The long-awaited results from 2 major randomized clinical trials of warfarin pharmacogenetics (i.e., COAG and EU-PACT: ClinicalTrial.gov identifiers NCT00839657 and NCT01119300, respectively) were recently published (1–2). These studies were aimed at demonstrating the clinical utility of guiding individual warfarin dosing by genotyping patients in order to attain a better control of anticoagulation over the first 4 weeks of therapy. To that end, the respective mean percentages of time within the therapeutic INR range (primary outcome) in the two arms of each study (i.e., treatments guided by a pharmacogenetic-driven regression model in one arm and the standard clinical algorithm in the other) were independently compared by the authors of these two studies after dosing patients, accordingly. The COAG trial findings suggested non-superiority (but also non-inferiority) of the pharmacogenetic-guided warfarin dosing method over standard clinical procedures to dosing in terms of ensuring the stability of patients on warfarin during the first 4 weeks of therapy.
Study authors also acknowledged that their pharmacogenetic algorithms showed significant differences of predictability across different ethnic groups and that these algorithms performed better at predicting the optimal maintenance doses for non-black patients than they did at predicting such doses for black patients. Because of such ethno-specific differences, a significant interaction between race and dosing strategy with respect to the primary outcome was found (p=0.003), after stratifying patients by race. Among individuals of African origin, the mean percentage of time on target was lower in the genotype-guided arm than it was among the same class of individuals belonging to the clinical group (35.2% vs. 43.5%; adjusted mean difference, −8.3%; p=0.01). Although not statistically significant, the primary outcome was slightly better among non-blacks in the genotype-guided arm of the study than it was among non-black members of the clinical group (49% vs. 46%; adjusted mean difference 2.8%, p=0.15). In the EU-PACT trial, in which 98.5% of the recruited patients were white, authors found a moderate but significantly better control of anticoagulation in the genotype-guided dosing group than was found in the other group (adjusted difference of 7%; p<0.001) (2). Based on these results, it could be concluded that both trials provide evidence of the need to develop and test pharmacogenetic-based, multiethnic dosing algorithms that are more comprehensive than those currently in use to draw valid conclusions and make adequate recommendations about the potential clinical utility of genotyping patients on warfarin, while also accounting for the admixture effect.
Linear regression models are data-driven and, hence, population-dependent. Algorithms derived from them are often valid only for the same population in which those algorithms were originally developed (i.e., in this case, mostly those populations composed of people of European descent) (3). That population specificity is the most likely explanation for the poor predictability found in African Americans in the COAG trial. It is now well recognized that these commonly used pharmacogenetic algorithms perform poorly when applied to people with substantial African heritage. Such poor performance is probably a consequence of omitting some genetic variants in the equations in question (e.g., CYP2C9*5, *6, *8 and *11), which variants have proven to be clinically relevant in Africans (4). Besides, common genetic variants included in these pharmacogenetic-guided regression models account only for sensitivity but not for resistant phenotypes. The generalizability of these regression models is questionable and, therefore, extrapolations to any different, barely characterized population with remarkable heterogeneity and admixture (e.g., African Americans, Amerindians or Hispanics) will be flawed. Strikingly, none of these trials has enrolled a significant portion of Hispanics. Our group has been working over the past 5 years on developing a novel admixture-adjusted, genotype-guided algorithm for warfarin dosing predictions in Caribbean Hispanics, mainly Puerto Ricans. While doing so, we have postulated the need for an admixture vector to improves predictability in admixed individuals of Caribbean descent, as highlighted by an ongoing study providing proof of concept, in which study the admixture vector explained ~6% of the variance in effective warfarin dose requirements after taking into account the contribution of other covariates (Figure 1). Our preliminary data demonstrates that an admixture-adjusted pharmacogenetic algorithm has significant potential in terms of its ability to serve the Caribbean Hispanic population. The admixture vector added to the algorithm is a composite of 3 individual ancestry proportions, as determined by a Bayesian clustering analysis (STRUCTURE software v2.3.4) of genotypes across multiple loci from an array of 384 physiogenomic (PG) markers of ancestry informative value. It is to represent the relative contribution of the 3 major historical groups that shaped the genetic background of the current Puerto Rican population (i.e., Europeans, West Africans and Taino Amerindians). All these PG markers were interrogated on each sampled individual, following a sound procedure of selection and validation, the details of which can be found elsewhere (5).
Figure 1.
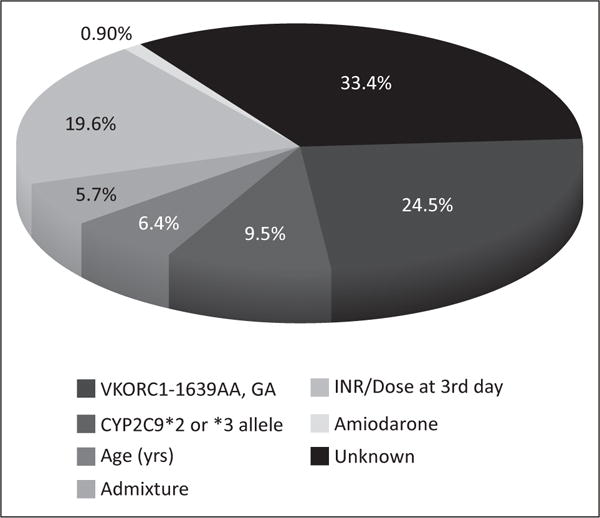
Percentage (%) of variance explained by covariates found to be significant predictors of warfarin dose variability in Puerto Ricans (i.e., genotypes, age, admixture, INR/dose on day 3 and amiodarone).
We also demonstrated a correlation between population sub-structure and warfarin-related genotypes and responses in Puerto Ricans (6). Briefly, when individual genotypes were visually overlaid atop a genetic-distance dendrogram, statistical analysis revealed a much higher frequency of the VKORC1-1639 “A” variant allele in the cluster representing Amerindians than in those clusters representing the rest of the population. We are currently performing deep sequencing analyses of samples from Puerto Ricans in order to identify the burden of unknown variants in the CYP2C9 and VKORC1 loci.
Human genetic diversity is a fundamental issue in the biomedical sciences, and is therefore relevant to population-based pharmacogenetic studies. The ability to identify relevant polymorphisms on pharmacogenes of interest is increased by considering multi-ethnic cohorts in admixture mapping-coupled association studies, population stratification and between- and within-population heterogeneity. Ancestral admixture proportions have to be quantified and further incorporated into the developed models in order to increase the accuracy of current predictions with regard to effective warfarin doses by expanding those models to incorporate mixed populations. Admixture is relevant to the clinical application of pharmacogenetics as multi-gene models are developed to predict dose variability. Previously, we have thoroughly discussed the need for admixture measurements in Caribbean Hispanics (5–7). Admixture measurements can be used as a reporter for the ethno-geographic genetic diversity that is not measured by specific polymorphisms on target pharmacogenes. Therefore, it can capture a significant proportion of the missing “genetic effect” in genotype-guided drug-prescribing algorithms. Moreover, failure to control for the effect of population stratification by admixture may give rise to confounder in future pharmacogenetic studies. A recent investigation into the effects of NQO1*2 and CYP4F2 V133M genotypes on warfarin dose requirements in Hispanic and African Americans successfully incorporated ancestry measurements to partially account for the observed variability in doses (8). The utility of genetic ancestry and admixture measures has been postulated previously, which serves to emphasize the need to be cautious when extrapolating genetic results from a homogeneous population to admixed ones (9).
The colonization of the islands in the Caribbean basin (i.e., Borikén [Puerto Rico], Cuba, Hispaniola [Haiti and the Dominican Republic) by European settlers, coming mainly from Spain, resulted in genetic mixture with local native populations. The populations of today’s Caribbean islands were also genetically enriched by a gene flow from West African natives, who were brought to America as slaves around the XVII to XVIII centuries, and to some extent by Native American admixture, as shown by the existence of the Arawak’s Taino mitochondrial DNA lineage in the contemporary population. We believe that the degree of admixture among Puerto Ricans, due to over 500 years of mixture with at least these 3 distant parental populations, contributed to a substantial enrichment of the population diversity (as represented by mosaic chromosomes). Likewise, it also contributes to its demographic stratification, which has been enhanced by demographic bottlenecks in New World settlements and geographic and cultural/religious/ethnic barriers, to name some factors. Because admixture proportions vary, both across the different regions of Puerto Rico and among individuals, as was earlier reported in a study that correlates ancestries to the locations of sugar mills and plantations on the island (10), such variations could have significantly affected the heterogeneity of the population. The population heterogeneity is of cardinal importance in pharmacogenetic studies, especially when rare genetic variants in susceptibility loci that underlie high-risk phenotypes are in play.
There are also important differences in linkage disequilibrium (LD) by ancestry that might have significant consequences for the association between genotypes and warfarin dosing across different populations. Differential association indicates that some common polymorphisms on warfarin pharmacogenes (i.e., those found to be highly prevalent in Caucasians) are in fact not causal SNPs but they rather are polymorphisms in LD with one or more functional variants (e.g., rs12777823 in African Americans and VKORC1-1639G>A in Asians) that occur specifically in a given population or ethnic group. Because of the complex and distinct pattern of mixture and recombination events that occurred in Caribbean Hispanics, it is expected that a significant reshuffling of this population’s haplotype blocks architecture has given rise to a unique LD structure in this population, with new putative markers or association signals arising from these events.
We firmly believe that the best approach for global pharmacogenetics is to guide warfarin dosing by using a pharmacogenetic-based algorithm that also accounts for the effect of admixture or ancestry proportions. Likewise, accounting for genetic variants that are important across ethnicities in warfarin pharmacogenetics would also be as effective as accounting for admixture to improve genotype-guided dosing in minorities. Future study designs must include admixture measures and novel variants so that the clinical utility of warfarin pharmacogenetics can be tested and proven. Only then will the full potential of genetically guided drug therapy to advance global health be realized. Otherwise, disparities in our ability to translate benefits from medical research findings to such medically underserved, minority populations as Caribbean Hispanics may, paradoxically, be exacerbated rather than mitigated by pharmacogenetics.
Acknowledgments
Financial Disclosures. Authors of this letter are funded in part by NIH grant #SC2HL110393 from the National Heart, Lung, and Blood Institute (NHLBI); the Research Centers in Minority Institutions (RCMI) grants from the National Center for Research Resources (NCRR) (award #2G12-RR003051) and the National Institute on Minority Health and Health Disparities (NIMHD) (award #8G12-MD007600); as well as grant #5R44GM085201-05 from the National Institute of General Medical Sciences (NIGMS) and grant #R01HS022304-01 from the Agency for Healthcare Research and Quality (AHRQ). G Ruaño is founder and President of Genomas, Inc.
Footnotes
Competing Interests Disclosures. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript, apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Contributor Information
Jorge Duconge, School of Pharmacy University of Puerto Rico Medical Sciences Campus, San Juan, PR.
Carmen l. Cadilla, School of Medicine University of Puerto Rico, Medical Sciences Campus, San Juan, PR.
Richard L. Seip, Genetics Research Center, Hartford Hospital, 67 Jefferson St., Hartford, CT 06106, USA.
Gualberto Ruaño, Genetics Research Center, Hartford Hospital, 67 Jefferson St., Hartford, CT 06106, USA.
References
- 1.Kimmel SE, French B, Kasner SE, et al. for the COAG Investigators A Pharmacogenetic versus a Clinical Algorithm for Warfarin Dosing. N Engl J Med. 2013;369:2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pirmohamed M, Burnside G, Eriksson N, et al. for the EU-PACT group A Randomized Trial of Genotype-Guided Dosing of Warfarin. N Engl J Med. 2013;369:2294–2303. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 Genotypes and Warfarin Dosing. Clin Pharmacol Ther. 2011;90:625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perera MA, Cavallari LH, Limdi NA, et al. Genetic variants associated with warfarin dose in African-American individuals: a genome-wide association study. Lancet. 2013;382:790–796. doi: 10.1016/S0140-6736(13)60681-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruaño G, Duconge J, Windemuth A, et al. Physiogenomic analysis of the Puerto Rican population. Pharmacogenomics. 2009;10:565–577. doi: 10.2217/pgs.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villagra D, Duconge J, Windemuth A, et al. CYP2C9 and VKORC1 genotypes in Puerto Ricans: A case for admixture-matching in clinical pharmacogenetic studies. Clin Chim Acta. 2010;411:1306–1311. doi: 10.1016/j.cca.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duconge J, Ruaño G. The Emerging Role of Admixture in the Pharmacogenetics of Puerto Rican Hispanics. J Pharmacogenomics Pharmacoproteomics. 2010;1(101) doi: 10.4172/2153-0645.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bress A, Patel SR, Perera MA, Campbell RT, Kittles RA, Cavallari LH. Effect of NQO1 and CYP4F2 genotypes on warfarin dose requirements in Hispanic Americans and African Americans. Pharmacogenomics. 2012;13:1925–1935. doi: 10.2217/pgs.12.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parra EJ, Kittles RA, Shriver MD. Implications of correlation between skin color and genetic ancestry for biomedical research. Nature Genetics. 2004;36(11 Suppl):S54–60. doi: 10.1038/ng1440. [DOI] [PubMed] [Google Scholar]
- 10.Via M, Gignoux CR, Roth LA, et al. History Shaped the Geographic Distribution of Genomic Admixture on the Island of Puerto Rico. PLoS One. 2011;6:e16513. doi: 10.1371/journal.pone.0016513. [DOI] [PMC free article] [PubMed] [Google Scholar]


