SUMMARY
Diverse effects of the microbiome on solid organ transplantation are beginning to be recognized. In allograft recipients, microbial networks are disrupted by immunosuppression, nosocomial and community-based infectious exposures, antimicrobial therapies, surgery, and immune processes. Shifting microbial patterns, including acute infectious exposures, have dynamic and reciprocal interactions with local and systemic immune systems. Both individual microbial species and microbial networks have central roles in the induction and control of innate and adaptive immune responses, in graft rejection, and in ischemia-reperfusion injury. Understanding the diverse interactions between the microbiome and the immune system of allograft recipients may facilitate clinical management in the future.
INTRODUCTION
Human microbial communities have diverse impacts on human physiology, including the development and maintenance of systemic immune function (1–3). Broad changes occur in the microbial flora of individuals undergoing solid organ transplantation (SOT) and hematopoietic stem cell transplantation (HSCT) via exposures to microbes, antimicrobial agents, vaccinations, infections, immunosuppression, surgery, and immune and inflammatory processes. These changes impact the outcomes of transplantation depending on the context in which they occur, including the nature, site, and intensity of infection and of immunosuppression. Several studies have examined alterations in allograft function after infection with individual viral and bacterial pathogens. This review examines the contributions of specific microbes in allograft function and the experimental and clinical data demonstrating the dynamic relationship that exists between the microbiome and systemic immune function relevant to transplantation.
Microorganisms in tissues and on barrier surfaces, including skin, airways, and gastrointestinal (GI) tract, are collectively termed the “microbiome” (4, 5). Microbial communities are distinct in different anatomic sites and under different clinical conditions (6–8). Exposures may be chronic, as for colonizing organisms, or acute, in association with infection of normally sterile sites or disruption of otherwise stable microbial flora. The gut microbiome comprises an estimated 1014 bacteria, fungi (including Candida species), and viruses (1, 9, 10). Viruses may alter host gene expression (e.g., endogenous retroviruses) or infect prokaryotic symbionts (e.g., bacteriophages) or eukaryotic cells as latent or productive infectious agents (e.g., herpesviruses) (11). In contrast to the case for bacteria, genetic sequencing and bioinformatics analyses of viral sequence diversity are relatively incomplete.
Many organisms comprising the human microbiome are unknown and often difficult to cultivate in vitro. Culture-independent nucleic acid sequencing tools and bioinformatics systems have revolutionized studies of microbial communities. Two major sequencing methods have been employed. Bacterial 16S ribosomal gene sequence data and fingerprinting methods (18S for fungi) resolve microbes to the taxon level and quantify the relative abundance of each species in a sample. Alternatively, next-generation sequencing or “shotgun” metagenomic sequencing uses direct sequencing of total DNA and has been used to examine viral sequences as well as genes in structural and metabolic pathways (12). Gene expression is further characterized with metatranscriptomic and metaproteomic analyses. With these tools and computational data analyses, the microbiome can be characterized in terms of specific organisms and relative abundance in anatomic sites and for changing populations over time with clinical syndromes and therapies.
SPECIFIC ORGANISMS CONTRIBUTE TO ALLOGRAFT REJECTION VIA INNATE AND ADAPTIVE IMMUNE MECHANISMS
In the face of the introduction of new organisms (“infections”), innate immune cells and proinflammatory mediators contribute to tissue injury and prime adaptive immune responses and/or may stimulate cross-reactive cellular alloimmunity. The relative contributions of each type of response to allograft injury are difficult to assess. “Heterologous immunity” describes memory T cell responses to previously encountered pathogens that cross-react with alloantigens (13). Prior infectious exposures may, in sum, enhance the risk for cross-reactive alloimmune responses. Thus, in a murine skin graft tolerance model, tolerance was more difficult to achieve in mice with immunity to a number of viruses (vaccinia virus [VV], herpes simplex virus [HSV], and vesicular stomatitis virus [VSV]) than in uninfected mice or mice infected with only one virus (14). Similarly, mice remotely infected with Leishmania major and undergoing tolerance induction for skin transplantation rejected grafts more rapidly than did uninfected controls. This effect was attributed to cross-reactive CD4 memory responses to Leishmania (15). Latent infections of B6 mice with murine gamma herpesvirus 68, a murine Epstein-Barr virus (EBV) homolog, accelerate BALB/c skin allograft rejection mediated by long-lasting viral antigen-specific CD8 memory responses (16, 17). In transplant recipients receiving calcineurin inhibitor-based immunosuppression, persistent alloimmune responses generated by chronic viral infections do not become “exhausted” and may contribute to graft injury (18–21). The lack of antiviral “exhaustion” may also occur with other viruses (cytomegalovirus [CMV] or EBV) capable of establishing latency with intermittent reactivation. The clinical significance of heterologous immunity requires further definition. Heterologous immune responses are typically restricted to single HLA molecules, limiting the breadth of these responses against HLA-diverse allografts (13, 22). Initial memory T cell infiltrates in rejecting allografts are not antigen specific, suggesting that heterologous immunity against the broad array of prior antigenic exposures rather than against single microbes might participate in allograft rejection (23). Microbial by-products, short-chain fatty acids, may modulate the balance between latent and lytic states of herpesviruses in the human host (24). Thus, interactions between bacterial and viral components of the microbiome may affect viral activation and the risk for alloimmune responses.
Microbial activation of innate immune responses has been implicated in acute and chronic allograft injury. In human transplant recipients with hypomorphic Toll-like receptor 4 (TLR4), an innate immune receptor, rates of bronchiolitis obliterans syndrome (BOS) in lung transplant recipients and of renal allograft rejection are reduced (25, 26). Inhaled lipopolysaccharide (LPS), a TLR4 agonist and mediator of sepsis syndromes, induces acute alloimmune lung injury in fully major histocompatibility complex (MHC)-mismatched murine bone marrow transplants but not in syngeneic recipients. Another innate immune activator, viral mimetic poly I·C, has also been shown to promote alloimmune lung injury (27, 28). In mice undergoing cardiac transplantation using a nondepletional (costimulatory blockade) tolerance-inducing regimen, early infection with Listeria monocytogenes produces allograft rejection (29, 30). Rejection occurs despite prior immunization against L. monocytogenes or treatment with ampicillin. In these animals, expression of type I interferons (IFNs) and bacterial expression of the virulence factor listeriolysin are required for cardiac allograft rejection (29, 30). Similarly, mice infected with Staphylococcus aureus reject skin allografts under a protocol that generally produces graft tolerance (31). In contrast, MyD88 knockout mice that lack a key signaling pathway of innate immune responses survive otherwise lethal challenges with heat-killed S. aureus and accept concurrent skin allografts, consistent with the role of innate immune responses to S. aureus in stimulating remote allograft rejection (31). In general, skin, lung, and intestinal transplants have been unsuccessful using tolerance induction based in costimulatory blockade. This may reflect TLR stimulation by commensal and environmental organisms (32).
Effects of viral infection on inflammation and graft function are complex and virus specific. Viral infection of grafts may increase allograft injury via innate immune mechanisms, possibly by increased expression of inflammatory mediators with recruitment of leukocytes (33). The Herpesviridae (e.g., CMV and EBV) are most often implicated in these processes, though hepatitis C virus (HCV) and pulmonary respiratory viral infections are also linked to allograft injuries (34–42). Posttransplantation, CMV has been implicated in endothelial inflammation, vasculopathy, and graft rejection (43). In vitro, CMV upregulates interleukin-8 (IL-8), transforming growth factor β (TGF-β), and other growth factors to increase fibrosis and inflammation (44, 45). Rat CMV (RCMV) upregulates genes associated with angiogenesis and wound repair with vasculopathy in rat cardiac allografts (46). Aggressive prophylaxis against CMV infection attenuates coronary artery vasculopathy in human cardiac allograft recipients and graft rejection in kidney recipients (47–49).
In addition to the effect of MHC-mismatched grafts on the intragraft efficacy of antiviral adaptive immune responses, CMV tends to suppress the host antiviral response (50). CMV reduces mobilization of monocytes and dendritic cells to inflammatory sites and decreases viral antigen presentation to T lymphocytes. CMV reduces expression of MHC class I receptors in infected host cells, facilitating immune evasion by decreasing antigen priming of T cells and cytolytic T cell responses (51). Murine cytomegalovirus (MCMV) downregulates the expression of natural killer (NK) cell receptors for MCMV, evading NK cell immune surveillance (51). The role of gamma-delta T cells at the interface of innate and adaptive immune mechanisms in the host response to CMV and in graft survival remains to be further defined (52). EBV similarly reduces expression of MHC class I and MHC class II receptors on infected cells, avoiding cellular immune responses (53). Both CMV and EBV encode a viral anti-inflammatory homolog of IL-10, further reducing host antiviral responses (54, 55). Allograft vasculopathy in murine cardiac transplant models (parental to F1) infected with lymphocytic choriomeningitis virus (LCMV) in the complete absence of T and B lymphocytes (RAG−/−) was found to be mediated by NK cells; depletion of NK cells abrogated vasculopathy (56). Thus, pathogen-specific effects on allograft survival relate to the timing, duration, and intensity of innate and adaptive responses (57).
DYSBIOSIS, INDUCTION THERAPIES, AND IMMUNE RECONSTITUTION IN ALLOGRAFT PATHOLOGY
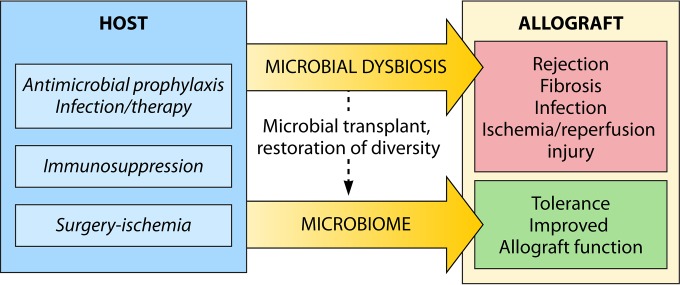
As opposed to the impact of specific infections common to the immunocompromised host, the effects of microbial networks on allograft function have only recently been explored. Early studies indicate bidirectional effects: allotransplantation induces microbial dysbiosis, and microbiome homeostasis has a key role in the control of allograft function (Fig. 1).
FIG 1.
Bidirectional effects of the microbiome in allotransplantation. The process of allotransplantation disrupts the composition of the microbiome through a variety of mechanisms, including surgery, immunosuppressive agents, and antimicrobial therapies. Microbial dysbiosis has been associated with the development of chronic rejection, injury from ischemia-reperfusion, and infection. Conversely, microbiome homeostasis is associated with improved allograft outcomes. The role of microbial manipulation (e.g., by fecal transplantation) as a therapeutic measure to improve allograft outcomes is under investigation.
Dysbiosis after Allotransplantation
Changes in the composition of the microbial profiles of pre- and posttransplant patients have been analyzed. The salivary microbiome in kidney and heart transplant recipients is disrupted compared with normal oral flora in favor of colonization with opportunistic pathogens, including Pseudomonas, Acinetobacter, and Enterobacteriaceae species (58). Such changes are termed “dysbiosis.” A comprehensive study of the blood, oral, urinary, and rectal microbiomes of kidney transplant patients before and after transplantation shows major shifts in composition by 1 month posttransplantation, with relative stability thereafter (59). In another study, the rectal microbiomes of five kidney transplant recipients in the first 90 days posttransplant showed significant increases in Bacteroides species and in species from the phylum Proteobacteria (60).
Some of these changes in the microbiome are due to the surgical process and some of these changes may be attributed to immunosuppressive regimens. For example, following small bowel transplants, analysis of ileal microbiome samples reveals inversion of the microbial composition from strict anaerobes to facultative anaerobes, likely due to the end ileostomy, which allows introduction of increased oxygen levels into the small bowel (61). Intestinal and hepatic ischemia-reperfusion injuries have been associated with a microbial dysbiosis and pathological predominance of Enterobacteriaceae (62).
Microbial Shifts in Induction Therapy and Immunosuppression
Allotransplantation often utilizes antibody-based T cell depletion at the time of transplantation, which is called “induction therapy.” Changed microbiome profiles have been identified in relation to the use of T cell-depleting agents (antithymocyte globulin), nondepleting therapies (basiliximab), early steroid withdrawal programs, or prolonged steroid use, but without statistical significance in small samples (62). All such studies are confounded by effects of perioperative and posttransplantation antimicrobial therapies. T cell-depleting therapies generally deplete central memory subsets, while effector memory and regulatory T cell (Treg) subsets persist (63). Immune reconstitution following depletion is shaped by antigen exposures and subsequent immunosuppressive regimens (64). Lymphopenia induces a compensatory repopulation of immune cells, termed “homeostatic proliferation,” which favors the emergence of memory T cells and may predispose to graft rejection. Rapid proliferation of lymphocytes is antigen specific, likely driven to a great degree by commensal bacterial antigens (65). A role for commensal organisms is suggested by the absence of postdepletional T cell proliferation in germfree, immunodeficient mice compared with conventionally raised mice (66, 67).
IMMUNOLOGIC CONSEQUENCES OF DYSBIOSIS AFTER ALLOTRANSPLANTATION: A CONCEPTUAL FRAMEWORK
How does the microbiome shape adaptive immune responses after allotransplantation? This concept is best considered in the context of recent data on the role of the localized microbiome in shaping systemic immune responses. T cell responses are classified according to surface markers and cytokine secretion patterns. Th1 responses include IFN-γ, Th2 responses include IL-4 and IL-5, and antimicrobial and proinflammatory Th17 responses are characterized by IL-17, IL-21, and IL-23 secretion. Tregs secrete IL-10. Subsets of Tregs include natural, thymus-derived Tregs with T cell receptors (TCRs) targeting self-antigens and induced Tregs (iTregs) derived from circulating CD4+ cells activated in the presence of antigen, TGF-β, and retinoic acid.
Select bacterial components mediate the maturation of mucosal and systemic T lymphocytes. For example, polysaccharide A (PSA) from Bacteroides fragilis mediates Th1-Th2 balance and development of invariant natural killer T (NKT) cells in the colonic lamina propria in germfree mice (68, 69). PSA also ameliorates murine colitis (70). In contrast, in gnotobiotic mice, Clostridium spp., “Candidatus Arthromitus,” and Gram-positive segmented filamentous bacteria (SFB) induce the development of proinflammatory Th1 and Th17 effector cells in the small intestinal lamina propria (71, 72). These effector cells participate in beneficial host defenses against GI pathogens, inducing the production of antimicrobial peptides and proinflammatory chemokines and cytokines via recruitment of neutrophils, macrophages, and dendritic cells. They may also participate in allograft rejection, particularly in the absence of tolerigenic iTregs.
In the lamina propria, iTreg TCRs are specific for intestinal microbial antigens (73). Under normal conditions, iTregs limit mucosal Th2 responses to commensal organisms and Th1 and Th17 responses to pathogenic bacteria, protecting the host from excessive tissue injury (74–76). Development of iTregs is stimulated by specific organisms, including capsular antigens of B. fragilis and a network of spore-forming clostridia (77, 78). Importantly, iTregs and Th17 cells share a developmental requirement for TGF-β signaling with the cofactor retinoic acid via the retinoic acid receptor-related orphan receptor alpha (RORα) and RORγ (79). In the presence of microbial antigens, generally those derived from noncommensals and proinflammatory cytokines, including IL-1β and IL-6, the action of retinoic acid is suppressed, iTreg development is blocked, and Tregs may be reprogrammed into IFN-γ- or IL-17-secreting effector T cells (Fig. 2) (80). In addition, antibacterial agents against Gram-positive bacteria have been shown to block local TLR and MyD88 innate immune signaling pathways that are essential for iTreg development (81, 82). Thus, recipient dysbiosis and/or antimicrobial agents, both common in allotransplantation, may alter iTreg phenotype and function toward proinflammatory adaptive immune responses. These shifts may predispose to the development of alloactive memory T cell responses, particularly after T cell depletional induction therapy.
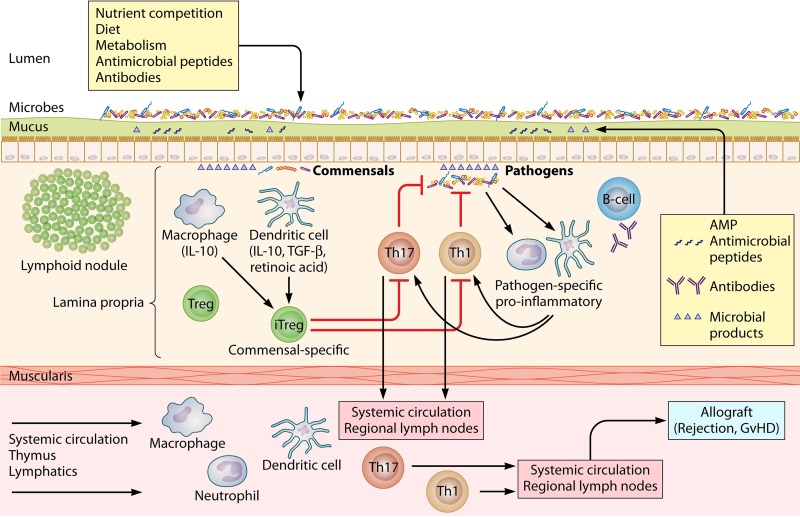
FIG 2.
The microbiome in alloimmunity. Under normal conditions, inflammatory responses to gastrointestinal flora are muted via both innate and adaptive immune mechanisms. Subsets of Tregs include both natural, thymus-derived cells with T cell receptors targeting self-antigens and induced Tregs (iTregs) derived from circulating CD4+ cells activated in the presence of microbial antigens, transforming growth factor β (TGF-β), and retinoic acid. In the presence of noncommensal microbial antigens, such as those from Clostridium spp. and Gram-positive segmented filamentous bacteria (SFB), and proinflammatory cytokines, iTreg development is blocked and Tregs may be reprogrammed into proinflammatory Th1 and Th17 effector cells. These effects may be blocked by antibacterial agents. Recipient dysbiosis and receipt of antimicrobial agents may shift iTreg development toward proinflammatory adaptive alloimmune responses. GvHD, graft-versus-host disease.
The microbiome also shapes relevant innate immune responses. In the intestine, molecules derived from gut microbes, including glycoproteins, LPS, and nucleic acids, are termed pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (MAMPs). MAMPs and PAMPs interact with the innate immune system via pattern recognition receptors (PRRs), including Toll-like receptors (TLRs) or Nod-like receptors (NLRs), on intestinal epithelial cells. Intestinal epithelial cells promote tolerigenic innate immune responses by conditioning intestinal dendritic cells to promote the development of regulatory T cell responses after these dendritic cells encounter antigen from commensal microorganisms (Fig. 2). Intestinal macrophages also respond to MAMP and PAMP signals via the development of inflammasomes, a complex of proteins that coordinate inflammatory processes. However, normal flora do not present antigen to intestinal macrophages as efficiently as newly introduced microbes in terms of the induction of inflammatory responses. Thus, innate immune responses to commensal organisms mediated by dendritic cells or by macrophages via dendritic cells are blunted (1, 2, 83, 84).
Innate immune responses to the microbiome have been explored in the context of liver transplantation. Under normal conditions, the liver contributes to containment of proinflammatory responses to commensal organisms (85). Conversely, in animal models of liver failure and in human studies of nonalcoholic steatohepatitis (NASH), liver disease has been associated with increased systemic immune responses to commensal microorganisms, possibly due to diminished filtration of bacteria and microbial products (85). Resident innate immune cells of the liver include natural killer (NK) cells, NKT cells, and macrophages (Kupffer cells); these interact with portal blood and participate in hepatic responses to ischemia-reperfusion (86). Germfree mice lack MAMPs in the portal circulation, and this absence of MAMPs appears to correlate with lower levels of hepatic leukocyte adhesion molecule expression and reduced numbers of Kupffer cells. Consistent with these studies, treatment of rats with polymyxin B prior to liver transplantation reduced intestinal levels of Enterobacteriaceae; this reduction was correlated with decreased portal circulation endotoxin levels, decreased hepatic Kupffer cell tissue factor activity, and decreased posttransplant hepatonecrosis (87). Thus, after ischemia-reperfusion, MAMPs may participate in recruitment of Kupffer cells to the liver allograft, contributing to graft injury after liver transplantation (88). This may reflect exaggerated inflammatory responses elicited by microbial flora in the portal circulation that upregulate expression of gastrointestinal TLRs and responses to danger-associated molecular patterns (DAMPs) from damaged cells after surgery.
MICROBIOME HOMEOSTASIS AMELIORATES ALLOGRAFT INJURY
Emerging evidence suggests that preservation of the pretransplant host microbiome in the posttransplantation period improves allograft outcomes. The persistence or resilience of the original pattern of the microbiome, notably in microbial diversity, through the process of allotransplantation is termed “microbiome homeostasis.” Microbiome homeostasis may promote tolerant host immune responses and avoid allograft injury. This has been best demonstrated in lung transplantation. In lung transplant patients undergoing protocol biopsies and bronchoalveolar lavage (BAL), de novo recipient colonization with Pseudomonas aeruginosa after transplantation was associated with development of early bronchiolitis obliterans syndrome (BOS), a form of chronic lung allograft injury (89). In another series, lung recipients with Firmicutes and Bacilli had more BOS than did those with Proteobacteria and Gammaproteobacteria (90). Willner et al. examined the relationship between lung transplant outcomes and the BAL fluid microbiome by 16S rRNA sequencing in a subset of cystic fibrosis patients. In those recipients who maintained the pretransplant colonizing flora in the posttransplant period, including those with Pseudomonas, there was a statistically significant decrease in BOS after lung transplantation (91). Conversely, colonization with new, pathogenic bacteria was associated with BOS (91).
Microbiome homeostasis may also ameliorate the effects of ischemia-reperfusion injury after transplantation. Short-chain fatty acids, the metabolic by-products of the intestinal microbiome, have been shown to attenuate ischemia-reperfusion injury (92). Short periods of hepatic ischemia followed by liver transplantation have been shown to ameliorate subsequent ischemic-reperfusion injury (93). This “ischemic preconditioning process” has been associated with restoration of pretransplant intestinal microbiota, including Clostridium and Bifidobacterium species. Amelioration of ischemia-reperfusion injury by ischemic preconditioning may be due to maintenance of normal intestinal flora, consistent with the potential importance of microbiome stability in prevention allograft injury. However, it is uncertain that the same effects would apply to organs not dependent upon the portal circulation.
POTENTIAL CLINICAL APPLICATIONS OF MICROBIOME MANAGEMENT IN ALLOTRANSPLANTATION
Manipulation of the microbiome may allow control of immune responses. In Pstip2cmo mice, spontaneous development of autoimmune osteomyelitis was prevented by diets rich in fat and cholesterol, with reductions in intestinal Prevotella levels and proinflammatory cytokines (94). Similarly, Lathrop et al. blocked the development of murine autoimmune arthritis using vancomycin treatment of SFB (73). These studies suggest that microbial modulation may durably impact proinflammatory immune responses. However, clinical applications of microbial manipulation are in their infancy. With respect to transplantation, the microbiome may serve as a marker of allograft health or a target for modulation of alloimmunity. Monitoring of the microbiome in intestinal transplant recipients may serve as a biomarker for allograft rejection; decrement of the Firmicutes population has been shown to be significantly correlated with acute rejection (95). However, similar associations between acute rejection and a specific microbiome composition(s) have not yet been identified in recipients of other organs. Recent studies in kidney transplant recipients did not demonstrate a clear correlation of the diversity in various microbiome sites with acute rejection events (59). Others have examined the microbiome profiles of small numbers of kidney transplant recipients with acute rejection and found significant dysbiosis following graft rejection compared with that in nonrejecting hosts (60). This analysis was confounded by the use of antimicrobial agents around the time of the diagnosis of rejection, possibly altering the microbial composition.
Loss of microbial diversity has implications for the risk for infections in the immunocompromised host. In stem cell transplantation, decreased microbial diversity and predominance of enterococcus in enteric flora preceded enterococcal bacteremia (6). Similarly, Fricke et al. demonstrated that decreased normal Firmicutes in rectal swabs in renal recipients preceded systemic infection of these hosts (59). Lee et al. reported an association between the absence of common intestinal microbiota, Bacteroides and Ruminococcus, and development of posttransplant diarrhea (60). In eight liver transplant recipients who did not preserve pretransplant intestinal microbiota, five developed posttransplant infections (96). Microbial shifts may prove to be useful diagnostic markers of acute rejection or impending infection in transplant patients; further studies are required to elucidate how to best use such data.
Modulation of the microbiome by fecal transplantation has demonstrated some success in the management of refractory Clostridium difficile infection both in immunocompetent and immunocompromised hosts; heightened surveillance for infection after fecal transplantation in immunocompromised hosts is recommended (97, 98). Specific microorganisms may contribute to resistance to C. difficile infection (99). Fecal transplantation may also assist in eradication of multidrug-resistant enteric flora (100) (101). Such changes in the composition of the microbiome will alter metabolism of antirejection immunosuppressants, including tacrolimus or cyclophosphamide (102, 103). As was noted, microbial by-products may affect the control of latency in herpesviral infections; these may achieve therapeutic applications if confirmed in clinical trials (30).
THE MICROBIOME IN CLINICAL ALLOTRANSPLANTATION
Complex interactions exist between the allograft recipient's microbiome, the immune system, and the allograft. There is emerging evidence that alteration of the pretransplant microbiome by the allotransplantation process results in poor clinical outcomes in both solid organ and stem cell recipients (6, 91). Viral and other new infections may contribute to detrimental immune responses to allografts. There are few studies that assess the impact of the networks of organisms and microbial products on immunity and graft function. Manipulation or reconstitution of the microbiome may be used to alter colonization patterns and as a diagnostic marker of impending posttransplant infections or of graft rejection. Therapeutic targets in the human microbiome may improve transplant outcomes in the future.
Biographies

Anoma Nellore, M.D., received her medical education at the University of Pennsylvania, followed by internal medicine training at the Hospital of the University of Pennsylvania and subsequent infectious diseases training at the combined Massachusetts General/Brigham and Women's program. She completed additional training in transplant infectious diseases at the Massachusetts General Hospital. Currently, she is an Assistant Professor at the University of Alabama at Birmingham, practicing transplant infectious diseases.

Jay A. Fishman, M.D., is Professor of Medicine at Harvard Medical School, Director of the Transplant Infectious Diseases and Compromised Host Program at the Massachusetts General Hospital (MGH), and Associate Director of the MGH Transplant Center. Dr. Fishman completed medical school at Johns Hopkins University School of Medicine, internal medicine training (1979 to 1981) and an Infectious Disease Fellowship at MGH (1981 to 1982), and a Fellowship in Molecular Biology and Genetics at MGH and Harvard Medical School (1982 to 1986). Dr. Fishman is a Fellow of the American College of Physicians and of the Infectious Disease Society of America. His laboratory investigates infections in xenotransplantation and viral pathogenesis in transplantation. He has a special interest in molecular diagnostics, biotechnology, and medical education. His leadership roles include past President of the American Society of Transplantation. He is a frequent contributor at international symposia and has received career achievement awards from the American Society of Transplantation and the Transplantation Society. Dr. Fishman has over 200 peer-reviewed publications.
REFERENCES
- 1.Maynard CL, Elson CO, Hatton RD, Weaver CT. 2012. Reciprocal interactions of the intestinal microbiota and immune system. Nature 489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belkaid Y, Hand TW. 2014. Role of the microbiota in immunity and inflammation. Cell 157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Surana NK, Kasper DL. 2014. Deciphering the tete-a-tete between the microbiota and the immune system. J Clin Invest 124:4197–4203. doi: 10.1172/JCI72332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. 2009. The NIH Human Microbiome Project. Genome Res 19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li K, Bihan M, Yooseph S, Methe BA. 2012. Analyses of the microbial diversity across the human microbiome. PLoS One 7:e32118. doi: 10.1371/journal.pone.0032118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, Perales MA, Jenq RR, van den Brink MR, Pamer EG. 2012. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. 2013. The role of the lung microbiome in health and disease. A National Heart, Lung, and Blood Institute workshop report. Am J Respir Crit Care Med 187:1382–1387. doi: 10.1164/rccm.201303-0488WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Human Microbiology Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huffnagle GB, Noverr MC. 2013. The emerging world of the fungal microbiome. Trends Microbiol 21:334–341. doi: 10.1016/j.tim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckburg PB. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virgin HW. 2014. The virome in mammalian physiology and disease. Cell 157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox MJ, Cookson WO, Moffatt MF. 2013. Sequencing the human microbiome in health and disease. Hum Mol Genet 22:R88–R94. doi: 10.1093/hmg/ddt398. [DOI] [PubMed] [Google Scholar]
- 13.Krummey SM, Ford ML. 2012. Heterogeneity within T cell memory: implications for transplant tolerance. Front Immunol 3:36. doi: 10.3389/fimmu.2012.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams AB, Williams MA, Jones TR, Shirasugi N, Durham MM, Kaech SM, Wherry EJ, Onami T, Lanier JG, Kokko KE, Pearson TC, Ahmed R, Larsen CP. 2003. Heterologous immunity provides a potent barrier to transplantation tolerance. J Clin Invest 111:1887–1895. doi: 10.1172/JCI200317477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pantenburg B, Heinzel F, Das L, Heeger PS, Valujskikh A. 2002. T cells primed by Leishmania major infection cross-react with alloantigens and alter the course of allograft rejection. J Immunol 169:3686–3693. doi: 10.4049/jimmunol.169.7.3686. [DOI] [PubMed] [Google Scholar]
- 16.Johannessen I, Crawford DH. 1999. In vivo models for Epstein-Barr virus (EBV)-associated B cell lymphoproliferative disease (BLPD). Rev Med Virol 9:263–277. [DOI] [PubMed] [Google Scholar]
- 17.Beus JM, Hashmi SS, Selvaraj SA, Duan D, Stempora LL, Monday SA, Cheeseman JA, Hamby KM, Speck SH, Larsen CP, Kirk AD, Kean LS. 2013. Heterologous immunity triggered by a single, latent virus in Mus musculus: combined costimulation- and adhesion-blockade decrease rejection. PLoS One 8:e71221. doi: 10.1371/journal.pone.0071221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi JS, Cox MA, Zajac AJ. 2010. T-cell exhaustion: characteristics, causes and conversion. Immunology 129:474–481. doi: 10.1111/j.1365-2567.2010.03255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, Kalia V, Subramaniam S, Blattman JN, Barber DL, Ahmed R. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Ou R, Zhang M, Huang L, Moskophidis D. 2008. Control of virus-specific CD8+ T-cell exhaustion and immune-mediated pathology by E3 ubiquitin ligase Cbl-b during chronic viral infection. J Virol 82:3353–3368. doi: 10.1128/JVI.01350-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wherry EJ. 2011. T cell exhaustion. Nat Immunol 12:492–499. [DOI] [PubMed] [Google Scholar]
- 22.Amir AL, D'Orsogna LJ, Roelen DL, van Loenen MM, Hagedoorn RS, de Boer R, van der Hoorn MA, Kester MG, Doxiadis II, Falkenburg JH, Claas FH, Heemskerk MH. 2010. Allo-HLA reactivity of virus-specific memory T cells is common. Blood 115:3146–3157. doi: 10.1182/blood-2009-07-234906. [DOI] [PubMed] [Google Scholar]
- 23.Schenk AD, Gorbacheva V, Rabant M, Fairchild RL, Valujskikh A. 2009. Effector functions of donor-reactive CD8 memory T cells are dependent on ICOS induced during division in cardiac grafts. Am J Transplant 9:64–73. doi: 10.1111/j.1600-6143.2008.02460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorres KL, Daigle D, Mohanram S, Miller G. 2014. Activation and repression of Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus lytic cycles by short- and medium-chain fatty acids. J Virol 88:8028–8044. doi: 10.1128/JVI.00722-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer SM, Burch LH, Davis RD, Herczyk WF, Howell DN, Reinsmoen NL, Schwartz DA. 2003. The role of innate immunity in acute allograft rejection after lung transplantation. Am J Respir Crit Care Med 168:628–632. doi: 10.1164/rccm.200303-447OC. [DOI] [PubMed] [Google Scholar]
- 26.Palmer SM, Burch LH, Mir S, Smith SR, Kuo PC, Herczyk WF, Reinsmoen NL, Schwartz DA. 2006. Donor polymorphisms in Toll-like receptor-4 influence the development of rejection after renal transplantation. Clin Transplant 20:30–36. doi: 10.1111/j.1399-0012.2005.00436.x. [DOI] [PubMed] [Google Scholar]
- 27.Kinnier CV, Martinu T, Gowdy KM, Nugent JL, Kelly FL, Palmer SM. 2011. Innate immune activation by the viral PAMP poly I:C potentiates pulmonary graft-versus-host disease after allogeneic hematopoietic cell transplant. Transpl Immunol 24:83–93. doi: 10.1016/j.trim.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garantziotis S, Palmer SM, Snyder LD, Ganous T, Chen BJ, Wang T, Cook DN, Schwartz DA. 2007. Alloimmune lung injury induced by local innate immune activation through inhaled lipopolysaccharide. Transplantation 84:1012–1019. doi: 10.1097/01.tp.0000286040.85007.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alegre ML, Chen L, Wang T, Ahmed E, Wang CR, Chong A. 2009. Antagonistic effect of toll-like receptor signaling and bacterial infections on transplantation tolerance. Transplantation 87:S77–S79. doi: 10.1097/TP.0b013e3181a2b90f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T, Chen L, Ahmed E, Ma L, Yin D, Zhou P, Shen J, Xu H, Wang CR, Alegre ML, Chong AS. 2008. Prevention of allograft tolerance by bacterial infection with Listeria monocytogenes. J Immunol 180:5991–5999. doi: 10.4049/jimmunol.180.9.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed EB, Wang T, Daniels M, Alegre ML, Chong AS. 2011. IL-6 induced by Staphylococcus aureus infection prevents the induction of skin allograft acceptance in mice. Am J Transplant 11:936–946. doi: 10.1111/j.1600-6143.2011.03476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Wang T, Zhou P, Ma L, Yin D, Shen J, Molinero L, Nozaki T, Phillips T, Uematsu S, Akira S, Wang CR, Fairchild RL, Alegre ML, Chong A. 2006. TLR engagement prevents transplantation tolerance. Am J Transplant 6:2282–2291. doi: 10.1111/j.1600-6143.2006.01489.x. [DOI] [PubMed] [Google Scholar]
- 33.Han Lee ED, Kemball CC, Wang J, Dong Y, Stapler DC Jr, Hamby KM, Gangappa S, Newell KA, Pearson TC, Lukacher AE, Larsen CP. 2006. A mouse model for polyomavirus-associated nephropathy of kidney transplants. Am J Transplant 6:913–922. doi: 10.1111/j.1600-6143.2006.01265.x. [DOI] [PubMed] [Google Scholar]
- 34.Spangenberg HC, Viazov S, Kersting N, Neumann-Haefelin C, McKinney D, Roggendorf M, von Weizsacker F, Blum HE, Thimme R. 2005. Intrahepatic CD8+ T-cell failure during chronic hepatitis C virus infection. Hepatology 42:828–837. doi: 10.1002/hep.20856. [DOI] [PubMed] [Google Scholar]
- 35.Nellore A, Fishman JA. 2011. NK cells, innate immunity and hepatitis C infection after liver transplantation. Clin Infect Dis 52:369–377. doi: 10.1093/cid/ciq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciuffreda D, Codarri L, Buhler L, Vallotton L, Giostra E, Mentha G, Morel P, Pantaleo G, Pascual M. 2010. Hepatitis C virus infection after liver transplantation is associated with lower levels of activated CD4(+)CD25(+)CD45RO(+)IL-7ralpha(high) T cells. Liver Transpl 16:49–55. doi: 10.1002/lt.21959. [DOI] [PubMed] [Google Scholar]
- 37.Carpentier A, Conti F, Stenard F, Aoudjehane L, Miroux C, Podevin P, Morales O, Chouzenoux S, Scatton O, Groux H, Auriault C, Calmus Y, Pancre V, Delhem N. 2009. Increased expression of regulatory Tr1 cells in recurrent hepatitis C after liver transplantation. Am J Transplant 9:2102–2112. doi: 10.1111/j.1600-6143.2009.02743.x. [DOI] [PubMed] [Google Scholar]
- 38.Gredmark S, Soderberg-Naucler C. 2003. Human cytomegalovirus inhibits differentiation of monocytes into dendritic cells with the consequence of depressed immunological functions. J Virol 77:10943–10956. doi: 10.1128/JVI.77.20.10943-10956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bando K, Paradis IL, Similo S, Konishi H, Komatsu K, Zullo TG, Yousem SA, Close JM, Zeevi A, Duquesnoy RJ, Manzetti J, Keenan RJ, Armitage JM, Hardesty RL, Griffith BP. 1995. Obliterative bronchiolitis after lung and heart-lung transplantation. An analysis of risk factors and management. J Thorac Cardiovasc Surg 110:4–13. (Discussion 110:13–14.) [DOI] [PubMed] [Google Scholar]
- 40.Grattan MT, Moreno-Cabral CE, Starnes VA, Oyer PE, Stinson EB, Shumway NE. 1989. Cytomegalovirus infection is associated with cardiac allograft rejection and atherosclerosis. JAMA 261:3561–3566. [PubMed] [Google Scholar]
- 41.Dzabic M, Rahbar A, Yaiw KC, Naghibi M, Religa P, Fellstrom B, Larsson E, Soderberg-Naucler C. 2011. Intragraft cytomegalovirus protein expression is associated with reduced renal allograft survival. Clin Infect Dis 53:969–976. doi: 10.1093/cid/cir619. [DOI] [PubMed] [Google Scholar]
- 42.Kumar D, Husain S, Chen MH, Moussa G, Himsworth D, Manuel O, Studer S, Pakstis D, McCurry K, Doucette K, Pilewski J, Janeczko R, Humar A. 2010. A prospective molecular surveillance study evaluating the clinical impact of community-acquired respiratory viruses in lung transplant recipients. Transplantation 89:1028–1033. doi: 10.1097/TP.0b013e3181d05a71. [DOI] [PubMed] [Google Scholar]
- 43.Koskinen P, Lemstrom K, Bruggeman C, Lautenschlager I, Hayry P. 1994. Acute cytomegalovirus infection induces a subendothelial inflammation (endothelialitis) in the allograft vascular wall. A possible linkage with enhanced allograft arteriosclerosis. Am J Pathol 144:41–50. [PMC free article] [PubMed] [Google Scholar]
- 44.Craigen JL, Yong KL, Jordan NJ, MacCormac LP, Westwick J, Akbar AN, Grundy JE. 1997. Human cytomegalovirus infection up-regulates interleukin-8 gene expression and stimulates neutrophil transendothelial migration. Immunology 92:138–145. doi: 10.1046/j.1365-2567.1997.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helantera I, Loginov R, Koskinen P, Tornroth T, Gronhagen-Riska C, Lautenschlager I. 2005. Persistent cytomegalovirus infection is associated with increased expression of TGF-beta1, PDGF-AA and ICAM-1 and arterial intimal thickening in kidney allografts. Nephrol Dial Transplant 20:790–796. doi: 10.1093/ndt/gfh714. [DOI] [PubMed] [Google Scholar]
- 46.Streblow DN, Kreklywich CN, Andoh T, Moses AV, Dumortier J, Smith PP, Defilippis V, Fruh K, Nelson JA, Orloff SL. 2008. The role of angiogenic and wound repair factors during CMV-accelerated transplant vascular sclerosis in rat cardiac transplants. Am J Transplant 8:277–287. doi: 10.1111/j.1600-6143.2007.02062.x. [DOI] [PubMed] [Google Scholar]
- 47.Potena L, Grigioni F, Magnani G, Lazzarotto T, Musuraca AC, Ortolani P, Coccolo F, Fallani F, Russo A, Branzi A. 2009. Prophylaxis versus preemptive anti-cytomegalovirus approach for prevention of allograft vasculopathy in heart transplant recipients. J Heart Lung Transplant 28:461–467. doi: 10.1016/j.healun.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 48.Lowance D, Neumayer HH, Legendre CM, Squifflet JP, Kovarik J, Brennan PJ, Norman D, Mendez R, Keating MR, Coggon GL, Crisp A, Lee IC. 1999. Valacyclovir for the prevention of cytomegalovirus disease after renal transplantation. International Valacyclovir Cytomegalovirus Prophylaxis Transplantation Study Group. N Engl J Med 340:1462–1470. [DOI] [PubMed] [Google Scholar]
- 49.Kliem V, Fricke L, Wollbrink T, Burg M, Radermacher J, Rohde F. 2008. Improvement in long-term renal graft survival due to CMV prophylaxis with oral ganciclovir: results of a randomized clinical trial. Am J Transplant 8:975–983. doi: 10.1111/j.1600-6143.2007.02133.x. [DOI] [PubMed] [Google Scholar]
- 50.Fishman JA. 2013. Overview: cytomegalovirus and the herpesviruses in transplantation. Am J Transplant 13(Suppl 3):1–8. doi: 10.1111/ajt.12002. [DOI] [PubMed] [Google Scholar]
- 51.Mocarski ES., Jr 2004. Immune escape and exploitation strategies of cytomegaloviruses: impact on and imitation of the major histocompatibility system. Cell Microbiol 6:707–717. doi: 10.1111/j.1462-5822.2004.00425.x. [DOI] [PubMed] [Google Scholar]
- 52.Couzi L, Pitard V, Moreau JF, Merville P, Dechanet-Merville J. 2015. Direct and indirect effects of cytomegalovirus-induced gammadelta T cells after kidney transplantation. Front Immunol 6:3. doi: 10.3389/fimmu.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ressing ME, Horst D, Griffin BD, Tellam J, Zuo J, Khanna R, Rowe M, Wiertz EJ. 2008. Epstein-Barr virus evasion of CD8(+) and CD4(+) T cell immunity via concerted actions of multiple gene products. Semin Cancer Biol 18:397–408. doi: 10.1016/j.semcancer.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 54.Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. 2000. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc Natl Acad Sci U S A 97:1695–1700. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki T, Tahara H, Narula S, Moore KW, Robbins PD, Lotze MT. 1995. Viral interleukin 10 (IL-10), the human herpes virus 4 cellular IL-10 homologue, induces local anergy to allogeneic and syngeneic tumors. J Exp Med 182:477–486. doi: 10.1084/jem.182.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Graham JA, Wilkinson RA, Hirohashi T, Chase CM, Colvin RB, Madsen JC, Fishman JA, Russell PS. 2009. Viral infection induces de novo lesions of coronary allograft vasculopathy through a natural killer cell-dependent pathway. Am J Transplant 9:2479–2484. doi: 10.1111/j.1600-6143.2009.02801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Floyd TL, Koehn BH, Kitchens WH, Robertson JM, Cheeseman JA, Stempora L, Larsen CP, Ford ML. 2011. Limiting the amount and duration of antigen exposure during priming increases memory T cell requirement for costimulation during recall. J Immunol 186:2033–2041. doi: 10.4049/jimmunol.1003015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diaz PI, Hong BY, Frias-Lopez J, Dupuy AK, Angeloni M, Abusleme L, Terzi E, Ioannidou E, Strausbaugh LD, Dongari-Bagtzoglou A. 2013. Transplantation-associated long-term immunosuppression promotes oral colonization by potentially opportunistic pathogens without impacting other members of the salivary bacteriome. Clin Vaccine Immunol 20:920–930. doi: 10.1128/CVI.00734-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fricke WF, Maddox C, Song Y, Bromberg JS. 2014. Human microbiota characterization in the course of renal transplantation. Am J Transplant 14:416–427. doi: 10.1111/ajt.12588. [DOI] [PubMed] [Google Scholar]
- 60.Lee JR, Muthukumar T, Dadhania D, Toussaint NC, Ling L, Pamer E, Suthanthiran M. 2014. Gut microbial community structure and complications after kidney transplantation: a pilot study. Transplantation 98:697–705. doi: 10.1097/TP.0000000000000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, Eisen JA. 2009. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci U S A 106:17187–17192. doi: 10.1073/pnas.0904847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang F, Li Q, Wang C, Tang C, Li J. 2012. Dynamic alteration of the colonic microbiota in intestinal ischemia-reperfusion injury. PLoS One 7:e42027. doi: 10.1371/journal.pone.0042027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gurkan S, Luan Y, Dhillon N, Allam SR, Montague T, Bromberg JS, Ames S, Lerner S, Ebcioglu Z, Nair V, Dinavahi R, Sehgal V, Heeger P, Schroppel B, Murphy B. 2010. Immune reconstitution following rabbit antithymocyte globulin. Am J Transplant 10:2132–2141. doi: 10.1111/j.1600-6143.2010.03210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruggenenti P, Perico N, Gotti E, Cravedi P, D'Agati V, Gagliardini E, Abbate M, Gaspari F, Cattaneo D, Noris M, Casiraghi F, Todeschini M, Cugini D, Conti S, Remuzzi G. 2007. Sirolimus versus cyclosporine therapy increases circulating regulatory T cells, but does not protect renal transplant patients given alemtuzumab induction from chronic allograft injury. Transplantation 84:956–964. doi: 10.1097/01.tp.0000284808.28353.2c. [DOI] [PubMed] [Google Scholar]
- 65.Tchao NK, Turka LA. 2012. Lymphodepletion and homeostatic proliferation: implications for transplantation. Am J Transplant 12:1079–1090. doi: 10.1111/j.1600-6143.2012.04008.x. [DOI] [PubMed] [Google Scholar]
- 66.Kieper WC, Troy A, Burghardt JT, Ramsey C, Lee JY, Jiang HQ, Dummer W, Shen H, Cebra JJ, Surh CD. 2005. Recent immune status determines the source of antigens that drive homeostatic T cell expansion. J Immunol 174:3158–3163. doi: 10.4049/jimmunol.174.6.3158. [DOI] [PubMed] [Google Scholar]
- 67.Min B, Yamane H, Hu-Li J, Paul WE. 2005. Spontaneous and homeostatic proliferation of CD4 T cells are regulated by different mechanisms. J Immunol 174:6039–6044. doi: 10.4049/jimmunol.174.10.6039. [DOI] [PubMed] [Google Scholar]
- 68.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 69.An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL. 2014. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 71.Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, Laufer TM, Ignatowicz L, Ivanov II. 2014. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal th17 cell differentiation. Immunity 40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. 2011. Peripheral education of the immune system by colonic commensal microbiota. Nature 478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. 2012. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Round JL, Mazmanian SK. 2010. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A 107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. 2011. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 77.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. 2007. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 80.Peck A, Mellins ED. 2010. Plasticity of T-cell phenotype and function: the T helper type 17 example. Immunology 129:147–153. doi: 10.1111/j.1365-2567.2009.03189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rivas MN, Koh YT, Chen A, Nguyen A, Lee YH, Lawson G, Chatila TA. 2012. MyD88 is critically involved in immune tolerance breakdown at environmental interfaces of Foxp3-deficient mice. J Clin Invest 122:1933–1947. doi: 10.1172/JCI40591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. 2008. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, Shaw MH, Kim YG, Nunez G. 2012. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol 13:449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hill DA, Artis D. 2010. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol 28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Balmer ML, Slack E, de Gottardi A, Lawson MA, Hapfelmeier S, Miele L, Grieco A, Van Vlierberghe H, Fahrner R, Patuto N, Bernsmeier C, Ronchi F, Wyss M, Stroka D, Dickgreber N, Heim MH, McCoy KD, Macpherson AJ. 2014. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci Transl Med 6:237ra266. [DOI] [PubMed] [Google Scholar]
- 86.Kolios G, Valatas V, Kouroumalis E. 2006. Role of Kupffer cells in the pathogenesis of liver disease. World J Gastroenterol 12:7413–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arai M, Mochida S, Ohno A, Arai S, Fujiwara K. 1998. Selective bowel decontamination of recipients for prevention against liver injury following orthotopic liver transplantation: evaluation with rat models. Hepatology 27:123–127. doi: 10.1002/hep.510270120. [DOI] [PubMed] [Google Scholar]
- 88.Corbitt N, Kimura S, Isse K, Specht S, Chedwick L, Rosborough BR, Lunz JG, Murase N, Yokota S, Demetris AJ. 2013. Gut bacteria drive Kupffer cell expansion via MAMP-mediated ICAM-1 induction on sinusoidal endothelium and influence preservation-reperfusion injury after orthotopic liver transplantation. Am J Pathol 182:180–191. doi: 10.1016/j.ajpath.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Botha P, Archer L, Anderson RL, Lordan J, Dark JH, Corris PA, Gould K, Fisher AJ. 2008. Pseudomonas aeruginosa colonization of the allograft after lung transplantation and the risk of bronchiolitis obliterans syndrome. Transplantation 85:771–774. doi: 10.1097/TP.0b013e31816651de. [DOI] [PubMed] [Google Scholar]
- 90.Poroyko V, Semenyuk E, Xu Z-P, Sperling A, Alegre M-L, Chong A, Garrity E, White S, Bhorade S. 2014. Post-transplant dynamics of the pulmonary microbiota. Ann Am Thorac Soc 11:S74. doi: 10.1513/AnnalsATS.201307-238MG. [DOI] [Google Scholar]
- 91.Willner DL, Hugenholtz P, Yerkovich ST, Tan ME, Daly JN, Lachner N, Hopkins PM, Chambers DC. 2013. Reestablishment of recipient-associated microbiota in the lung allograft is linked to reduced risk of bronchiolitis obliterans syndrome. Am J Respir Crit Care Med 187:640–647. doi: 10.1164/rccm.201209-1680OC. [DOI] [PubMed] [Google Scholar]
- 92.Andrade-Oliveira V, Amano MT, Correa-Costa M, Castoldi A, Felizardo RJ, de Almeida DC, Bassi EJ, Moraes-Vieira PM, Hiyane MI, Rodas AC, Peron JP, Aguiar CF, Reis MA, Ribeiro WR, Valduga CJ, Curi R, Vinolo MA, Ferreira CM, Camara NO. 2015. Gut bacteria products prevent AKI induced by ischemia-reperfusion. J Am Soc Nephrol 26:1877–1888. doi: 10.1681/ASN.2014030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ren Z, Cui G, Lu H, Chen X, Jiang J, Liu H, He Y, Ding S, Hu Z, Wang W, Zheng S. 2013. Liver ischemic preconditioning (IPC) improves intestinal microbiota following liver transplantation in rats through 16s rDNA-based analysis of microbial structure shift. PLoS One 8:e75950. doi: 10.1371/journal.pone.0075950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lukens JR, Gurung P, Vogel P, Johnson GR, Carter RA, McGoldrick DJ, Bandi SR, Calabrese CR, Walle LV, Lamkanfi M, Kanneganti TD. 2014. Dietary modulation of the microbiome affects autoinflammatory disease. Nature 516:246–249. doi: 10.1038/nature13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oh PL, Martinez I, Sun Y, Walter J, Peterson DA, Mercer DF. 2012. Characterization of the ileal microbiota in rejecting and nonrejecting recipients of small bowel transplants. Am J Transplant 12:753–762. doi: 10.1111/j.1600-6143.2011.03860.x. [DOI] [PubMed] [Google Scholar]
- 96.Lu H, He J, Wu Z, Xu W, Zhang H, Ye P, Yang J, Zhen S, Li L. 2013. Assessment of microbiome variation during the perioperative period in liver transplant patients: a retrospective analysis. Microb Ecol 65:781–791. doi: 10.1007/s00248-013-0211-6. [DOI] [PubMed] [Google Scholar]
- 97.Friedman-Moraco RJ, Mehta AK, Lyon GM, Kraft CS. 2014. Fecal microbiota transplantation for refractory Clostridium difficile colitis in solid organ transplant recipients. Am J Transplant 14:477–480. doi: 10.1111/ajt.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 99.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Singh R, van Nood E, Nieuwdorp M, van Dam B, ten Berge IJ, Geerlings SE, Bemelman FJ. 2014. Donor feces infusion for eradication of extended spectrum beta-lactamase producing Escherichia coli in a patient with end stage renal disease. Clin Microbiol Infect 20:O977–O978. doi: 10.1111/1469-0691.12683. [DOI] [PubMed] [Google Scholar]
- 101.Stripling J, Kumar R, Baddley JW, Nellore A, Dixon P, Howard D, Ptacek T, Lefkowitz EJ, Tallaj JA, Benjamin WH Jr, Morrow CD, Rodriguez JM. 2015. Loss of vancomycin-resistant Enterococcus fecal dominance in an organ transplant patient with Clostridium difficile colitis after fecal microbiota transplant. Open Forum Infect Dis 2:ofv078. doi: 10.1093/ofid/ofv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee JR, Muthukumar T, Dadhania D, Taur Y, Jenq RR, Toussaint NC, Ling L, Pamer E, Suthanthiran M. 2015. Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS One 10:e0122399. doi: 10.1371/journal.pone.0122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, Schlitzer A, Ginhoux F, Apetoh L, Chachaty E, Woerther PL, Eberl G, Berard M, Ecobichon C, Clermont D, Bizet C, Gaboriau-Routhiau V, Cerf-Bensussan N, Opolon P, Yessaad N, Vivier E, Ryffel B, Elson CO, Dore J, Kroemer G, Lepage P, Boneca IG, Ghiringhelli F, Zitvogel L. 2013. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]