Abstract
Meiotic crossovers (COs) are tightly regulated to ensure that COs on the same chromosome are distributed far apart (crossover interference, COI) and that at least one CO is formed per homolog pair (CO homeostasis). CO formation is controlled in part during meiotic double-strand break (DSB) creation in Caenorhabditis elegans, but a second level of control must also exist because meiotic DSBs outnumber COs. We show that the anti-recombinase RTEL-1 is required to prevent excess meiotic COs, probably by promoting meiotic synthesis-dependent strand annealing. Two distinct classes of meiotic COs are increased in rtel-1 mutants, and COI and homeostasis are compromised. We propose that RTEL-1 implements the second level of CO control by promoting noncrossovers.
Homologous recombination repair of meiotic DNA double-strand breaks (DSBs) is regulated to ensure the correct number and placement of meiotic crossovers (COs). One CO per chromosome ensures that homologous chromosomes are held together, can orient toward opposite spindle poles, and thereby segregate correctly at the first meiotic division. Crossover interference (COI) ensures appropriate distribution of COs among chromosomes because the formation of one CO reduces the likelihood of other COs occurring nearby. Meiotic COI is “complete” in Caenorhabditis elegans: Only a single CO occurs on each chromosome (1, 2). COI is regulated in part by the condensin I complex, which limits meiotic DSB formation (3). Because the average number of meiotic DSBs per chromosome is 2.1 (3), and only one of these is repaired as a CO, a second tier of CO control must exist downstream of meiotic DSB formation that channels about half of all DSBs into noncrossovers (NCOs). However, the proteins involved in generating a meiotic CO versus NCO are not well understood.
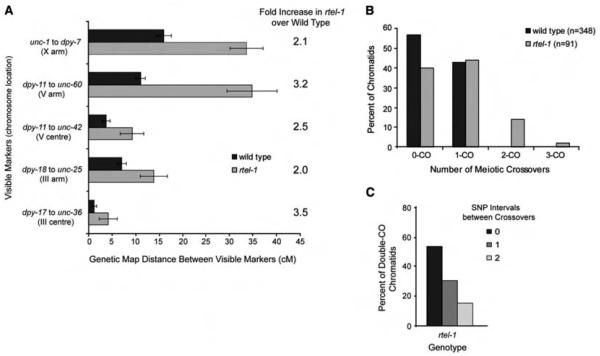
Human RTEL1 (and C. elegans RTEL-1, by homology) negatively regulates recombination by disassembling D loop–recombination intermediates during DNA repair (4). If RTEL1 acts similarly on meiotic recombination intermediates, it could be the key protein required to execute NCOs by promoting meiotic synthesis-dependent strand annealing (SDSA). By genetic measurements, recombination in C. elegans rtel-1 mutants was significantly increased in five genetic intervals on three chromosomes, including both chromosome center and arm regions (Fig. 1A and table S1) (4, 5). We used five snip–single-nucleotide polymorphisms (snip-SNPs) distributed along 80% of the X chromosome to track recombination events (Fig. 2B). Of wild-type chromatids, 43% had a single CO, and no double COs were observed, as expected (3) (Fig. 1B) [normally, 50% of chromatids have a single CO, as one CO occurs per homolog pair (6)]. In rtel-1 mutants, single, double, and triple CO chromatids occurred at a frequency of 44, 14, and 2%, respectively (Figs. 1B and 2B). The number of chromatids with COs in rtel-1 mutants was significantly different from that of wild type (P = 1.68e−10), which indicated that complete COI is defective in the absence of RTEL-1. The 44% of single CO chromatids in rtel-1 mutants is consistent with the observation that 38% of bivalents receive only one meiotic DSB (3). The number of double-CO chromatids in rtel-1 mutants is also greater than triple-CO chromatids (Figs. 1B and 2B), consistent with the reported distribution of meiotic DSBs (3). Therefore, it is possible that all DSBs in rtel-1 mutants become COs. Examination of the relative positions of COs on double-CO chromatids suggests that there is no interference between multiple COs in rtel-1 mutants, because double COs occur in adjacent SNP intervals at a frequency that does not differ from random (P = 1.00) (Fig. 1C and fig. S1). This lack of interference agrees with the idea that all DSBs in rtel-1 mutants become COs.
Fig. 1.
Recombination is increased in multiple chromosomal regions in rtel-1 mutants. (A) Recombination as measured by genetic map distance (in centimorgans) between pairs of marker genes in wild-type and rtel-1 mutants. Error bars are 95% CI. (B) Percentage of total chromatids (n) with no CO or single, double, or triple COs in wild-type and rtel-1 mutants. (C) Number of SNP intervals occurring between COs on double-CO chromatids.
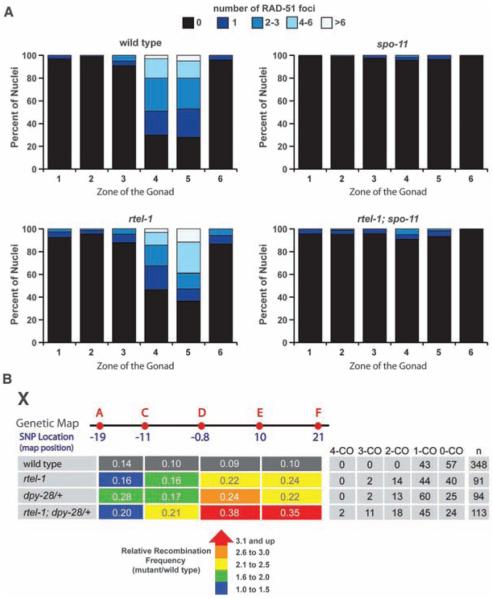
Fig. 2.
DSBs are similar in wild-type and rtel-1 mutants; rtel-1 and dpy-28/+ mutations have an additive effect on CO frequency. (A) Number of RAD-51 foci in 100 nuclei in each zone of the gonad for each genotype. Zones are as previously defined (17). (B) Recombination frequenciesmeasured between SNPs A to C, C to D, D to E, and E to F in each genotype. Relative recombination frequencies in each mutant compared with wild type are by color: blue, 1.0- to 1.5-fold; green, 1.6- to 2.0-fold; yellow, 2.1- to 2.5-fold; orange, 2.6- to 3.0-fold; and red, 3.1-fold increase or greater. The table at right shows the percentage of total chromatids (n) with no COs or single, double, triple, or quadruple COs.
Condensin I complex mutants, such as dpy-28, alter the distribution and increase the overall number of COs that occur on each chromosome because of an increase in meiotic DSB formation (3, 7). To determine whether the increased COs seen in rtel-1 mutants reflect an elevation in recombination precursors (as does dpy-28), we measured the number of DSBs generated in rtel-1 mutants using RAD-51 protein as a marker (3). rtel-1 mutants had slightly more RAD-51 foci, consistent with RTEL-1 having a role in disassembling unstable D loop–repair intermediates (4). The distribution of RAD-51 foci in rtel-1 mutants, as well as dependence on the DSB formation protein SPO-11, was similar to that of wild type (Fig. 2A and fig. S2). Meiotic DSB formation was also quantified in wild-type and rtel-1 mutants subject to RNA interference (RNAi) with rad-54, which stalls recombination intermediates after RAD-51 loading in yeast (8) and C. elegans (3). No significant difference in RAD-51 foci was observed in rad-54 RNAi–treated wild-type and rtel-1 mutants (average wild type, 11.8 ± 3.2 SD; rtel-1, 12.2 ± 4.1 SD). Elevated levels of meiotic DSBs generated in the dpy-28 mutant are sufficient to partially rescue him-17 mutants, which are deficient in meiotic DSB formation (7). Combining the rtel-1 mutation with either the spo-11 or him-17 mutations did not rescue the spo-11 or him-17 mutant phenotypes (fig. S3). Unlike dpy-28 mutants, the elevated COs in rtel-1 mutants cannot be explained by an increase in CO precursors, which suggests a breakdown at a second level of CO control distinct from DSB formation.
Because RTEL-1 and DPY-28 appear to control CO formation by different means, we examined the effect on recombination of combining rtel-1 and dpy-28 mutations. In dpy-28(s939)/+ mutants, 60, 13, and 2% of chromatids showed single, double, and triple COs, respectively (3) (Fig. 2B). When rtel-1 mutation was combined with dpy-28(s939)/+, quadruple COs were observed in 2% of rtel-1; dpy-28/+ mutant chromatids (but were not observed in either single mutant), and 45, 18, and 11% of rtel-1; dpy-28/+ mutant chromatids had single, double, and triple COs, respectively (Fig. 2B). Furthermore, 44% of the total measured intervals had a CO in rtel-1; dpy-28/+ animals, whereas 32% had a CO in dpy-28/+, and 25% hada CO in rtel-1 mutants. In the rtel-1; dpy-28/+ mutant, the total CO frequency was significantly different from that of either single mutant (P = 0.00014 for rtel-1 versus rtel-1; dpy-28; P = 0.0097 for dpy-28/+ versus rtel-1; dpy-28/+). Double COs in rtel-1; dpy-28/+ double mutants occurred in adjacent SNP intervals at a frequency that did not differ from random (P = 0.824), which suggested no residual interference (fig. S1). These data indicate an additive effect of combining dpy-28/+ and rtel-1 mutations on meiotic CO frequency, which supports the hypothesis that RTEL-1 and DPY-28 regulate CO formation through distinct mechanisms. These data also reinforce the idea that all DSBs are converted to COs in the absence of RTEL-1.
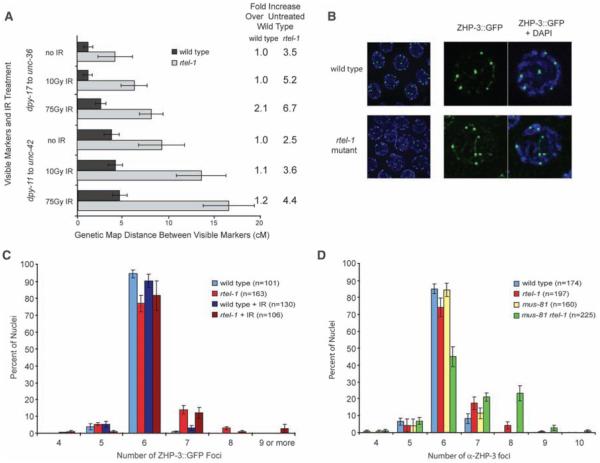
Meiotic CO homeostasis in yeast maintains COs at the expense of NCOs under conditions where meiotic DSBs are decreased (9, 10). Conversely, beyond the single “obligate” CO per chromosome, most extra DSBs in C. elegans appear to be channeled into NCO pathways, such as SDSA. Given that mutation in dpy-28 causes an increase in meiotic DSBs (3, 7), the additive effect of rtel-1 and dpy-28 mutations on meiotic CO frequency suggested that RTEL-1 may function to maintain homeostasis when there are extra DSBs. If additional meiotic DSBs generated by treatment with ionizing radiation (IR) are repaired predominantly through CO pathways in rtel-1 mutants, this would lead to increased numbers of COs. Treating rtel-1 mutants with 10 or 75 Gy of IR resulted in a large, dose-dependent increase in COs, up to 6.7-fold over untreated wild-type animals in the intervals measured, whereas relatively small increases in recombination were observed in wild-type animals after IR (Fig. 3A, fig. S2, and table S2). These data indicate that homeostasis is compromised in the absence of RTEL-1.
Fig. 3.
Recombination increases greatly in rtel-1 mutants after IR treatment but ZHP-3::GFP foci do not; ZHP-3 foci are increased in mus-81 rtel-1 mutants. (A) Recombination as measured by genetic map distance between pairs of marker genes for two intervals with no IR or 10 or 75 Gy IR in wildtype and rtel-1 mutants. Fold increase in recombination is comparedwith untreated wild type. Error bars are 95% CI. (B) ZHP-3::GFP foci (green) and 4′,6′-diamidino-2-phenylindole (DAPI)–stained (blue) in wildtype and rtel-1 mutants at meiotic diplotene. (C) Percentage of total nuclei (n) with the indicated number of ZHP-3::GFP foci in wild-type and rtel-1 mutants without treatment and 24 hours after 75Gy IR. Error bars are SEM. (D) Percentage of total nuclei (n) with the indicated number of anti–ZHP-3 foci in each genotype. Error bars are SEM.
During C. elegans meiosis, the ZHP-3 protein becomes restricted to recombination foci; one focus per chromosome and six spots per nucleus are observed in wild type (11). In wild-type nematodes, only 1% of nuclei had greater than six ZHP-3::GFP (ZHP-3 marked with green fluorescent protein) foci, whereas in rtel-1 mutants, 18% of nuclei had more than six ZHP-3 foci (Fig. 3, B and C). This was a significant, but relatively small, increase compared with the increased COs observed by our genetic and snip-SNP methods. As recombination greatly increases in rtel-1 mutants after IR, if ZHP-3 marks all COs, a large increase in ZHP-3 foci should occur after 75 Gy IR. However, ZHP-3 foci remained unchanged in wild-type animals and rtel-1 mutants after IR (Fig. 3C) [supporting online material (SOM) text]. These data support the hypothesis that ZHP-3 marks only a subset of COs and imply that two classes of meiotic CO events are elevated in rtel-1 mutants.
In Saccharomyces cerevisiae, two classes of COs exist: (i) Those that predominate are dependent on the ZMM proteins (Zip1-4, Mer3, Msh4, and Msh5) and exhibit COI; and (ii) a second class includes those that are independent of the ZMMs, require Mms4 and Mus81, and do not exhibit COI (12, 13). Until now, C. elegans was thought to have only class I COs (14, 15). We found COs in the rtel-1 mutant to fall into two classes: ZHP-3–associated obligate-type COs and COs produced by repair events not associated with ZHP-3 occurring in the absence of rtel-1. A reliance of COs not associated with ZHP-3 on MUS-81 in rtel-1 mutants is consistent with the synthetic embryonic lethality of mus-81 rtel-1 double mutants (4). Excess COs generated in wild type after x-ray are similarly dependent on MUS-81 (fig. S4 and SOM text). In mus-81 rtel-1 double mutants, RAD-51 foci persist (4), and ZHP-3 restriction to foci is delayed or does not occur in 65% of nuclei (SOM text), which suggests defects in intermediate processing that may account for the synthetic lethality. In doublemutant nuclei where ZHP-3 foci are observed, foci are increased compared with wild type and with either single mutant (Fig. 3D and SOM text), which indicates that more obligate-type COs are formed in the absence of both rtel-1 and MUS-81 (fig. S7).
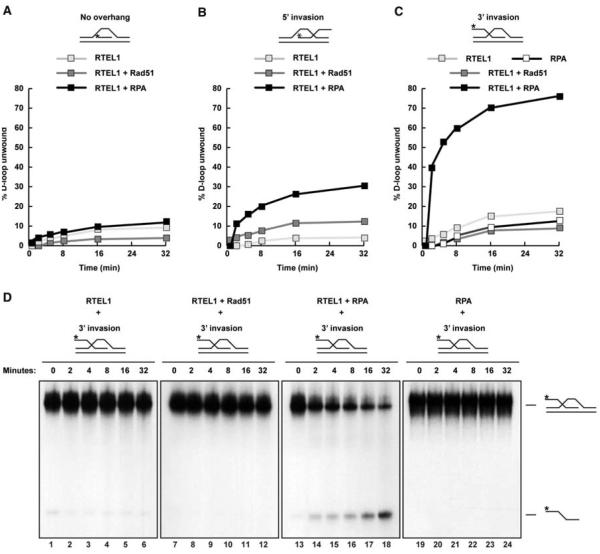
NCO repair of meiotic DSBs in S. cerevisiae is thought to occur mainly through the SDSA pathway (16). A key step in SDSA is the disruption of the D loop joint molecule. Human RTEL1 can disrupt a preformed D loop in vitro, but the mechanism of RTEL1 action is not well understood (4). We considered whether D loop unwinding might require species-specific interactions with RAD51. However, RTEL1 efficiently disrupted D loops preformed by using the E. coli RAD51 homolog RecA and single-stranded DNA (ssDNA) binding protein SSB (fig. S5). To determine whether RTEL1 exhibits structural preference for D loops, we generated three substrates: a 3′ ssDNA invasion with a 5′ overhang, a 5′ ssDNA invasion with a 3′ overhang, and a substrate with no overhang. When incubated with RAD51 and replication protein A (RPA), RTEL1 preferentially disrupted the 3′ invasion D loop but showed negligible activity toward D loop substrates with either 5′ invasion or no overhang (Fig. 4, A to C, and fig. S6). Efficient unwinding of the 3′ invasion D loop required incubation with RPA (Fig. 4D). Thus, RTEL1 activity in vitro is consistent with a role in displacing transient strand invasion events in vivo. Taken together, our data support a role for RTEL-1 in meiotic SDSA that enforces COI by preventing further DSBs (beyond the obligate CO) from becoming COs.
Fig. 4.
RTEL1 preferentially dissociates D loops with 3′ invasion and is dependent on RPA. D loop substrates with (A)nooverhang, (B) 5′ invasion, or (C) 3′ invasion. Shown is quantification of the percentage of D loop unwound over time. (D) Time course of RTEL1 activity toward a D loop substrate with 3′ invasion alone or with the indicated proteins. The fastest migrating band is the displaced radiolabeled (*) ssDNA probe; the slower species is the D loop substrate.
In summary, our data support the hypothesis that RTEL-1 regulates meiotic recombination and CO homeostasis in C. elegans by physically dissociating strand invasion events and thereby promotes NCO repair by meiotic SDSA (fig. S7). Two levels of meiotic CO control have now been identified in C. elegans: regulation of DSB number and distribution by the condensin I complex (3) and execution of NCOs by RTEL-1.
Supplementary Material
Acknowledgments
This work was supported by Cancer Research UK (S.J.B. and S.C.W.) and the Canadian Institute of Health Research (A.M.R.). B.J.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/327/5970/1254/DC1
Materials and Methods
SOM Text
Figs. S1 to S7
Tables S1 and S2
References
References and Notes
- 1.Hillers KJ, Villeneuve AM. Curr. Biol. 2003;13:1641. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 2.Wood W. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; NY; 1988. [Google Scholar]
- 3.Mets DG, Meyer BJ. Cell. 2009;139:73. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber LJ, et al. Cell. 2008;135:261. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Materials and methods are available as supporting material on Science Online.
- 6.Meneely PM, Farago AF, Kauffman TM. Genetics. 2002;162:1169. doi: 10.1093/genetics/162.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai CJ, et al. Genes Dev. 2008;22:194. doi: 10.1101/gad.1618508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krogh BO, Symington LS. Annu. Rev. Genet. 2004;38:233. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 9.Martini E, Diaz RL, Hunter N, Keeney S. Cell. 2006;126:285. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen SY, et al. Dev. Cell. 2008;15:401. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhalla N, Wynne DJ, Jantsch V, Dernburg AF, Hawley RS. PLoS Genet. 2008;4:e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bishop DK, Zickler D. Cell. 2004;117:9. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- 13.Hollingsworth NM, Brill SJ. Genes Dev. 2004;18:117. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Muse T, Boulton SJ. Chromosome Res. 2007;15:607. doi: 10.1007/s10577-007-1146-x. [DOI] [PubMed] [Google Scholar]
- 15.Zetka M. Genome Dyn. 2009;5:43. doi: 10.1159/000166618. [DOI] [PubMed] [Google Scholar]
- 16.McMahill MS, Sham CW, Bishop DK. PLoS Biol. 2007;5:e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colaiácovo MP, et al. Dev. Cell. 2003;5:463. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.