In the structure of the naphthoquinone derivative 2-hydroxy-3-(2-methylprop-1-en-1-yl)naphthalene-1,4-dione, the molecules form a centrosymmetric cyclic dimer through intermolecular O—H⋯O hydrogen bonds which, together with intermolecular C—H⋯O hydrogen bonds and weak π–π ring interactions, give rise to an overall two-dimensional structure.
Keywords: crystal structure, naphthoquinone derivative, molecular conformation, hydrogen bonding
Abstract
In the structure of the title compound, C14H12O3, the substituent side chain, in which the H atoms of both methyl groups are disordered over six equivalent sites, lies outside of the plane of the naphthalenedione ring. The ring-to-chain C—C—C—C torsion angles are 50.7 (3), −176.6 (2) and 4.9 (4)°. An intramolecular methyl–hydroxy C—H⋯O hydrogen bond is present. In the crystal, molecules are primarily connected by intermolecular O—H⋯O hydrogen bonds, forming a centrosymmetric cyclic dimer motif [graph set R 2 2(10)]. Also present is a weak intermolecular C—H⋯O hydrogen bond linking the dimers and a weak π–π ring interaction [ring centroid separation = 3.7862 (13) Å], giving layers parallel to (10-3).
Chemical context
Naphthoquinone compounds exhibit several biological activities, being utilized for the treatment of parasitic diseases (Salas et al., 2008 ▸) some types of cancer (Tonholo et al., 1998 ▸) and cardiovascular disease (Silva & Torres, 2013 ▸). The compound in this study, 2-hydroxy-3-(2-metilprop-1-enol)naphthalene-1,4-dione, C14H12O3, is a naphthoquinone derivative and the structure is reported herein.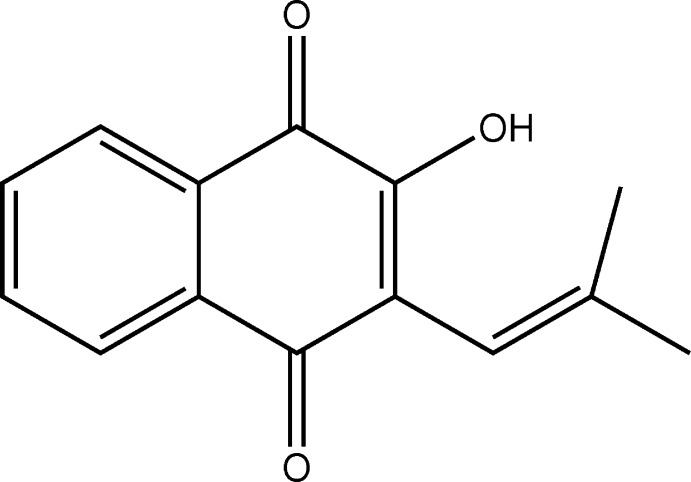
Structural commentary
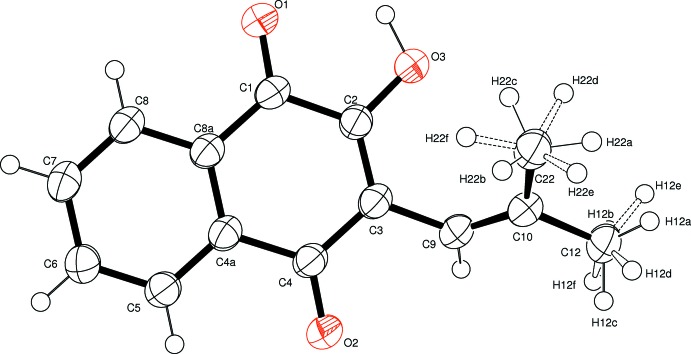
The molecular structure of the title compound is shown in Fig. 1 ▸. In this structure the side chain is rotated out of the plane of the naphthalenedione ring, with torsion angles C2—C3—C9—C10, C3—C9—C10—C12 and C3—C9—C10—C22 of 50.7 (3), −176.6 (2) and 4.9 (4)°, respectively. Present also in the molecule is an intramolecular methyl C22⋯O3 [2.959 (3) Å; see Table 1 ▸] and a short O3⋯O1 contact [2.665 (2) Å]. When compared with other analogous structures in the literature, e.g. 2-chloro-3-(4-chlorobenzamido)-1,4-naphthoquinone (Brandy et al., 2009 ▸), it is observed that the title compound has similar conformational features with respect to the side chain, which lies out of the naphthoquinone plane.
Figure 1.
Molecular conformation and atom-numbering scheme, with non-H atoms drawn at the 50% probability level. The H atoms of the rotationally disordered methyl groups are shown as six equivalent half-occupancy sites.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H1O3⋯O1i | 0.97 (3) | 1.93 (3) | 2.770 (2) | 143 (3) |
| C7—H7⋯O2ii | 0.93 | 2.43 | 3.339 (3) | 164 |
| C22—H22C⋯O3 | 0.96 | 2.21 | 2.959 (3) | 134 |
Symmetry codes: (i) 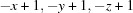 ; (ii)
; (ii) 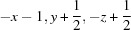 .
.
Supramolecular features
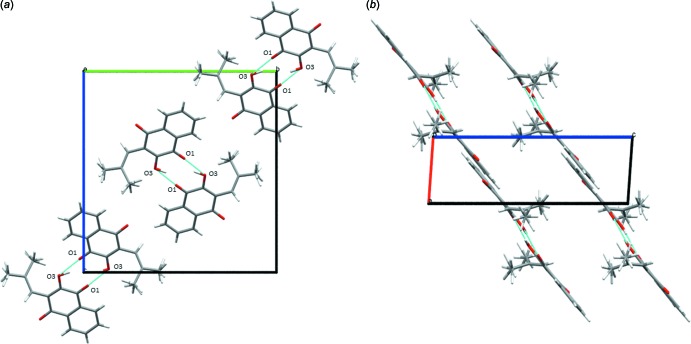
In the crystal, the molecules are connected by classic intermolecular O3—H⋯O1i hydrogen bonds (Table 1 ▸), forming a centrosymmetric cyclic dimer [graph set 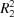 (10)] (Bernstein et al., 1995 ▸) (Fig. 2 ▸
a). Also present in the structure is a weak intermolecular C7—H⋯O2ii hydrogen bond [3.339 (3) Å], linking the dimers and a weak π–π ring interaction between the benzene and quinone ring moieties of the parent ring system [ring centroid separation Cg⋯Cg
iii = 3.7862 (13) Å; symmetry code: (iii) x + 1, y, z], giving layers parallel to (10
(10)] (Bernstein et al., 1995 ▸) (Fig. 2 ▸
a). Also present in the structure is a weak intermolecular C7—H⋯O2ii hydrogen bond [3.339 (3) Å], linking the dimers and a weak π–π ring interaction between the benzene and quinone ring moieties of the parent ring system [ring centroid separation Cg⋯Cg
iii = 3.7862 (13) Å; symmetry code: (iii) x + 1, y, z], giving layers parallel to (10 ) (Figs. 2 ▸
b and 3 ▸).
) (Figs. 2 ▸
b and 3 ▸).
Figure 2.
The centrosymmetric dimers formed from the O3—H⋯O1i hydrogen bonds, viewed (a) along a and (b) along b. For symmetry code (i), see Table 1 ▸.
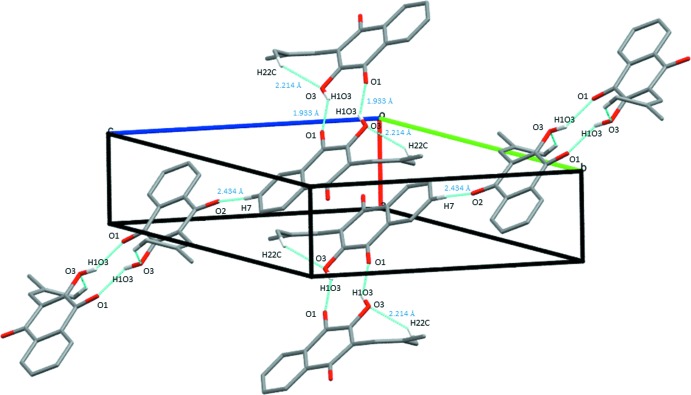
Figure 3.
The crystal packing in the unit cell, showing intra- and intermolecular interactions as dashed lines.
Database survey
A search of the Cambridge Structural Database (Groom & Allen, 2014 ▸) revealed the presence of 40 structures containing the 2-hydroxynaphthalene-1,4-dione core moiety. There were 787 structures which possess the naphthalene-1,4-dione moiety. There are structures similar to the title compound, whichvary depending on the oxidant used in the synthesis.
Synthesis and crystallization
The compound was obtained through to the lapachol oxidation product as can be seen in the scheme below (Hooker, 1936 ▸). The sample was subjected to an ethyl acetate solution at 301 K for crystallization.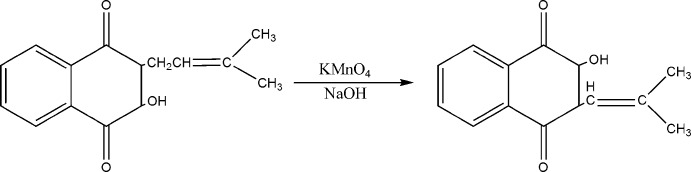
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The O3-bound H atom was located in a difference Fourier map and was freely refined. The remaining H atoms were positioned geometrically with aromatic C—H = 0.93 Å and U iso(H) = 1.2U eq(C). Rotational disorder was identified in the hydrogen atoms of the methyl carbon atoms C12 and C22 and these were included in the refinement over six equivalent 60° sites with 50% occupation, with C—H = 0.96 Å and U iso(H) = 1.5U eq(C).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C14H12O3 |
| M r | 228.24 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 293 |
| a, b, c (Å) | 4.3564 (2), 16.4069 (8), 15.8598 (7) |
| β (°) | 94.793 (2) |
| V (Å3) | 1129.62 (9) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.09 |
| Crystal size (mm) | 0.14 × 0.11 × 0.10 |
| Data collection | |
| Diffractometer | Nonius KappaCCD |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 4661, 2585, 1802 |
| R int | 0.041 |
| (sin θ/λ)max (Å−1) | 0.650 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.061, 0.191, 1.03 |
| No. of reflections | 2585 |
| No. of parameters | 158 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.31, −0.30 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015024755/zs2357sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015024755/zs2357Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015024755/zs2357Isup3.cml
CCDC reference: 1444109
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
UFAL, IQB, LabCriMM, CNPq and FAPEAL are acknowledged for support. We thank Professor Dr Antonio Ventura Pinto (in memorium) for his collaboration in the works of this research group, specifically for the synthesis of the title compound.
supplementary crystallographic information
Crystal data
| C14H12O3 | F(000) = 480 |
| Mr = 228.24 | Dx = 1.342 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 2659 reflections |
| a = 4.3564 (2) Å | θ = 1.0–27.5° |
| b = 16.4069 (8) Å | µ = 0.09 mm−1 |
| c = 15.8598 (7) Å | T = 293 K |
| β = 94.793 (2)° | Block, red |
| V = 1129.62 (9) Å3 | 0.14 × 0.11 × 0.10 mm |
| Z = 4 |
Data collection
| Nonius KappaCCD diffractometer | 1802 reflections with I > 2σ(I) |
| Radiation source: Enraf-Nonius FR590 | Rint = 0.041 |
| Graphite monochromator | θmax = 27.5°, θmin = 2.6° |
| Detector resolution: 9 pixels mm-1 | h = −5→5 |
| CCD rotation images, thick slices scans | k = −19→21 |
| 4661 measured reflections | l = −20→20 |
| 2585 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.061 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.191 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0946P)2 + 0.4119P] where P = (Fo2 + 2Fc2)/3 |
| 2585 reflections | (Δ/σ)max < 0.001 |
| 158 parameters | Δρmax = 0.31 e Å−3 |
| 0 restraints | Δρmin = −0.30 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O3 | 0.3690 (4) | 0.37038 (10) | 0.48362 (10) | 0.0407 (4) | |
| O1 | 0.2205 (4) | 0.52143 (9) | 0.43377 (9) | 0.0404 (4) | |
| O2 | −0.3406 (4) | 0.27382 (9) | 0.26721 (10) | 0.0481 (5) | |
| C10 | 0.0940 (5) | 0.19603 (13) | 0.47932 (13) | 0.0395 (5) | |
| C9 | 0.0749 (5) | 0.23006 (12) | 0.40272 (13) | 0.0386 (5) | |
| H9 | 0.1126 | 0.1961 | 0.3578 | 0.046* | |
| H1O3 | 0.448 (7) | 0.424 (2) | 0.5005 (19) | 0.073 (9)* | |
| C4A | −0.3114 (5) | 0.41650 (12) | 0.28476 (13) | 0.0349 (5) | |
| C8A | −0.1689 (5) | 0.48205 (13) | 0.32860 (13) | 0.0348 (5) | |
| C2 | 0.1441 (5) | 0.38047 (12) | 0.42115 (13) | 0.0351 (5) | |
| C1 | 0.0733 (5) | 0.46622 (12) | 0.39675 (13) | 0.0350 (5) | |
| C5 | −0.5333 (5) | 0.43122 (14) | 0.21845 (13) | 0.0400 (5) | |
| H5 | −0.6268 | 0.3879 | 0.1885 | 0.048* | |
| C3 | 0.0007 (5) | 0.31557 (12) | 0.38155 (12) | 0.0358 (5) | |
| C4 | −0.2235 (5) | 0.33077 (13) | 0.30859 (13) | 0.0369 (5) | |
| C6 | −0.6151 (5) | 0.51093 (14) | 0.19709 (14) | 0.0426 (5) | |
| H6 | −0.7628 | 0.5207 | 0.1524 | 0.051* | |
| C8 | −0.2532 (5) | 0.56203 (13) | 0.30691 (14) | 0.0386 (5) | |
| H8 | −0.1584 | 0.6056 | 0.3362 | 0.046* | |
| C7 | −0.4789 (5) | 0.57607 (13) | 0.24159 (14) | 0.0413 (5) | |
| H7 | −0.5389 | 0.6291 | 0.2277 | 0.05* | |
| C12 | 0.1899 (6) | 0.10858 (13) | 0.49043 (15) | 0.0475 (6) | |
| H12A | 0.1919 | 0.0939 | 0.5491 | 0.071* | 0.5 |
| H12B | 0.3923 | 0.1015 | 0.4718 | 0.071* | 0.5 |
| H12C | 0.0468 | 0.0744 | 0.4575 | 0.071* | 0.5 |
| H12D | 0.2288 | 0.086 | 0.4365 | 0.071* | 0.5 |
| H12E | 0.0283 | 0.0784 | 0.5138 | 0.071* | 0.5 |
| H12F | 0.3738 | 0.1055 | 0.5281 | 0.071* | 0.5 |
| C22 | 0.0189 (6) | 0.23815 (14) | 0.55869 (14) | 0.0452 (6) | |
| H22A | 0.0507 | 0.2013 | 0.6056 | 0.068* | 0.5 |
| H22B | −0.1923 | 0.2554 | 0.553 | 0.068* | 0.5 |
| H22C | 0.1503 | 0.2848 | 0.5684 | 0.068* | 0.5 |
| H22D | −0.0449 | 0.2931 | 0.5457 | 0.068* | 0.5 |
| H22E | 0.1981 | 0.2389 | 0.5983 | 0.068* | 0.5 |
| H22F | −0.1445 | 0.2095 | 0.583 | 0.068* | 0.5 |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O3 | 0.0435 (9) | 0.0323 (8) | 0.0450 (9) | −0.0013 (6) | −0.0051 (6) | −0.0002 (7) |
| O1 | 0.0456 (9) | 0.0318 (8) | 0.0431 (8) | −0.0041 (6) | −0.0005 (6) | −0.0022 (6) |
| O2 | 0.0651 (11) | 0.0308 (8) | 0.0460 (9) | −0.0033 (7) | −0.0096 (7) | −0.0026 (7) |
| C10 | 0.0438 (12) | 0.0302 (10) | 0.0439 (12) | −0.0025 (8) | −0.0013 (9) | −0.0004 (9) |
| C9 | 0.0462 (12) | 0.0282 (10) | 0.0409 (11) | 0.0009 (9) | 0.0011 (9) | −0.0027 (9) |
| C4A | 0.0422 (11) | 0.0292 (10) | 0.0340 (10) | −0.0011 (8) | 0.0061 (8) | 0.0012 (8) |
| C8A | 0.0398 (11) | 0.0305 (11) | 0.0346 (10) | −0.0016 (8) | 0.0057 (8) | −0.0001 (8) |
| C2 | 0.0384 (11) | 0.0314 (11) | 0.0355 (10) | 0.0003 (8) | 0.0035 (8) | 0.0009 (8) |
| C1 | 0.0398 (11) | 0.0288 (10) | 0.0367 (10) | −0.0019 (8) | 0.0055 (8) | −0.0034 (8) |
| C5 | 0.0486 (13) | 0.0343 (11) | 0.0368 (11) | −0.0025 (9) | 0.0015 (9) | −0.0004 (9) |
| C3 | 0.0431 (11) | 0.0292 (10) | 0.0355 (10) | −0.0006 (8) | 0.0064 (8) | −0.0006 (8) |
| C4 | 0.0453 (12) | 0.0299 (10) | 0.0357 (11) | −0.0025 (9) | 0.0040 (9) | −0.0011 (8) |
| C6 | 0.0500 (13) | 0.0383 (12) | 0.0391 (11) | 0.0013 (9) | 0.0003 (9) | 0.0039 (9) |
| C8 | 0.0460 (12) | 0.0293 (10) | 0.0410 (11) | −0.0005 (8) | 0.0062 (9) | −0.0002 (8) |
| C7 | 0.0493 (12) | 0.0312 (11) | 0.0436 (11) | 0.0027 (9) | 0.0060 (9) | 0.0055 (9) |
| C12 | 0.0651 (15) | 0.0315 (11) | 0.0447 (12) | 0.0020 (10) | −0.0024 (10) | 0.0011 (9) |
| C22 | 0.0587 (14) | 0.0346 (11) | 0.0420 (12) | 0.0006 (10) | 0.0033 (10) | 0.0011 (9) |
Geometric parameters (Å, º)
| O3—C2 | 1.344 (3) | C3—C4 | 1.472 (3) |
| O3—H1O3 | 0.97 (4) | C6—C7 | 1.387 (3) |
| O1—C1 | 1.230 (2) | C6—H6 | 0.93 |
| O2—C4 | 1.228 (2) | C8—C7 | 1.387 (3) |
| C10—C9 | 1.333 (3) | C8—H8 | 0.93 |
| C10—C22 | 1.496 (3) | C7—H7 | 0.93 |
| C10—C12 | 1.501 (3) | C12—H12A | 0.96 |
| C9—C3 | 1.472 (3) | C12—H12B | 0.96 |
| C9—H9 | 0.93 | C12—H12C | 0.96 |
| C4A—C5 | 1.389 (3) | C12—H12D | 0.96 |
| C4A—C8A | 1.398 (3) | C12—H12E | 0.96 |
| C4A—C4 | 1.498 (3) | C12—H12F | 0.96 |
| C8A—C8 | 1.398 (3) | C22—H22A | 0.96 |
| C8A—C1 | 1.469 (3) | C22—H22B | 0.96 |
| C2—C3 | 1.361 (3) | C22—H22C | 0.96 |
| C2—C1 | 1.485 (3) | C22—H22D | 0.96 |
| C5—C6 | 1.390 (3) | C22—H22E | 0.96 |
| C5—H5 | 0.93 | C22—H22F | 0.96 |
| C2—O3—H1O3 | 108.3 (18) | C10—C12—H12C | 109.5 |
| C9—C10—C22 | 124.9 (2) | H12A—C12—H12C | 109.5 |
| C9—C10—C12 | 120.2 (2) | H12B—C12—H12C | 109.5 |
| C22—C10—C12 | 114.91 (19) | C10—C12—H12D | 109.5 |
| C10—C9—C3 | 127.1 (2) | H12A—C12—H12D | 141.1 |
| C10—C9—H9 | 116.5 | H12B—C12—H12D | 56.3 |
| C3—C9—H9 | 116.5 | H12C—C12—H12D | 56.3 |
| C5—C4A—C8A | 119.67 (19) | C10—C12—H12E | 109.5 |
| C5—C4A—C4 | 120.09 (19) | H12A—C12—H12E | 56.3 |
| C8A—C4A—C4 | 120.23 (18) | H12B—C12—H12E | 141.1 |
| C4A—C8A—C8 | 120.21 (19) | H12C—C12—H12E | 56.3 |
| C4A—C8A—C1 | 119.46 (19) | H12D—C12—H12E | 109.5 |
| C8—C8A—C1 | 120.32 (19) | C10—C12—H12F | 109.5 |
| O3—C2—C3 | 121.45 (19) | H12A—C12—H12F | 56.3 |
| O3—C2—C1 | 115.56 (18) | H12B—C12—H12F | 56.3 |
| C3—C2—C1 | 122.95 (19) | H12C—C12—H12F | 141.1 |
| O1—C1—C8A | 122.31 (19) | H12D—C12—H12F | 109.5 |
| O1—C1—C2 | 119.00 (18) | H12E—C12—H12F | 109.5 |
| C8A—C1—C2 | 118.68 (18) | C10—C22—H22A | 109.5 |
| C4A—C5—C6 | 119.8 (2) | C10—C22—H22B | 109.5 |
| C4A—C5—H5 | 120.1 | H22A—C22—H22B | 109.5 |
| C6—C5—H5 | 120.1 | C10—C22—H22C | 109.5 |
| C2—C3—C4 | 118.61 (19) | H22A—C22—H22C | 109.5 |
| C2—C3—C9 | 123.83 (19) | H22B—C22—H22C | 109.5 |
| C4—C3—C9 | 117.35 (18) | C10—C22—H22D | 109.5 |
| O2—C4—C3 | 120.66 (19) | H22A—C22—H22D | 141.1 |
| O2—C4—C4A | 119.58 (18) | H22B—C22—H22D | 56.3 |
| C3—C4—C4A | 119.76 (18) | H22C—C22—H22D | 56.3 |
| C7—C6—C5 | 120.7 (2) | C10—C22—H22E | 109.5 |
| C7—C6—H6 | 119.6 | H22A—C22—H22E | 56.3 |
| C5—C6—H6 | 119.6 | H22B—C22—H22E | 141.1 |
| C7—C8—C8A | 119.7 (2) | H22C—C22—H22E | 56.3 |
| C7—C8—H8 | 120.2 | H22D—C22—H22E | 109.5 |
| C8A—C8—H8 | 120.2 | C10—C22—H22F | 109.5 |
| C6—C7—C8 | 119.9 (2) | H22A—C22—H22F | 56.3 |
| C6—C7—H7 | 120 | H22B—C22—H22F | 56.3 |
| C8—C7—H7 | 120 | H22C—C22—H22F | 141.1 |
| C10—C12—H12A | 109.5 | H22D—C22—H22F | 109.5 |
| C10—C12—H12B | 109.5 | H22E—C22—H22F | 109.5 |
| H12A—C12—H12B | 109.5 | ||
| O1—C1—C2—O3 | 0.2 (3) | O2—C4—C4A—C5 | 2.6 (3) |
| O1—C1—C2—C3 | −177.5 (2) | O2—C4—C4A—C8A | −176.8 (2) |
| C8A—C1—C2—O3 | 179.69 (19) | C3—C4—C4A—C5 | −177.0 (2) |
| C8A—C1—C2—C3 | 2.0 (3) | C3—C4—C4A—C8A | 3.5 (3) |
| O1—C1—C8A—C4A | 175.2 (2) | C4—C4A—C5—C6 | 179.6 (2) |
| O1—C1—C8A—C8 | −3.9 (3) | C8A—C4A—C5—C6 | −1.0 (3) |
| C2—C1—C8A—C4A | −4.3 (3) | C4—C4A—C8A—C1 | 1.6 (3) |
| C2—C1—C8A—C8 | 176.6 (2) | C4—C4A—C8A—C8 | −179.4 (2) |
| O3—C2—C3—C4 | −174.50 (19) | C5—C4A—C8A—C1 | −177.9 (2) |
| O3—C2—C3—C9 | 0.2 (3) | C5—C4A—C8A—C8 | 1.2 (3) |
| C1—C2—C3—C4 | 3.1 (3) | C4A—C5—C6—C7 | −0.4 (3) |
| C1—C2—C3—C9 | 177.8 (2) | C5—C6—C7—C8 | 1.5 (3) |
| C2—C3—C4—O2 | 174.5 (2) | C6—C7—C8—C8A | −1.3 (3) |
| C2—C3—C4—C4A | −5.9 (3) | C7—C8—C8A—C1 | 179.0 (2) |
| C9—C3—C4—O2 | −0.5 (3) | C7—C8—C8A—C4A | −0.1 (3) |
| C9—C3—C4—C4A | 179.15 (19) | C3—C9—C10—C12 | −176.6 (2) |
| C2—C3—C9—C10 | 50.7 (3) | C3—C9—C10—C22 | 4.9 (4) |
| C4—C3—C9—C10 | −134.6 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H1O3···O1i | 0.97 (3) | 1.93 (3) | 2.770 (2) | 143 (3) |
| C7—H7···O2ii | 0.93 | 2.43 | 3.339 (3) | 164 |
| C22—H22C···O3 | 0.96 | 2.21 | 2.959 (3) | 134 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x−1, y+1/2, −z+1/2.
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Brandy, Y., Butcher, R. J., Adesiyun, T. A., Berhe, S. & Bakare, O. (2009). Acta Cryst. E65, o64. [DOI] [PMC free article] [PubMed]
- Enraf–Nonius (2001). Kappa CCD Operation Manual. Enraf–Nonius, Delft, The Netherlands.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R. & Allen, F. H. (2014). Angew. Chem. Int. Ed. 53, 662–671. [DOI] [PubMed]
- Hooker, S. C. (1936). J. Am. Chem. Soc. 58, 1168–1173.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, Editors C. W. Carter & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Salas, C., Tapia, R. A., Ciudad, K., Armstrong, V., Orellana, M., Kemmerling, U., Ferreira, J., Maya, J. D. & Morello, A. (2008). Bioorg. Med. Chem. 16, 668–674. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Silva, A. K. Soares e, de Oliveira Cipriano Torres, D., Santos Rocha, S. W., dos Santos Gomes, F. O., dos Santos Silva, B., Donato, M. A. M., Raposo, C., Santos, A. C. O., de Lima, M. do C. A., Galdino, S. L., da Rocha Pitta, I., de Souza, J. R. B. & Peixoto, C. A. (2013). Cardiovascular Pathol. 22, 81–90. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Tonholo, J. L. R., Freitas, L. R., de Abreu, F. C., Azevedo, D. C., Zani, C. L., de Oliveira, A. B. & Goulart, M. O. F. (1998). J. Braz. Chem. Soc. 9, 163–169.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015024755/zs2357sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015024755/zs2357Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015024755/zs2357Isup3.cml
CCDC reference: 1444109
Additional supporting information: crystallographic information; 3D view; checkCIF report