Abstract
Background
MicroRNA (miRNA) is a small, non-coding RNA molecule which plays a role in the carcinogenesis and progression of cancers. Abnormal expression of miRNA in plasma has been found in some patients with malignant tumors.
Material/Methods
This study was conducted to investigate the expression of miRNA-30a in plasma of patients with non-small cell lung cancer (NSCLC). The plasma miRNA-30a in 87 patients with NSCLC, 20 patients with benign lung diseases, and 76 healthy subjects were measured by real-time PCR. The diagnostic value of miRNA-30a in NSCLC was evaluated via the ROC curve method.
Results
Plasma miRNA-30a level was significantly higher in the NSCLC group compared with benign control and healthy control groups (P<0.01). No statistically significant difference was found in the expression level of miRNA-30a among various clinical pathologic features in NSCLC. ROC curve analysis showed that the specificity and sensitivity cut-off points were at 61.0% and 84.3% for NSCLC. The specificity and sensitivity values were 54.9% and 94.4%, respectively, in the analysis based on in-patients only.
Conclusions
All these results suggest that plasma miRNA-30a measurement may be a novel and noninvasive method for NSCLC preliminary screening and differential diagnosis.
MeSH Keywords: Carcinoma, Non-Small-Cell Lung; MicroRNAs; Plasma
Background
MicroRNA (miRNA) is a small, non-coding RNA molecule found in eukaryotes, containing about 22 nucleotides, which plays a negative role in transcriptional and post-transcriptional regulation of gene expression [1]. Some studies concluded that over 30% of the proteins in the human body are regulated by miRNA via several mechanisms [2]. This indicates miRNA may be important regulatory factors in many diseases. Recent research has shown that miRNAs not only regulate various biological processes, but are also associated with the carcinogenesis and progression of diverse human cancers [3–5]. MiRNA-30a, a member of the miR-30 family, located at human chromosome 6q13, has 2 mature forms: miR-30a-3p and miRNA-30a-5p [6]. Increasing evidence confirms the close relationship between miRNA-30a expression level and numerous types of human cancers [7,8], but the exact role of miRNA-30a in cancers remains controversial. Some research suggests that miRNA-30a may act as a tumor suppressor, but other studies suggest it acts as an oncogene.
Lung cancer, a common malignant lung tumor, has become the major cause of cancer-related death worldwide [9]. It is divided into 2 broad classes: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC accounts for 80% of human lung cancer, and the 5-year survival rate is only 15%. Uncontrolled cell growth and multitudinous metastasis of lung cancer lead to high morbidity and mortality. Distinguishing the significant molecular alterations at the early stage of lung cancer contributes to early diagnosis, and thus to early treatment, which may have the potential to reduce the mortality of lung cancer. Currently, the diagnosis of lung cancer mainly depends on clinical symptoms, sputum cytology, immunological examinations, bronchoscopy, percutaneous needle lung biopsy, and many common tumor markers. Because lung cancer screening cannot normally be conducted in high-risk populations and in those lacking symptoms at the early stage, early diagnosis and timely treatment for lung cancer are still challenges for clinical practice.
Tumor markers are biomarkers found in various body fluids or body tissues, which can help detect the presence and development of tumors. Each tumor marker has a high diagnostic value in 1 or several specific tumor types, such as alpha fetoprotein (AFP) in hepatic carcinoma diagnosis. At present, the following tumor markers are used for lung cancer diagnosis and screening: cytokeratin 19 fragment (CYFRA21-1), carcinoembryonic antigen (CEA), neuron-specific enolase (NSE), pro-gastrin-releasing peptide (ProGRP), carbohydrate antigen 125 (CA125), and carbohydrate Antigen 199 (CA199). Due to insufficient sensitivity and specificity of these common tumor markers [10,11], new biomarkers with a high diagnostic value for lung cancer diagnosis are needed.
MiRNA-30a may be used as a tumor biomarker in lung cancer diagnosis, as some researches [7,8] found its expression was remarkably downregulated in lung cancer tissue. Because invasive examination or surgery must be performed to obtain the lung cancer tissue, it is almost impossible to use the miRNA-30a in the lung tissue as a biomarker for clinical lung cancer screening. Furthermore, after invasive examination or surgery, precise diagnostic results will be provided by pathological examination, while the biomarker from the tissue is not necessary for cancer diagnosis. Therefore, tumor markers in plasma or sputum have the advantage of convenience compared with those in the lung tissue. This study aimed to investigate the expression pattern of miRNA-30a in the plasma of NSCLC patients, and also to evaluate the potential of plasma miRNA-30a as a novel tumor biomarker in NSCLC diagnosis.
Material and methods
Patients
This study was approved by the Medical Ethics Committee of the West China Hospital, Sichuan University. Three patients with lung cancers who had undergone surgical resection in the Department of Thoracic Surgery at West China Hospital, Sichuan University, were included in array analysis. All the patients were males with a history of cigarette smoking. The average age was 59±5.57 years. All the tissues were confirmed as adenocarcinoma with TNM stage I and II. Tumor tissues and para-carcinoma normal tissues were collected after the resection of lung tissue. Samples were stored at −80°C until use.
A total of 99 plasma samples were collected from lung cancer patients who underwent surgical resection with histologically verified lung cancer in the Department of Thoracic Surgery at West China Hospital, Sichuan University between 2012 and 2014. The patients consisted of 68 males and 31 females and the mean age at surgery was 60.73±9.42 years. Pathological types included 52 cases of adenocarcinoma, 35 cases of squamous cell carcinoma (SCC), 6 cases of unclassified cancers, and the other 6 cases were small cell lung cancer (SCLC). According to the 7th Lung Cancer TNM Classification and Staging System, revised by the International Union Against Cancer (UICC) in 2009, intra-operative and postoperative pathology indicated 56 cases with stages I and II and 37 with stages III and IV cancer. Twenty patients (mean age 62.3±8.78, 11 males, 9 females) with benign lung nodules, such as tuberculosis, inflammatory pseudotumor, pulmonary cystitis and aspergillosis, verified by pathology, served as benign controls. Seventy-six healthy persons without obvious disease who had health examinations in our hospital were also included as healthy controls, with a mean age of 51.5±8.26 years. There were 47 males and 29 females in the healthy control group. All plasma samples of controls were collected during the same period.
Plasma specimen collection
To avoid the impacts of diet, physiological activities, and physical activities on the sample, all blood specimens were collected between 7 and 8 AM after fasting for at least 8 h. We collected 3–5 ml of venous blood into an EDTA-anticoagulant tube (BD, Franklin Lakes, NJ, USA), and processed by centrifugation at 2000 rpm for 8 min. Plasma was transferred to a fresh tube (Axygen Scientific, Union City, CA, USA) and stored at −80°C until use.
RNA extraction and reverse transcriptase
Total RNA was extracted from plasma using the Trizol reagent (Invitrogen, Carlsbad, CA, USA), and then the RNA pellet was dissolved in 30 ul of RNAase free water and stored at −80°C for use. The purity and concentration of RNA was determined by spectrophotometer. Reverse transcription was conducted with Thermo Scientific Revert Aid Reverse kit (Thermo, Waltham, MA, US) and cDNA was collected after the transcription. Total RNA of lung tissue was extracted using Trizol (Invitrogen) and RNeasy mini kit (QIAGEN) according to the manufacturer’s instructions.
miRNA array
MiRCURY LNA™ microRNA Arrays (v.10.0, Exiqon) were used in array analysis. The samples were labeled using the miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon) and hybridized on the miRCURY™ LNA Array (Exiqon). Scanning was performed with the Axon GenePix 4000B microarray scanner. GenePix pro software (V6.0) was used to read the raw intensity of the image. The intensity of the green signal was calculated after background subtraction and 4 replicated spots of each probe on the same slide were averaged. The Median Normalization Method was used to obtain Normalized Data and Normalized Data=(Foreground-Background)/median. Unsupervised hierarchical clustering and correlation analysis were performed on the miRNA expression profiling. The software produced the scatter plot and the corresponding correlation R-value.
Quantitative Real-Time PCR
Quantitative real-time PCR (qRT-PCR) was performed on the IQ5 Real-time PCR system (Bio-Rad, Hercules, CA, USA) under the following conditions: incubated at 95°C for 3 min followed by 40 cycles of 10 s at 95°C, 15 s at 59°C, and 72°C for 10 s. The relative transcript levels of miRNA-30a were calculated using the 2−ΔCT method. U6-snRNA acted as an external control, and all primers were synthesized by RiboBio Co. (Guangzhou, China).
Statistical analysis
Data analyses were performed using SPSS version 19.0 (SPSS, Inc., Chicago, IL). The data are expressed as means±SEMs. ANOVA, Kruskal-Wallis Test, multiple linear regression analysis, and the unpaired t test were used to distinguish the differences between groups. P<0.05 was considered as a statistically significant difference with 2-tailed tests. ROC curve was used to analyze the diagnosis value, including AUC, cut-off point, sensitivity, and specificity.
Results
Array results
To investigate the different expression of miRNA in lung cancer tissues, the miRNA expression profile was analyzed with array in 3 lung adenocarcinoma patients. Six miRNAs were downregulated in tumor tissue compared with para-carcinoma normal tissues (Table 1, p<0.05). No miRNA was found significantly upregulated in tumor tissue compared with para-carcinoma normal tissues. Correlation coefficient matrix analysis showed that most of the miRNA expression levels remained almost the same in tumor tissues compared with para-carcinoma normal tissues (Table 2). This was further demonstrated by the Heat Map and Unsupervised Hierarchical Clustering (Figure 1).
Table 1.
The results of miRNA expression levels in tumor tissue and para-carcinoma normal tissues. Six miRNAs were down regulated in lung cancer tissue compared with para-carcinoma normal tissues. And no miRNAs were up regulated.
| Down Regulated microRNAs | ||
|---|---|---|
| Name | t/p average | P-value |
| Has-miR-30a | 0.3334 | 0.0389 |
| hsa-miR-320 | 0.5935 | 0.1033 |
| miRPlus_42487 | 0.2267 | 0.0143 |
| miRPlus_42526 | 0.4296 | 0.0370 |
| hsa-miR-193a-5p | 0.6579 | 0.0548 |
| ebv-miR-BART8* | 0.5623 | 0.1419 |
| hsa-miR-638 | 0.5363 | 0.0729 |
| hsa-miR-129-5p | 0.3460 | 0.0205 |
| miRPlus_42745 | 0.6315 | 0.0354 |
| hsa-miR-801 | 0.6073 | 0.2302 |
| ebv- -BHRF1-2 | 0.3991 | 0.0417 |
Table 2.
Correlation coefficient matrix analysis of miRNA expression in lung cancer tissues and para-carcinoma normal tissues. Most miRNA expression levels in tumor tissues remained the same compared with para-carcinoma normal tissues.
| Correlation coefficient matrix | ||||||
|---|---|---|---|---|---|---|
| 016–0807p | 016–0807t | 017–0807p | 017–0807t | 018–0807p | 018–0807t | |
| 016–0807p | 1.0000 | 0.6673 | / | / | / | / |
| 016–0807t | 0.6673 | 1.0000 | / | / | / | / |
| 017–0807p | / | / | 1.0000 | 0.7758 | / | / |
| 017–0807t | / | / | 0.7758 | 1.0000 | / | / |
| 018–0807p | / | / | / | / | 1.0000 | 0.9664 |
| 018–0807t | / | / | / | / | 0.9664 | 1.0000 |
Figure 1.
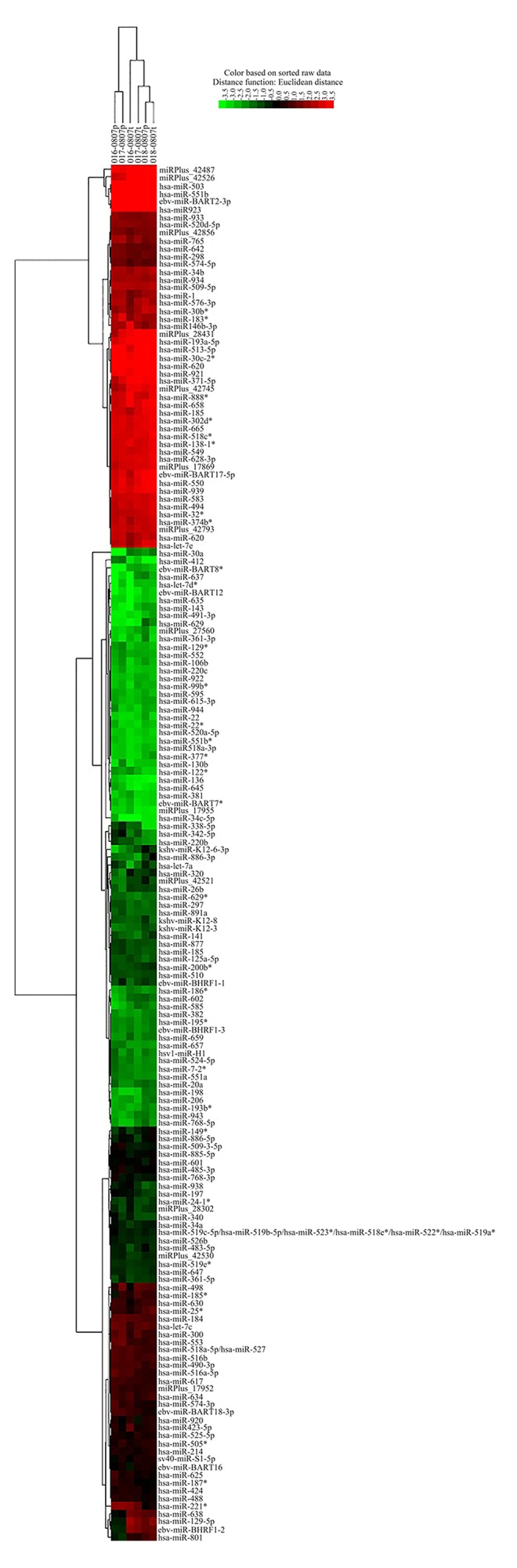
Heat map and unsupervised hierarchical clustering.
Plasma miRNA-30a in three groups
As miRNA-30a was one of the most downregulated miRNAs in our array data, we investigated the expression levels of miRNA-30a in plasmas of NSCLC patients using qRT-PCR, compared with benign controls and healthy controls. To our surprise, the level of miRNA-30a was significantly increased in the NSCLC group compared to the benign and healthy control groups (P<0.01, Figure 2A).
Figure 2.
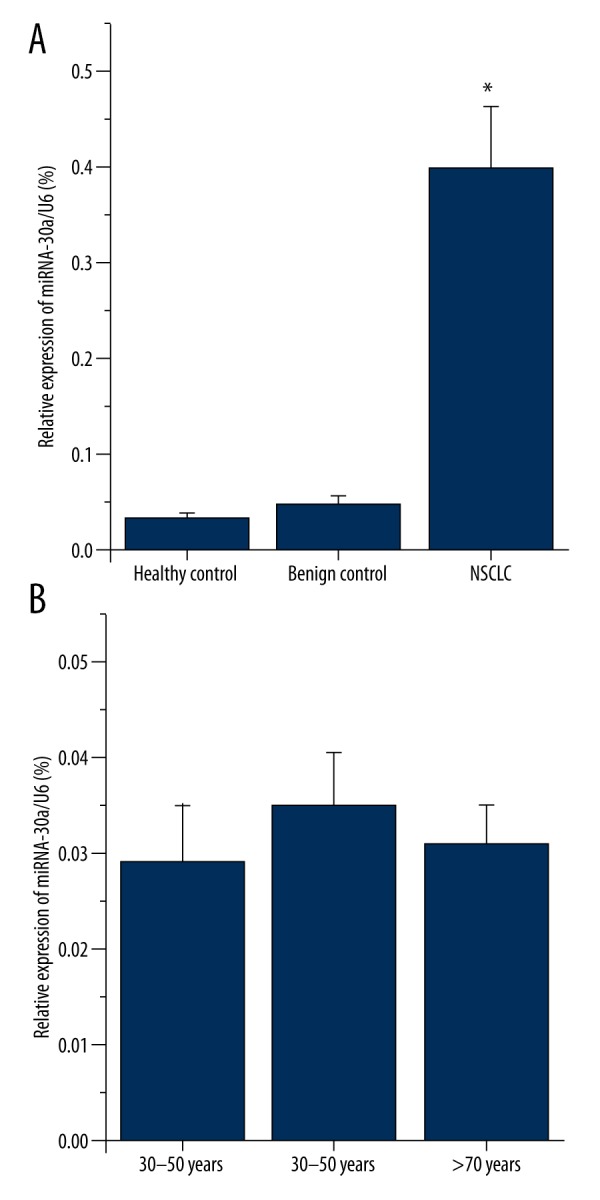
Plasma miRNA-30a relative expression level in three groups. (A) miRNA-30a were up-regulated in NSCLC group, as detected by qRT-PCR (n=76 in healthy control, n=20 in benign control, and n=87 in NSCLC group). (B)There was no significant difference in miRNA-30a expression level between three age groups of healthy control (n=36 of 30-50 years, n=31 of 51-79 years, n=9 of over 70 years, P>0.05, determined by Student-Newman-Keuls test). Graph represents means ±SEMs. * P<0.01, versus control, values determined by the Student t test.
We found that there was a significant difference in age between the healthy controls and the other 2 groups (P<0.05) during the data analysis. To evaluate if the age impacted our result authenticity, we grouped the healthy controls to 3 age grades (30–50 years, 51–70 years, and >70 years) and further analyzed the data based on the age groups. As shown in Figure 2B, there is no significant difference in the plasma miRNA-30a between these 3 age groups (Figure 2B). We also compared plasma miRNA-30a level in the control included subjects who had the age matched to the patients group and higher plasma mrRNA-30a also found in lung cancer group (data not shown).
The relationship between miRNA-30a expression and histological types
All lung cancer patients except unclassified cases were divided into 3 groups according to histological type: lung adenocarcinoma, lung squamous cell cancer (SCC), and small cell lung cancer (SCLC). There were no significant differences in the plasma miRNA-30a levels among these 3 groups (Figure 3).
Figure 3.
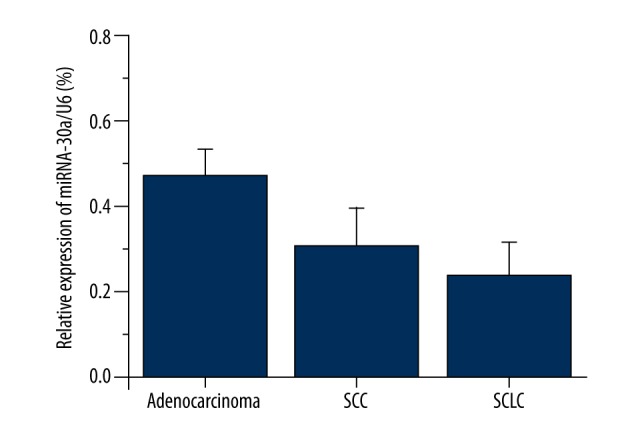
PlasmamiRNA-30a levels in different histological types. Plasma miRNA-30a expression was measured via qRT-PCR (n=52 of adenocarcinoma, n=35 of SCC, n=6 of SCLC), data represent means ±SEMs. No Statistical difference was found among three groups (P>0.05, using the Student-Newman-Keuls test). SCC – lung squamous cell cancer, SCLC – small cell lung cancer.
The correlation between plasma miRNA-30a and NSCLC clinicopathologic features
To investigate the possible relationships between plasma miRNA-30a levels and NSCLC clinicopathologic features, we used multiple linear regression analysis to analyze the data based on the following factors: age, gender, smoking history, pathological type, TNM classification, lymph node metastasis, and distant metastasis (the assignment of independent variable is shown in Table 3). Then, as shown in Table 4, no statistically significant correlation was found between miRNA-30a levels and these characteristics.
Table 3.
Independent variable assignment.
| Independent variable | Assignment |
|---|---|
| Age | Raw value |
| Gender | Male=1, Female=2 |
| Smoking history | No=1, Yes=2 |
| Pathological type | Adenocarcinoma=1, SCC=2, SCLC=3 |
| TNM classification | I=1, II=2, III=3, IV=4 |
| Lymph node metastasis | N0=1, N1–3=2 |
| Distant metastasis | M0=1, M1=2 |
Table 4.
The results of multiple linear regression analysis.
| Independent variable | B | t value | P value |
|---|---|---|---|
| Age | 0.03 | 0.39 | 0.698 |
| Gender | 0.071 | 0.452 | 0.693 |
| Smoking history | 0.251 | 1.768 | 0.083 |
| Pathological type | −0.241 | −1.755 | 0.085 |
| TNM classification | −0.145 | −1.127 | 0.264 |
| Lymph node metastasis | 0.046 | 1.794 | 0.078 |
| Distant metastasis | 0.031 | 0.097 | 0.923 |
F=1.884; p=0.097; R2=0.554; B – unstandardized coefficients.
Diagnostic value analysis of miRNA-30a levels using Receiver Operating Characteristic curves (ROC curve)
Pathologic result, as the criterion standard for diagnosis, was used to evaluate the value of miRNA-30a level as the test variable in the diagnosis of NSCLC. ROC curve analysis showed that the Area Under the Curve (AUC) of ROC was 0.727, indicating the miRNA-30a level may be a valuable index in diagnosing NSCLC (Figure 4).
Figure 4.
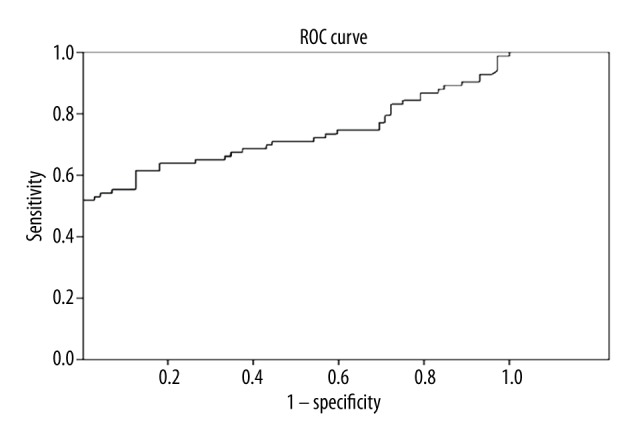
Receiver operating characteristic (ROC) curve for miRNA-30a. The area under the ROC was 72.7% (Standard error was 4.2%, 95% confidence interval was 64.5–81.0%)
Diagnosis cut-off point
After analyzing the ROC curve of miRNA-30a expression, the top Youden index was selected as the cut-off point for NSCLC diagnosis [12], depending on the sensitivities and specificities of different cut-off points. As indicated in Table 5, the best diagnostic cut-off value of miRNA-30a levels in plasma stood at 0.061 (sensitivity and specificity were 61.0 and 84.3%, respectively).
Table 5.
Diagnosis cut-off point for NSCLC.
| Test variable | Cut-off | TPR | TNR | LR+ | LR− | YI |
|---|---|---|---|---|---|---|
| miRNA-30a Level | 0.061 | 61.0% | 84.3% | 3.89 | 0.46 | 45.3% |
Cut-off – cut-off point; TPR – true positive rate (sensitivity); TNR – true negative rate (specificity); LR+ – positive likelihood ratio; LR− – negative likelihood ratio; YI – Youden index.
The results based on inpatients only
The data above (Figure 4, Table 5) were calculated with both the benign control group and the healthy control group. In fact, tumor biomarker tests are applied not only to screening asymptomatic and high-risk populations, but also to differential diagnosis for lung cancer. Although there was no statistically significant difference in miRNA-30a expression levels between the healthy group and the benign group, we re-analyzed the data excluding the healthy group. The results may be more useful for use in clinical practice. The best diagnostic cut-off value was 0.095 with 54.9% and 94.4% sensitivity and specificity, respectively (Figure 5, Table 6).
Figure 5.
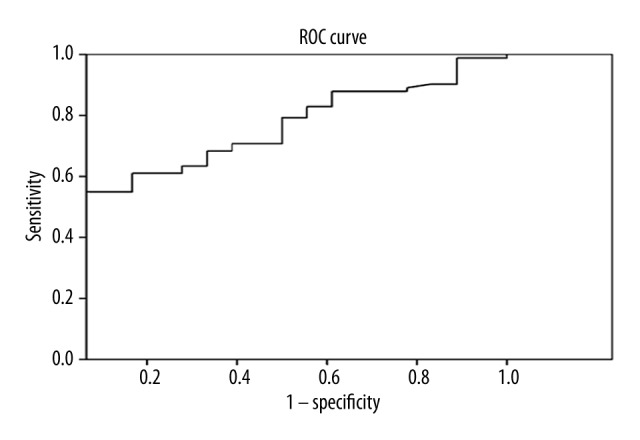
ROC curve of the diagnostic value of miRNA-30a in NSCLC based on in-patients only. The area under the ROC was 72.7% (Standard error was 6.2%, 95% confidence interval 60.4–84.9%)
Table 6.
Diagnosis cut-off point for NSCLC base on in-patients only.
| Test variable | Cut-off | TPR | TNR | LR+ | LR− | YI |
|---|---|---|---|---|---|---|
| miRNA-30a Level | 0.095 | 54.9% | 94.4% | 9.80 | 0.48 | 49.3% |
Cut-off – cut-off point; TPR – true positive rate (sensitivity); TNR – true negative rate (specificity); LR+ – positive likelihood ratio; LR− – negative likelihood ratio; YI – Youden index.
Discussion
Although several new tumor biomarkers have been studied recently [13], biomarkers from minor invasive procedures should be more useful in the screening and early diagnosis of lung cancer. In this study, we first investigated the expression profile of miRNA in lung cancer tissue with arrays. Array data suggested that miRNA-30a level was downregulated in lung cancer tissues compared with para-carcinoma normal tissue. Then, we measured this miRNA in the plasma. To our surprise, the average level of miRNA-30a was dramatically increased in plasma of NSCLC patients compared with both the benign control group and the healthy group (P<0.01). This suggests that miRNA-30a may be used as a biomarker in NSCLC diagnosis. We are unsure whether the sample size influenced the difference of plasma miRNA-30a level between the benign and healthy groups (P>0.05). The average level of miRNA-30a in the benign group was higher than that in the healthy group, implying miRNA-30a may play a role in various pulmonary diseases. In addition, no significant correlation was found between the miRNA-30a level and NSCLC clinicopathologic features, including histological type, age, gender, smoking history, TNM classification, lymph node metastasis, distant metastasis, and clinical stage.
Baffa et al. [14] found that subtypes of the miRNA-30 family (miRNA-30a, 30d, and 30e) were remarkably downregulated in metastatic cancerous tissues, and similar results were reported in the following tumor tissues: kidney [15], lung [16,17], leukemia [18], and breast [19]. Furthermore, miRNA-30a was a markedly and frequently decreased miRNA compared to other members of the miRNA-30 family [14,20,21]. Those phenomena imply that miRNA-30a may restrain the generation and progress of tumors. At present, most authors consider miRNA-30a as a tumor suppressor, inhibiting the malignant phenotype of tumors [8,18,20,21], including lung cancer.
Although the mechanism of miRNA-30a as a tumor suppressor in lung cancer is still not entirely clear, 2 possibilities have been suggested. Wu et al. [22] found that the miRNA-30 family negatively regulates the expression of Ubc9 (Ubiquitin Conjugating Enzyme), the sole E2-conjugating enzyme for SUMOylation, which has been proven to possess an oncogenic function, and then inhibit its carcinogenesis in lung cancer. Kumarswamy et al. [8] found miRNA-30a targets the 3′-UTR of Snai1 and reduces its expression, up-regulates the E-cadherin (epithelial marker) expression, and correlates positively with N-cadherin (mesenchymal marker) expression, resulting in a decrease of the EMT (epithelial to mesenchymal transition), finally inhibiting invasion and metastasis of NSCLC. Due to its function of carcinostasis, the decrease of miRNA-30a may lead to tumorigenesis in NSCLC. Thus, significantly downregulated miRNA-30a in cancer tissues compared with para-carcinoma normal tissue may promote NSCLC.
In addition to the mechanisms described above, unknown mechanisms may affect the interaction between NSCLC and miRNA-30a. In this study we measured the plasma miRNA-30a in 3 different groups (NSCLC group, benign control, and healthy control). Unexpectedly, we found plasma miRNA-30a expression level increased significantly in NSCLC patients compared with benign and healthy controls (P<0.01). This suggests to us that miRNA-30a may be involved in NSCLC via unknown mechanisms outside the lung. It is possible that NSCLC follows the abnormal downregulation of miRNA-30a in lung tissue, leading to the increase of miRNA-30a in plasma to restrain the metastasis of existing lung cancer in some way. The role of increased plasma miRNA-30a in NSCLC needs to be investigated further.
Further analysis was performed to evaluate the diagnostic value of miRNA-30a via ROC curve [23]. According to the clinical requirements, we analyzed the data with the following 2 conditions: including and excluding the healthy group. The AUC of ROC in the 2 conditions was similar, at about 72.2%, which shows miRNA-30a expression level has a moderate accuracy in NSCLC diagnosis. We then compared the cut-off value-related data for the 2 conditions, and a high specificity (94.4%) was observed in the inpatient group, indicating miRNA-30a testing may be a potential candidate in lung cancer differential diagnosis for inpatients. Many studies [24–31] have revealed a positive correlation between levels of the most common tumor markers (such as CEA, CYFRA21-1, NSE, and SCC) and clinical stage of NSCLC patients. These may indicate false-negative results in early diagnosis, causing insufficient detection or diagnosis at an early stage. On the contrary, plasma miRNA-30a expression has a higher stability and less interference compared with traditional tumor markers, because no significant difference was found in patients with different clinical and pathological characteristics. This implies miRNA-30a may be helpful for preliminary NSCLC screening. As Figure 3 shows, trends in mean plasma miRNA-30a expression in the 3 lung cancer pathological types were different (adenocarcinoma >SCC>SCLC), although no statistically significant difference was found among the 3 types of pathology in our study. It is difficult to completely rule out the influence of sample size on this result. Further study should be conducted to investigate the role of miRNA-30a in the diagnosis of different pathology types of lung cancer, as other tumor markers do [24–26].
The sample sizes of the 3 groups are imbalanced in this pilot study. Fewer patients in some groups resulted in difficulty for further analysis. Also, we analyzed the diagnostic value based on NSCLC only, because of the insufficient sample size of the SCLC group. Further study with sufficient and balanced samples is required to evaluate the differential diagnosis and prognosis for NSCLC.
Conclusions
In summary, all these results show that plasma miRNA-30a measurement may be a novel and noninvasive method for preliminary NSCLC screening and differential diagnosis. miRNA-30a may be a useful bio-marker in NSCLC diagnosis.
Footnotes
Source of support: National Natural Science Foundation of China, 30871118, 30971325, 81270129, 81470268 (FL)
Conflict of interest
None.
References
- 1.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Reviews Genet. 2007;8(2):93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 2.Berezikov E, van Tetering G, Verheul M, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16(10):1289–98. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 4.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–50. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Shell S, Park SM, Radjabi AR, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104(27):11400–5. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103(7):2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumarswamy R, Mudduluru G, Ceppi P, et al. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer. 2012;130(9):2044–53. doi: 10.1002/ijc.26218. [DOI] [PubMed] [Google Scholar]
- 9.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CancerJ Clin. 2006;56(2):106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 10.Cedres S, Nunez I, Longo M, Martinez P, et al. Serum tumor markers CEA, CYFRA21-1, and CA-125 are associated with worse prognosis in advanced non-small-cell lung cancer (NSCLC) Clin Lung Cancer. 2011;12(3):172–79. doi: 10.1016/j.cllc.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 11.Yu D, Du K, Liu T, Chen G. Prognostic value of tumor markers, NSE, CA125 and SCC, in operable NSCLC Patients. Int J Mol Sci. 2013;14(6):11145–56. doi: 10.3390/ijms140611145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bantis LE, Nakas CT, Reiser B. Construction of confidence regions in the ROC space after the estimation of the optimal Youden index-based cut-off point. Biometrics. 2014;70(1):212–23. doi: 10.1111/biom.12107. [DOI] [PubMed] [Google Scholar]
- 13.Adam J, Planchard D, Marabelle A, et al. PD-L1 expression: An emerging biomarker in non-small cell lung cancer. Ann Pathol. 2016;36(1):94–102. doi: 10.1016/j.annpat.2015.11.004. [in French] [DOI] [PubMed] [Google Scholar]
- 14.Baffa R, Fassan M, Volinia S, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219(2):214–21. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 15.Heinzelmann J, Henning B, Sanjmyatav J, et al. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J Urol. 2011;29(3):367–73. doi: 10.1007/s00345-010-0633-4. [DOI] [PubMed] [Google Scholar]
- 16.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Chen YY, Li SQ, et al. Expression of miR-148/152 family as potential biomarkers in non-small-cell lung cancer. Med Sci Monit. 2015;21:1155–61. doi: 10.12659/MSM.892940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon-McIver A, East P, Mein CA, et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One. 2008;3(5):e2141. doi: 10.1371/journal.pone.0002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan LX, Huang XF, Shao Q, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14(11):2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47(3):897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 21.Yu Y, Cao L, Yang L, et al. microRNA 30A promotes autophagy in response to cancer therapy. Autophagy. 2012;8(5):853–55. doi: 10.4161/auto.20053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu F, Zhu S, Ding Y, et al. MicroRNA-mediated regulation of Ubc9 expression in cancer cells. Clin Cancer Res. 2009;15(5):1550–57. doi: 10.1158/1078-0432.CCR-08-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moses LE, Shapiro D, Littenberg B. Combining independent studies of a diagnostic-test into a summary roc curve – data-analytic approaches and some additional considerations. Stat Med. 1993;12(14):1293–316. doi: 10.1002/sim.4780121403. [DOI] [PubMed] [Google Scholar]
- 24.Molina R, Filella X, Auge JM, et al. Tumor markers (CEA, CA 125, CYFRA 21-1, SCC and NSE) in patients with non-small cell lung cancer as an aid in histological diagnosis and prognosis. Comparison with the main clinical and pathological prognostic factors. Tumour Biol. 2003;24(4):209–18. doi: 10.1159/000074432. [DOI] [PubMed] [Google Scholar]
- 25.Okada M, Sakamoto T, Nishio W, et al. Characteristics and prognosis of patients after resection of nonsmall cell lung carcinoma measuring 2 cm or less in greatest dimension. Cancer. 2003;98(3):535–41. doi: 10.1002/cncr.11530. [DOI] [PubMed] [Google Scholar]
- 26.Barak V, Goike H, Panaretakis KW, Einarsson R. Clinical utility of cytokeratins as tumor markers. Clin Biochem. 2004;37(7):529–40. doi: 10.1016/j.clinbiochem.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Fluss R, Faraggi D, Reiser B. Estimation of the Youden Index and its associated cutoff point. Biom J. 2005;47(4):458–72. doi: 10.1002/bimj.200410135. [DOI] [PubMed] [Google Scholar]
- 28.Stieber P, Hatz R, Holdenrieder S, et al. National Academy of Clinical Biochemistry (NACB) guidelines for the use of tumor markers in lung cancer. Tumor Biol. 2006:27. [Google Scholar]
- 29.Alm El-Din MA, Farouk G, Nagy H, et al. Cytokeratin-19 fragments, nucleosomes and neuron-specific enolase as early measures of chemotherapy response in non-small cell lung cancer. Int J Biol Markers. 2012;27(2):e139–46. doi: 10.5301/JBM.2012.9141. [DOI] [PubMed] [Google Scholar]
- 30.Lin XF, Wang XD, Sun DQ, et al. High serum CEA and CYFRA21-1 levels after a two-cycle adjuvant chemotherapy for NSCLC: Possible poor prognostic factors. Cancer Biol Med. 2012;9(4):270–73. doi: 10.7497/j.issn.2095-3941.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S, Lee CY, Kim DJ, et al. Pathologic correlation of serum carcinoembryonic antigen and cytokeratin 19 fragment in resected nonsmall cell lung cancer. Korean J Thoracic Cardiovasc Surg. 2013;46(3):192–96. doi: 10.5090/kjtcs.2013.46.3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]


