In C6H6N2O2·H2O, the N-hydroxypicolinamide molecule adopts a strongly flattened conformation. O—H⋯O interactions and π–π stacking interactions between the pyridine rings organize the crystal components into columns extending along the b axis while N—H⋯N hydrogen bonds link these columns into a two-dimensional framework parallel to (100).
Keywords: crystal structure, hydroxamic acids, hydrogen bonds, π–π stacking interactions
Abstract
The crystal structure of the title compound, C6H6N2O2·H2O, consists of N-hydroxypicolinamide and water molecules connected through O—H⋯O and N—H⋯N hydrogen bonds. The O—H⋯O interactions and π–π stacking interactions between the pyridine rings [centroid–centroid distance = 3.427 (1) Å] organize the components into columns extending along the b axis and the N—H⋯N hydrogen bonds link these columns into a two-dimensional framework parallel to (100). The N-hydroxypicolinamide molecule adopts a strongly flattened conformation and only the O—H group H atom deviates significantly from the molecule best plane. The dihedral angle between the hydroxamic group and the pyridine ring is 5.6 (2)°. The conformation about the hydroxamic group C—N bond is Z and that about the C—C bond between the pyridine and hydroxamic groups is E.
Chemical context
Hydroxamic acids (HA) are weak organic acids with the general formula R—C(=O)—NH—OH. HA can exist as keto and imino(enol) tautomers with two isomers, E and Z, for each form, and in the zwitterionic form (see Scheme below). They have found broad application in coordination chemistry due to their diversity and comparatively facile synthesis (Świątek-Kozłowska et al., 2000 ▸; Dobosz et al., 1999 ▸). In addition, they exhibit biological activities related to their enzyme-inhibitory properties (Marmion et al., 2013 ▸). HAs are widely used in coordination and supramolecular chemistry as scaffolds in the preparation of metallacrowns (Seda et al., 2007 ▸; Jankolovits et al., 2013 ▸; Safyanova et al., 2015 ▸) and as building blocks of coordination polymers (Gumienna-Kontecka et al., 2007 ▸; Golenya et al., 2014 ▸; Pavlishchuk et al., 2010 ▸, 2011 ▸). 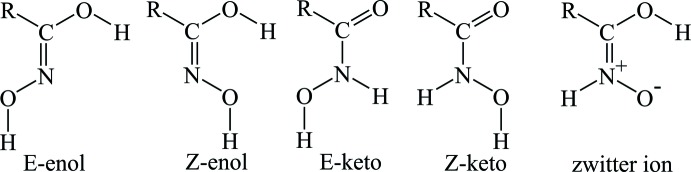
N-Hydroxypicolinamide, known also as picoline-2-hydroxamic acid (o-PicHA), has been used extensively for the synthesis of polynuclear complexes, especially in the synthesis of diverse metallacrowns (Stemmler et al., 1999 ▸; Seda et al., 2007 ▸; Jankolovits et al., 2013 ▸; Golenya et al., 2012 ▸; Gumienna-Kontecka et al., 2013 ▸). Presently, the Cambridge Structural Database (Groom & Allen, 2014 ▸) contains more than 20 entries of coordination compounds based on N-hydroxypicolinamide.
Our interest in N-hydroxypicolinamide stems also from the fact that in the course of synthesis of the title and related compounds from 2-picolinic acid esters (Hynes, 1970 ▸), the products are frequently contaminated with impurities that result from hydrolysis of the ester or hydroxamic groups to the carboxylic group. Structural information about the title compound will be helpful in controlling the purity of the synthesised ligand by powder diffraction.
Structural commentary
The molecular structure of the title compound is presented in Fig. 1 ▸. The crystal structure of the title compound consists of an N-hydroxypicolinamide molecule in the Z-keto tautomeric form in agreement with the C=O and C—N bond lengths [1.234 (2) and 1.325 (2) Å, respectively] and a water molecule. The N-hydroxypicolinamide molecule adopts a strongly flattened conformation and only the O—H group H atom deviates significantly from the molecular best plane. The maximum deviation from this plane for non-hydrogen atom is 0.083 (1) Å for O1 and the hydroxyl group H2 atom is displaced from the mean plane by 0.80 (1) Å in the direction of the water molecule. The dihedral angle between the hydroxamic group and the pyridine ring is 5.6 (2)°. The configuration about the hydroxamic group C—N bond is Z and that about the C—C bond between the pyridine and hydroxamic groups is E [torsion angles O2—N2—C6—O1 −0.4 (3)°, N1—C1—C6—O1 175.6 (2)°].
Figure 1.
The asymmetric unit of the title compound, with displacement ellipsoids drawn at the 50% probability level. H atoms are shown as spheres of arbitrary radius. The dashed line indicates a hydrogen bond.
Supramolecular features
The molecular components of the title compound are connected by O—H⋯O and N—H⋯N hydrogen bonds (Table 1 ▸) into a two-dimensional framework parallel to (100) (Fig. 2 ▸). The O—H⋯O interactions and π–π stacking interactions between the pyridine rings [centroid–centroid distance 3.427 (1) Å] organize the crystal components into columns extending along the b axis while the N—H⋯N hydrogen bonds link these columns into a two-dimensional framework parallel to (100) (Fig.2).
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2⋯O1W | 0.82 | 1.86 | 2.656 (2) | 163 |
| N2—H2A⋯N1i | 0.86 | 2.31 | 3.010 (2) | 139 |
| O1W—H1WA⋯O1ii | 0.85 | 2.14 | 2.976 (2) | 168 |
| O1W—H1WB⋯O1iii | 0.85 | 1.94 | 2.788 (2) | 173 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Figure 2.
A packing diagram of the title compound. Hydrogen bonds are indicated by dashed lines. H atoms not involved in hydrogen bonding have been omitted for clarity.
Database survey
A search of the Cambridge Structural Database (Version 5.36, last update February 2015; Groom & Allen, 2014 ▸) revealed two crystal structures of isomeric pyridine hydroxamic acids and the crystal structure of 2,6-pyridinedihydroxamic acid (Golenya et al., 2007 ▸; Makhmudova et al., 2001 ▸; Griffith et al., 2008 ▸).
Synthesis and crystallization
The title compound was obtained by the reaction of methyl 2-picolinate and hydroxylamine in methanol solution according to a reported procedure (Hynes, 1970 ▸). Colorless crystals suitable for X-ray diffraction were obtained from a methanol solution by slow evaporation at room temperature (yield 79%).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The crystal was modelled as a non-merohedral twin with the volume ratio of two twin domains refined at 89:19. The C—H, N—H and O—H hydrogen atoms of the organic molecule were found from the difference Fourier maps but for further calculations they were positioned geometrically and constrained to ride on their parent atoms with C—H = 0.93 Å, N—H = 0.86 Å and O—H = 0.82 Å, and with U iso = 1.2U eq(C,N) or U iso = 1.5U eq(O). The H atoms of the water molecule were located in the difference Fourier maps, the O—H distances standardized to 0.85 Å and refined in riding-model approximation with U iso(H) = 1.5U eq(O).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C6H6N2O2·H2O |
| M r | 156.14 |
| Crystal system, space group | Monoclinic, C2/c |
| Temperature (K) | 298 |
| a, b, c (Å) | 18.7471 (13), 3.8129 (4), 20.4813 (17) |
| β (°) | 100.570 (7) |
| V (Å3) | 1439.2 (2) |
| Z | 8 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.12 |
| Crystal size (mm) | 0.4 × 0.4 × 0.1 |
| Data collection | |
| Diffractometer | Agilent Xcalibur, Sapphire3 |
| Absorption correction | Multi-scan (CrysAlis PRO; Agilent, 2013 ▸) |
| T min, T max | 0.476, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 2491, 1401, 1053 |
| R int | 0.037 |
| (sin θ/λ)max (Å−1) | 0.617 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.053, 0.143, 0.99 |
| No. of reflections | 1401 |
| No. of parameters | 102 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.19, −0.25 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015024706/gk2650sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015024706/gk2650Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015024706/gk2650Isup3.cml
CCDC reference: 1444026
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
Financial support from the European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreement PIRSES-GA-2013–611488 is gratefully acknowledged. KAO acknowledges for the DAAD fellowship (Leonhard-Euler-Programm).
supplementary crystallographic information
Crystal data
| C6H6N2O2·H2O | Dx = 1.441 Mg m−3 |
| Mr = 156.14 | Melting point: 393 K |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 18.7471 (13) Å | Cell parameters from 893 reflections |
| b = 3.8129 (4) Å | θ = 4.1–29.0° |
| c = 20.4813 (17) Å | µ = 0.12 mm−1 |
| β = 100.570 (7)° | T = 298 K |
| V = 1439.2 (2) Å3 | Plate, clear colourless |
| Z = 8 | 0.4 × 0.4 × 0.1 mm |
| F(000) = 656 |
Data collection
| Agilent Xcalibur, Sapphire3 diffractometer | 1401 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 1053 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.037 |
| Detector resolution: 16.1827 pixels mm-1 | θmax = 26.0°, θmin = 3.3° |
| ω scans | h = −22→22 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2013) | k = −4→4 |
| Tmin = 0.476, Tmax = 1.000 | l = −24→24 |
| 2491 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.053 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.143 | H-atom parameters constrained |
| S = 0.99 | w = 1/[σ2(Fo2) + (0.0751P)2] where P = (Fo2 + 2Fc2)/3 |
| 1401 reflections | (Δ/σ)max < 0.001 |
| 102 parameters | Δρmax = 0.19 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refined as a 2-component twin. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.52649 (8) | 0.3700 (4) | 0.43254 (7) | 0.0468 (5) | |
| O2 | 0.40179 (7) | 0.5190 (5) | 0.35109 (7) | 0.0506 (5) | |
| H2 | 0.3916 | 0.6429 | 0.3808 | 0.076* | |
| N1 | 0.59448 (9) | 0.8106 (5) | 0.30306 (8) | 0.0366 (5) | |
| N2 | 0.46909 (9) | 0.6163 (5) | 0.33806 (8) | 0.0410 (5) | |
| H2A | 0.4722 | 0.7305 | 0.3025 | 0.049* | |
| C1 | 0.59810 (10) | 0.6575 (5) | 0.36271 (9) | 0.0323 (5) | |
| C2 | 0.66231 (11) | 0.6152 (6) | 0.40743 (11) | 0.0428 (6) | |
| H2B | 0.6628 | 0.5119 | 0.4487 | 0.051* | |
| C3 | 0.72601 (11) | 0.7308 (6) | 0.38915 (13) | 0.0519 (7) | |
| H3 | 0.7703 | 0.7030 | 0.4178 | 0.062* | |
| C4 | 0.72296 (11) | 0.8863 (6) | 0.32857 (13) | 0.0482 (6) | |
| H4 | 0.7650 | 0.9656 | 0.3154 | 0.058* | |
| C5 | 0.65653 (12) | 0.9234 (6) | 0.28737 (11) | 0.0434 (6) | |
| H5 | 0.6549 | 1.0327 | 0.2465 | 0.052* | |
| C6 | 0.52864 (10) | 0.5321 (6) | 0.38079 (9) | 0.0332 (5) | |
| O1W | 0.39862 (8) | 0.9488 (5) | 0.45255 (8) | 0.0501 (5) | |
| H1WA | 0.4308 | 1.0938 | 0.4455 | 0.075* | |
| H1WB | 0.4202 | 0.8351 | 0.4861 | 0.075* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0447 (8) | 0.0657 (11) | 0.0287 (8) | 0.0009 (7) | 0.0035 (6) | 0.0147 (7) |
| O2 | 0.0332 (8) | 0.0832 (13) | 0.0350 (9) | −0.0121 (8) | 0.0051 (6) | 0.0062 (8) |
| N1 | 0.0370 (9) | 0.0465 (11) | 0.0267 (9) | −0.0051 (8) | 0.0069 (7) | −0.0036 (8) |
| N2 | 0.0307 (9) | 0.0670 (13) | 0.0252 (9) | −0.0038 (8) | 0.0048 (7) | 0.0107 (9) |
| C1 | 0.0344 (10) | 0.0371 (11) | 0.0252 (10) | 0.0012 (8) | 0.0047 (8) | −0.0061 (8) |
| C2 | 0.0369 (11) | 0.0543 (14) | 0.0347 (12) | 0.0052 (9) | 0.0000 (9) | −0.0030 (11) |
| C3 | 0.0322 (11) | 0.0640 (17) | 0.0554 (16) | 0.0028 (11) | −0.0027 (10) | −0.0116 (13) |
| C4 | 0.0351 (11) | 0.0569 (15) | 0.0552 (15) | −0.0091 (10) | 0.0156 (10) | −0.0148 (12) |
| C5 | 0.0435 (12) | 0.0538 (15) | 0.0348 (12) | −0.0062 (11) | 0.0121 (9) | −0.0047 (11) |
| C6 | 0.0359 (11) | 0.0409 (12) | 0.0224 (10) | −0.0012 (9) | 0.0043 (8) | −0.0013 (9) |
| O1W | 0.0408 (8) | 0.0687 (11) | 0.0404 (9) | 0.0044 (8) | 0.0068 (7) | 0.0163 (8) |
Geometric parameters (Å, º)
| O1—C6 | 1.234 (2) | C2—C3 | 1.387 (3) |
| O2—N2 | 1.387 (2) | C2—H2B | 0.9300 |
| O2—H2 | 0.8200 | C3—C4 | 1.367 (3) |
| N1—C5 | 1.334 (3) | C3—H3 | 0.9300 |
| N1—C1 | 1.344 (3) | C4—C5 | 1.378 (3) |
| N2—C6 | 1.325 (2) | C4—H4 | 0.9300 |
| N2—H2A | 0.8600 | C5—H5 | 0.9300 |
| C1—C2 | 1.382 (3) | O1W—H1WA | 0.8503 |
| C1—C6 | 1.496 (3) | O1W—H1WB | 0.8499 |
| N2—O2—H2 | 109.5 | C4—C3—H3 | 120.4 |
| C5—N1—C1 | 117.26 (17) | C2—C3—H3 | 120.4 |
| C6—N2—O2 | 119.60 (16) | C3—C4—C5 | 118.9 (2) |
| C6—N2—H2A | 120.2 | C3—C4—H4 | 120.6 |
| O2—N2—H2A | 120.2 | C5—C4—H4 | 120.6 |
| N1—C1—C2 | 123.08 (19) | N1—C5—C4 | 123.4 (2) |
| N1—C1—C6 | 117.54 (16) | N1—C5—H5 | 118.3 |
| C2—C1—C6 | 119.38 (18) | C4—C5—H5 | 118.3 |
| C1—C2—C3 | 118.2 (2) | O1—C6—N2 | 122.15 (18) |
| C1—C2—H2B | 120.9 | O1—C6—C1 | 122.66 (17) |
| C3—C2—H2B | 120.9 | N2—C6—C1 | 115.17 (17) |
| C4—C3—C2 | 119.2 (2) | H1WA—O1W—H1WB | 102.7 |
| C5—N1—C1—C2 | 0.3 (3) | C3—C4—C5—N1 | −1.0 (4) |
| C5—N1—C1—C6 | 179.67 (17) | O2—N2—C6—O1 | −0.4 (3) |
| N1—C1—C2—C3 | −1.2 (3) | O2—N2—C6—C1 | −178.54 (17) |
| C6—C1—C2—C3 | 179.4 (2) | N1—C1—C6—O1 | 175.6 (2) |
| C1—C2—C3—C4 | 1.0 (3) | C2—C1—C6—O1 | −5.0 (3) |
| C2—C3—C4—C5 | 0.0 (4) | N1—C1—C6—N2 | −6.2 (3) |
| C1—N1—C5—C4 | 0.8 (3) | C2—C1—C6—N2 | 173.20 (18) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···O1W | 0.82 | 1.86 | 2.656 (2) | 163 |
| N2—H2A···N1i | 0.86 | 2.31 | 3.010 (2) | 139 |
| O1W—H1WA···O1ii | 0.85 | 2.14 | 2.976 (2) | 168 |
| O1W—H1WB···O1iii | 0.85 | 1.94 | 2.788 (2) | 173 |
Symmetry codes: (i) −x+1, y, −z+1/2; (ii) x, y+1, z; (iii) −x+1, −y+1, −z+1.
References
- Agilent (2013). CrysAlis PRO. Agilent Technologies, Yarnton, England.
- Dobosz, A., Dudarenko, N. M., Fritsky, I. O., Głowiak, T., Karaczyn, A., Kozłowski, H., Sliva, T. Yu. & Świątek-Kozłowska, J. (1999). J. Chem. Soc. Dalton Trans. pp. 743–750.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Golenya, I. A., Gumienna-Kontecka, E., Boyko, A. N., Haukka, M. & Fritsky, I. O. (2012). Inorg. Chem. 51, 6221–6227. [DOI] [PubMed]
- Golenya, I. A., Gumienna-Kontecka, E., Haukka, M., Korsun, O. M., Kalugin, O. N. & Fritsky, I. O. (2014). CrystEngComm, 16, 1904–1918.
- Golenya, I. A., Haukka, M., Fritsky, I. O. & Gumienna-Kontecka, E. (2007). Acta Cryst. E63, o1515–o1517.
- Griffith, D., Chopra, A., Müller-Bunz, H. & Marmion, C. (2008). Dalton Trans. 48, 6933–6939. [DOI] [PubMed]
- Groom, C. R. & Allen, F. H. (2014). Angew. Chem. Int. Ed. 53, 662–671. [DOI] [PubMed]
- Gumienna-Kontecka, E., Golenya, I. A., Dudarenko, N. M., Dobosz, A., Haukka, M., Fritsky, I. O. & Świątek-Kozłowska, J. (2007). New J. Chem. 31, 1798–1805.
- Gumienna-Kontecka, E., Golenya, I. A., Szebesczyk, A., Haukka, M., Krämer, R. & Fritsky, I. O. (2013). Inorg. Chem. 52, 7633–7644. [DOI] [PubMed]
- Hynes, J. B. (1970). J. Med. Chem. 13, 1235–1237. [DOI] [PubMed]
- Jankolovits, J., Kampf, J. W. & Pecoraro, V. L. (2013). Inorg. Chem. 52, 5063–5076. [DOI] [PubMed]
- Makhmudova, N. K., Kadyrova, Z. Ch., Del’yaridi, E. A. & Sharipov, Kh. T. (2001). Russ. J. Org. Chem. 37, 866–868.
- Marmion, C. J., Parker, J. P. & Nolan, K. B. (2013). Comprehensive Inorganic Chemistry II: From Elements to Applications, edited by J. Reedijk & K. Poeppelmeier, Vol. 3, pp. 684–708. Amsterdam: Elsevier.
- Pavlishchuk, A. V., Kolotilov, S. V., Zeller, M., Shvets, O. V., Fritsky, I. O., Lofland, S. E., Addison, A. W. & Hunter, A. D. (2011). Eur. J. Inorg. Chem. pp. 4826–4836.
- Pavlishchuk, A. V., Kolotilov, S. V., Zeller, M., Thompson, L. K., Fritsky, I. O., Addison, A. W. & Hunter, A. D. (2010). Eur. J. Inorg. Chem. pp. 4851–4858.
- Safyanova, I. S., Golenya, I. A., Pavlenko, V. A., Gumienna-Kontecka, E., Pekhnyo, V. I., Bon, V. V. & Fritsky, I. O. (2015). Z. Anorg. Allg. Chem. 641, 2326–2332.
- Seda, S. H., Janczak, J. & Lisowski, J. (2007). Eur. J. Inorg. Chem. pp. 3015–3022.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Stemmler, A. J., Kampf, J. W., Kirk, M. L., Atasi, B. H. & Pecoraro, V. L. (1999). Inorg. Chem. 38, 2807–2817. [DOI] [PubMed]
- Świątek-Kozłowska, J., Fritsky, I. O., Dobosz, A., Karaczyn, A., Dudarenko, N. M., Sliva, T. Yu., Gumienna-Kontecka, E. & Jerzykiewicz, L. (2000). J. Chem. Soc. Dalton Trans. pp. 4064–4068.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015024706/gk2650sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015024706/gk2650Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989015024706/gk2650Isup3.cml
CCDC reference: 1444026
Additional supporting information: crystallographic information; 3D view; checkCIF report




