Abstract
The single-stranded nucleic acid binding protein KHSRP (KH-Type Splicing Regulatory Protein) modulates RNA life and gene expression at various levels. KHSRP controls important cellular functions as different as proliferation, differentiation, metabolism and response to infectious agents. We summarize and discuss experimental evidence providing a potential link between changes in KHSRP expression/function and human diseases including neuromuscular disorders, obesity, type II diabetes, and cancer.
Keywords: KHSRP, RNA-binding protein, neuromuscular disorders, inflammation, obesity, cancer
KHSRP (also known as KSRP) is a single-stranded nucleic acid binding protein independently discovered by Levens and Black laboratories1,2. Davis-Smyth and coworkers described KHSRP (named FBP2 in their paper) as a factor interacting with an enhancer element upstream of the c-myc oncogene promoter1. Min and coworkers reported that KHSRP is a component of a multi-protein complex that binds to a splicing enhancer element of c-src pre-mRNA2. The official name of the protein (KHSRP, for KH-Type Splicing Regulatory Protein, used in this review) reflects both the presence in the molecule of four distinct hnRNPK-Homology (KH) domains and its splicing function.
In the last twenty years additional roles of KHSRP in the post-transcriptional control of gene expression have been discovered. Considering that several recent reviews have exhaustively addressed the mechanisms by which KHSRP exerts its numerous functions3-6, this review is mainly focused on how KHSRP-dependent regulation of RNA metabolism affects distinct cell functions in different tissues and can impact on pathological conditions. However, for scholarly reasons we briefly summarize below the molecular functions of KHSRP that have been most extensively investigated and that will be cited in the following Sections of this review.
1. Implication of KHSRP in pre-mRNA splicing
Biochemical studies performed more than fifteen years ago revealed that KHSRP is a component of a multiprotein complex (also including hnRNPF, hnRNPH, and Polypyrimidine Tract Binding Protein) that binds specifically to a G+U-rich intronic splicing enhancer element downstream of the neuron-specific SRC N1 exon and is required for proper splicing2. Subsequently, Russo and coworkers observed that KHSRP, together with hnRNPH and nucleophosmin, interacts with intron 3 of human RPL3 gene thus promoting the expression of an alternative isoform of RPL37.
2. Implication of KHSRP in mRNA decay
Upon the initial reports, KHSRP has been extensively studied as a factor required for rapid decay of inherently labile transcripts3,4. The AU-rich element (ARE) is the landmark cis-acting motif responsible for rapid mRNA decay in mammalian cells and can be found in the 3’ untranslated regions (UTRs) of many short-lived transcripts3,4. We and others have shown that KHSRP interacts with AREs and recruits the RNA exosome (and other nucleases) on labile transcripts that encode cytokines and transcriptional regulators of cell identity and fate thus promoting their decay3,4.
3. Implication of KHSRP in microRNA biogenesis
Expanding the array of KHSRP functions on RNA metabolism, the protein has been proved to promote maturation of select microRNAs (miRNAs) from precursors. KHSRP binds to the terminal loop (enriched in G nucleotides) of a cohort of miRNA precursors and interacts with both Drosha and Dicer complexes in nuclear and cytoplasmic compartments, respectively3. The miRNAs whose maturation is favored by KHSRP exert important functions in controlling cell proliferation, immune response, metabolic homeostasis and cell fate determination in response to extracellular stimuli3,5,6. We and others proposed that the terminal loop of miRNA precursors constitutes a pivotal structure where miRNA processing co-activators (such as KHSRP) and miRNA processing co-repressors function in a coordinated way to convey proliferating and differentiating cues into changes of miRNA expression3,5,6.
4. Implication of KHSRP-long noncoding RNA interaction in gene expression control
We have recently reported that long non-coding (nc) RNAs (lncRNAs) represent an unanticipated class of RNAs that interact with KHSRP8. KHSRP directly binds to H19 lncRNA in undifferentiated multipotent mesenchymal C2C12 cells and this interaction favors KHSRP-mediated destabilization of labile mRNAs such as myogenin8. AKT1 and AKT2 activation, which occurs during the early steps of myogenesis, induces KHSRP dismissal from H19 lncRNA and, as a consequence, myogenin mRNA is stabilized while KHSRP is repurposed to promote maturation of myogenic miRNAs, thus favoring myogenic differentiation8. Our unpublished data indicate that other lncRNAs associate with KHSRP in different cell types. This allows to hypothesize that the regulatory role played by H19 in C2C12 cells might be operated by different lncRNAs in other cell types.
Table I summarizes the diverse molecular functions of KHSRP and how they are affected by post-translational modifications.
Table I.
Molecular functions of KHSRP in different cellular contexts.
| Molecular function |
Biological outcome |
Cell type / tissue | Post- translational modification |
Physiopathological context |
References |
|---|---|---|---|---|---|
|
mRNA
decay |
FOS, IL2, TNF mRNA stabilization |
HeLa, HT1080 | — | — | 9 |
|
mRNA
decay |
Myog,
Cdkn1a mRNA stabilization |
C2C12 multipotent mesenchymal |
MAPK14 | Myogenesis | 27 |
|
mRNA
decay |
IL8, CXCL2,
CXCL3 and others mRNA stabilization |
HeLa | MAPK14 | — | 10 |
|
mRNA
decay |
Ifna, Ifnb
mRNA stabilization |
Fibroblasts, macrophages Ksrp−/− mice |
— | Response to viral infections |
12 |
|
mRNA
decay |
Il1b, Tnf
mRNA stabilization |
Astrocytes Ksrp−/−
mice |
— | Inflammatory response |
13 |
|
mRNA
decay |
NOS2A, IL8,
TNF mRNA stabilization |
DLD-1 colon carcinoma |
MAPK14 | Inflammatory response |
15, 16 |
|
mRNA
decay |
Utrophin
mRNA stabilization |
C2C12, mouse muscle |
MAPK14, AKT |
Myogenesis, Duchenne Muscular Atrophy |
35 |
|
mRNA
decay |
Gap43
mRNA stabilization |
Neurons Ksrp−/−
mice |
— | Axonal outgrowth | 50 |
|
mRNA
decay |
Per2 mRNA stabilization |
Liver Ksrp−/− mice | — | Non-alchoolic liver steatosis |
62 |
|
mRNA
translation |
Il1b, Tnf
mRNA translation |
Astrocytes Ksrp−/−
mice |
— | Inflammatory response |
13 |
|
mRNA
translation |
Il6, Il23a, Tnf and other mRNA translation |
HeLa | MAPK14 | — | 17 |
|
mRNA
translation |
EV71 viral RNA |
RD rhabdomyosarcoma cells |
MAPK14 | Viral infection | 23, 24 |
|
miRNA
maturation |
Let-7 family, miR-16, miR-20b, and others |
U2OS, HeLa | ATM | Cell proliferation, DNA damage response |
66, 67 |
|
miRNA
maturation |
miR-155 targeting Il1b, Il12b, Cxcl11 and other mRNAs |
Mouse macrophages |
— | Inflammatory response |
19 |
|
miRNA
maturation |
miR-150 targeting Prdm16, Ppargc1a |
iWAT Ksrp−/− mice | — | Control of body adiposity |
29 |
|
miRNA
maturation |
miR- 198/FSTL1 |
Human keratinocytes |
— | Wound healing (non- healing diabetic ulcers?) |
33 |
|
miRNA
maturation |
miR-145 targeting Foxo1, Cgi58 |
eWAT Ksrp−/− mice | — | Control of lipolysis (obesity?) |
57 |
|
mRNA
decay + miRNA maturation + interaction with lncRNA |
Myog, miR- 1, miR-133, miR-206 (myomiRs), lncRNA H19 |
C2C12 multipotent mesenchymal |
AKT | Cell fate determination |
8, 31, 37 |
| unknown | unknown | Glioblastoma cells | — | Cell migration | 68 |
| unknown | unknown | Glioblastoma cells | MAPK14, MAPKAPK2 |
Cell cycle progression |
69 |
| unknown | unknown | Hepatoma cells | — | Cell motility | 70,71 |
1. Role of KHSRP in the immune response
The expression of cytokines and chemokines undergoes a finely tuned control. Deregulated levels of these immune modulators can cause or aggravate many pathological conditions of high social impact such as auto-immune diseases, neurodegenerative disorders and cancer. Thus, it is conceivable that several layers of transcriptional and post-transcriptional control exist to maintain homeostatic balance of immune modulators. Post-transcriptional control of cytokines and chemokines expression is operated through distinct mechanisms involving i) specific RNA elements (including the AREs) that control mRNA decay and/or its translation ii) miRNA-mediated gene silencing. KHSRP controls the expression of several immune modulators by acting at both these post-transcriptional levels.
After our initial observation that KHSRP associates with the RNA exosome to control rapid decay of FOS, TNF and IL2 mRNAs9, a report from Winzen and coworkers identified a number of modulators of the immune response as KHSRP mRNA targets in a pull-down-based screening (among them IL8, CXCL2, and CXCL3)10. They also reported that cell stimulation with IL1 reduced the interaction of KHSRP with IL8 mRNA leading to mRNA stabilization10,11. The analysis of KHSRP knockout mice revealed that IFNA and IFNB are up-regulated in cells derived from Khsrp−/− mice because of enhanced mRNA half-life and this regulation explains the robust response of Khsrp−/− mice to viral infections (see Section 2)12. Further, Li and coworkers observed that the expression of TNF and IL1B significantly increased in astrocytes derived from Khsrp−/− mice when compared to wild-type littermate13. Considering that IFNs and cytokines play a crucial role in the pathogenesis of autoimmune diseases14, the elucidation of KHSRP function in restraining their expression in immune cells may lead to the development of therapeutic strategies for these pathologies.
NOS2A (also known as iNOS) is an important component of the inflammatory response pathway that is encoded by an unstable mRNA whose expression is modulated by cytokines15. Linker and coworkers reported that ZFP36 (also known as TTP, an RNA-binding protein (RBP) that usually promotes decay of ARE-containing labile mRNAs) enhances NOS2A mRNA stability by interacting with KHSRP and hijacking the KHSRP-exosome complex away from NOS2A mRNA in colorectal adenocarcinoma cells15. More recently, the same group has reported that KHSRP represents a pivotal element for the anti-inflammatory function of resveratrol, a natural phenol produced by plants in response to injury or upon invasion by bacteria or fungi16. Resveratrol has been proved to have beneficial effects on cancer or cardiovascular diseases15. Resveratrol shortens the half-life of the transcripts encoding NOS2A, IL8 and TNF, by directly interacting with KHSRP and preventing the MAPK14 (also known as MAPK p38)-mediated inhibition of KHSRP decay promoting activity15. These findings underline the relevance of Thr692 phosphorylation by MAPK14 in the regulation of KHSRP function (see Section 4) and hint to KHSRP modulation as a target of therapeutic intervention in inflammatory diseases.
KHSRP can negatively control gene expression also by down-regulating ARE-containing mRNA translation. Dhamija and coworkers reported that as many as 50 mRNAs that are targets of KHSRP are enriched in polysomes after KHSRP knock-down and among these mRNAs are those encoding IL6, IL23A, and TNF17. Also studies performed in Khsrp−/− astrocytes support a role of KHSRP in TNF and IL1B translation13.
We and others have reported on the role of KHSRP as a factor required for optimal biogenesis of select miRNAs from their primary transcripts6. Among these, miR-155 is able to regulate inflammatory and immune responses through down-regulation of multiple targets18. We showed that KHSRP promotes miR-155 biogenesis in murine macrophages in response to lipopolysaccharide treatment19. Gene expression analysis of cells in which either KHSRP was knocked-down or miR-155 was silenced showed that the expression of a large number of transcripts encoding cytokines and chemokines, such as IL1B, IL12B, and CXCL11, was significantly affected19. Remarkably, the effect of KHSRP silencing was reverted by forced expression of mature miR-15519.
Patients suffering from granulomatosis with polyangiitis (Wegener's granulomatosis), eosinophilic granulomatosis with polyangiitis (Churg-Strauss syndrome), and microscopic polyangiitis express anti-endothelial cell antibodies that cause endothelial cell dysfunction20. Recently, Regent and coworkers observed that anti-KHSRP antibodies are present in 50% of patients suffering for these diseases20. This finding suggests a possible implication of KHSRP in endothelial cell physiology and pathology. Although intriguing, the relevance of pathological conditions dependent on the presence of anti-KHSRP autoantibodies deserves further confirmation.
2. Role of KHSRP in viral infections
Mammalians respond to viral invasion and replication through specialized innate immune sensors in cells that detect viral components and trigger downstream signaling pathways that can ultimately result in the activation of systemic immune response5. Several innate immune sensors recognize cytoplasmic viral RNA and lead to the production of Interferons that, in turn, trigger various antiviral pathways aimed at halting viral replication and spread21. In the last few years several studies have revealed that viral RNA, interacting with cellular RBP, can affect their physiological functions thus impacting on cellular metabolism of various RNAs22.
Studies from Lin and coworkers revealed that KHSRP, competing with factors that enhance IRES (internal ribosome entry site)-mediated translation of enterovirus 71 (EV71), down-regulates IRES-dependent EV71 translation23. This effect is achieved through direct interaction of KHSRP with the IRES present in the 5’UTR of EV71. Interestingly, more recent studies from the same laboratory demonstrated that EV71 infection induces KHSRP cleavage through multiple mechanisms, including caspase activation, proteasome activity, and autophagy induction24. Thus, enterovirus-induced cleavage of KHSRP reduces the level of a negative regulator of viral protein translation envisaging a negative feedback loop. Further, the study from Chen and coworkers also demonstrated that the truncated form of KHSRP lacking the C-terminal domain (KHSRP1-503), instead of being a negative regulator, becomes a positive regulator of EV71 IRES-mediated translation. The C-terminal domain lacking in KHSRP1-503 includes Thr692, an amino acid that undergoes phosphorylation by MAPK14. We and others reported that Thr692 phosphorylation impairs KHSRP decay-promoting activity on a variety of target mRNAs6,11. It would be interesting to directly verify whether Thr692 phosphorylation influences also the IRES modulating activity of KHSRP.
Avian influenza virus (AIV) often results in a virus-induced cytokine deregulation called “cytokine storm" typically characterized by the presence of elevated levels of pro-inflammatory cytokines and IFNs25. Recently, Liu and coworkers reported that KHSRP expression levels are strongly induced by the low pathogenic H9N2 viral strain in a spontaneously immortalized cell line of chicken embryo fibroblasts while it is not regulated by the highly pathogenic H5N1 viral strain infection26. The subtype-dependent host response observed in this study offers new insights into the potential roles of KHSRP in the control and modulation of replication and virulence of different subtypes or strains of avian influenza A virus26. Further, considering that KHSRP has been implicated in MAPK14 signaling pathway and that the regulation of this pathway has been reported to be associated with host defense to AIV infection, it would be interesting to investigate whether KHSRP phosphorylation by MAPK14 occurs during AIV infection6,11,16.
Taking advantage of Khsrp knock-out mice, the Chen laboratory described an additional mechanism by which KHSRP controls susceptibility to viral infection in mammalians. These animals display enhanced type I IFN levels in response to viral infections when compared to wild-type littermate12. Specifically, Khsrp-deleted mice proved to be refractory to herpes simplex virus 1 and vesicular stomatitis virus infection as a consequence of enhanced expression of IFNs12. This observation has been recently extended to human cytomegalovirus infected cells where viral replication was significantly reduced in cells in which KHSRP has been silenced28.
3. KHSRP controls cell fate determination and tissue remodeling
In response to developmental and differentiative cues, cells activate a network of signaling proteins, transcription factors, epigenetic modifiers, ncRNAs, and RNA-binding proteins that modulate functional and combinatorial interactions at the genomic, epigenomic, transcriptomic, and proteomic levels. These, ultimately, culminate in the activation or the repression of gene expression resulting in cell fate determination and tissue remodeling. On this regard, in the last few years KHSRP has emerged as a regulator of cell state transitions able to orient cell fate decisions operating at different levels6.
Starting from the evidence that mice lacking Khsrp gene display reduced body adiposity, Chou and coworkers demonstrated that KHSRP favors brown-like transformation in subcutaneous/inguinal white adipose tissue (iWAT)29. Targeted deletion of Khsrp enhances brown fat-selective gene expression in iWAT through an elevation of the levels of some important regulators of the thermogenic program such as PRDM16, PPARGC1A, and PPARA29. Importantly, KHSRP absence results in down-regulation of miR-150 due to its defective maturation from precursors29. Overexpression of Prdm16 and Ppargc1a, that are direct targets of miR-150, leads to brown-like transformation of iWAT29. It is known that brown adipose tissue oxidizes fatty acids for heat generation and energy expenditure. Thus, promoting brown-like transformation in WAT is a promising strategy for combating obesity. Therefore, modulation of KHSRP-dependent miR-150 maturation from precursors could potentially lead to the development of therapeutic tools against obesity and metabolic disorders (see also Section 6)29.
Multipotent mesenchymal cell differentiation is subject to intense regulation via integrated signaling networks that orchestrate changes in gene expression leading to commitment toward specific cell lineages30. We have extensively studied multipotent mesenchymal C2C12 cells for their potential to differentiate into myofibers upon serum withdrawal (see Section 4) or into osteoblasts upon addition to the culture media of BMP/TGF-β ligands31. In a recent study we have demonstrated that SMAD proteins, mediators of BMP/TGF-β ligands, regulate KHSRP function31. SMADs interact with KHSRP and impair its ability to bind to primary myogenic miRNAs (usually referred to as myomiRs) and to promote their maturation. Prevention of myomiR maturation is instrumental to promote the differentiation of C2C12 cells into osteoblasts31. In line with this observation, KHSRP silencing causes a reshaping of the transcriptome largely overlapping that produced by BMP/SMAD signaling activation in C2C12 cells31. The observation that myomiR re-expression in KHSRP-silenced cells is sufficient to enhance myogenin expression and to abrogate the osteoblastic phenotype, allowed us to propose that the most critical consequence of KHSRP silencing in C2C12 cells is the impairment of myomiR maturation and this is sufficient to direct C2C12 cell fate decision31.
TGF-β orchestrates wound healing by regulating cell proliferation, differentiation, extracellular matrix production, and immune modulation32. In the early phases of wound healing, TGF-β is induced and, among other effects, down-regulates KHSRP levels33. Sundaram and coworkers have demonstrated that, in normal epidermis, KHSRP promotes biogenesis of miR-198 whose primary miRNA is located in the 3’UTR of follistatin-like 1 (FSTL1) transcript33. As a consequence of KHSRP down-regulation, miR-198 maturation from precursors is abrogated and the expression of FSTL1 is enhanced. Importantly, FSTL1 stimulates keratinocyte migration whereas miR-198 expression produces the opposite effect33. Thus, regulation of KHSRP expression by TGF-β is required to switch the post-transcriptional control of FSTL1/miR-198 expression to a “wound healing-mode”33. In their paper, Sundaram and coworkers point to the consequences of the failure of the FSTL1/miR-198 switch in non-healing chronic diabetic ulcers where miR-198 levels persist high, FSTL1 expression is absent, and keratinocyte migration —as well as wound healing— fail to occur33. Thus, the importance of KHSRP in the pathogenesis of diabetic ulcers deserves further investigation.
4. Role of KHSRP in myogenesis, muscle functionality, and neuromuscular diseases
Experimental evidence published in the last years demonstrated that post-transcriptional control of gene expression plays a central role in myogenesis and RBPs are essential gatekeepers able to ensure proper progression of myoblasts through the myogenic differentiation program34.
Several studies have explored KHSRP role during normal myogenesis, a complex process in which at least two enzymatic signaling cascades, MAPK14 and PI3K/AKT, are activated and play a pivotal regulatory role6 (see also Section 3).
We initially showed that the MAPK14 phosphorylates KHSRP at Thr692 in differentiating myoblasts27. This, in turn, impairs KHSRP ability to associate with ARE-containing transcripts encoding myogenin and CDKN1A (also known as p21) and results in stabilization and enhanced steady-state expression of the mRNAs encoding these two important regulators of myogenesis. More recently, Amirouche and coworkers reported that MAPK14-mediated phosphorylation of KHSRP impairs its mRNA decay function thus favoring the expression of utrophin35. Due to its high degree of sequence identity with dystrophin and its ability to associate with members of the “dystrophin-associated protein complex”, utrophin has been proposed as an excellent substitute to dystrophin itself with high potential to alleviate the dystrophic pathology35. Importantly, either KHSRP knock-down or activation of MAPK14 in a mouse model of Duchenne muscular dystrophy (mdx mice) significantly increased utrophin expression35. Therefore, modulating either KHSRP expression or its function could be envisaged as a novel strategy to treat dystrophic disease36.
We also found that phosphorylation by AKT1 and AKT2 promotes KHSRP-dependent maturation of myomiRs in the early phases of myogenic differentiation of C2C12 cells37. The analysis of the decay of unstable transcripts (e.g. myogenin) combined with the analysis of myomiR maturation from precursors enabled us to conclude that PI3K/AKT signaling activation inhibits the ability of KHSRP to promote decay of myogenin mRNA while favors KHSRP ability to promote myomiR maturation37. More recently, we found that H19 lncRNA favors the mRNA decay promoting function of KHSRP via a “molecular scaffold” mechanism and prevents, at the same time, KHSRP ability to interact with specific miRNA precursors and to favor their processing into the mature forms8. Remarkably, the cytoplasmic association of KHSRP with lncRNA H19 is abrogated by PI3K/AKT signaling activation8.
Adult skeletal muscle regeneration in response to injury is a complex process in which PI3K/AKT signaling activation has an essential role (reviewed in ref. 38) and has been linked to induction of myomiR expression39. We have explored the role of KHSRP in muscle regeneration and found that the induction of myomiRs occurring during muscle regeneration is impaired in Khsrp−/−mice due to maturation blockade37. Interestingly, similarly to MAPK14, also PI3K/AKT signaling activation in mdx mice muscle is beneficial, since enhances expression of utrophin and promotes muscle regeneration35. Thus, it is possible to hypothesize that regulation of KHSRP functions by both MAPK14 and PI3K/AKT is required for the process of muscle regeneration.
Tadesse and coworkers reported that KHSRP expression is down-regulated in spinal cord tissues from mild spinal muscular atrophy (SMA) model mice, and this correlates with increased Cdkn1a mRNA levels thus suggesting the involvement of exaggerated stabilization of specific KHSRP mRNA targets in the etiology of SMA40.
Over the last years, converging lines of evidence indicate that the expression of miR-206 is altered in several neuromuscular disorders39. miR-206 is essential for muscle regeneration and able to delay progression of dystrophic disease in mdx mice41. We have reported that miR-206 maturation from precursors is severely impaired in C2C12 cells depleted of KHSRP as well as in regenerating muscle derived from Khsrp−/− mice37. In a mouse model of amyotrophic lateral sclerosis (ALS), a neurodegenerative disease characterized by loss of motor neurons and degeneration of target muscle fibers, deficiency of miR-206 accelerates disease progression42. Williams and coworkers proposed that miR-206 delays progression of ALS by stimulating compensatory regeneration of neuromuscular synapses targeting histone deacetylase 4 and fibroblast growth factors42.
Based on the above observations, it is tempting to speculate that manipulation of KHSRP function in order to enhance its ability to promote myomiR maturation and/or affect its mRNA decay promoting function on select mRNAs could contribute to the treatment of degenerative neuromuscular disorders.
5. Role of KHSRP in neurons
Post-transcriptional regulatory mechanisms that control gene expression have been shown to be important for virtually all stages of assembling neural networks, from neurite guidance, branching, and growth to synapse morphogenesis and function43.
Although KHSRP is known to be expressed in both neurons and glial cells44,45, its function in the nervous system is now beginning to be understood. After the initial characterization of KHSRP as a splicing factor in neurons2, two homologs of the human protein were identified in neurons and shown to participate in mRNA transport: the chicken zipcode-binding protein 2 (ZBP2), a protein that is required for localizing β-actin mRNA to growth cones46, and rat MARTA1, a protein that mediates the transport of Map2 mRNA to dendrites47. Also, KHSRP is located in RNA granules in peripheral nerve axons, where it interacts with —and it is regulated by— SMN, a RBP required for motor neuron survival40. In dorsal root ganglion (DRG)-like neuronal cells and cortical neurons, KHSRP co-localizes with the X-chromosome-linked intellectual disability protein (PQBP1) which is a component of neuronal RNA granules and regulates the formation of stress granules48. Furthermore, the recent finding that KHSRP is present together with Dicer and some pre-miRNAs in distal DRG axons suggests its involvement in localized pre-miRNA processing49.
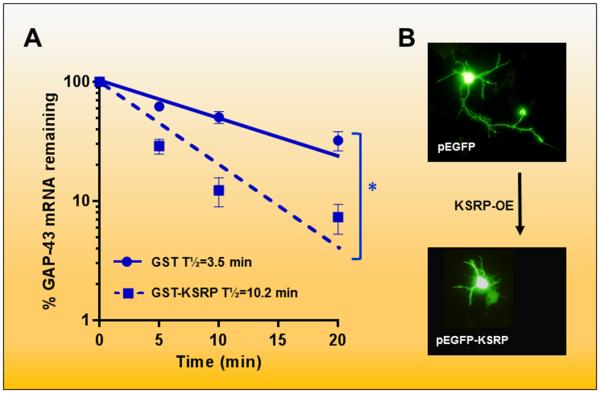
Our studies indicate that KHSRP, by controlling mRNA stability, affects axonal outgrowth during neuronal development50. As shown in Figure 1A, KHSRP is able to destabilize the mRNA encoding the neuronal growth associated protein GAP4350. Notably, KHSRP competes with the neuronal stabilizing factor ELAVL4 (also known as HuD) which is known to increase Gap43 mRNA stability and axonal elongation during neuronal development51-55. Overexpression of either full length KHSRP (Figure 1B) or a deletion mutant that lacks its nuclear localization signal was found to impair axonal outgrowth in hippocampal neurons, whereas overexpression of a mutant protein without the KH4 domain did not have any effects50. Further, overexpression of Gap43 mRNA with axonal targeting sequences in the 3’UTR, but not an mRNA restricted to the cell body, is able to rescue axonal elongation in the presence of an excess of KHSRP55. Finally, depletion of KHSRP in hippocampal neurons led to a 2-3 fold increase in Gap43 expression and to enhanced axonal length50. Altogether these findings suggest that some of the effects of KHSRP on neuronal differentiation are mediated by localized regulation of mRNA stability.
Figure 1. KSRP decreases GAP-43 mRNA stability and axonal outgrowth.
(A) As shown by Bird and coworkers47, addition of recombinant KSRP to S100 extracts from Ksrp -/- mouse brains decreases the half-life of the GAP-43 mRNA. Decay curves show the average results of 3 separate decay experiments fitted with a single rate exponential decay curve. *p<0.05. The effect of KSRP required both the presence of the KH4 domain in the protein and the ARE in the GAP-43 3’ UTR. (B) Overexpression (OE) of KSRP in primary hippocampal neuron cultures impairs axonal elongation. This process is reversed by either KSRP knockdown (KD) or by OE of a GAP-43 mRNA with a 3’ UTR targeting sequence for axonal localization47.
6. Role of KHSRP in lipid metabolism
Rapid changes in mRNA levels are critical for altering the pattern of protein expression in response to various metabolic stimuli in order to maintain cellular homeostasis56. miRNAs and RBPs that controls mRNA decay have been recently reported to be major regulators of glucose and lipid homeostasis under physiologal conditions and in the presence of metabolic disorders, such as type 2 diabetes and obesity.
We have recently provided evidence that a KHSRP/miR-145 axis can be viewed as an important negative regulator of lipolysis in epididymal white adipose tissue (eWAT)57. It is well known that increasing lipolysis in WAT causes an elevation in fatty acid utilization and energy expenditure, thus protecting against obesity58. During fast, catecholamines cause elevation of cyclic AMP levels and activation of protein kinase A resulting in stimulation of lipolysis while in the fed state lipolysis is inhibited by insulin59. Transcriptional activation of the gene Pnpla2 (which encodes the lipase ATGL) by forkhead box O1 (FOXO1) and other factors enhances lipolysis in adipocytes60. Further, ATGL activity is modulated through interaction with the co-activator CGI5861. We have shown that lipolysis is increased in Khsrp−/− eWAT due to enhanced expression of the genes encoding the lipases ATGL and HSL (hormone sensitive lipase) as well as of FOXO1 and CGI5857. From a mechanistic point of view, the expression of miR-145 is decreased in Khsrp−/− eWAT as a consequence of impaired pri-miR-145 processing57. Reduction of miR-145 expression in eWAT of Khsrp−/− mice leads to up-regulation of FOXO1 and CGI58, thereby enhancing lipolysis.
More recently, the Chen laboratory has provided evidence that KHSRP can impinge on lipid homeostasis through a distinct additional mechanism62. In the liver of Khsrp−/− mice the expression of Per2 (encoding a transcriptional repressor which belongs to the core component of the circadian clock) is significantly increased leading to an altered circadian clock. This leads to a reduction in liver triglycerides (TG) content in mutant mice, which become protected from diet-induced hepatic steatosis. Elevation of Per2 mRNA levels depends on increased mRNA stability in the absence of KHSRP and results in down-regulation of important modulators of lipid metabolism. As a consequence, Khsrp−/− mice exhibit reduced lipogenesis and TG content62. Interestingly, the consequences of KHSRP deletion on Per2 mRNA levels are evident only in the liver but not in adipose tissue, suggesting a tissue-restricted role of KHSRP in controlling Per2 mRNA stability. Non-alcoholic liver steatosis is the most common form of chronic liver disease associated with obesity, type 2 diabetes, and insulin resistance63,64. These observations point to KHSRP as a critical factor in governing hepatic lipid metabolism and it can be viewed as an unpredicted potential therapeutic target to control liver steatosis.
7. Role of KHSRP in cancer
In this Section we summarize the experimental evidence that links altered KHSRP expression or function to cell transformation. Further, in the last subsection we propose that KHSRP gene mutations detected in human tumors target specific protein domains (KH3 and KH4 as well as the putative nuclear localization signal [NLS]) and hypothesize a link between oncogenic mutations and protein functions.
1. Implication in the response to DNA damage
Ataxia-telangiectasia mutated kinase (ATM) plays an essential role in the maintenance of genome stability regulating the function of a large number of proteins in order to facilitate cell cycle checkpoints, to promote DNA repair, and to control many other aspects of physiological responses in the event of DNA double-strand breaks65. Overall, ATM activation results in a dramatic change in the gene-expression program that can be ascribed, in part, to regulation of miRNA expression65. A study from Zhang and coworkers demonstrated that the induction of a large population of miRNAs by ATM is controlled at post-transcriptional level as suggested by lack of significant changes in pri-miRNA transcription levels after induction of DNA damage66. Interestingly, all the miRNAs whose maturation was shown to be favored by KHSRP in a previous study67 were induced in ATM-expressing cells upon DNA damage while ATM knock-out abrogated this effect66. Accordingly, KHSRP silencing abolished DNA damage-induced upregulation of the same cohort of mature miRNAs66. Zhang and coworkers extended this observation demonstrating that KHSRP is phosphorylated by ATM and that this phosphorylation enhances the interaction of the protein with a subset of pri-miRNAs as well as its maturation-promoting activity66. These results suggest that KHSRP can play a role in tumorigenesis participating in the signaling cascade that regulates cellular response to DNA damage.
2. Role in tumors
KHSRP knock-down enhances the migratory capability of glioblastoma multiforme (GBM) cells as revealed by a siRNA screening68. This study, performed by Yang and coworkers, also demonstrated that KHSRP down-regulation induces the formation of multifocal GBM in a mouse model64. Interestingly, high expression levels of KHSRP were observed in patients who survived longer after surgery suggesting that KHSRP may be used as a novel prognostic marker for GBM patients68. A recent independent study from Boucas and coworkers confirmed that high KHSRP transcript levels are associated with increased overall survival of patients suffering from GBM and suggested that this might result from a better response to therapy in patients expressing high KHSRP levels69. This last study also demonstrated that KHSRP is highly regulated in response to genotoxic stress through the MAPK14/MAPKAPK2 (the latter also known as MK2) signaling module with cells deficient for MAPK14/MAPKAPK2 showing an overall altered interaction between KHSRP and target mRNAs69.
Differently from GBM, silencing of KHSRP in hepatocellular carcinoma cells represses migration. Zubaidah and coworkers as well as Malz and coworkers independently reported that the coordinated activation of FUBP1 (a nucleic acids binding protein whose KH domains displays high sequence homology with KHSRP) and of KHSRP represents a pro-tumorigenic mechanism promoting cell proliferation (in the case of FUBP1) and cell motility (in the case of KHSRP) of human liver cancer cells70,71.
From these studies it is evident that the consequences of altered levels of KHSRP are different in distinct human tumors and may reflect cell-restricted functions of KHSRP that could depend on the participation of the protein in distinct multi-protein complexes and to its ability to interact with different targets in different cells. Although intriguing, the above results necessitate both mechanistic exolanations and confirmation on a broader scale.
3. KHSRP gene translocation and mutations in cancer
Malouf and coworkers recently identified human KHSRP gene as a novel partner of TFE3 in Translocation Renal Cell Carcinoma (TRCC) tumors72. TFE3 is a transcription factor that transactivates expression of genes downstream of TGF-β signaling73. The translocation-generated fusion protein detected in TRCC tumors includes almost the entire KHSRP protein at its C-terminus and the TFE3 protein at the N-terminus. Remarkably, both the helix-loop-helix domain of TFE3 and the KH domains of KHSRP are present in the fusion protein72. The study also revealed that genes differentially spliced between TRCC and other renal cell carcinoma types were enriched for TFE3 targets, suggesting a putative role for KHSRP-regulated RNA splicing events in kidney carcinogenesis72. It will be interesting to experimentally verify whether transcripts whose alternative splicing is affected by TFE3-KHSRP fusion protein play a role either in the pathogenesis or in the natural history of TRCC and whether the KHSRP moiety in the fusion protein plays a role in the modulation of alternative splicing events.
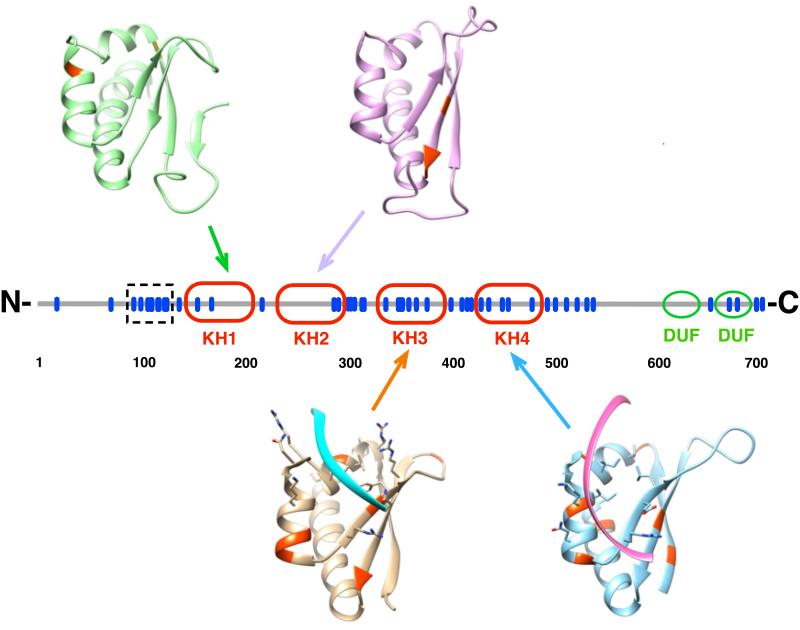
We sought to gain insight into a potential role of missense mutations of human KHSRP gene in cancer and, to this purpose, we performed a metadata analysis of identified KHSRP mutations in public available databases (Bordo et al., unpublished). Fifty-one amino acid substitutions are reported in the COSMIC database (catalogue of somatic mutations in cancer74), 16 are reported in the Intogen database75 while other 16 are reported in Biomuta database76. The merging of the three datasets resulted in 53 distinct amino acid mutations affecting KHSRP. The distribution of these mutations shows that 20 of them involve amino acids belonging to one of the four KH domains, two involve one of the two domains of unknown function (DUFs, as defined in Pfam77) and 31 instances map on the remaining regions (Figure 2). Among the KH domains, KH3 and KH4 appear to be more subject to mutations. We mapped mutations affecting KH domains on the three-dimensional structure of the corresponding KH domain and found that the majority (16 out of 19) involves either α-helices or β-strands (Figure 2). Thus, it is likely that these amino acid substitutions influence the structural stability of the corresponding KH domain. No positional bias is observed concerning the putative nucleotide binding groove of the KH domains (Figure 2) while the linker regions connecting KH2-KH3 and KH3-KH4 host each five amino acid substitutions. Notably, although the nuclear localization signal (NLS) of KHSRP has not been unambiguously defined, 9 amino acid substitutions localize to a region spanning amino acid positions 90-120, predictably hosting the NLS of the protein. Our metadata analysis also showed that 16 mutations associate with cancer75. Interestingly, although KHSRP does not possess the features of a driver gene for the switch to cancer, there is an increased frequency of mutations (that affect KH3) in Small Cell Lung Carcinoma, with a PAM (Protein Affecting Mutation) of about 5%, a figure that is about four times higher than that of other cancer types75.
Figure 2. Missense point mutations in KHSRP.
The primary sequence of KHSRP is represented as a straight line in the center, with the positions of the known missense point mutations indicated by blue bars. The position of the KH domains and that of the two domains of unknown function (DUF, as from Pfam73) is shown. The region spanning amino acids 90-120 containing a cluster of nine distinct mutations and putatively involved in nuclear localization is highlighted with a dotted box. The three-dimensional structures of the four KH domains are also represented as ribbon diagrams in the same orientation with respect to the RNA binding groove, with the position of the mutated residues highlighted in red. The experimentally determined position of the bound RNA is shown for KH3 and KH4. The Protein Data Bank codes for the four KH domains are 2opu (KH1), 2opv (KH2), and 1j4w (KH3, KH4).
8. Conclusive remarks
Among the cellular functions of KHSRP that we have described in this review, some have been more extensively investigated and broadly accepted by the scientific community. These include mRNA decay promoting function and miRNA maturation induction that have been confirmed by many laboratories in different cellular contexts and have gained the support of data derived from the analysis of knock-out mice. Conversely, although based on solid biochemical data, the pre-mRNA alternative splicing control operated by KHSRP has not been supported by in vivo experiments and clearly deserves further investigations. Similarly, the KHSRP-dependent control of mRNA translation as well as the lncRNA-mediated regulation of KHSRP activity need to be further confirmed in additional experimental settings.
As a matter of fact, given its proven ability to interact with a variety of RNA targets in distinct cellular contexts due to its modular structure, KHSRP is able to regulate a broad spectrum of cellular functions. More work is needed to understand from a mechanistic point of view the role of KHSRP in the context of large ribonucleoprotein complexes comprising other RBPs, enzymes, adaptor proteins, and ncRNAs.
Finally, results derived from the analysis of knock-out mice phenotype suggest that KHSRP deletion is favorable for certain aspects of mice physiology (Sections 2 and 6) stressing the fact that KHSRP expression and function need to be tightly regulated to ensure tissue homeostasis. Therefore, further studies are required to systematically investigate how KHSRP expression and function can be finely modulated in response to a variety of agents in the context of normal and pathological conditions.
ACKNOWLEDGMENTS
The experimental work performed in the Gene Expression Laboratory of IRCCS AOU San Martino-IST and cited in this article has been supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC; IG 10090, IG 15195; to R.G.) and Ministero della Salute (129/RF-2010-2306205; to R.G.) while the experimental work performed at the University of New Mexico was supported by the NIH grant 1R01NS089633 (to N.P-B.). The funding sources had no involvement in the study design; in the collection, analysis and interpretation of data; in the writing of reports; and in the decision to submit articles for publication. The authors would like to apologize for not being able to cite all of the primary literature due to space limitations.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Davis-Smyth T, Duncan RC, Zheng T, Michelotti G, Levens D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J Biol Chem. 1996;271:31679–31679. doi: 10.1074/jbc.271.49.31679. doi: 10.1074/jbc.271.49.31679. [DOI] [PubMed] [Google Scholar]
- 2.Min H, Turck CW, Nikolic JM, Black DL. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1023. doi: 10.1101/gad.11.8.1023. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 3.Gherzi R, Chen CY, Trabucchi M, Ramos A, Briata P. The role of KSRP in mRNA decay and microRNA precursor maturation. Wiley Interdiscip Rev RNA. 2010;1:230–239. doi: 10.1002/wrna.2. doi: 10.1002/wrna.2. [DOI] [PubMed] [Google Scholar]
- 4.Briata P, Chen CY, Ramos A, Gherzi R. Functional and molecular insights into KSRP function in mRNA decay. Biochim Biophys Acta. 2013;1829:689–694. doi: 10.1016/j.bbagrm.2012.11.003. doi: 10.1016/j.bbagrm.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 5.King PH, Chen CY. Role of KSRP in control of type I interferon and cytokine expression. Interferon Cytokine Res. 2014;34:267–267. doi: 10.1089/jir.2013.0143. doi: 10.1089/jir.2013.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gherzi R, Chen CY, Ramos A, Briata P. KSRP controls pleiotropic cellular functions. Semin Cell Dev Biol. 2014;34:2–2. doi: 10.1016/j.semcdb.2014.05.004. doi: 10.1016/j.semcdb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Russo A, Catillo M, Esposito D, Briata P, Pietropaolo C, Russo G. Autoregulatory circuit of human rpL3 expression requires hnRNP H1, NPM and KHSRP. Nucleic Acids Res. 2011;39:7576–7576. doi: 10.1093/nar/gkr461. doi: 10.1093/nar/gkr461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giovarelli M, Bucci G, Ramos A, Bordo D, Wilusz CJ, Chen CY, Puppo M, Briata P, Gherzi R. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc Natl Acad Sci U S A. 2014;111:E5023–E5028. doi: 10.1073/pnas.1415098111. doi: 10.1073/pnas.1415098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gherzi R, Lee KY, Briata P, Wegmüller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–571. doi: 10.1016/j.molcel.2004.05.002. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Winzen R, Thakur BK, Dittrich-Breiholz O, Shah M, Redich N, Dhamija S, Kracht M, Holtmann H. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol Cell Biol. 2007;27:8388–8388. doi: 10.1128/MCB.01493-07. doi: 10.1128/MCB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiedje C, Holtmann H, Gaestel M. The role of mammalian MAPK signaling in regulation of cytokine mRNA stability and translation. J Interferon Cytokine Res. 2014;34:220–220. doi: 10.1089/jir.2013.0146. doi: 10.1089/jir.2013.0146. [DOI] [PubMed] [Google Scholar]
- 12.Lin WJ, Zheng X, Lin CC, Tsao J, Zhu X, Cody JJ, Coleman JM, Gherzi R, Luo M, Townes TM, Parker JN, Chen CY. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Mol Cell Biol. 2011;31:3196–3196. doi: 10.1128/MCB.05073-11. doi:10.1128/MCB.05073-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Lin WJ, Chen CY, Si Y, Zhang X, Lu L, Suswam E, Zheng L, King PH. KSRP: a checkpoint for inflammatory cytokine production in astrocytes. Glia. 2012;60:1773–1773. doi: 10.1002/glia.22396. doi: 10.1002/glia.22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagy G, Huszthy PC, Fossum E, Konttinen Y, Nakken B, Szodoray P. Selected Aspects in the Pathogenesis of Autoimmune Diseases. Mediators Inflamm. 2015;2015:351732. doi: 10.1155/2015/351732. doi: 10.1155/2015/351732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linker K, Pautz A, Fechir M, Hubrich T, Greeve J, Kleinert H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4813. doi: 10.1093/nar/gki797. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bollmann F, Art J, Henke J, Schrick K, Besche V, Bros M, Li H, Siuda D, Handler N, Bauer F, et al. Resveratrol post-transcriptionally regulates pro-inflammatory gene expression via regulation of KSRP RNA binding activity. Nucleic Acids Res. 2014;42:12555–12555. doi: 10.1093/nar/gku1033. doi: 10.1093/nar/gku1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhamija S, Kuehne N, Winzen R, Doerrie A, Dittrich-Breiholz O, Thakur BK, Kracht M, Holtmann H. Interleukin-1 activates synthesis of interleukin-6 by interfering with a KH-type splicing regulatory protein (KSRP)-dependent translational silencing mechanism. J Biol Chem. 2011;286:33279–33279. doi: 10.1074/jbc.M111.264754. doi: 10.1074/jbc.M111.264754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253:146–146. doi: 10.1111/imr.12057. doi: 10.1111/imr.12057. [DOI] [PubMed] [Google Scholar]
- 19.Ruggiero T, Trabucchi M, De Santa F, Zupo S, Harfe BD, McManus MT, Rosenfeld MG, Briata P, Gherzi R. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009;23:2898–2898. doi: 10.1096/fj.09-131342. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 20.Régent A, Lofek S, Dib H, Bussone G, Tamas N, Federici C, Broussard C, Guillevin L, Mouthon L. Identification of target antigens of anti-endothelial cell antibodies in patients with anti-neutrophil cytoplasmic antibody-associated vasculitides: a proteomic approach. Clin Immunol. 2014;153:123–123. doi: 10.1016/j.clim.2014.03.020. doi: 10.1016/j.clim.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 21.Gürtler C, Bowie AG. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21:413–413. doi: 10.1016/j.tim.2013.04.004. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charley PA, Wilusz J. Sponging of cellular proteins by viral RNAs. Curr Opin Virol. 2014;9:14–14. doi: 10.1016/j.coviro.2014.09.001. doi: 10.1016/j.coviro.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin JY, Li ML, Shih SR. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 2009;37:47–47. doi: 10.1093/nar/gkn901. doi: 10.1093/nar/gkn901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen LL, Kung YA, Weng KF, Lin JY, Horng JT, Shih SR. Enterovirus 71 infection cleaves a negative regulator for viral internal ribosomal entry site-driven translation. J Virol. 2013;87:3828–3828. doi: 10.1128/JVI.02278-12. doi: 10.1128/JVI.02278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1203. doi: 10.1038/nm1477. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu AL, Li YF, Qi W, Ma XL, Yu KX, Huang B, Liao M, Li F, Pan J, Song MX. Comparative analysis of selected innate immune-related genes following infection of immortal DF-1 cells with highly pathogenic (H5N1) and low pathogenic (H9N2) avian influenza viruses. Virus Genes. 2015;50:189–189. doi: 10.1007/s11262-014-1151-z. doi: 10.1007/s11262-014-1151-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briata P, Forcales SV, Ponassi M, Corte G, Chen CY, Karin M, Puri PL, Gherzi R. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol Cell. 2005;20:891–891. doi: 10.1016/j.molcel.2005.10.021. doi: 0.1016/j.molcel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 28.Chai F, Li HY, Wang W, Zhu XJ, Li Y, Wang S, Guo L, Zhang LK, Xiao G. Subcellular quantitative proteomic analysis reveals host proteins involved in human cytomegalovirus infection. Biochim Biophys Acta. 2015;1854:967–967. doi: 10.1016/j.bbapap.2015.04.016. doi: 10.1016/j.bbapap.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 29.Chou CF, Lin YY, Wang HK, Zhu X, Giovarelli M, Briata P, Gherzi R, Garvey WT, Chen CY. KSRP ablation enhances brown fat gene program in white adipose tissue through reduced miR-150 expression. Diabetes. 2014;63:2949–2949. doi: 10.2337/db13-1901. doi: 10.2337/db13-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asakura A, Komaki M, Rudnicki M. Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation. 2001;68:245–245. doi: 10.1046/j.1432-0436.2001.680412.x. doi: 10.1046/j.1432-0436.2001.680412.x. [DOI] [PubMed] [Google Scholar]
- 31.Pasero M, Giovarelli M, Bucci G, Gherzi R, Briata P. Bone morphogenetic pro-tein/SMAD signaling orients cell fate decision by impairing KSRP-dependent microRNA maturation. Cell Rep. 2012;2:1159–1159. doi: 10.1016/j.celrep.2012.10.020. doi: 10.1016/j.celrep.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez H, Patel SB, Pastar I. The Role of TGFβ Signaling in Wound Epithelialization. Adv Wound Care. 2014;3:482–482. doi: 10.1089/wound.2013.0466. doi: 10.1089/wound.2013.0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundaram GM, Common JE, Gopal FE, Srikanta S, Lakshman K, Lunny DP, Lim TC, Tanavde V, Lane EB, Sampath P. ‘See-saw’ expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature. 2013;495:103–103. doi: 10.1038/nature11890. doi: 10.1038/nature11890. [DOI] [PubMed] [Google Scholar]
- 34.Apponi LH, Corbett AH, Pavlath GK. RNA-binding proteins and gene regulation in myogenesis. Trends Pharmacol Sci. 2011;32:652–652. doi: 10.1016/j.tips.2011.06.004. doi:10.1016/j.tips.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amirouche A, Tadesse H, Lunde JA, Bélanger G, Côté J, Jasmin BJ. Activation of p38 signaling increases utrophin A expression in skeletal muscle via the RNA-binding protein KSRP and inhibition of AU-rich element-mediated mRNA decay: implications for novel DMD therapeutics. Hum Mol Genet. 2013;22:3093–3093. doi: 10.1093/hmg/ddt165. doi: 10.1093/hmg/ddt165. [DOI] [PubMed] [Google Scholar]
- 36.Ruegg UT. Pharmacological prospects in the treatment of Duchenne muscular dystrophy. Curr Opin Neurol. 2013;26:577–577. doi: 10.1097/WCO.0b013e328364fbaf. doi: 0.1097/WCO.0b013e328364fbaf. [DOI] [PubMed] [Google Scholar]
- 37.Briata P, Lin WJ, Giovarelli M, Pasero M, Chou CF, Trabucchi M, Rosenfeld MG, Chen CY, Gherzi R. PI3K/AKT signaling determines a dynamic switch between distinct KSRP functions favoring skeletal myogenesis. Cell Death Differ. 2012;19:478–478. doi: 10.1038/cdd.2011.117. doi: 10.1038/cdd.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013;280:4294–4294. doi: 10.1111/febs.12253. doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- 39.Mitchelson KR, Qin WY. Roles of the canonical myomiRs miR-1, -133 and -206 in cell development and disease. World J Biol Chem. 2015;6:162–162. doi: 10.4331/wjbc.v6.i3.162. doi: 10.4331/wjbc.v6.i3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tadesse H, Deschênes-Furry J, Boisvenue S, Côté J. KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum Mol Genet. 2008;17:506–524. doi: 10.1093/hmg/ddm327. doi: 10.1093/hmg/ddm327. [DOI] [PubMed] [Google Scholar]
- 41.Ma G, Wang Y, Li Y, Cui L, Zhao Y, Zhao B, Li K. MiR-206, a key modulator of skeletal muscle development and disease. Int J Biol Sci. 2015;11:345–345. doi: 10.7150/ijbs.10921. doi: 10.7150/ijbs.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1549. doi: 10.1126/science.1181046. doi: 0.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loya CM, Van Vactor D, Fulga TA. Understanding neuronal connectivity through the post-transcriptional toolkit. Genes Dev. 2015;29:625–625. doi: 10.1101/gad.1907710. doi: 10.1101/gad.1907710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snee M, Kidd GJ, Munro TP, Smith R. RNA trafficking and stabilization elements associate with multiple brain proteins. J Cell Sci. 2002;115:4661–4661. doi: 10.1242/jcs.00137. doi: 10.1242/jcs.00137. [DOI] [PubMed] [Google Scholar]
- 45.Lu JY, Schneider RJ. Tissue Distribution of AU-rich mRNA-binding Proteins Involved in Regulation of mRNA Decay. J Biol Chem. 2004;279:12974–12974. doi: 10.1074/jbc.M310433200. doi: 10.1074/jbc.M310433200. [DOI] [PubMed] [Google Scholar]
- 46.Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J Cell Biol. 156:41–51. doi: 10.1083/jcb.200105133. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rehbein M, Wege K, Buck F, Schweizer M, Richter D, Kindler S. Molecular characterization of MARTA1, a protein interacting with the dendritic targeting element of MAP2 mRNAs. J Neurochem. 2002;82:1039–1039. doi: 10.1046/j.1471-4159.2002.01058.x. doi: 10.1046/j.1471-4159.2002.01058.x. [DOI] [PubMed] [Google Scholar]
- 48.Kunde SA, Musante L, Grimme A, Fischer U, Müller E, Wanker EE, Kalscheuer VM. The X-chromosome-linked intellectual disability protein PQBP1 is a component of neuronal RNA granules and regulates the appearance of stress granules. Hum Mol Genet. 2011;20:4916–4916. doi: 10.1093/hmg/ddr430. doi: 10.1093/hmg/ddr430. [DOI] [PubMed] [Google Scholar]
- 49.Kim HH, Kim P, Phay M, Yoo S. Identification of precursor microRNAs within distal axons of sensory neuron. J Neurochem. 2015;134:193–193. doi: 10.1111/jnc.13140. doi: 10.1111/jnc.13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bird CW, Gardiner AS, Bolognani F, Tanner DC, Chen CY, Lin WJ, Yoo S, Twiss JL, Perrone-Bizzozero N. KSRP modulation of GAP-43 mRNA stability restricts axonal outgrowth in embryonic hippocampal neurons. PLoS One. 2013;8:e79255. doi: 10.1371/journal.pone.0079255. doi: 10.1371/journal.pone. 0079255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mobarak CD, Anderson KD, Morin M, Beckel-Mitchener A, Rogers SL, Furneaux H, King P, Perrone-Bizzozero NI. The RNA-binding protein HuD is required for GAP-43 mRNA stability, GAP-43 gene expression, and PKC-dependent neurite outgrowth in PC12 cells. Mol Biol Cell. 2000;11:3191–3191. doi: 10.1091/mbc.11.9.3191. doi: 10.1091/mbc.11.9.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson KD, Sengupta J, Morin M, Neve RL, Valenzuela CF, Perrone-Bizzozero NI. Overexpression of HuD accelerates neurite outgrowth and increases gap-43 mRNA expression in cortical neurons and retinoic acid-induced embryonic stem cells in vitro. Exp Neurol. 2001;168:250–258. doi: 10.1006/exnr.2000.7599. doi: 10.1006/exnr.2000.7599. [DOI] [PubMed] [Google Scholar]
- 53.Bolognani F, Tanner DC, Nixon S, Okano HJ, Okano H, Perrone-Bizzozero NI. Coordinated expression of HuD and GAP-43 in hippocampal dentate granule cells during developmental and adult plasticity. Neurochem Res. 2007;32:2142–2142. doi: 10.1007/s11064-007-9388-8. [DOI] [PubMed] [Google Scholar]
- 54.Perrone-Bizzozero N, Bolognani F. Role of HuD and other RNA-binding proteins in neural development and plasticity. J Neurosci Res. 2002;68:121–121. doi: 10.1002/jnr.10175. doi: 10.1002/jnr.10175. [DOI] [PubMed] [Google Scholar]
- 55.Perrone-Bizzozero NI, Tanner DC, Mounce J, Bolognani F. Increased expression of axogenesis-related genes and mossy fibre length in dentate granule cells from adult HuD overexpressor mice. ASN Neuro. 2011;3:259–259. doi: 10.1042/AN20110015. doi: 10.1042/AN20110015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim W, Lee EK. Post-transcriptional regulation in metabolic diseases. RNA Biol. 2012;9:772–780. doi: 10.4161/rna.20091. doi: 10.4161/rna.20091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin YY, Chou CF, Giovarelli M, Briata P, Gherzi R, Chen CY. KSRP and MicroRNA 145 are negative regulators of lipolysis in white adipose tissue. Mol Cell Biol. 2014;34:2339–2339. doi: 10.1128/MCB.00042-14. Doi: 10.1128/MCB.00042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahmadian M, Duncan RE, Sul HS. The skinny on fat: lipolysis and fatty acid utilization in adipocytes. Trends Endocrinol Metab. 2009;20:424–424. doi: 10.1016/j.tem.2009.06.002. doi: 10.1016/j.tem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duncan RE, Ahmadian M, Jaworski K, Sarkadi-Nagy E, Sul HS. Regulation of lipolysis in adipocytes. Annu Rev Nutr. 2007;27:79–79. doi: 10.1146/annurev.nutr.27.061406.093734. doi: annurev.nutr.27.061406.093734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manteiga S, Choi K, Jayaraman A, Lee K. Systems biology of adipose tissue metabolism: regulation of growth, signaling and inflammation. Wiley Interdiscip. Rev Syst Biol Med. 2013;5:425–425. doi: 10.1002/wsbm.1213. doi: 10.1002/wsbm.1213. [DOI] [PubMed] [Google Scholar]
- 61.Lass A, Zimmermann R, Oberer M, Zechner R. Lipolysis-a highly regulated multienzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res. 2011;50:14–14. doi: 10.1016/j.plipres.2010.10.004. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chou CF, Zhu X, Lin YY, Gamble KL, Garvey WT, Chen CY. KSRP is critical in governing hepatic lipid metabolism through controlling Per2 expression. J Lipid Res. 2015;56:227–227. doi: 10.1194/jlr.M050724. doi: 10.1194/jlr.M050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abdelmalek MF, Diehl AM. Nonalcoholic fatty liver disease as a complication of insulin resistance. Med Clin North Am. 2007;91:1125–1125. doi: 10.1016/j.mcna.2007.06.001. doi: 10.1016/j.mcna.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 64.Lewis JR, Mohanty SR. Nonalcoholic fatty liver disease: a review and update. Dig Dis Sci. 2010;55:560–578. doi: 10.1007/s10620-009-1081-0. doi: 10.1016/j.plipres.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 65.Shiloh Y. ATM: expanding roles as a chief guardian of genome stability. Exp Cell Res. 2014;329:154–154. doi: 10.1016/j.yexcr.2014.09.002. doi: 0.1016/j.yexcr.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 66.Zhang X, Wan G, Berger FG, He X, Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell. 2011;41:371–371. doi: 10.1016/j.molcel.2011.01.020. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1010. doi: 10.1038/nature08025. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang J, Fan J, Li Y, Li F, Chen P, Fan Y, Xia X, Wong ST. Genome-wide RNAi screening identifies genes inhibiting the migration of glioblastoma cells. PLoS One. 2013;8:e61915. doi: 10.1371/journal.pone.0061915. doi: 10.1371/journal.pone.0061915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boucas J, Fritz C, Schmitt A, Riabinska A, Thelen L, Peifer M, Leeser U, Nuernberg P, Altmueller J, Gaestel M, et al. Label-free protein-RNA interactome analysis identifies Khsrp signaling downstream of the p38/Mk2 kinase complex as a critical modulator of cell cycle progression. PLoS One. 2015;10:e0125745. doi: 10.1371/journal.pone.0125745. doi: 10.1371/journal.pone.0125745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zubaidah RM, Tan GS, Tan SB, Lim SG, Lin Q, Chung MC. 2-D DIGE profiling of hepatocellular carcinoma tissues identified isoforms of far upstream binding protein (FUBP) as novel candidates in liver carcinogenesis. Proteomics. 2008;8:5086–5086. doi: 10.1002/pmic.200800322. doi: 10.1002/pmic.200800322. [DOI] [PubMed] [Google Scholar]
- 71.Malz M, Weber A, Singer S, Riehmer V, Bissinger M, Riener MO, Longerich T, Soll C, Vogel A, Angel P, et al. Overexpression of far upstream element binding proteins: a mechanism regulating proliferation and migration in liver cancer cells. Hepatology. 2009;50:1130–1130. doi: 10.1002/hep.23051. doi: 10.1002/hep.23051. [DOI] [PubMed] [Google Scholar]
- 72.Malouf GG, Su X, Yao H, Gao J, Xiong L, He Q, Compérat E, Couturier J, Molinié V, Escudier B, et al. Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin Cancer Res. 2014;20:4129–4129. doi: 10.1158/1078-0432.CCR-13-3036. doi: 10.1158/1078-0432.CCR-13-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nijman SM, Hijmans EM, El Messaoudi S, van Dongen MM, Sardet C, Bernards R. A functional genetic screen identifies TFE3 as a gene that confers resistance to the anti-proliferative effects of the retinoblastoma protein and transforming growth factor-beta. J Biol Chem. 2006;281:21582–21582. doi: 10.1074/jbc.M602312200. doi: 10.1074/jbc.M602312200. [DOI] [PubMed] [Google Scholar]
- 74.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–811. doi: 10.1093/nar/gku1075. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, Jene-Sanz A, Santos A, Lopez-Bigas N. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods. 2013;10:1081–1081. doi: 10.1038/nmeth.2642. doi: 10.1038/nmeth.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu TJ, Shamsaddini A, Pan Y, Smith K, Crichton DJ, Simonyan V, Mazumder R. A framework for organizing cancer-related variations from existing databases, publications and NGS data using a High-performance Integrated Virtual Environment (HIVE) Database (Oxford) 2014;25:bau022. doi: 10.1093/database/bau022. doi: 10.1093/database/bau022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–230. doi: 10.1093/nar/gkt1223. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]