Summary
We characterize the interaction of RecA with membranes in vivo and in vitro and demonstrate that RecA binds tightly to the anionic phospholipids (aPLs) cardiolipin (CL) and phosphatidylglycerol (PG). Using computational models, we identify two regions of RecA that interact with PG and CL: 1) the N-terminal helix and 2) loop L2. Mutating these regions decreased the affinity of RecA to PG and CL in vitro. Using 3D-super-resolution microscopy (3D-SIM), we demonstrate that depleting Escherichia coli PG and CL altered the localization of RecA foci and hindered the formation of RecA filament bundles. In consequence, E. coli cells lacking aPLs fail to initiate a robust SOS response after DNA damage, indicating that the membrane acts as a scaffold for nucleating the formation of RecA filament bundles and plays an important role in the SOS response.
Introduction
RecA repairs DNA damage by performing homologous recombination and is the primary mediator of the DNA damage (SOS) response in Escherichia coli. RecA binds ATP, polymerizes on exposed, single-stranded DNA (ssDNA), and creates nucleoprotein filaments that facilitate DNA repair through a mechanism that entails homology searching and matching of complementary strands. RecA nucleoprotein filaments subsequently cleave the LexA repressor and initiate the SOS response leading to the downstream transcription of ~40 genes that facilitate DNA repair. After the process of DNA repair is complete, RecA filaments hydrolyze ATP and dissociate from DNA.
A translational fusion of green fluorescent protein (GFP) to the C-terminus of RecA (RecA-GFP) localizes into polar foci in E. coli (Renzette et al., 2005). These foci, referred to as ‘storage structures’, are reminiscent of the RecA aggregates observed in vitro (Brenner et al., 1988; Story et al., 1992). These foci do not form in RecA mutants unable to bind ATP, highlighting the importance of the ATP-bound form of RecA in the stability of these aggregates and mirroring observations from in vitro studies (Brenner et al., 1988; Egelman and Stasiak, 1988). When DNA is damaged, RecA-GFP redistributes from foci into long filament-like structures that span the chromosome and are referred to as bundles (Lesterlin et al., 2014; Renzette and Sandler, 2008). Lesterlin et al., demonstrated that these laterally associated bundles of RecA filaments that extend from the site of a double-stranded DNA break (DSB), assemble along the length of the cell in a step that appears to be connected to the search for DNA sequence homology. These bundles have been observed in vivo in E. coli by electron microscopy (Levin-Zaidman et al., 2000). Interestingly, the majority of RecA protein within these bundles (hereafter referred to as ‘filament bundles’) appears to not be associated with DNA (Lesterlin et al., 2014). They are occluded from the nucleoid and positioned close to the inner membrane (IM), suggesting the possibility of membrane-RecA interactions during DNA repair. The association of RecA with membranes during SOS was reported nearly three decades ago in E. coli lysates (Garvey et al., 1985), and yet very little is known about this interaction (Papanastasiou et al., 2013), its relevance to RecA location and function, and its role in cell physiology.
In this study, we test the hypothesis that the interaction of RecA with membranes is important for its activity in vivo. We use experimental and computational methods to characterize the binding of RecA to anionic phospholipids (aPLs), identify the RecA interfaces responsible, and demonstrate that this interaction decreases the ATPase activity of RecA in vitro. Further, we observe that RecA is mislocalized in E. coli mutants unable to produce aPLs and the SOS response in these mutants is significantly reduced. We show that the formation of RecA filament bundles is decreased in mutants lacking aPLs and this impairment in filament bundle formation hinders the progress of the SOS response. Our results indicate that aPLs stabilize polarly localized ‘storage forms’ of RecA, and also provide a scaffold for the formation of RecA filament bundles that facilitate the SOS response after DNA damage. These observations provide a context for the physiological significance of RecA-membrane interactions in E. coli and highlight an important role of the IM in the cellular response to DNA damage.
Results
RecA binds aPLs in vitro
To determine the affinity of RecA to PLs, we purified recombinant RecA from E. coli and measured the affinity of RecA to zwitterionic PLs (phosphatidylethanolamine [PE], phosphatidylcholine [PC]) and aPLs (phosphatidylglycerol [PG], cardiolipin [CL]) using liposome co-floatation assays. We quantified the amount of E. coli RecA that floats with liposomes and found that RecA displays a specific and high affinity interaction with the aPLs, PG and CL (Fig 1A–D) and a weaker association with PE (all found in E. coli IM), but no association with PC (not found in E. coli IM).
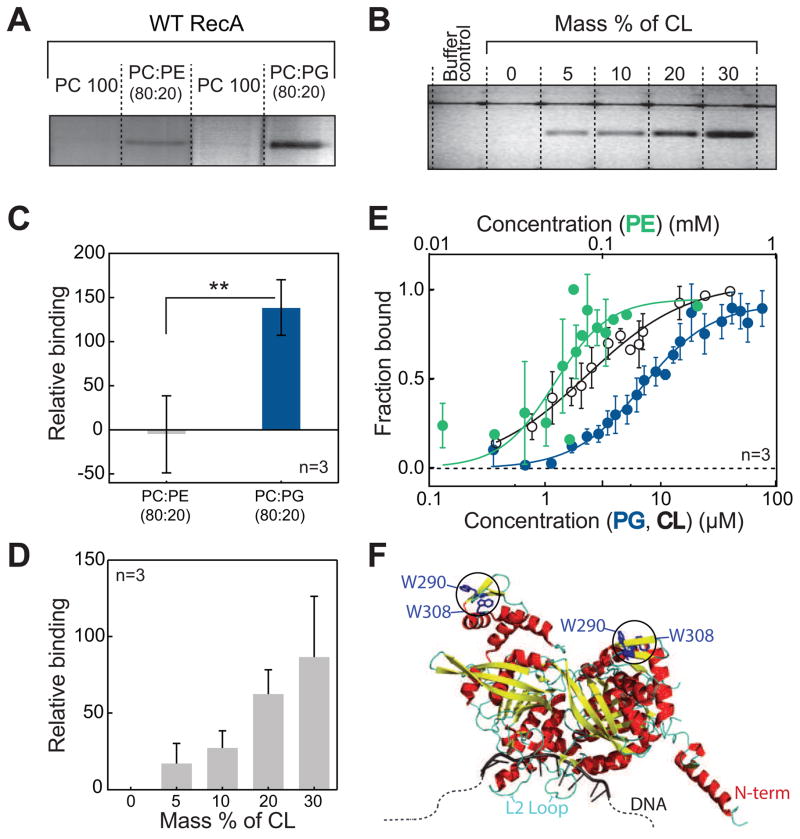
Figure 1. E. coli RecA interacts specifically and with high affinity to anionic phospholipids.
(A and C) Co-flotation assays of RecA with PE and PG demonstrate that RecA binds to anionic PLs with high affinity. ** p<0.05 indicates statistical significance calculated by a two-tailed t test. (B and D) The binding of RecA with CL increases proportionally with increasing CL concentration. (E) Langmuir binding isotherms of RecA with PE, PG, and CL demonstrate binding parameters as follows: PE (Kd = 52.9 ± 8.6 μM, Hill coefficient = 2.8 ± 1.1 and R2=0.7), PG (Kd = 6.9 ± 1.2 μM, Hill coefficient = 1.5 ± 0.3 and R2 = 0.93), and CL (Kd = 2.4 ± 0.4 μM, Hill coefficient = 1.0 ± 0.1 and R2 = 0.91). (F) Crystal structure (PDB ID 3CMU) of a dimer of RecA showing the position of the two Trp residues used to quantify binding affinities. All error bars indicate SD.
We quantitatively determined apparent dissociation constants (Kds) for PG, CL, and PE by utilizing the inherent fluorescence of tryptophans (Trp) at positions 290 and 309 in RecA (Fig 1F). The fluorescence intensity and wavelength of maximum emission of Trp residues reflect changes in the chemical microenvironment within proteins (Vivian and Callis, 2001) that may arise in response to protein-membrane binding events and can be used to quantify these interactions. We found the interaction of RecA with PG, CL and PE-containing liposomes is coupled to a change in the environment around the Trps that alters the intensity of their fluorescence emission. By titrating in different concentrations of aPLs and fitting the change in Trp fluorescence intensity to a Langmuir isotherm, we determined apparent Kds for RecA binding to PE (53 μM), PG (7 μM) and CL (2.5 μM) (Fig 1E). Remarkably, the values for PG and CL are within an order of magnitude of the Kd of RecA to ssDNA (0.8–2.5 μM) reported by others (Britt et al., 2011; Menetski and Kowalczykowski, 1985) and measured by us (0.7 μM; Fig S3B, D). This high affinity interaction of RecA to PG and CL suggests that RecA may spend a significant proportion of time on the IM when not bound to ssDNA.
Molecular dynamics simulations identify key membrane-binding interfaces in RecA
To predict the binding orientation, depth of insertion, and affinity of RecA to the membrane, we combined implicit and explicit membrane simulations. Explicit membrane simulations describe protein/solvent/lipid interactions at an atomistic level and are generally more accurate; however, these simulations are too computationally expensive for binding free energy calculations that involve a relatively large protein such as RecA and are not suited for identifying the residues/motifs that are important in membrane binding. Our strategy was to start with implicit membrane simulations and test the predicted binding conformations further using explicit membrane simulations. Implicit membrane simulations can be used to compute semi-quantitative binding free energies and predict the effect of mutations on binding, which can be tested experimentally.
We collected a binding energy map (Fig 2A) using GBSW-GCS and a non-flexible RecA protein and found two low energy configurations. This data is qualitatively consistent with the two binding conformations obtained from flexible MD simulations (Fig 2C, D). The binding interfaces of the two binding conformations (Fig 2B) are largely consistent and involve inserting the largely hydrophobic DNA binding loop L2 (I195 to N205) and the N-terminal α-helix, which contains both hydrophobic and charged residues (AIDENKQKAL- ‘N10’) into the membrane. Therefore, RecA binding to the anionic membrane involves the interplay of both electrostatic interactions and hydrophobic insertion, which was confirmed further by explicit membrane simulations (Fig S1).
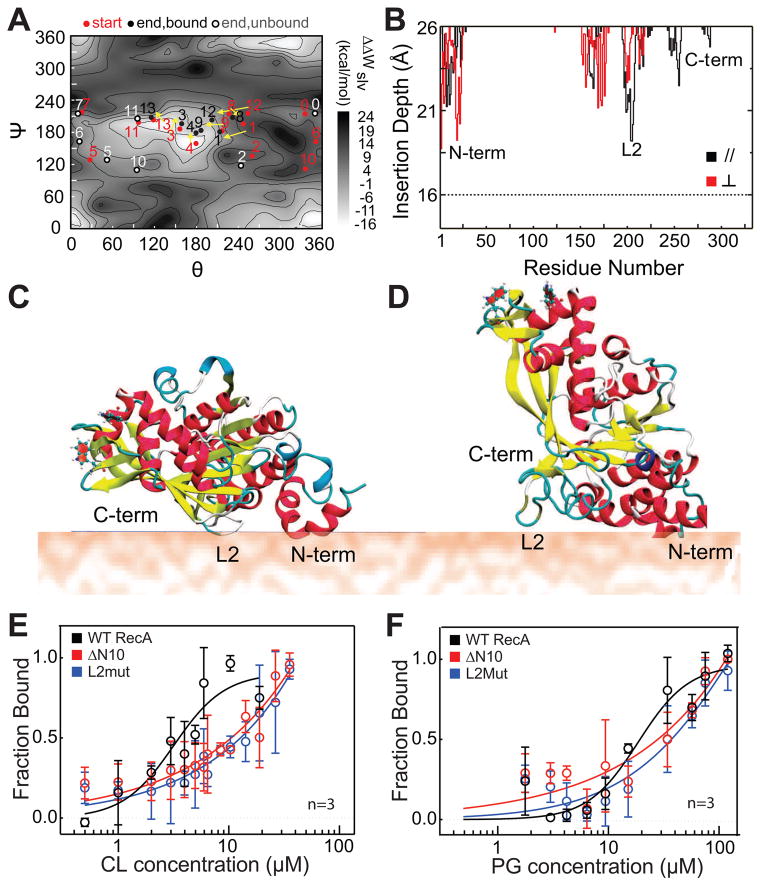
Figure 2. L2 and N10 regions form the binding interface of RecA with anionic PLs.
(A) 14 different trajectories are shown in dots (starting points are red, and ending points are black if RecA is bound and grey if RecA is unbound) on a solvation PMF (ΔWslv) scan map of the interaction energy between RecA and the membrane. Trajectories of RecA- bound to the membrane are connected with arrows. (B) A plot of the average insertion depth of RecA into the membrane calculated from the two typical binding trajectories observed. The insertion depth for the N-terminus, DNA binding loop L2, and C- terminus regions of RecA are labeled. (C) A cartoon depicting the structure of the membrane-bound orientation observed in trajectory 4 (shown in panel A), denoted as the parallel ‘//’ orientation. (D) A cartoon depicting the structure of the membrane- bound orientation observed in trajectory 13, denoted as the perpendicular ‘ ⊥ ‘ orientation. (E and F) Langmuir binding isotherms of N10 and L2 mutants of RecA compared to WT. The N10 and L2 regions of RecA are important for its interaction with PG and CL, and mutating these regions of RecA weakens its binding to anionic PLs. Error bars indicate SD. Also see Fig S1, Fig S2 and Table S1.
Similar to our previous observations, the vertical differential solvation free energies (ΔΔWslv) of RecA binding to the membrane correlate with ΔFbind and overestimate this parameter (Zhang et al., 2014). The calculated binding affinity for the parallel binding conformation of RecA to the membrane is substantially stronger than the perpendicular mode by ~5 kcal/mol. We evaluated the parallel and perpendicular conformation for the mutants in which one binding motif (either the N-terminal helix or loop L2) was altered. Our calculations (Table 1) suggest that altering either motif leads to a significant (~50%) reduction in the binding free energy and is qualitatively in agreement with our experimental binding measurements (Fig 2E,F and Table S1). The data suggests that both motifs are likely to be involved in binding of RecA to the membrane.
Table 1.
Binding free energy (in kcal/mol) calculations using the GBSW-GCS model for WT RecA and mutants to an anionic membrane.
| System a | Mutations | ΔΔWslv b | ΔFbind c |
|---|---|---|---|
| WT // | - | −21.3 ± 2.3 | −9.6 ± 0.9 |
| WT ⊥ | - | −13.6 ± 2.3 | −5.0 ± 0.5 |
| Mut1 // | Δ10N | −19.2 ± 4.0 | −5.3 ± 0.6 |
| Mut1 ⊥ | −10.8 ± 1.4 | −2.3 ± 0.1d | |
| Mut2 // | I195A, R196A, K198A, I199A, V201A, F203A | −15.2 ± 6.7 | −4.7 ± 2.9 |
| Mut2 ⊥ | −9.2 ± 1.3 | −0.8 ± 0.1 |
. // and ⊥ refer to the parallel and perpendicular binding conformations (see Fig 2C,D), respectively.
. The vertical differential solvation free energy of the protein at the membrane surface vs. in bulk solution. As discussed in (Zhang et al., 2014), this quantity correlates with binding free energy when the system does not undergo significant conformational transitions upon binding.
. Binding free energy of the protein calculated using the protocol established in (Zhang et al., 2014) based on a specific thermodynamic cycle and the Bennett Acceptance Ratio (BAR) approach (Bennett, 1976).
. Mut1 for the perpendicular orientation leads to a 1.6 kcal/mol less binding free energy change than the parallel orientation. This is because the weakly perpendicularly bound RecA adjusts Loop L2 insertion to compensate the truncation of N-terminal amino acids to remain as the bound state. Meanwhile for parallel orientation, the change in insertion depth is negligible.
Verification of membrane-binding regions in vitro
To verify our computational results, we purified two RecA mutants: 1) a mutant lacking the 10 N-terminal amino acids (ΔN10); and 2) a mutant containing the substitutions I195A, R196A, K198A, I199A, V201A and F203A (L2mut); circular dichroism data indicated that both mutants were folded correctly (Fig S2A). We performed Trp fluorescence binding assays of these mutants with CL- and PG-containing liposomes (Fig 2E,F) and found both mutants showed a > 3-fold reduction in binding (Table S1). To confirm that the wild-type (WT) and mutant proteins were not partially denatured by the presence of aPLs, we performed guanidine hydrochloride (GuHCl) titrations in the presence and absence of 70 μM PG liposomes (~10X the Kd for WT RecA). We observed that all three proteins denatured completely at a GuHCl concentration of 3.75 M. The CD spectra in the presence of PG liposomes showed no significant differences when compared to the spectra in the absence of liposomes and at all GuHCl concentrations tested (Fig S2B-D). These results confirm our computational predictions that the N10 and L2 regions contribute significantly to and form the interface for RecA-membrane interactions. The N-terminus of RecA has been hypothesized to play a role in the assembly/disassembly of RecA-ssDNA filaments (Mikawa et al., 1995) and the L2 loop is implicated in ssDNA binding and ATPase (Voloshin et al., 2000). Previous work has demonstrated that mutations in both of these regions lead to recombination negative phenotypes in vivo (Table S2). The L2 loop is evolutionarily conserved across many bacterial species (Karlin and Brocchieri, 1996). Interestingly, only 4 residues within the 10 N-terminal amino acids are stringently conserved (K6, K8, A9 and L10). We wondered whether the interaction of RecA with aPLs affects the functions related to these regions 1) monomer-monomer interactions (ΔN10) and 2) ssDNA binding (L2).
aPLs inhibit ATPase within RecA nucleoprotein filaments
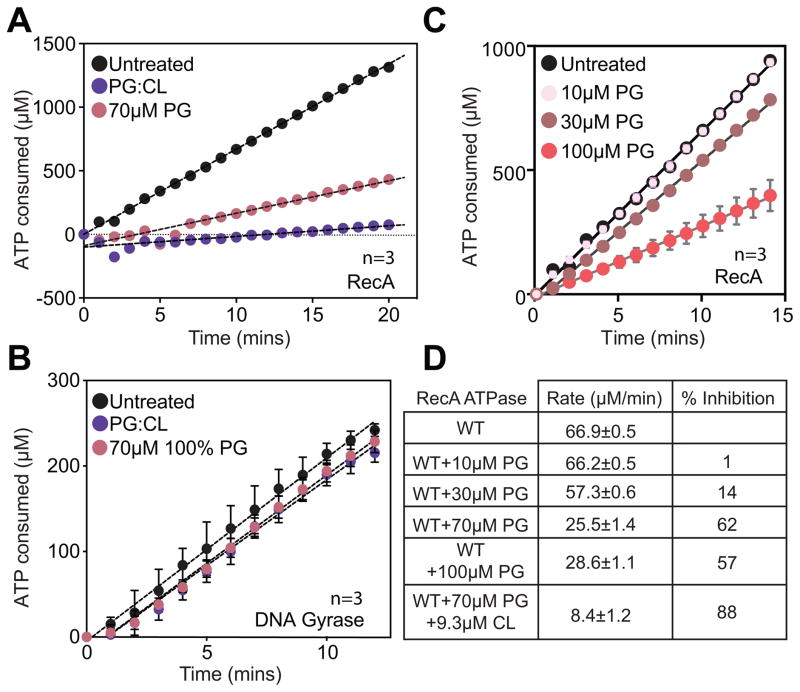
We performed in vitro assays to determine whether interactions with PG/CL perturb RecA binding to ssDNA, strand exchange, and/or ATPase activities. All assays of RecA activity in vitro require Mg2+ ions to facilitate ATP binding. The zwitterionic PL, PC, forms a ternary complex with DNA when Mg2+ ions are present and confounds results (Kuvichkin et al., 2009). To circumvent this limitation, we avoided the use of PC/PE, and utilized 100% PG and a mixture of 80% PG+20% CL at 10X the measured Kds. We measured the DNA-dependent ATPase rate of RecA. We pre-incubated RecA simultaneously with aPLs and ssDNA prior to the addition of ATP to initiate the reaction. This experimental setup enabled us to mimic in vivo conditions where both aPLs and ssDNA (after DNA is damaged) are available simultaneously. We observed that ATPase was inhibited in the presence of CL and PG (Fig 3A, Fig 3D). In identical experimental conditions—controlled for order of addition of different components as well as concentrations of aPLs—we found ssDNA binding to RecA was unaffected (Fig S3A,C), and strand exchange progressed to completion (Fig S3E-G). We also titrated PG into ATPase reactions to confirm that rates of ATPase decrease as a function of PG concentration (Fig 3C,D). This set of experiments revealed four important conclusions: 1) the presence of aPLs did not alter the affinity of RecA to ssDNA, which is consistent with Kd measurements indicating that ssDNA has a higher affinity to RecA than PG or CL; 2) the inhibition of ATPase did not affect the ability of RecA to perform homologous recombination, which is congruent with proposed models that ATPase is required for dissociation of RecA nucleoprotein filaments and is not required for the process of recombination; 3) the reduction in ATPase activity was not due to a reduction in ATP binding, as a decrease in ATP binding would impair strand exchange due to the inability to form RecA filaments; and 4) the RecA protein was not denatured in the presence of aPLs as it remained able to perform DNA strand exchange. To confirm that the effect of aPLs on RecA’s ATPase activity is specific, we performed a control experiment with aPLs at identical concentrations with another DNA-dependent ATPase, E. coli DNA gyrase (Rajendram et al., 2014). We pre-incubated gyrase with aPLs and DNA prior to the addition of ATP to initiate the reaction. In contrast to RecA, we observed no decrease in ATPase by gyrase when aPLs were present (Fig 3B). These observations indicate that PG and CL produce a specific effect on RecA ATPase but do not alter ATP/DNA binding. We hypothesized that aPLs inhibit dissociation of RecA nucleoprotein filaments by inhibiting ATPase and provide a mechanism for the formation of stable nucleoprotein filaments.
Figure 3. ATPase assays with aPLs for RecA and DNA Gyrase.
(A) Anionic PLs inhibit DNA dependent ATP hydrolysis by RecA. (B) Negative control with DNA gyrase DNA-dependent ATP hydrolysis indicates that the inhibition of RecA ATP hydrolysis is due to a specific interaction of anionic PLs with RecA. (C) Titration of PG liposomes show that ATPase rates are inhibited as a function of concentration of PG. (D) Absolute ATPase rates calculated from fits to the curves above. Error bars indicate SD. Also see Fig S3.
RecA mislocalizes in the absence of aPLs
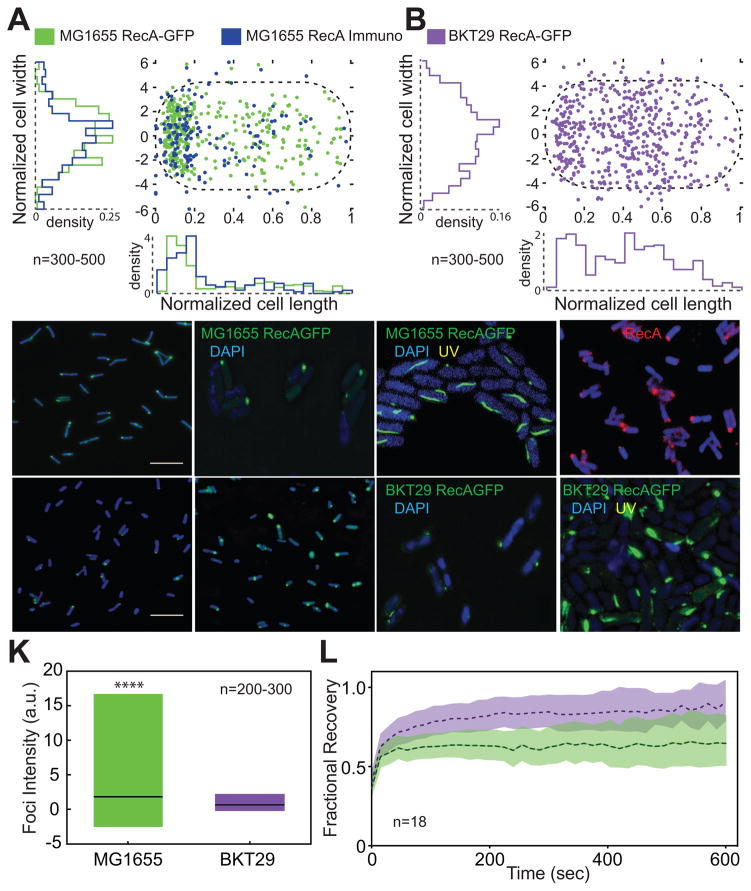
aPLs localize preferentially to the poles of E. coli (Oliver et al., 2014; Renner and Weibel, 2011). One hypothesis is that aPLs act as landmarks to recruit proteins to cell poles. Due to the reduction in RecA ATPase in the presence of aPLs, we wondered whether depleting aPLs (E. coli BKT29 (Tan et al., 2012)) would alter the localization of RecA-GFP foci that are observed at the E. coli poles. Consistent with this hypothesis, previous studies have shown that mutants of RecA (K72A) that do not bind ATP display a decrease in foci formation (Renzette and Sandler, 2008). We transduced RecA-GFP described previously (Renzette et al., 2005) into E. coli MG1655 and analyzed RecA-GFP localization and found RecA-GFP foci localized predominantly at the poles (Fig 4A,C,D). ~50% of cells had a single focus and ~70% of these cells displayed a unipolar distribution. When multiple foci were present (~5% of cells), at least one focus was present at a pole. As fluorescent protein tags can cause artifacts in protein localization, we confirmed these results by quantifying the subcellular localization of RecA in E. coli using immunofluorescence labeling. We expressed RecA with a C-terminal epitope tag (Xpress™ DLYDDDDK) from a low-copy pBAD plasmid induced with arabinose and quantified foci distribution. We observed a predominantly polar distribution correlating very closely with that of RecA-GFP (Fig 4A,F) indicating polarly localized RecA-GFP foci in E. coli are not artifacts.
Figure 4. RecA foci localize to the poles of E. coli and mislocalize in the absence of anionic PLs.
(A) Quantification of the position of RecA in WT cells using RecA-GFP and immunofluorescence labeling. (B) RecA-GFP foci mislocalize in the absence of aPLs (strain BKT29). (C) Representative image of RecA-GFP in WT cells (cells are false colored blue). (D) 3D-SIM images show that RecA foci are located on the periphery of the nucleoid (visualized in blue by DAPI staining) in WT. (E) We observe the formation of RecA filament bundles following UV treatment that were positioned proximal to the nucleoid. (F) A representative image of immunofluorescence of RecA in WT cells. (G and H) Representative images of RecA-GFP foci in BKT29 using epifluorescence microscopy (cells are false colored blue) and (I) 3D-SIM with DAPI. (J) Images of UV-treated BKT29 cells using 3D-SIM and labeled with DAPI demonstrating short, ellipsoidal filament bundles. (K) BKT29 cells have foci that display a significantly lower fluorescence intensity compared to WT. Plot displays distribution of foci intensity from min to max, with black line indicating mean. **** Indicates p<0.001 calculated using a two-tailed t-test. (L) FRAP recovery curves for RecA-GFP foci for WT and BKT29. Shaded area indicates 95% confidence intervals. BKT29 displays a larger mobile fraction of RecA compared to WT, indicating a higher turnover rate of protein within the foci. This result indicates that anionic PLs have a stabilizing effect on RecA within the cell. Also see Fig S4, S5 and S7.
Next, we quantified the localization of RecA-GFP in E. coli BKT29 (Fig 4B,G,H,I) and observed that the distribution of foci was altered, with a >50% reduction in the incidence of polar foci compared to wild-type (WT) cells. Strain E. coli BKT29 contains suppressor mutations that stabilize cells depleted of aPLs. To account for the effect of these suppressor mutations, we tested strain E. coli UE53, which contains only these mutations, as a background control (Fig S7A,E) and found there were no significant differences in RecA localization between UE53 and MG1655. Our observations are consistent with the hypothesis that aPLs stabilize RecA structures at the poles. These foci were not associated with the nucleoid as observed by DAPI staining (Fig 4D,I). In contrast, RecA bundles formed in MG1655 and BKT29 after UV damage, associated with the periphery of the nucleoid (Fig 4E,J). RecA bundles in BKT29 appear morphologically distinct to those we observed in MG1655.
We also analyzed the localization of the ΔN10 and L2mut proteins in vivo using immunofluorescence (Fig S4A-D). Of the two mutants, L2mut cells had the largest decrease in polar RecA foci; however, this result may not be significant because both L2mut and ΔN10 showed very weak foci formation in vivo (<10% incidence). We tested whether weak foci formation is a result of alterations in protein expression by performing western blots of cell lysates expressing WT, L2mut, and ΔN10. L2mut and WT RecA expressed to similar levels, whereas ΔN10 expressed at 60–70% of WT level (Fig S4E, F). The absence of foci in ΔN10 may be attributed to the decrease in DNA-less oligomers observed with mutations in this region (Eldin et al., 2000) or due to the slight decrease in protein expression. The involvement of L2 in the oligomeric states of RecA is less clear, however mutations immediately flanking the L2-loop have been implicated in ATP binding and RecA oligomerization (Kelley and Knight, 1997; Logan et al., 1997).
As ATPase activity within the RecA filament is required for filament disassembly and aPLs inhibited ATPase in vitro, we explored whether the slower rates of ATPase activity for membrane-associated RecA stabilized these structures in vivo. If correct, RecA-GFP foci in BKT29 cells should contain fewer RecA molecules than WT cells. We found the magnitude of the fluorescence intensity of RecA foci was significantly lower in BKT29 than in WT (Fig. 4K), whereas strain UE53 showed no significant difference (Fig S7K). This suggests an increase in RecA turnover or a decrease in the accumulation of RecA in foci in the absence of PG and CL, which we probed using fluorescence recovery after photobleaching (FRAP) experiments in these cells (Movie S1, S2). ~60% of the RecA-GFP in WT,UE53 and BKT29 cells was dynamic (Fig 4L, S5A-D,S7H), further indicating that RecA-GFP foci are not misfolded aggregates of protein in inclusion bodies which do not recover after photobleaching (Winkler et al., 2010). The recovery curves were composed of two distinct populations of molecules- a steep recovery by a fast-moving population (similar to free RecA diffusion) and a shallow recovery (faster than protein synthesis but slower than diffusion) characteristic of a reaction-dominant recovery (Fig S5C). In WT cells, ~26% of the RecA-GFP exchanged rapidly (Fig 4L, S5D), referred to as the fast-moving fraction (C2), whereas in BKT29 cells C2 was ~36% (Fig S5A,B,D). C2 had recovery kinetics and koff values in the effective diffusion regime (Fig S5C,D), indicating that there were fewer free RecA-GFP molecules available in WT compared to BKT29. The remainder of the mobile RecA-GFP fraction in both strains corresponded to a slow-moving reaction-dominant fraction (C1), which is significantly reduced in BKT29 (~21%) compared to WT (~37%) (Fig S5A,B,D). C1 also displayed drastically decreased koff rates for BKT29 (0.001s−1) compared to WT (0.00011s−1). Because the total % of recovered fluorescence does not vary significantly for both strains, the increase of free RecA in C2 of BKT29 corresponds closely with the decrease of RecA in C1. This relationship is also reflected in the ratio of k1on/k1off for BKT29 of 0.49 (Fig S5D), which indicates a high turnover rate of RecA within the C1 fraction and release of these molecules into the free RecA pool. These observations support a model in which aPLs decrease the ATPase activity of RecA, stabilize the protein in a self-associated state, and promote the nucleation of RecA monomers.
aPLs promote stable and efficient SOS response
In vitro, LexA cleavage is efficient in the presence of extended RecA nucleoprotein filaments (Craig and Roberts, 1980). In vivo, Lesterlin et al., demonstrated that bundles of RecA form only after DNA damage but are occluded from the nucleoid with ~>85% of RecA within the bundles observed to be DNA-free. In this model, RecA proteins nucleate on exposed ssDNA at the site of damage and form a filament. This single filament is then sandwiched by multiple RecA filaments to produce bundles, which do not associate with DNA. It was also observed that RecA was not exchanged within these bundles suggesting that they are highly stable. A RecA mutant (E38K, K72R) that binds ssDNA and forms extended filaments but does not hydrolyze ATP, constitutively initiates the SOS response in vivo (Gruenig et al., 2008). This observation implies that RecA filaments that do not dissociate by hydrolyzing ATP are capable of producing an enhanced SOS response. We hypothesized that alterations in membrane composition will destabilize RecA filament bundles in vivo and impact the kinetics of the SOS response.
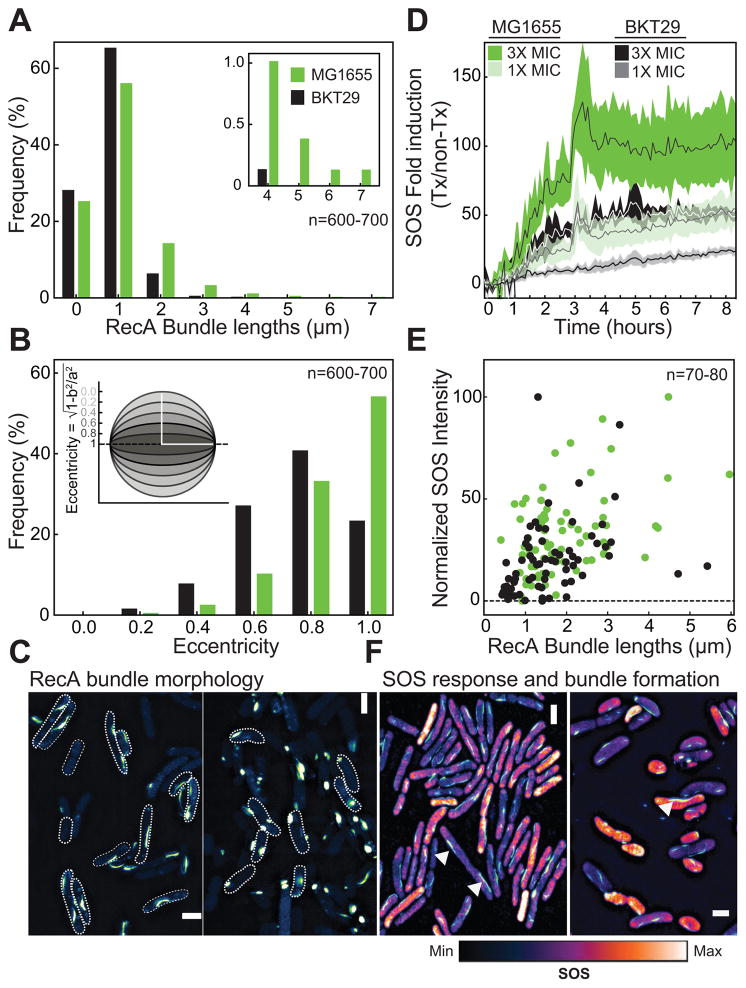
We analyzed the distribution of filament bundle lengths and morphology in WT and BKT29 cells using 3D-SIM (Fig 5A,C). We treated growing cell cultures with 10ng/mL norfloxacin (~1X minimal inhibitory concentration (MIC)) at 0.1–0.2 OD and continued growth for a further 1.5 h, after which cells were imaged. We found that WT and UE53 cells had a broad distribution of bundle lengths, with ~19% of bundles > 2 μm in length (Fig 5A, Fig S7I). In BKT29 cells, only 6% of the bundles were > 2 μm in length with the longest bundles we observed being ~4 μm compared to 7 μm for WT cells (Fig 5A). We quantified bundle morphology in both strains using a metric for characterizing the deviation of bundle architecture from a sphere (eccentricity). Filament bundles that are spherical (i.e. foci-like) have an eccentricity of 0, whereas those that deviate away from a sphere (i.e. a straight line) have an eccentricity approaching 1. In BKT29 cells, bundles of all lengths were largely spherical and non-linear, unlike the morphology observed during DNA repair. ~60% of all bundles in WT and UE53 cells were linear whereas only 20% of bundles were linear for BKT29 (Fig 5B, S7G). Both the length and morphology analyses indicate that BKT29 is largely unable to form RecA filament bundles. This observation confirms the function of aPLs as a scaffold for the nucleation and stabilization of RecA bundles.
Figure 5. Analysis of SOS at a single cell level in the absence of anionic PLs.
(A) Measurement of RecA bundle lengths 1.5 h after treatment with 1X MIC of norfloxacin shows that in WT (MG1655), >20% of bundles are >2 μm. These RecA bundles are 15% longer than in BKT29 in which ~85% RecA bundles are typically <1.5 μm. (B) ~60% of bundles display a straight morphology (eccentricity = 1) in WT compared to ~20% in BKT29. (C) Representative SIM images showing bundles in WT, and punctate, ellipsoidal RecA- GFP structures in BKT29. (D) Using a sulA-promoter fusion to GFP, we observed that BKT29 has a reduced SOS response compared to WT when treated with 1X MIC of norfloxacin. This difference is also observed at 3X MIC. Shaded area represents SD of three biological replicates, with line and mean. (E) The length of RecA bundles within the cell is positively correlated to the degree of SOS induced as observed by a recN-mCherry promoter fusion expressed with RecA-GFP. Shaded area indicates 95% confidence intervals of a linear regression fit. (F) Representative SIM image showing the overlay of SOS intensity with RecA bundles for WT and BKT29. White arrows indicate RecA bundles. Most cells show increased SOS (orange-white) when long bundles are present. The image displays the maximum intensity within one frame, which is a subset of the population of cells analyzed. Some outliers exist for BKT29 (seen in the accompanying images) in which short filaments have high levels of SOS and are visible in the plot. Because cells are also normalized by total area, longer cells with SOS intensities equal to shorter cells will have an overall higher SOS response. Cells that are blue indicate a lower level of SOS. Also see Fig S6 and Fig S7.
To test whether the inability to form RecA filament bundles affected the SOS response in BKT29, we used a plate-reader based assay and measured kinetics of SOS induction in strains of E. coli with WT RecA treated with 1X and 3X MIC norfloxacin (for WT, UE53, and BKT29) using a plasmid encoded copy of the SulA promoter located upstream of super-folder GFP (sfGFP). Norfloxacin produces DSBs that induce the SOS response and are repaired by RecA mediated recombination. We measured the fold-increase of sfGFP expression from the sulA promoter in untreated and norfloxacin treated cells normalized by cell density, and observed a significant decrease in the amount of SOS induction after DNA damage in BKT29 cells (Fig 5D).
We also performed a survival analysis by monitoring colony-forming units after 3 h of treatment with 10ng/mL norfloxacin. ~60% of MG1655 RecA-GFP survived treatment whereas <5% of ΔrecA survived. MG1655 RecA-GFP is at least partially functional in repairing DSBs in the conditions that we tested (Fig S6A). Interestingly, BKT29 had lower survival compared to WT (~60%) confirming that these cells are impaired in repairing DNA damage. Defects in RecA-GFP foci and bundles were unique to BKT29 and did not appear in UE53, the parent strain of BKT29 that contains the suppressor mutations necessary for aPL deletion (Fig S7A-K).
We explored the possibility that the defect in RecA bundle formation in BKT29 may be the reason for the decreased SOS response observed in this strain. We monitored the level of SOS expression in WT and BKT29 at the single cell level while simultaneously measuring the lengths of RecA bundles formed. To obtain bundles of comparable lengths between WT and BKT29, we treated BKT29 with norfloxacin for an additional 1 h (for a total of 2.5 h). We found that the mean intensity of SOS observed in individual cells increased in proportion with bundle lengths observed for WT and BKT29 (Fig 5E,F). We determined the mean intensity by dividing the total intensity of each cell by the area and plotting it as a fraction of the maximum intensity observed within the population. This indicates BKT29 cells that eventually form long bundles are capable of producing the SOS response. However, BKT29 cells are slow to form RecA bundles with lengths comparable to those in WT cells and therefore display a decreased SOS response as a population.
Discussion
The role of RecA in recombination repair is critical and during normal growth and replication, the nucleation of RecA on exposed ssDNA or double stranded DNA can impede cellular processes. Our data is consistent with a model in which the IM stabilizes polymeric RecA structures at the poles and prevents its association with DNA during normal cellular growth and division. Induction of DNA damage (by UV or norfloxacin) redistributed RecA-GFP from the poles to the region of E. coli cells where the chromosome is located and where RecA formed visible bundles. The mechanisms underlying this process of RecA redistribution are currently unknown, but may be driven by the affinity of freely diffusing RecA to ssDNA.
CL and PG stabilize RecA structures without affecting the binding of ssDNA to RecA and altering its strand exchange activity in vitro. In contrast, CL and PG inhibited RecA ATPase significantly. ATPase is not required for RecA to catalyze strand pairing (Menetski et al., 1990); instead the hydrolysis of ATP has been appropriated to the disassembly of RecA filaments into monomers and facilitating strand pairing of heterologous chromosomal regions (Cox, 2007). Extended RecA nucleoprotein filaments are required for efficient SOS induction in vivo, however ATPase is not required (Gruenig et al., 2008). These observations—framed within the context of the results presented in this study—imply that aPLs may also stabilize RecA bundles formed after the induction of DSBs by reducing the rate of ATPase within RecA filaments. In support of this hypothesis, we observed a dramatic decrease in the SOS response in cells lacking aPLs.
Our combined experimental and computational approach elucidated the membrane-binding domains in RecA. These regions are conserved across multiple species of bacterial RecA and mutational variation within these regions is infrequently tolerated in E. coli (Karlin and Brocchieri, 1996). The stringent conservation of amino acids in these positions has been attributed to the involvement of these regions in: 1) strengthening the association between RecA monomers; and 2) interacting with ssDNA and catalyzing the hydrolysis of ATP. The placement of membrane-association domains of RecA at regions that are essential for its activity may provide a mechanism to reduce the probability that RecA will lose the ability to bind aPLs by genetic drift. The dominant negative phenotype associated with mutations at these regions made it impossible for us to manipulate these regions to observe the effects of membrane association on DNA repair. Regardless, the positions of the membrane binding interfaces may provide insight into the modulation of ATPase activity within the RecA filament by aPLs. The N-terminal 10 amino acids of RecA have been hypothesized to participate in monomer-monomer contacts important for polymerization. The N-terminus (residues 1–33) has been suggested to play a role in RecA assembly-disassembly kinetics, specifically the coupling of a helix-coil transition in this region to RecA filament disassembly (Mikawa et al., 1995). Thus, stabilization provided by the membrane may prevent this structural transition and enable the formation of stable RecA bundles. Interestingly, Bacillus subtilis RecA—which possesses an N-terminus that is not similar to E. coli—does not form RecA foci under normal growth, implicating the importance of this region for RecA self-association. The loop L2 (amino acids 195–209), and specifically F203 makes direct contact with ssDNA in vitro (Malkov and Cameriniotero, 1995). Q194 and R196 are involved in catalyzing the hydrolysis of ATP. L2 has been demonstrated to mediate conformational changes associated with ATP binding to interaction with ssDNA and is subsequently involved in coupling DNA dependent ATPase to strand exchange. It is possible that inhibition of DNA dependent ATPase by aPLs occurs by blocking communication of the conformational changes associated with strand pairing by L2 to ATPase. Lesterlin et al. demonstrated that most of the RecA within bundles are not associated with DNA. This observation implies that the L2 loop is freely available for interaction with the IM when RecA is present as bundles.
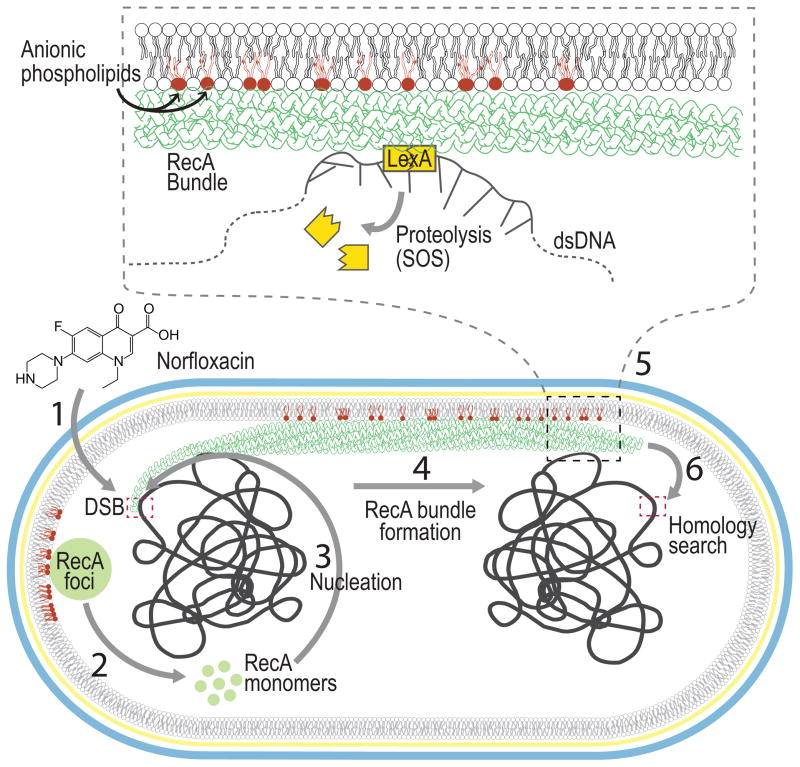
To summarize, we have demonstrated that the IM is a key component for the nucleation and formation of RecA bundles in E. coli. We propose a model (Fig 6) in which RecA is nucleoid occluded and bound to aPLs at the poles of E. coli under normal conditions in ‘storage structures’ that turn over at a constant rate. Damage to DNA that produces exposed ssDNA drives RecA nucleation due to high affinity RecA-ssDNA interactions. ssDNA bound RecA produces filament bundles in an attempt to search for DNA homology utilizing the IM as a scaffold for stability. This study sheds light on an exciting new dimension in RecA biochemistry and provides an example of yet another cytoplasmic protein that relies on its interaction with the bacterial membrane for spatial control of its activity and cellular function.
Figure 6. Model for the role of the inner membrane in RecA filament bundle formation.
(1) Norfloxacin treatment of E. coli cells produces double stranded breaks (DSBs). (2) RecA protein which undergoes exchange between the cytoplasm and RecA foci, (3) nucleates on the processed DSB due to its high affinity to ssDNA (4) and forms RecA bundles associated with the IM (6) which extend across the cell searching for homology. (5) At some point during this search for homology, contact between the RecA bundle and LexA facilitates the autocleavage of LexA leading to the SOS response. RecA bundles are a stylized version of those observed by electron microscopy (Yu and Egelman, 1992).
Experimental Procedures
Strain Construction
E. coli strain BKT29 (ΔaraBAD,rcsF::mini-Tn10cam,λ−, ΔclsABC, ΔymdB::kan, ΔpgsA,lpp-2) was transformed with plasmid pCP20 (containing FLP recombinase) and grown overnight at 30 °C to remove the kanamycin (kan) resistance cassette (kanS). pCP20 was cured from strain BKT29 by growing at 43 °C. To generate chromosomal RecA-GFP fusions, BKT29 kanS, E. coli MG1655, and UE53 (ΔaraBAD,rcsF::mini-Tn10cam, λ−,lpp-2) were subjected to P1 transduction using a phage P1 lysate grown on strain ss3041 (ygaD1::kan recAo1403 recA4136::gfp-901. recAo1403 ). recAo1403 refers to a single point mutation (T to A) in the operator/promoter region of recA that causes a two-fold higher level of basal expression. Gfp-901 refers to mut-2 with the additional monomeric mutation A209T, which reduces aggregation that cause the formation of aberrant foci. For assays measuring SOS induction, plasmids pUA66-sulA promoter-GFP (pSulA-GFP), and pUA66-GFP were transformed into WT MG1655, UE53 and BKT29 kanS. Plasmid precN-mCherry was constructed with a recN promoter fused to mCherry on a pBR322 backbone and transformed into BKT29 RecA-GFP and MG1655 RecA-GFP to monitor SOS. This plasmid was SOS-inducible under norfloxacin treatment (Fig S6B).
Quantifying RecA binding to liposomes by measuring the fluorescence quenching of tryptophan
To determine an apparent Kd of RecA to aPLs, we measured changes in the fluorescence intensity of Trp290 and 309 during the titration of RecA with liposomes. We fixed the excitation of a PTI Quantamaster 300 spectrofluorimeter (Birmingham, NJ) at λ= 295 and collected fluorescence data at 37 °C. We titrated liposomes consisting of 100% PC (negative control), 100% PG, 50:50% PC:PE, and 80:20% PC:CL into purified E. coli RecA (0.7 μM) in 1.4 mL of R buffer (see SI) and mixed the suspension thoroughly using a pipette. We incubated mixtures for 5 min at 37 °C and imaged the fluorescence emission by scanning from λ=300–400 nm. We corrected measurements of fluorescence intensity for the dilution of RecA during titration. We also accounted for scattering of light by liposomes and corrected our fluorescence measurements by subtracting values of fluorescence that we measured for liposome-only titrations into buffer. The titration of 100% PC liposomes into RecA did not produce an appreciable change in fluorescence intensity at λ=300–400 nm, and thus we calculated the binding constants for PC:CL and PC:PE liposomes to RecA using only the concentration of CL or PE respectively. We calculated the fraction of RecA bound to liposomes using the equation: where Fadj is the fluorescence intensity corrected for dilution and scattering, and Fmax and Fmin are the maximum and minimum fluorescence intensities we measured at λ=320 nm. We calculated the free lipid concentrations for CL, PG and PE using the equation: . We plotted the values we calculated for B against the free lipid concentration of CL, PG and PE to obtain Langmuir binding isotherms and fit the data to the modified one-site specific binding equation: .
Implicit Membrane Simulations
For the implicit membrane model, we use the generalized Born model with a simple switching function (GBSW) (Im et al., 2003), augmented with the Gouy-Chapman-Stern (GCS) model (Barrat and Hansen, 2003; Stern, 1924) for the anionic membrane surface. The salt concentration was set to 0.02 M NaCl, and parameters in the GCS model were chosen based a membrane with 27% PG and 73% PC. To identify favorable binding orientations, we performed a binding energy scan by rotating a rigid RecA monomer—constructed based on the crystal structure, PDB ID 3CMT—with its center of mass fixed at 25 Å away from the membrane center. Guided by this binding energy surface (Fig 2A), we used 14 independent GBSW-GCS molecular dynamics trajectories (~10 ns each) without imposing any structural restraints on RecA. Each trajectory started with a different orientation. Some of the trajectories led to dissociation of RecA from the membrane surface, while the rest converged to two distinct binding conformations that featured similar binding interfaces (Fig 2B) and had different orientations of the C-terminal domain; we refer to these two orientations as the parallel (Fig 2C) and perpendicular (Fig 2D) binding conformations. These conformations were present in subsequent explicit membrane simulations on a time scale of ~110 ns.
We computed the binding free energies of the two binding conformations using the GBSW-GCS model and the thermodynamic cycle described recently (Zhang et al., 2014), which demonstrated that this methodology accurately predicts the binding free energy of peptides with membranes. We tested several mutants that were designed based on the observed RecA/membrane binding interface, which involves mainly the DNA binding loop L2 and the N-terminal helix; to a lesser extent, the C-terminal domain also contributes to binding in the parallel binding conformation. We computed the binding free energies of all of these mutants (Table 1).
For more information on experimental methods, please see Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Steven Sandler for E. coli strain ss3041 and Brandon Tan and Ziqiang Guan for E. coli strain BKT29. We are grateful to Angela Gruber, Anjon Audhya, Alessandro Senes, Elizabeth Wood, and Darrell McCaslin for input, experimental protocols, and reagents. This research was supported by the National Science Foundation under Grant No. DMR-1121288, National Institutes of Health Grant 1DP2OD008735-01 (to D.B.W.), National Institutes of Health Grant GM32335 (to M.M.C.), a Mao Wisconsin Distinguished Graduate Fellowship (to M.R.), and grants BIR-9512577 (NSF) and S10 RR13790 (NIH) (to the Biophysics Instrumentation Facility).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barrat J-L, Hansen J-P. Basic concepts for simple and complex liquids. New York: Cambridge University Press; 2003. [Google Scholar]
- Brenner SL, Zlotnick A, Griffith JD. RecA protein self-assembly. Multiple discrete aggregation states. J Mol Biol. 1988;204:959–972. doi: 10.1016/0022-2836(88)90055-1. [DOI] [PubMed] [Google Scholar]
- Britt RL, Chitteni-Pattu S, Page AN, Cox MM. RecA K72R filament formation defects reveal an oligomeric RecA species involved in filament extension. J Biol Chem. 2011;286:7830–7840. doi: 10.1074/jbc.M110.194407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM. Motoring along with the bacterial RecA protein. Nat Rev Mol Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- Craig NL, Roberts JW. E. coli recA protein-directed cleavage of phage lambda repressor requires polynucleotide. Nature. 1980;283:26–30. doi: 10.1038/283026a0. [DOI] [PubMed] [Google Scholar]
- Egelman EH, Stasiak A. Structure of helical RecA-DNA complexes. II. Local conformational changes visualized in bundles of RecA-ATP gamma S filaments. J Mol Biol. 1988;200:329–349. doi: 10.1016/0022-2836(88)90245-8. [DOI] [PubMed] [Google Scholar]
- Eldin S, Forget AL, Lindenmuth DM, Logan KM, Knight KL. Mutations in the N-terminal region of RecA that disrupt the stability of free protein oligomers but not RecA-DNA complexes. J Mol Biol. 2000;299:91–101. doi: 10.1006/jmbi.2000.3721. [DOI] [PubMed] [Google Scholar]
- Garvey N, St John AC, Witkin EM. Evidence for RecA protein association with the cell membrane and for changes in the levels of major outer membrane proteins in SOS-induced Escherichia coli cells. J Bacteriol. 1985;163:870–876. doi: 10.1128/jb.163.3.870-876.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenig MC, Renzette N, Long E, Chitteni-Pattu S, Inman RB, Cox MM, Sandler SJ. RecA-mediated SOS induction requires an extended filament conformation but no ATP hydrolysis. Mol Microbiol. 2008;69:1165–1179. doi: 10.1111/j.1365-2958.2008.06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im W, Lee MS, Brooks CL., 3rd Generalized born model with a simple smoothing function. J Comput Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- Karlin S, Brocchieri L. Evolutionary conservation of RecA genes in relation to protein structure and function. J Bacteriol. 1996;178:1881–1894. doi: 10.1128/jb.178.7.1881-1894.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JA, Knight KL. Allosteric regulation of RecA protein function is mediated by Gln194. J Biol Chem. 1997;272:25778–25782. doi: 10.1074/jbc.272.41.25778. [DOI] [PubMed] [Google Scholar]
- Kuvichkin VV, Danev RS, Shigematsu H, Nagayama K. DNA-induced aggregation and fusion of phosphatidylcholine liposomes in the presence of multivalent cations observed by the cryo-TEM technique. J Membr Biol. 2009;227:95–103. doi: 10.1007/s00232-008-9146-0. [DOI] [PubMed] [Google Scholar]
- Lesterlin C, Ball G, Schermelleh L, Sherratt DJ. RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature. 2014;506:249–253. doi: 10.1038/nature12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Zaidman S, Frenkiel-Krispin D, Shimoni E, Sabanay I, Wolf SG, Minsky A. Ordered intracellular RecA-DNA assemblies: a potential site of in vivo RecA-mediated activities. Proc Natl Acad Sci U S A. 2000;97:6791–6796. doi: 10.1073/pnas.090532397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan KM, Skiba MC, Eldin S, Knight KL. Mutant RecA proteins which form hexamer-sized oligomers. J Mol Biol. 1997;266:306–316. doi: 10.1006/jmbi.1996.0751. [DOI] [PubMed] [Google Scholar]
- Malkov VV, Cameriniotero RD. Photocross-Links between Single-Stranded-DNA and Escherichia-Coli Reca Protein Map to Loops L1 (Amino-Acid-Residues 157–164) and L2 (Amino-Acid-Residues 195–209) J Biol Chem. 1995;270:30230–30233. doi: 10.1074/jbc.270.50.30230. [DOI] [PubMed] [Google Scholar]
- Menetski JP, Bear DG, Kowalczykowski SC. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc Natl Acad Sci U S A. 1990;87:21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J Mol Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- Mikawa T, Masui R, Ogawa T, Ogawa H, Kuramitsu S. N-terminal 33 amino acid residues of Escherichia coli RecA protein contribute to its self-assembly. J Mol Biol. 1995;250:471–483. doi: 10.1006/jmbi.1995.0391. [DOI] [PubMed] [Google Scholar]
- Oliver PM, Crooks JA, Leidl M, Yoon EJ, Saghatelian A, Weibel DB. Localization of Anionic Phospholipids in Escherichia coli Cells. J Bacteriol. 2014;196:3386–3398. doi: 10.1128/JB.01877-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanastasiou M, Orfanoudaki G, Koukaki M, Kountourakis N, Sardis MF, Aivaliotis M, Karamanou S, Economou A. The Escherichia coli peripheral inner membrane proteome. Mol Cell Proteomics. 2013;12:599–610. doi: 10.1074/mcp.M112.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendram M, Hurley KA, Foss MH, Thornton KM, Moore JT, Shaw JT, Weibel DB. Gyramides prevent bacterial growth by inhibiting DNA gyrase and altering chromosome topology. ACS Chem Biol. 2014;9:1312–1319. doi: 10.1021/cb500154m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner LD, Weibel DB. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc Natl Acad Sci U S A. 2011;108:6264–6269. doi: 10.1073/pnas.1015757108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzette N, Gumlaw N, Nordman JT, Krieger M, Yeh SP, Long E, Centore R, Boonsombat R, Sandler SJ. Localization of RecA in Escherichia coli K-12 using RecA-GFP. Mol Microbiol. 2005;57:1074–1085. doi: 10.1111/j.1365-2958.2005.04755.x. [DOI] [PubMed] [Google Scholar]
- Renzette N, Sandler SJ. Requirements for ATP binding and hydrolysis in RecA function in Escherichia coli. Mol Microbiol. 2008;67:1347–1359. doi: 10.1111/j.1365-2958.2008.06130.x. [DOI] [PubMed] [Google Scholar]
- Stern O. Zur theorie der elektrolytischen doppelschicht. Z Elektrochem Angew P. 1924;30:508–516. [Google Scholar]
- Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- Tan BK, Bogdanov M, Zhao J, Dowhan W, Raetz CR, Guan Z. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc Natl Acad Sci U S A. 2012;109:16504–16509. doi: 10.1073/pnas.1212797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian JT, Callis PR. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloshin ON, Wang L, Camerini-Otero RD. The homologous pairing domain of RecA also mediates the allosteric regulation of DNA binding and ATP hydrolysis: a remarkable concentration of functional residues. J Mol Biol. 2000;303:709–720. doi: 10.1006/jmbi.2000.4163. [DOI] [PubMed] [Google Scholar]
- Winkler J, Seybert A, Konig L, Pruggnaller S, Haselmann U, Sourjik V, Weiss M, Frangakis AS, Mogk A, Bukau B. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 2010;29:910–923. doi: 10.1038/emboj.2009.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Egelman HE. Direct Visualization of Dynamics and Co-operative Conformational Changes within RecA filaments that appear to be associated with the hydrolysis of adenosine 5’-O-(3-thiotriphosphate) J Mol Biol. 1992;225:193–216. doi: 10.1016/0022-2836(92)91036-o. [DOI] [PubMed] [Google Scholar]
- Zhang LL, Yethiraj A, Cui Q. Free energy calculations for the peripheral binding of proteins/peptides to an anionic membrane. 1. Implicit Membrane Models. J Chem Theory Comput. 2014;10:2845–2859. doi: 10.1021/ct500218p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.