with the 10% and 6% obtained with singly positive selection (Fig. 4F). These results suggest that the LeX+/C1qR1+ fraction was the most highly enriched source of NSCs.
Visualization of NSCs
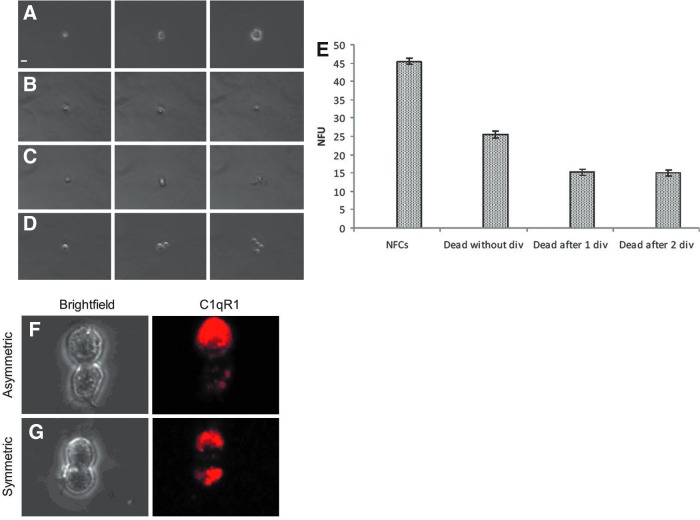
Eighty percent of the neurospheres formed from the LeX+/C1qR1+ cells, under clonal conditions, were tripotent (and therefore NSCs), suggesting that 8 of every 10 cells, which form neurospheres, would be an NSC. We next used time-lapse microscopy to observe NSCs forming neurospheres as distinct from NPs at the single-cell level. LeX+/C1qR1+ cells were seeded at clonal density and followed for 5 days. Fifty-five percent of cells died before forming neurospheres and there were three distinct classes of cells that died: (1) cells that died without division (25%) (Fig. 5A), (2) cells that died after dividing once (15%) (Fig. 5B), and (3) cells that died after dividing twice (15%) (Fig. 5C). We next used C1qR1 to label cells after 24 h of culture where cells on average had divided once. We followed these doublets over 5 days, forming neurospheres using time-lapse microscopy. We observed two types of doublets: (1) where C1qR1 intensity was predominantly in one cell (asymmetric distribution) (Fig. 5F) and (2) where C1qR1 intensity was divided equally in the two cells (symmetric distribution) (Fig. 5G). Of 61 cells observed forming neurospheres, 57 (93%) had asymmetric distribution of C1qR1.
FIG. 5.
Time-lapse imaging of neurosphere formation. LeX+/C1qR1+ cells were seeded at clonal density in a 96-well plate and imaged from the single-cell stage for 5 days. (A) Cells forming neurospheres. (B) Cells dying without division. (C) Cells dying after one division and (D) cells dying after two divisions. (E) Quantification of data presented in (A–D). Scale bar = 10 μm (Mean ± SEM; n = 5). (F, G) LeX+/C1qR1+ cells were seeded at clonal density and left for 24 h. Cells were stained with LeX (or C1qR1), doublets imaged for LeX distribution, and scored for neurosphere formation after 5 days.
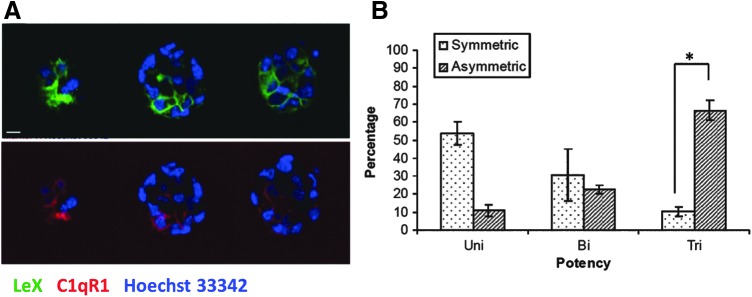
To explore asymmetric division further, we labeled neurospheres grown under clonal conditions for 5 days with LeX and C1qR1, and then imaged them using three-dimensional confocal imaging (Fig. 6A). We found two distributions of label: symmetric and asymmetric. Individual neurospheres were then differentiated and a multipotency assay carried out. Seventy percent of neurospheres that had asymmetric distribution of label were tripotent (Fig. 6B). Taken together, these data suggest that NSCs in neurosphere culture divide asymmetrically and can be visualized with surface markers such as LeX and C1qR1.
FIG. 6.
Distribution of LeX and C1qR1 in neurospheres. (A) Day 5 neurospheres derived from LeX+/C1qR1+ cells and grown under clonal conditions were labeled with LeX (green) or C1qR1 (red) antibodies and imaged on an laser scanning confocal microscope (LSCM) to reveal distribution patterns. Slices from one three-dimensional stack are shown. Quantification of the fluorescence intensity of LeX in two different neurospheres revealed a difference in fluorescence intensity between bright cells relative to other cells within the same neurosphere. (B) Differentiated neurospheres were labeled with lineage-specific markers, O4, βIII tubulin, and GFAP, and the potency of individual neurospheres was scored (Mean ± SEM; 60 neurospheres, n = 4; *P < 0.01).
Single-cell mRNA profiling of LeX+/C1qR1+ and LeX+/C1qR1− populations
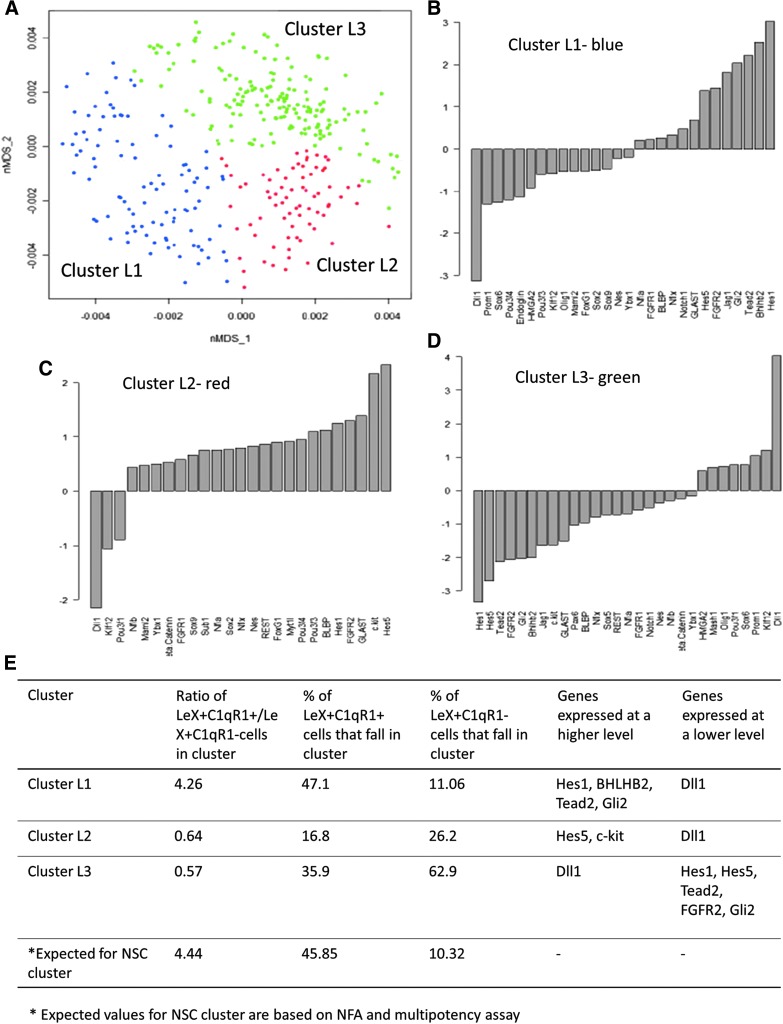
To identify the molecular signature of the NSCs present within the LeX+/C1qR1+ cells, we profiled the mRNA levels of 48 genes in parallel, at the single-cell level. These 48 genes included genes involved in major signaling pathways (Notch, Shh, Wnt), classes of genes such as POU factors and basic helix-loop-helix factors and other transcription factors, surface markers, and epigenetic regulators, which are closely associated with NSCs (Supplementary Table S1). We profiled 172 cells each from LeX+/C1qR1+ and LeX+/C1qR1− populations, combined the expression data, and clustered the cells using nMDS and Mclust. Three clusters were obtained, although not distinct—cluster I, II, and III (Fig. 7A). Expression bar plots showing the genes that were significantly expressed at a higher or lower level in one cluster compared with the other two clusters combined were derived (Fig. 7B).
FIG. 7.
Single-cell messenger RNA (mRNA) profiling of LeX+/C1qR1+ and LeX+/C1qR1− cells. (A) Forty-eight-dimensional mRNA profiling data were compressed to two dimensions by nonmetric multidimensional scaling (nMDS). Axes for the two dimensions are labeled as nMDS_1 and nMDS_2. Model-based clustering (Mclust) was then used to cluster cells based on the mRNA profile of 48 genes. Three clusters were derived—I, II, and III. Each point represents one cell. (B–D) Bar plots showing log2-fold change in gene expression for genes that show significant change in expression (P < 0.05) in a cluster in comparison with the remaining clusters combined. (E) The ratio of LeX+/C1qR1+ cells to LeX+/C1qR1− cells and the percentage of LeX+/C1qR1+ cells and LeX+/C1qR1− cells in each cluster were calculated. The genes that show significant changes in expression in each cluster are shown. Passage 2 cells were used for single-cell mRNA profiling.
The LeX+/C1qR1+ population has a 4.44-fold higher NSC percentage than the LeX+/C1qR1− population at clonal density. If all the NSCs from the LeX+/C1qR1+ and LeX+/C1qR1− populations were to fall in one cluster (ie, the NSC cluster), the ratio of LeX+/C1qR1+ to LeX+/C1qR1− cells in this cluster would be 4.44. We therefore checked the ratio of LeX+/C1qR1+ to LeX+/C1qR1− cells in the three clusters and found that cluster L1 had a ratio of 4.26 compared with clusters L2 and L3, which had a ratio of 0.64 and 0.57, respectively (Fig. 7E). Further analysis revealed that 47.1% of all LeX+/C1qR1+ cells and 11.1% of all LeX+/C1qR1− cells analyzed fell in cluster L1 (Fig. 7E). These percentages fit well with the percentage of NSCs in the LeX+/C1qR1+ (45.85%) and LeX+/C1qR1− cells (10.32%) derived from the NFA and multipotency assay (Fig. 7E). Taken together, cluster L1 represents an NSC cluster.
The probability that cluster L1 is formed by chance was determined. Random clustering of LeX+/C1qR1+ and LeX+/C1qR1− cells was performed 1,000 times to form a cluster of 100 cells (number of cells in cluster L1). The ratio of LeX+/C1qR1+ to LeX+/C1qR1− cells in the cluster formed was calculated each time. A distribution curve showing the probability of obtaining a cluster with specific LeX+/C1qR1+-to-LeX+/C1qR1− ratios was constructed (Supplementary Fig. S2). From the distribution curve, the probability of obtaining a cluster with LeX+/C1qR1+-to-LeX+/C1qR1− ratio of 4.26 by chance is not statistically significant (Supplementary Fig. S2A). Similar analysis for clusters L2 and L3 also showed that the probability of obtaining these clusters by chance is not statistically significant (Supplementary Fig. S2B, C).
Discussion
C1qR1, also known as C1qRp, a collectin receptor and AA4 antigen in rodents, mediates enhanced phagocytosis by monocytes and macrophages upon interaction with soluble defense collagens [32]. It is a type I transmembrane glycoprotein that was originally identified as a B-cell lineage marker [33]. C1qR1 has also been identified as a near marker of multipotent hematopoietic progenitors that can give rise to all mature blood cells [34]. It has been used to enrich human HSCs [35]. C1qR1 is highly expressed in cytoplasmic vesicles in endothelial cells, platelets, and cells of myeloid origin, such as monocytes and neutrophils. Besides, C1qR1-positive cells are also detected in adult spleen and fetal liver [36].
C1qR1+ cells derived from neurosphere culture were enriched for proliferating cells, with high self-renewal capacity and multipotency. Investigation of primary brain material revealed that C1qR1 colocalized with Nestin in the VZ and SVZ. Furthermore, C1qR1+ primary cells from brain were enriched for NFUs, self-renewal capacity, and multipotency. Our analysis using clonal neurosphere formation, followed by single neurosphere differentiation, allowed us to calculate an NSC frequency for unsorted and selected populations. C1qR1+ cells gave an NSC frequency of 5.8%. This is in the same range as NSC frequencies reported for LeX+ cells [18]. Next, we determined whether double selection might improve the enrichment of NSCs by C1qR1. Cells were sorted for C1qR1 with either LeX or Prom1 and compared for NSC frequency and gene expression. Interestingly, LeX+/C1qR1+ cells gave an NSC frequency 46% significantly higher than the other selection combinations used. LeX+/C1qR1+ cells expressed significantly higher levels of Hes1 and Hes5, reporters of the Notch signaling pathway, compared with the unsorted populations, which is consistent with recent work on CBF1 [37]. Importantly, under clonal conditions, 80% of the LeX+/C1qR1+ cells formed neurospheres that were tripotent, suggesting that the double selected cells are very useful to investigate the cell biology of NSCs. We observed that some of the double selected cells did not form neurospheres and died after one or two divisions. We tried a range of experiments to increase neurosphere formation, including the addition of CM, ApoE, CSPG, and shh, but none of these conditions were successful.
Time-lapse analysis following neurosphere formation was carried out under conditions optimized for cell survival. It was not possible to use cytosolic or nuclear labels to follow neurosphere formation as the majority of cells died. Interestingly, we found that cell surface labels such as LeX, C1qR1, and Prom1 do not affect the viability of cells and could be used to follow neurosphere formation. Our time-lapse microscopy with LeX and C1qR1 data suggest that NSCs divide asymmetrically in contrast to NPs. Furthermore, labeling of neurospheres after 5–6 days of growth with either LeX or C1qR1 suggests that asymmetric distribution of label was indicative of NSCs. We did observe neurospheres with intermediate distributions of LeX or C1qR1 label, which were difficult to categorize as symmetric or asymmetric distributions. This may explain why the correlation of asymmetric distribution of label with tripotency was 76% in neurospheres lower than that observed with single-cell analysis. Further improvement of quantitation of label intensity will be required. Nonetheless, these experiments are helpful in distinguishing NSCs from NPs.
Single-cell mRNA profiling on LeX+/C1qR1+ and LeX+/C1qR1− cells revealed an NSC cluster. The heatmap for the NSC cluster showed that the NSCs are heterogeneous in terms of gene expression (Supplementary Fig. S3). The mRNA profile of the NSC cluster showed strong expression of genes involved in self-renewal, proliferation, and repression of differentiation, which are important characteristics of NSCs. Specifically, the NSC cluster showed high expression of genes in the Notch (Notch1, Jag1, Hes1, and Hes5) and Shh (Gli2) signaling pathways, two major signaling pathways involved in NSC self-renewal. The NSC cluster also showed high expression of growth factor receptors, FGFR1 and FGFR2, indicating high proliferative capacity. High expression of known NSC or RGC markers such as GLAST and BLBP further supports characteristics of the NSC cluster. Dll1, a Notch ligand, was the gene with the lowest expression in the NSC cluster. This mimics NSCs in vivo, which have low surface expression of Dll1 so as to minimize activation of Notch receptors on surrounding NPs in a process known as lateral inhibition. Klf12, which is expressed at a low level in early developmental cells [5], and Olig1 [38–40] and Sox9 [41], which are involved in oligodendrocyte differentiation, are expressed at a low level in the NSC cluster, indicating repression of differentiation.
The mRNA profile of the NSC cluster identified genes, which may be important for NSC function and behavior. Tead2 was highly expressed in the NSC cluster. Strong expression of Tead2 is also observed in the VZ/SVZ of the developing cortex where NSCs reside (Allen Brain Atlas). It has been reported that transgenic mice deficient in Tead2 display neural tube defects [42]. Tead2 expression is negatively regulated by the Hippo signaling pathway. The Hippo signaling pathway is well studied in Drosophila and has been shown to inhibit cell proliferation and organ growth [43,44]. High expression of Tead2 in NSCs suggests that the Hippo signaling pathway is modulated in NSCs to enable proliferation and, when activated, could control NSC differentiation. Further work is necessary to reveal the importance of the Notch, Shh, and Hippo pathways in NSC behavior.
Conclusion
In conclusion, we have identified a novel NSC marker, C1qR1. We have used C1qR1 to purify NSCs, which allowed us to visualize formation of neurospheres from NSCs and identify a molecular signature for NSCs. The molecular signature reasserts the self-renewal and proliferative capacity and undifferentiated state of NSCs. From the molecular signature, we propose that the Hippo signaling pathway plays an important role in NSC function and behavior.
Supplementary Material
Acknowledgments
The authors thank the Biological Resource Centre for maintenance of mice. This work was funded by the Agency for Science, Technology and Research, Singapore. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Disclosure Statement
The authors indicate no potential conflicts of interest.
References
- 1.Lindvall O. and Kokaia Z. (2004). Recovery and rehabilitation in stroke: stem cells. Stroke 35:2691–2694 [DOI] [PubMed] [Google Scholar]
- 2.Parker MA, Anderson JK, Corliss DA, Abraria VE, Sidman RL, Park KI, Teng YD, Cotanche DA. and Snyder EY. (2005). Expression profile of an operationally-defined neural stem cell clone. Exp Neurol 194:320–332 [DOI] [PubMed] [Google Scholar]
- 3.Reynolds BA. and Weiss S. (1992). Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255:1707–1710 [DOI] [PubMed] [Google Scholar]
- 4.Davis AA. and Temple S. (1994). A self-renewing multipotential stem cell in embryonic rat cerebral cortex. Nature 372:263–266 [DOI] [PubMed] [Google Scholar]
- 5.Narayanan G, Poonepalli A, Chen J, Sankaran S, Hariharan S, Yu YH, Robson P, Yang H. and Ahmed S. (2012). Single-cell mRNA profiling identifies progenitor subclasses in neurospheres. Stem Cells Dev 21:3351–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M. and Matsuzaki F. (2008). Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development 135:3113–3124 [DOI] [PubMed] [Google Scholar]
- 7.Morrison SJ, White PM, Zock C. and Anderson DJ. (1999). Prospective identification, isolation by flow cytometry, and in vivo self-renewal of multipotent mammalian neural crest stem cells. Cell 96:737–749 [DOI] [PubMed] [Google Scholar]
- 8.Rietze RL, Valcanis H, Brooker GF, Thomas T, Voss AK. and Bartlett PF. (2001). Purification of a pluripotent neural stem cell from the adult mouse brain. Nature 412:736–739 [DOI] [PubMed] [Google Scholar]
- 9.Nagato M, Heike T, Kato T, Yamanaka Y, Yoshimoto M, Shimazaki T, Okano H. and Nakahata T. (2005). Prospective characterization of neural stem cells by flow cytometry analysis using a combination of surface markers. J Neurosci Res 80:456–466 [DOI] [PubMed] [Google Scholar]
- 10.Ida M, Shuo T, Hirano K, Tokita Y, Nakanishi K, Matsui F, Aono S, Fujita H, Fujiwara Y, Kaji T. and Oohira A. (2006). Identification and functions of chondroitin sulfate in the milieu of neural stem cells. J Biol Chem 281:5982–5991 [DOI] [PubMed] [Google Scholar]
- 11.Hamanoue M, Matsuzaki Y, Sato K, Okano HJ, Shibata S, Sato I, Suzuki S, Ogawara M, Takamatsu K. and Okano H. (2009). Cell surface N-glycans mediated isolation of mouse neural stem cells. J Neurochem 110:1575–1584 [DOI] [PubMed] [Google Scholar]
- 12.Nakatani Y, Yanagisawa M, Suzuki Y. and Yu RK. (2010). Characterization of GD3 ganglioside as a novel biomarker of mouse neural stem cells. Glycobiology 20:78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capela A. and Temple S. (2002). LeX/ssea-1 is expressed by adult mouse CNS stem cells, identifying them as nonependymal. Neuron 35:865–875 [DOI] [PubMed] [Google Scholar]
- 14.Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV, Tsukamoto AS, Gage FH. and Weissman IL. (2000). Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci U S A 97:14720–14725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corti S, Nizzardo M, Nardini M, Donadoni C, Locatelli F, Papadimitriou D, Salani S, Del Bo R, Ghezzi S, et al. (2007). Isolation and characterization of murine neural stem/progenitor cells based on Prominin-1 expression. Exp Neurol 205:547–562 [DOI] [PubMed] [Google Scholar]
- 16.Gan HT, Tham M, Hariharan S, Ramasamy S, Yu YH. and Ahmed S. (2011). Identification of ApoE as an autocrine/paracrine factor that stimulates neural stem cell survival via MAPK/ERK signaling pathway. J Neurochem 117:565–578 [DOI] [PubMed] [Google Scholar]
- 17.Tham M, Ramasamy S, Gan HT, Ramachandran A, Poonepalli A, Yu YH. and Ahmed S. (2010). CSPG is a secreted factor that stimulates neural stem cell survival possibly by enhanced EGFR signaling. PLoS One 5:e15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis SA, Rietze RL, Deleyrolle L, Wagey RE, Thomas TE, Eaves AC. and Reynolds BA. (2008). Enumeration of neural stem and progenitor cells in the neural colony-forming cell assay. Stem Cells 26:988–996 [DOI] [PubMed] [Google Scholar]
- 19.Kim M. and Morshead CM. (2003). Distinct populations of forebrain neural stem and progenitor cells can be isolated using side-population analysis. J Neurosci 23:10703–10709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurvansuu J, Zhao Y, Leung DS, Boulaire J, Yu YH, Ahmed S. and Wang S. (2008). Transmembrane protein 18 enhances the tropism of neural stem cells for glioma cells. Cancer Res 68:4614–4622 [DOI] [PubMed] [Google Scholar]
- 21.Huang CH, Sankaran S, Racoceanu D, Hariharan S. and Ahmed S. (2012). Online 3-D tracking of suspension living cells imaged with phase-contrast microscopy. IEEE Trans Biomed Eng 59:1924–1933 [DOI] [PubMed] [Google Scholar]
- 22.Reynolds BA. and Weiss S. (1996). Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol 175:1–13 [DOI] [PubMed] [Google Scholar]
- 23.Weiss S, Dunne C, Hewson J, Wohl C, Wheatley M, Peterson AC. and Reynolds BA. (1996). Multipotent CNS stem cells are present in the adult mammalian spinal cord and ventricular neuroaxis. J Neurosci 16:7599–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss S, Reynolds BA, Vescovi AL, Morshead C, Craig CG. and van der Kooy D. (1996). Is there a neural stem cell in the mammalian forebrain? Trends Neurosci 19:387–393 [DOI] [PubMed] [Google Scholar]
- 25.Nakamura Y, Sakakibara S, Miyata T, Ogawa M, Shimazaki T, Weiss S, Kageyama R. and Okano H. (2000). The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J Neurosci 20:283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohtsuka T, Sakamoto M, Guillemot F. and Kageyama R. (2001). Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J Biol Chem 276:30467–30474 [DOI] [PubMed] [Google Scholar]
- 27.Capela A. and Temple S. (2006). LeX is expressed by principle progenitor cells in the embryonic nervous system, is secreted into their environment and binds Wnt-1. Dev Biol 291:300–313 [DOI] [PubMed] [Google Scholar]
- 28.Lee A, Kessler JD, Read TA, Kaiser C, Corbeil D, Huttner WB, Johnson JE. and Wechsler-Reya RJ. (2005). Isolation of neural stem cells from the postnatal cerebellum. Nat Neurosci 8:723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zappone MV, Galli R, Catena R, Meani N, De Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R, Ottolenghi S. and Nicolis SK. (2000). Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development 127:2367–2382 [DOI] [PubMed] [Google Scholar]
- 30.Lendahl U, Zimmerman LB. and McKay RD. (1990). CNS stem cells express a new class of intermediate filament protein. Cell 60:585–595 [DOI] [PubMed] [Google Scholar]
- 31.Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, Ueda S, Uchiyama Y, Noda T. and Okano H. (2002). RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci U S A 99:15194–15199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim TS, Park M, Nepomuceno RR, Palmarini G, Winokur S, Cotman CA, Bengtsson U. and Tenner AJ. (2000). Characterization of the murine homolog of C1qR(P): identical cellular expression pattern, chromosomal location and functional activity of the human and murine C1qR(P). Mol Immunol 37:377–389 [DOI] [PubMed] [Google Scholar]
- 33.McKearn JP, Baum C. and Davie JM. (1984). Cell surface antigens expressed by subsets of pre-B cells and B cells. J Immunol 132:332–339 [PubMed] [Google Scholar]
- 34.Jordan CT, McKearn JP. and Lemischka IR. (1990). Cellular and developmental properties of fetal hematopoietic stem cells. Cell 61:953–963 [DOI] [PubMed] [Google Scholar]
- 35.Danet GH, Luongo JL, Butler G, Lu MM, Tenner AJ, Simon MC. and Bonnet DA. (2002). C1qRp defines a new human stem cell population with hematopoietic and hepatic potential. Proc Natl Acad Sci U S A 99:10441–10445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dean YD, McGreal EP, Akatsu H. and Gasque P. (2000). Molecular and cellular properties of the rat AA4 antigen, a C-type lectin-like receptor with structural homology to thrombomodulin. J Biol Chem 275:34382–34392 [DOI] [PubMed] [Google Scholar]
- 37.Mizutani K, Yoon K, Dang L, Tokunaga A. and Gaiano N. (2007). Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449:351–355 [DOI] [PubMed] [Google Scholar]
- 38.Kabrun N, Buhring HJ, Choi K, Ullrich A, Risau W. and Keller G. (1997). Flk-1 expression defines a population of early embryonic hematopoietic precursors. Development 124:2039–2048 [DOI] [PubMed] [Google Scholar]
- 39.Ling V. and Neben S. (1997). In vitro differentiation of embryonic stem cells: immunophenotypic analysis of cultured embryoid bodies. J Cell Physiol 171:104–115 [DOI] [PubMed] [Google Scholar]
- 40.Maire CL, Wegener A, Kerninon C. and Nait Oumesmar B. (2010). Gain-of-function of Olig transcription factors enhances oligodendrogenesis and myelination. Stem Cells 28:1611–1622 [DOI] [PubMed] [Google Scholar]
- 41.Petrenko O, Beavis A, Klaine M, Kittappa R, Godin I. and Lemischka IR. (1999). The molecular characterization of the fetal stem cell marker AA4. Immunity 10:691–700 [DOI] [PubMed] [Google Scholar]
- 42.Kaneko KJ, Kohn MJ, Liu C. and DePamphilis ML. (2007). Transcription factor TEAD2 is involved in neural tube closure. Genesis 45:577–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halder G. and Johnson RL. (2011). Hippo signaling: growth control and beyond. Development 138:9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, Wu S, Barrera J, Matthews K. and Pan D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122:421–434 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.