Abstract
Escherichia coli O111 is an emerging non-O157:H7 serotype of Shiga toxin-producing E. coli (STEC). We previously reported that outbreak and environmental, but not sporadic-case, strains of STEC O111 share a distinct aggregation phenotype (M. E. Diodati, A. H. Bates, M. B. Cooley, S. Walker, R. E. Mandrell, and M. T. Brandl, Foodborne Pathog Dis 12:235−243, 2015, http://dx.doi.org/10.1089/fpd.2014.1887). We show here the natural occurrence of nonaggregative variants in single STEC O111 strains. These variants do not produce curli fimbriae and lack RpoS function but synthesize cellulose. The deletion of csgBAC or rpoS in an aggregative outbreak strain abolished aggregate formation, which was rescued when curli biogenesis or RpoS function, respectively, was restored. Complementation of a nonaggregative variant with RpoS also conferred curli production and aggregation. These observations were supported by Western blotting with an anti-CsgA antibody. Immunomicroscopy revealed that curli were undetectable on the cells of the nonaggregative variant and the RpoS mutant but were present in large quantities in the intercellular matrix of the assemblages formed by aggregative strains. Sequence analysis of rpoS in the aggregative strain and its variant showed a single substitution of threonine for asparagine at amino acid 124. Our results indicate that the multicellular behavior of STEC O111 is RpoS dependent via positive regulation of curli production. Aggregation may confer a fitness advantage in O111 outbreak strains under stressful conditions in hydrodynamic environments along the food production chain and in the host, while the occurrence of nonaggregative variants may allow the cell population to adapt to conditions benefiting a planktonic lifestyle.
INTRODUCTION
The global burden of Shiga toxin-producing Escherichia coli (STEC) infection is estimated to be 2.8 million cases of acute illnesses annually worldwide (1), and non-O157 STEC infections result in nearly 113,000 illnesses yearly in the United States (2). STEC O111 is among the six most commonly reported non-O157 STEC serogroups (3). It is the most prevalent STEC serogroup in Europe (4) and the third most important STEC serogroup in the United States, causing 19% of the illness cases in 2000 to 2010 (3). Outbreaks of STEC O111 illness also have been reported in Australia (5), Canada (6), and Japan (7). Many of the reported cases of STEC O111 infection in the United States are of sporadic illness. However, this serogroup has caused more than 15 well-known outbreaks in the United States since 1999 (8–10).
We have previously assembled a collection of environmental and clinical strains of E. coli O111 from diverse sources and investigated various genotypic and phenotypic characteristics of these strains to gain a better understanding of the epidemiology and biology of this serogroup. Our study revealed that many environmental STEC O111 strains isolated from diverse sources over time and from various geographical locations are genotypically identical, suggesting that these strains circulate in the environment and may contaminate the food chain (11). The environmental and outbreak strains in this collection were similar in their virulence determinants, and all displayed a strong aggregative phenotype characterized by the settlement of large bacterial assemblages in broth culture with agitation; in contrast, the sporadic case strains, which are strains isolated from single patients that were not part of a recognized outbreak, were nonaggregative (11).
Autoaggregation is caused by bacterial cell-cell interactions that lead to the production of a range of multicellular assemblages and, in some cases, may cause flocculation and settling of the cells in static liquid suspensions. A number of bacterial surface factors that are involved in E. coli interactions with host cells and virulence are also implicated in autoaggregation (12). These adhesins, which mediate the attachment of E. coli cells to one another and to human epithelial cells, include type I fimbriae, type IV bundle-forming pili, AAF/I and AAF/II aggregative adherence fimbriae, and the autotransporter proteins TibA and Ag43 (12). In 2011, a strain of STEC O104:H4 that harbored the AAF virulence and aggregative adherence determinants of enteroaggregative E. coli (EAEC) caused a large outbreak in Germany (13), calling attention to bacterial aggregation as an important factor in epidemics of foodborne disease. The ability to form aggregates also may increase the tolerance of human pathogens to physicochemical stresses in host and in nonhost environments. Expression of the self-aggregative adhesins Ag43 and TibA in E. coli promotes the formation of single- and mixed-species biofilms (14, 15), and Ag43 and type IV bundle-forming pili enhance resistance to antimicrobial agents in E. coli (16, 17), thus supporting the view that autoaggregation may be an important mechanism for bacterial survival.
Given the role of aggregation in the pathogenicity and environmental fitness of E. coli and our previous observation that STEC O111 outbreak and environmental strains aggregate in culture, whereas sporadic strains are nonaggregative (11), we sought to identify the factors involved in this aggregative behavior in order to further our understanding of STEC O111 epidemiology. Because STEC O111 aggregation strongly correlated with high levels of curli fimbrial production and RpoS function in our previous study (11), we assessed the role of these fimbriae and sigma RpoS-mediated regulation in the aggregation of this important enteric pathogen. Curli promote initial adhesion to abiotic and biotic surfaces, including epithelial cells, as well as cell-cell interactions during biofilm formation (18). The expression of curli operons is regulated by environmental signals via RpoS and at least five transcription factors (19, 20), allowing transcription to respond to a number of different stimuli, including pH, temperature, low osmolarity, and nutrient limitation (20, 21).
In the study described herein, we obtained evidence that curli fimbriae are required for the autoaggregative behavior of STEC O111 in liquid culture and that this phenomenon depends on the presence of functional RpoS. Furthermore, we show that the polymorphic nature of this aggregative phenotype is linked to the regulation of curli synthesis by RpoS and that the nonaggregative variants of STEC O111 arise naturally due the presence of RpoS-defective mutants in a given cell population. This heterogeneity in aggregative behavior may impart to STEC O111 the adaptability required to persist in the environment along the food production chain and reach suitable hosts.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study are listed in Tables 1 and 2. All strains were cultured in Luria-Bertani half-salt (5 g NaCl/liter) (LBHS) broth or Luria-Bertani no-salt (0 g NaCl) (LBNS) broth when appropriate and incubated at 28°C. LBHS broth was amended as appropriate with kanamycin (30 μg/ml), gentamicin (15 μg/ml), or chloramphenicol (20 μg/ml).
TABLE 1.
List of previously described strains and plasmids used in this study
| E. coli serogroup and strain name or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Serogroups and strains | ||
| O111 | ||
| ECRC 00.1441 | O111:H8 isolate from cow (feces); USA (WI); 2008 | 11 |
| ECRC 00.1932 | O111:H8 isolate from cow (feces); USA (PA); 2008 | 11 |
| RM9131 | O111:H8 isolate from cow (feces); USA (CA); 2009 | 11 |
| RM9975 | O111:H7 isolate from crow (intestines); USA (CA); 2009 | 11 |
| BE99-1161 | O111:H8 clinical isolate associated with ice and salad-linked outbreak; USA (TX); 1999 | 11 |
| K6807 | O111:H8 clinical isolate associated with salad bar-linked outbreak; USA (OK); 2008 | 11 |
| SD134209 | O111:H8 clinical isolate associated with person-to-person outbreak; USA (SD); 2009 | 11 |
| O157:H7 | ||
| EDL933 | Clinical isolate associated with hamburger-linked outbreak; USA (MI); 1982 | 55 |
| RM6607R | Clinical isolate associated with hamburger-linked outbreak; USA; 1993 | 56 |
| K-12 | ||
| MG1655 | General laboratory strain | ATCC 29425 |
| Plasmids | ||
| pBBR1MCS-5 | Broad-range cloning vector; Gmr | 23 |
| pMBcsgO111 | pBBR1MCS-5 carrying the csgBAC operon and native promoter of MB975; Gmr | This study |
| pMB682 | pBBR1MCS-5 carrying the cellulose operon of Salmonella enterica serovar Typhimurium SL1344; Gmr | 28 |
| pXQ29 | pBBR1MCS-5 carrying the rpoS gene and native promoter of RM6067W; Gmr | 29 |
For strains, the description is followed by the location and year of isolation.
TABLE 2.
List of strains isolated or constructed in this study and their relevant phenotypic characteristics
| Strain name | Description | Phenotypea |
|||
|---|---|---|---|---|---|
| Aggregation | Curli | Cellulose | RpoS | ||
| Isolated variants of E. coli O111 | |||||
| MB975 | Aggregative isolate of outbreak strain SD134209 | P | +++ | A | +++ |
| MB971 | Nonaggregative variant of MB975 | A | − | P | − |
| MB1005 | Aggregative isolate of outbreak strain K6807 | P | +++ | A | ++ |
| MB1006 | Nonaggregative variant of K6807 | A | − | P | − |
| MB1108 | Aggregative isolate of environmental strain ECRC 00.1441 | P | +++ | A | ++ |
| MB1109 | Nonaggregative variant of MB1108 | A | − | P | − |
| MB1110 | Aggregative isolate of environmental strain ECRC 00.1932 | P | +++ | A | + |
| MB1111 | Nonaggregative variant of MB1110 | A | − | P | − |
| MB1117 | Aggregative isolate of outbreak strain BE99-1161 | P | +++ | A | + |
| MB1118 | Nonaggregative variant of MB1117 | A | − | P | − |
| Mutants and transformants of E. coli O111 SD134209 | |||||
| MB1044 | MB975 ΔcsgBAC::kan (Kmr) | A | − | A | +++ |
| MB1054 | MB1044/pMBcsgO111 | P | +++ | A | +++ |
| MB1062 | MB971/pMBcsgO111 | A | − | P | − |
| MB1064 | MB971/pBBR1MCS-5 | A | − | P | − |
| MB1066 | MB975/pBBR1MCS-5 | P | +++ | A | +++ |
| MB1068 | MB1044/pBBR1MCS-5 | A | − | A | +++ |
| MB1096 | MB971/pXQ29 | P | ++ | A | +++ |
| MB1191 | MB975 ΔrpoS::cat (Cmr) | A | − | P | − |
| MB1192 | MB1191/pXQ29 | P | +++ | A | ++ |
| Non-O111 strains used as controls | |||||
| EDL933 | E. coli O157:H7 | A | + | A | ++ |
| RM6607R | E. coli O157:H7 RcsB− | A | ++ | A | + |
| MG1655 | E. coli K-12 | A | + | A | + |
Aggregation, curli production, cellulose synthesis, and RpoS function were assayed in glass culture tubes with LBHS broth on CRI plates, on calcofluor white plates, and with iodine staining. +++, ++, +, and −, strong, medium, weak, and undetectable production of curli or RpoS function, respectively; P or A, presence or absence of aggregation and cellulose synthesis.
Strain construction.
Deletion mutants of MB975 that are defective in curli fimbriae (MB1044) or RpoS (MB1191) were generated via lambda Red-mediated recombination using pKD4 or pKD3 (22) by replacing csgBAC or rpoS with a kanamycin or chloramphenicol resistance cassette, respectively. The deletions of csgBAC and rpoS were verified by PCRs. The PCR primers used for mutagenesis and to verify deletion of the target genes in the mutants are listed in Table S1 in the supplemental material. The absence of the curli structural subunits was confirmed also by streaking of the mutants onto Congo red indicator (CRI) plates and observing white colonies, by immunomicroscopy, and by Western blot analysis using an anti-CsgA antibody (see below). The loss of RpoS was confirmed with an iodine-based assay, which is described below.
The curli complementation plasmid (pMBcsgO111) was constructed by PCR cloning of csgBAC and ligation into HinDIII- and KpnI-restricted pBBR1MCS-5, a medium-copy-number broad-host-range plasmid (23), using primers for the amplification of each clone that harbored a HinDIII or KpnI site at its 5′ end. Primers used for the construction of pMBcsgO111 are listed in Table S1 in the supplemental material. The presence of the clone on pBBR1MCS-5 was verified by restriction digestion.
Aggregation in liquid culture.
Strains were grown in borosilicate Kimax test tubes (Kimble Chase, Vineland, NJ) in 5 ml LBHS broth and incubated to early stationary phase at 28°C with 25-rpm rotary agitation in a rolling drum. Aggregation at the bottom of the culture tube was observed directly during agitated incubation and did not require static conditions to allow for settling of the aggregates over time. For determination of aggregation over time, cells were grown as described above and absorbance at 600 nm was measured at regular time intervals. When the maximum absorbance had been reached after 5.5 h, aliquots of the cultures also were diluted 1:10 in separate tubes to allow for more accurate absorbance measurements and to better assess the progression of aggregative behavior.
Isolation of nonaggregative variants.
Aggregative and nonaggregative variants of single O111 outbreak strains and environmental strains were isolated by growing cultures at 37°C (nonaggregating conditions) in LBHS broth and dilution plating onto LBHS agar. Twenty-five to 50 colonies were chosen, inoculated into glass tubes, and grown under agitation at 28°C to test for aggregation. Nonaggregative variants were selected for further analysis.
Phenotypic tests on growth agar.
Strains were screened for curli fimbrial production on CRI plates (LB no-salt agar with 40 μg/ml Congo red dye and 10 μg/ml Coomassie brilliant blue), on which curli-positive colonies are red, and for cellulose production on LB low-salt (0.5g/liter NaCl) agar containing 0.025% calcofluor white M2R with Evans blue dye (Fluka; Sigma-Aldrich, St. Louis, MO), on which cellulose-positive colonies have bright fluorescence under UV exposure, based on previously described methods (24, 25). The presence of functional RpoS in the strains was detected by staining glycogen in the colonies on LBHS agar with iodine, based on a modification of the method of King et al. (26) that we described previously (11).
Western blot analysis.
The purified polyclonal anti-curli peptide antibodies were prepared by Biomatik Custom Antibody Service (Ontario, Canada). Two peptides of 14 amino acids (aa) each were selected and designed based on the sequence of csgA from E. coli K-12. The sequence of these peptides is identical to that of CsgA in E. coli O111 and E. coli O157:H7. The polyclonal IgG antibody was derived from two rabbits immunized with the two synthesized peptides. Western blot analysis was carried out as previously described (27, 28), with a few modifications. Overnight cultures (5 ml) in LBHS or LBNS broth with agitation at 28°C were centrifuged, and the cells were resuspended in 100 μl sterile H2O. Twenty microliters of each bacterial cell suspension was added to 150 μl 95% formic acid and vortexed for 1 min to depolymerize the curli fibers. The suspensions were incubated for 40 min and then dried in a Savant DNA120 SpeedVac concentrator (Thermo Scientific, Waltham, MA) for 2 h at high temperature. The samples were solubilized by the addition of 150 μl 2× Tricine sample buffer (Novex; Life Technologies, Grand Island, NY) containing 2.5% mercaptoethanol, vortexed for 1 min, and then sonicated for 5 min in a water bath sonicator. The samples were heated at 100°C for 3 min for analysis by gel electrophoresis.
The samples were resolved on 10 to 20% Tricine gels (Novex; Life Technologies, Grand Island, NY). The proteins were electrotransferred onto Novex 0.2-μm-pore-size nitrocellulose membranes in a Bio-Rad Trans-Blot SD semidry transfer cell (Bio-Rad, Hercules, CA). The membranes were blocked with a bovine serum albumin (BSA)–Tris-buffered saline (TBS)–Tween 20 solution (1% BSA, 10 mM Tris-HCl, 150 mM NaCl, 5 mM MgCl2, 0.05% Tween 20 [pH 7.4]). The membranes were probed with the polyclonal anti-curli peptide antibodies described above for the detection of curli, and mouse anti-DnaK monoclonal antibody (LifeSpan BioSciences, Seattle, WA) as an internal control for normalization of the protein concentrations across the samples. The membranes were then incubated with alkaline phosphatase-labeled goat anti-rabbit and rabbit anti-mouse secondary antibodies for CsgA and DnaK, respectively (Thermo Scientific, Waltham, MA). The immunoblots were visualized with the SigmaFast BCIP-NBT (5-bromo-4-chloro-3-indolyl phosphate–nitroblue tetrazolium) precipitating substrate kit (Sigma-Aldrich, St. Louis, MO).
Confocal scanning microscopy.
Confocal laser scanning microscopy was performed to determine the presence of curli fimbriae in O111 cells grown in LBHS broth. A small aliquot of bacterial aggregates or of cells sampled from the bottom of the culture tubes in the case of nonaggregative strains was resuspended in sterile water, spotted onto microscope slides, and air dried. The slides were rinsed in double-distilled water (ddH2O) and then incubated in BSA-TBS-Tween 20 solution to eliminate nonspecific binding of the antibody to glass. The bacterial cells were fluorescently stained with Hoechst 33342 nucleic acid stain (Life Technologies, Grand Island, NY) at 100 μg/ml for 40 min. The curli proteins were labeled by incubation in polyclonal anti-CsgA antibody diluted in BSA-TBS-Tween 20 solution followed with goat anti-rabbit secondary antibody labeled with Alexa Fluor 488 (Life Technologies). The cells were observed under a Leica SP5 scanning confocal microscope at 405-nm and 488-nm excitation wavelengths. The signals from the Hoechst stain and the anti-CsgA antibody were assigned the pseudocolors blue and green, respectively.
Sequence analysis.
The sequences of the curli operons, RpoS, and the rpoS promoter regions were extracted from the E. coli O111 (strains MB971, MB975, RM9131, and RM9975) and O157:H7 (strain RM6607R; curli operon only) draft genomes or the complete genomes of E. coli K-12 strain MG1655 (GenBank accession number NC_000913), E. coli O111:H− strain 11128 (accession number AP010960), and E. coli O157:H7 strains Sakai, EDL933, and TW14359 (accession numbers NC_002695, NC_002655, and NC_013008, respectively). Each set of sequences was aligned using the ClustalW method in the MegAlign module of Lasergene (v. 8.0; DNAStar, Madison, WI). Mutations within the Csg proteins were determined using either the Lasergene EditSeq module or manual curation of MegAlign alignments.
RESULTS
STEC O111 has aggregative behavior in culture.
The environmental and outbreak STEC O111 strains in this study, which we described previously (11), exhibited aggregative behavior when cultured with agitation at 28°C. We isolated naturally occurring nonaggregative variants from the same agar plate as that of the stock of the original isolate for STEC O111 strains from cow feces in Wisconsin and Pennsylvania and from clinical patients in three different outbreaks in the United States (Table 2; Fig. 1). A systematic approach for the isolation of nonaggregative variants of other strains in our O111 collection was not pursued. Strain MB971 is 1 of 10 nonaggregative variants out of a total of 25 colonies of STEC O111 outbreak strain SD143209, suggesting that such variants possibly occur in a cell population at high frequency; one of the colonies of SD143209 that displayed aggregative behavior in liquid culture was isolated to further investigate this phenotype, and the isolate was named MB975. The nonaggregative variants of the other strains listed in Table 2 were isolated at frequencies of approximately 12 to 28%. Thus, these STEC O111 variants may arise at various frequencies in different strains, depending on environmental conditions. Once isolated from the original culture stock in our collection, the aggregative and nonaggregative isolates were observed to be very stable with this phenotype under our laboratory conditions.
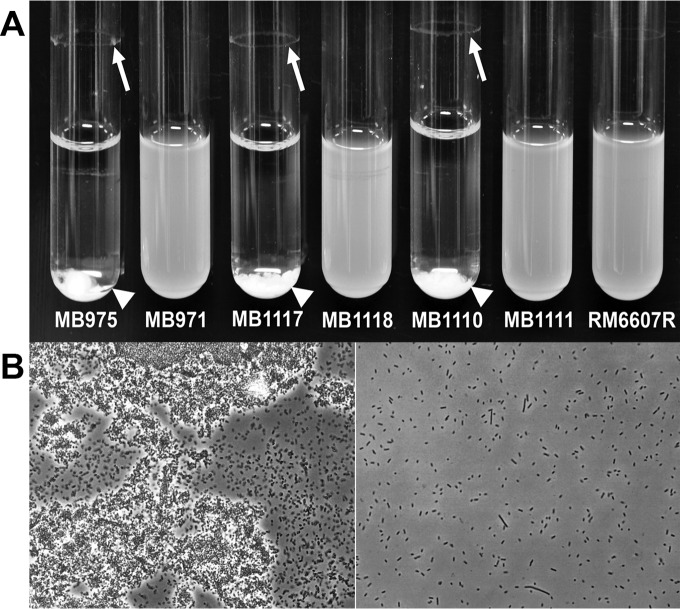
FIG 1.
Aggregative phenotype of STEC O111 cells grown to stationary phase in LBHS broth in glass tubes at 28°C for 24 h in a rotary shaker. (A) Photograph of cultures of aggregative outbreak (MB975 and MB1117) and environmental (MB1110) O111 strains and their respective natural nonaggregative variants (MB971, MB1118, and MB1111, respectively) and of E. coli O157:H7 RM6607R. Aggregates formed at the bottom of the tubes (arrowheads), resulting in a clear suspension in the environmental and outbreak O111 cultures. The natural variants and RM6607R remained in suspension. Arrows point to biofilm rings at the liquid-air interface. (B) Representative phase-contrast images of the cells at the bottom of the culture tubes for aggregative (MB975) (left) and nonaggregative (MB971) (right) O111 variant strains. Large dense clusters of cells are visible for the aggregative strains, but only single cells are present in the other cultures. Magnification, ×400.
The aggregative behavior of MB975 and other STEC O111 strains of various origins was evidenced by the production of large and dense cell clusters accumulating at the bottom of the culture tubes during incubation with agitation on a rolling drum, concomitant with the formation of a biofilm ring on the tube surface at the liquid-air interface (Fig. 1). Over time during incubation, the planktonic cells were depleted from the suspension and the number and size of aggregates increased. This aggregation was observed in LB broth with various NaCl concentrations (0 to 14.5%), in M9 glucose minimal medium, and in glass, polystyrene, and polypropylene culture tubes (data not shown). However, aggregation was not observed under static conditions or at 37°C. Figure 1 illustrates that, in contrast, STEC O157:H7 strain RM6607R, which caused an outbreak linked to hamburger meat in 1993, did not aggregate under the conditions tested in this study.
The cell-cell interaction observed for STEC O111 appears to be distinct from that of enteroaggregative E. coli (EAEC), for which aggregates were not detected under our culture conditions at the macroscopic or the microscopic level but which formed a thin biofilm coating on the inner surface of the culture tube (data not shown). Preliminary data indicate also that the EAEC pAA-encoded aggregative determinants were not present in the genomes of four aggregative STEC O111 strains that we sequenced, including strain MB975 (unpublished data).
Aggregation is stationary phase dependent.
A time course of aggregation during the growth of strain MB975 in LBHS broth was obtained by measuring the absorbance of the suspension over time at a λ of 600 nm (Fig. 2). Because maximum absorbance had been reached after 5.5 h, the optical density at 600 nm (OD600) of the cultures diluted 1:10 in separate tubes was measured also after this time in order to better determine the start and progression of aggregative behavior (Fig. 2, inset). This approach revealed that the planktonic cells of the two outbreak strains MB975 and MB1005 began to be depleted from the suspension during entry of the culture into early stationary phase and that aggregation increased thereafter until most cells had formed large assemblages, settling at the bottom of the tube. In contrast, the absorbance measurements of the cultures of the nonaggregative variant strain MB971 and of STEC O157:H7 strain RM6607R continued to increase slightly after this time as the cells entered stationary phase and remained in suspension.
FIG 2.
Culture-phase-dependent aggregation: OD600 time course of cultures in LBHS broth at 28°C with agitation in glass tubes. Absorbance is illustrated for the aggregative outbreak strains MB975 and MB1005, the nonaggregative variant of MB975 (MB971), and the O157:H7 outbreak strain (RM6607R). Error bars represent standard deviations for three replicate samples, and the experiment was repeated three times. (Inset) Because the maximum absorbance had been reached after 5.5 h, cultures were diluted 1:10 in separate tubes to determine the start and progression of aggregative behavior.
Phase-contrast microscopy of the cells in MB975 cultures over time clearly showed that small aggregates were present starting in early stationary phase and were larger in full stationary phase (Fig. 3). At the latter time, a decrease in the density of single cells in suspension was also evident, as well as a considerable increase in the size of the cell clusters at the bottom of the tube; this process continued until nearly all cells were located in the large assemblages that settled at the bottom of the tube, as observed in the overnight cultures (Fig. 3).
FIG 3.
Phase-contrast micrographs of MB975 cells over time in the late log, early stationary, full stationary, and overnight phases of culture in LBHS broth under agitation at 28°C. The upper and lower panels show cells retrieved from the suspension and from the bottom of the culture tubes, respectively. Note the formation of small cell clusters starting in early stationary phase (dashed circles) and their increase in size and number with concurrent depletion of the planktonic cells from the suspension over time, leading to the settlement of large aggregates at the bottom of the tubes. Magnification, ×400.
Aggregation depends on curli production.
Aggregation was positively correlated with the presence of curli fimbriae in STEC O111 strains, as assayed on CRI plates (Table 2) (11). It was noted also that the environmental and outbreak STEC O111 aggregative strains tested in this study formed darker-red colonies on these plates than the curli-producing STEC O157:H7 strains EDL933 and RM6607R, indicating higher levels of curli production in the aggregative O111 strains. In contrast, the indicator plates revealed that the nonaggregative variants had lost the ability to produce curli fimbriae and lacked RpoS function but gained the ability to synthesize cellulose (Table 2). The csgBAC operon, which codes for the structural curli subunits, was deleted in strain MB975. The resulting mutant, strain MB1044, and its derivative, strain MB1068 (MB1044 transformed with the empty cloning vector pBBR1MCS-5), were impaired in curli synthesis and in aggregation (Table 2; Fig. 4 and 5). The csgBAC operon was cloned with its native promoter into pBBR1MCS-5 (pMBcsgO111), and both phenotypes were rescued by transformation of the mutant with pMBcsgO111, to yield strain MB1054 (Table 2; Fig. 4 and 5). It is noteworthy that the complemented mutant, strain MB1054, aggregated faster than strain MB1066 (the parental strain MB975 transformed with the empty cloning vector pBBR1MCS-5), as was evidenced by the earlier decrease in the OD600 of the suspension at the start of stationary phase (Fig. 4A). Transformation of the nonaggregative variant MB971 with pMBcsgO111 (strain MB1062) did not impart to this strain the ability to aggregate (Table 2), providing the first evidence that the nonaggregative variant lacks factors (other than the genes coding for curli subunits per se) that are required for aggregation to occur.
FIG 4.
(A) OD600 time course of aggregation in strains MB1066, MB1068, MB1054, and MB1064, described below. (Inset) See the explanation in the legend to Fig. 2. (B) Phenotypes of MB1066 (MB975/pBBR1MCS-5), MB1068 (MB975 ΔcsgBAC::kan/pBBR1MCS-5), MB1054 (MB975 ΔcsgBAC::kan/pMBcsgO111), MB1064 (MB971/pBBR1MCS-5), MB1096 (MB971/pXQ29), and MB1191 (MB975 ΔrpoS::cat) in cultures in LBHS broth in glass tubes with agitation at 28°C. WT, wild type.
FIG 5.

Western blots of formic acid-treated whole-cell lysates of MB1066 (MB975/pBBR1MCS-5) (lane 1), MB1068 (MB975 ΔcsgBAC::kan/pBBR1MCS-5) (lane 2), MB1054 (MB975 ΔcsgBAC::kan/pMBcsgO111) (lane 3), MB1064 (MB971/pBBR1MCS-5) (lane 4), MB1096 (MB971/pXQ29) (lane 5), MB1191 (MB975 ΔrpoS::cat) (lane 6), and MB1192 (MB975 ΔrpoS::cat/pXQ29) (lane 7) cells grown in LBHS broth in glass tubes and probed with anti-DnaK antibody (top) and polyclonal anti-curli peptide (CsgA) antibody (bottom). The mass standards are shown on the left.
In order to obtain further evidence for the role of curli in aggregation of STEC O111, the presence of curli protein on cells grown in liquid culture was assessed by Western blotting and immunomicroscopy with anti-curli peptide antibody. A Western blot of formic acid-treated whole lysates of cells grown in LBHS broth showed that the aggregative parental strain and its complemented curli-defective mutant produced large amounts of curli protein, as evidenced by the presence of a 17-kDa band corresponding to the CsgA subunit (20), whereas the protein was undetected in the curli deletion strain and the naturally nonaggregative variant (Fig. 5). Confocal imaging of STEC O111 strains grown in LBHS broth to the stationary phase and probed with Alexa Fluor 488-labeled anticurli antibody showed a strong green fluorescent signal for the presence of curli on the cell surface of the aggregative strain and large masses of the protein in the intercellular matrix of the aggregates (Fig. 6A). In contrast, no antibody signal was detected on the cells of the curli-deficient mutant (Fig. 6B). Likewise, the signal was absent on the cells of the nonaggregative variant MB971 (data not shown).
FIG 6.
Representative confocal image overlays of O111 MB1066 (MB975/pBBR1MCS-5) (A), MB1068 (MB975 ΔcsgBAC::kan/pBBR1MCS-5) (B), and MB1096 (MB971/pXQ29) (C) cells grown in LBHS broth in glass tubes. Curli fimbriae were tagged with Alexa Fluor 488-labeled polyclonal anti-curli peptide (CsgA) antibody (green fluorescence), and cells were stained with Hoechst 33342 dye (blue fluorescence). Bars, 5 μm.
RpoS regulation of aggregation via curli production.
Because of a positive correlation in STEC O111 between aggregation and functional RpoS based on the iodine plate assay (Table 2) (11) and the knowledge that the expression of the curli operons depends on RpoS in E. coli K-12 and Salmonella enterica (20, 24), the nonaggregative and RpoS-negative variant MB971 was transformed with pXQ29. This plasmid carries the rpoS gene of E. coli O157:H7 expressed from its own promoter (29). The presence of this functional RpoS gene in the resulting strain, MB1096, complemented the variant for RpoS function, curli production, and aggregation and suppressed cellulose synthesis (Table 2; Fig. 4B, 5, and 6C). An rpoS deletion mutant of MB975 was constructed. This mutant (strain MB1191) lacked not only RpoS function but also curli production, recovered the ability to synthesize cellulose, as revealed by indicator plate assays and Western blot analysis, and displayed a nonaggregative phenotype (Table 2; Fig. 4B and 5). The RpoS mutant, MB1191, was fully complemented for RpoS function, and thus for curli production and aggregation, by transformation with pXQ29 (resulting in strain MB1192) (Table 2; Fig. 5).
Comparative aggregation and production of curli in E. coli O111, O157:H7, and K-12.
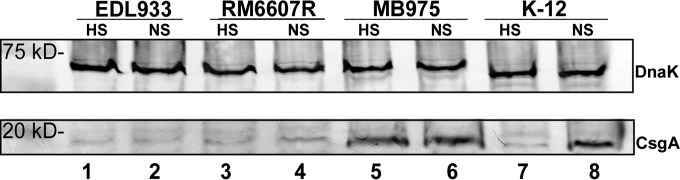
The nonpathogenic strain E. coli K-12 and two STEC O157:H7 strains were tested for aggregation under our culture conditions. None of these strains displayed an aggregative phenotype in LBHS culture at 28°C, despite the presence of functional RpoS and curli production, based on indicator plate assays (Table 2). However, strain K-12, but not O157:H7 EDL933 or O157:H7 RM6607R, aggregated under LBNS broth growth conditions, although the K-12 aggregates appeared much smaller than those of any aggregative STEC O111 strain that we investigated (data not shown). Because the transcription of curli operons is known to be induced in low/no-salt conditions in E. coli and S. enterica (20, 21), comparative curli production in LBHS and LBNS by strains MB975, K-12, RM6607R, and EDL933 was assessed by Western blotting. This revealed that the non-O111 strains discussed above produced minimal quantities of curli in LBHS broth compared with the large amounts produced by MB975 in both LBHS and LBNS broth (Fig. 7). Although the LBNS conditions appeared to induce greater production of CsgA in K-12 and the O157:H7 strains, this increase was substantial only in the K-12 strain.
FIG 7.
Western blots of formic acid-treated whole lysates of cells grown in LBHS (HS) and LBNS (NS) broth. (Lanes 1 and 2) E. coli O157:H7 EDL933 in LBHS and LBNS broth, respectively; (lanes 3 and 4) E. coli O157:H7 RM6607R in LBHS and LBNS broth, respectively; (lanes 5 and 6) E. coli O111 MB975 in LBHS and LBNS broth, respectively; (lanes 7 and 8) E. coli K-12 MG1655 in LBHS and LBNS broth, respectively. Cells were probed with anti-DnaK antibody (top) and polyclonal anti-curli peptide (CsgA) antibody (bottom). Mass standards are shown on the left.
Comparative sequence analysis of E. coli O111, O157:H7, and K-12.
In order to further understand the differences in ability to aggregate between RpoS- and curli-positive strains of E. coli, the nucleotide sequences of the csgGFEDBAC curli loci were extracted from the draft and complete genomes of eight E. coli strains and aligned. These sequences represent five STEC O111 strains (11128, as well as MB971, MB975, RM9131, and RM9975 [unpublished data]), three STEC O157:H7 strains (EDL933, Sakai, and TW14359), and E. coli K-12 (MG1655). Analysis of the alignments indicated that the csgGFEDBAC loci were identical in all of the above-named O111 strains. The majority of the single nucleotide polymorphisms (SNPs) identified among all strains were synonymous substitutions. The amino acid sequences of CsgF, CsgE, and CsgB were identical between all eight strains (data not shown). However, some SNPs were missense mutations. Relative to K-12's sequence, in CsgG, TW14539 has a P76T mutation, in CsgD, the O111 and O157 strains have S19P and S110A mutations, and in CsgC, the O111 strains contain a D82E mutation, the O157 strains contain a V24M mutation, and both serotypes contain a T3A mutation. The majority of the csg variation was identified within the gene encoding the major structural curli subunit CsgA, the comparative amino acid sequence analysis of which is illustrated for all eight strains in Fig. 8. Relative to K-12, all of the O111-specific SNPs were synonymous. However, five O157-specific missense mutations were identified. Although four of these were either within or directly adjacent to the repeat units, none altered the conserved residues within the repeat unit. It is noteworthy that one additional glycine residue was present at position 31 in the N-terminal secretion-associated domain of csgA in all three E. coli O157:H7 strains (Fig. 8). Finally, the 656-bp intergenic region between csgD and csgB contained 14 SNPs; 5 were O157 specific and 4 were O111 specific. None were present within either the csgD or csgB promoter.
FIG 8.
Alignment of E. coli CsgA sequences. Amino acid alignments were performed using the ClustalW method in MegAlign (Lasergene v. 12.0; DNAStar, Madison, WI). The amino acid differences between O157 strains and O111/K-12 strains are boxed and marked with asterisks. Sec, secretory signal sequence (57); N22, N-terminal domain necessary for secretion through the outer membrane (57); R1 to R5, repeat units within CsgA (57, 58). Amino acid residues conserved between the repeat units (58) are shaded.
Importantly, we observed also that the RpoS sequence of the nonaggregative O111 variant MB971 differs from that of MB975 by the substitution of threonine for asparagine at amino acid (aa) 124 (data not shown); the rpoS promoter region sequences in the two strains were identical. The RpoS sequence of MB975 was identical to those of the other three O111 strains named above but differed from that of O157:H7 EDL933 at aa 19 and aa 21, from that of O157:H7 TW14359 at aa 263, and from that of K-12 at aa 33 (data not shown).
DISCUSSION
Our study investigated the distinctive aggregative behavior displayed by all of the 22 outbreak and environmental STEC O111 strains in our collection under agitated, but not static, culture conditions and at 28°C, but not 37°C. We reported previously that this phenomenon, which positively correlated with curli fimbrial production and RpoS function and negatively with cellulose synthesis based on the results from indicator plate assays, was not observed in any of the 16 sporadic case strains that we studied (11). Curli fimbriae are highly stable self-polymerizing fibers (30) that are involved in the attachment of E. coli to a variety of abiotic and biotic surfaces, including human cells and plants (31–34). In addition, curli fimbriae play a role in cell-cell interactions and biofilm architecture in E. coli (18, 35, 36). The O111 curli-deficient mutant generated by deletion of csgBAC and its complementation confirmed that curli fimbriae are required for the aggregative phenomenon observed in this serogroup. Furthermore, immunomicroscopy with fluorescently labeled anticurli antibody revealed the clear presence of large amounts of the fiber subunits in the clusters formed by aggregative strains in culture. Curli fiber remained undetected on the cells of the nonaggregative strains.
The critical role of curli in O111 aggregation provides an explanation for the RpoS-dependent regulation of this behavior, since the transcription of the csg operons is under the positive control of RpoS in E. coli K-12 and S. enterica (20, 24). Uhlich et al. reported that in many strains of STEC O157:H7, the inability to produce curli correlated with mutations in rpoS (37). Thus, it appears that regulation of curli expression in STEC O111 is similar to that in K-12 and STEC O157:H7, with the exception of certain strains, such as those implicated in the spinach-linked outbreak in 2006, for which the csg operons are expressed in an RpoS− background (29). The onset of O111 aggregation during entry into the stationary phase of growth, as reflected by the subsequent continuous decrease in the OD600 of the culture suspension and the formation of increasingly larger cell clusters, likely resulted from the induction of curli synthesis by this stationary-phase sigma factor (38). The early start of aggregation in the complemented csgBAC deletion mutant further supports the important role of curli in this multicellular behavior, since the greater synthesis of curli subunits due to the multiple copy numbers of the pBBR1-derived complementation plasmid may have accelerated cell-cell interactions.
The aggregation of O111 in culture was observed in tubes of various compositions and surface properties as well as in flasks but required gentle agitation and was the most pronounced in borosilicate tubes; silanization of the borosilicate tubes to neutralize their surface charge prevented aggregation (data not shown). Based on the results of this study, we hypothesize that as the culture approaches stationary phase, O111 cells produce sufficient curli to mediate cell-to-surface adhesion, which is promoted by agitation of the culture and leads to the formation of a biofilm at the liquid-air interface. This increase in the density of curli fibers on the cell surface also prompts cell-cell interactions. The role of curli fibers in the formation of intercellular bundles that allow for the stable association of curli-overproducing E. coli K-12 has been reported (18). This intercellular adhesion may also prevail at the liquid-air interface during the production of a dense multilayered biofilm on the substratum, part of which may slough off due to culture agitation. This continued process may contribute to depletion of the planktonic cells from the suspension and to the formation of large cell clusters that settle at the bottom of the tube. The cell-cell interactions that underlie aggregation in our system are distinct from those involving the aggregative adherence fimbriae of EAEC (39), which do not form cell clusters but rather coat the tube inner surface with a thin film (data not shown). Moreover, they are not likely to involve aggregation factors such as Ag43, which mediates autoaggregation under static conditions, since O111 cells produce aggregates only in agitated culture (40, 41). However, the aggregation in O111 appears very similar to that described briefly by Vidal et al. during culture of an E. coli K-12 OmpR mutant that produces high levels of curli (35). The OmpR sequences are identical in K-12 and the O111 strains MB975 and MB971 (data not shown). Our study extensively investigated the role of curli in the aggregative behavior of E. coli cells in suspension, a common phenotype among STEC O111 strains, with the exception of sporadic case strains, which generally lack a functional RpoS and are curli deficient (11).
In E. coli K-12 and S. enterica, high osmolarity represses csgD expression and therefore prevents activation of the curli operons (42–44). In light of the dependence of STEC O111 aggregation on curli production and the strong aggregation of these strains irrespective of the NaCl content in LB broth, the regulation of curli by the osmotic potential of the milieu does not appear to predominate in this serogroup. STEC O157:H7 strains EDL933 and RM6607R, a high-curli-producing strain on CRI plates due to a mutation in the global regulator RcsB (45), do not form aggregates in LB broth containing 0.5% (LBHS) or 0% (LBNS) NaCl. In contrast, E. coli K-12, which grew to very homogeneous suspensions in LBHS broth, formed numerous small cell clusters that depleted the planktonic cell suspension over time during incubation in LBNS broth. Western blot analysis with anti-CsgA antibody revealed that these differences were likely caused, at least partly, by a lack of significant induction of curli production under low osmolarity in liquid culture in the STEC O157:H7 strains, whereas curli were indeed more abundant in K-12 aggregative cultures in LBNS broth than in its nonaggregative cultures in LBHS broth. As expected based on aggregative behavior, the antibody signal for curli was very strong under both of the latter conditions, as well as in M9 glucose cultures (data not shown), in O111 MB975 cells.
The csgDEFG promoter is subject to one of the most complex regulation mechanisms in E. coli (46). The intergenic region between this operon and csgBAC, which are divergently transcribed, is the site of interaction with several stress-sensing transcription factors (19). Comparative sequence analysis of the csg operons in strains K-12, STEC O157:H7, and STEC O111 showed that the intergenic region between csgD and csgB contains five O157-specific- and four O111-specific SNPs. None of these are located within the csgB promoter or the csgD promoter, as was reported previously for curli variants in STEC O157:H7 (47), but it is not clear whether these SNPs affect interactions with the various transcription factors that come into play in this region. Furthermore, compared with K-12, the O111 and O157:H7 strains have an S19P and an S110A mutation, respectively, in CsgD, the activator of the curli structural operon. Overall, the effect of these missense mutations on the relative abundances of curli produced by the strains and, therefore, of their aggregative behavior in response to various environmental stimuli remains to be defined.
Curli-driven cell-cell and cell-surface adhesion may be modulated also by chemical differences in the polymeric fiber per se, brought about by the amino acid sequences of curli structural subunits. It is noteworthy that the sequence of the major curli subunit CsgA has the most variation among the strains investigated in our study. While the SNPs in O111 csgA are synonymous to those of K-12, five SNPs, including an additional glycine residue, are missense mutations specific to O157:H7. Curlin is rich in glycine repeats that may afford the fiber greater flexibility and may be involved in protein-protein interactions (20). Thus, besides the altered regulation of curli production in the STEC O157:H7 strains investigated in this study compared with that in STEC O111 strains, the additional glycine residue in O157:H7 CsgA may affect the protein-protein interactions required for effective cell-cell aggregation in these strains. However, although curli fimbriae are required for aggregation, one cannot discount the possibility that other surface factors may contribute to the interactions involved in this phenotype.
Despite the predominance of aggregative behavior in STEC O111, nonaggregative variants within the population of a given outbreak or environmental strain were isolated in our study at a considerable rate. The curli-negative and cellulose-positive phenotype of these variants is likely due generally to their impaired RpoS function, according to the indicator plate assay, since introduction of functional RpoS in variant MB971 restored curli production, suppressed cellulose synthesis, and also imparted aggregation. Additionally, the csg operons and their intergenic regions are identical in O111 strain MB975 and its variant MB971. Thus, the substitution of a threonine residue for an asparagine at aa 124 in MB971 likely renders the sigma factor nonfunctional and abolishes expression of the csg operon. In an E. coli water isolate, a nonsynonymous mutation to a tyrosine residue also at aa 124 resulted in attenuated RpoS function (48). It is unclear whether the negative correlation of aggregation and cellulose synthesis in O111 is simply related to the divergent regulation of curli and cellulose production by RpoS in this serogroup or whether the absence of cellulose actually promotes more efficient cell-cell interactions since the overproduction of cellulose in nonpathogenic E. coli was shown to inhibit curli-mediated adhesion to surfaces (49). The observation that strain MB975 retained its full aggregative behavior despite the constitutive production of high levels of cellulose imparted by transformation with pMB682, which carries the full cellulose biosynthesis operon of S. enterica (28), suggests that the interference of cellulose fibers with aggregation does not take place in this strain.
RpoS is a global regulator of stationary-phase- and stress-related genes, and therefore, RpoS polymorphism is the source of considerable phenotypic diversity in bacteria. RpoS mutants have been isolated at high frequency in a population of STEC O157:H7 strains from the environment, from bagged spinach, and from clinical patients associated with a spinach-linked outbreak in 2006 (29, 50). The diversification of RpoS function may allow for the adaptation of E. coli cells to shifting environmental conditions and the selection of cells with altered phenotypes. The SPANC (self-preservation and nutritional competence) hypothesis of King et al. postulates a trade-off between high-stress resistance (RpoS+) and greater nutrient scavenging capability (RpoS−) within a given population of cells (26). The ability to form aggregates is an important mechanism of protection in a diverse range of bacterial species (51–53), including during pathogenesis, as aggregated cells resist various host defenses more efficiently than individual bacteria (54). Hence, the RpoS-based intrastrain heterogeneity in STEC O111 may ensure that a given population at large has sufficient protection against external stresses via formation of multicellular clusters, whereas the planktonic state of the nonaggregative variant cells may afford them increased ability to seek substrates under nutrient-poor conditions. This phenotypic diversity likely promotes the survival of STEC O111 strains in hydrodynamic environments along the food production chain and in the human host, potentially leading to outbreaks, while the inability to aggregate may hamper the overall survival of sporadic-case O111 strains and their spread to epidemic proportions.
Supplementary Material
ACKNOWLEDGMENT
We thank Michael K. Fleming for technical help with the graphics.
Funding Statement
The USDA Agricultural Research Service (ARS) provided funding to Michelle E. Diodati, Anne H. Bates, William G. Miller, Michelle Q. Carter, Yaguang Zhou, and Maria T. Brandl under CRIS project 2030-42000-046-00D.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03935-15.
REFERENCES
- 1.Majowicz SE, Scallan E, Jones-Bitton A, Sargeant JM, Stapleton J, Angulo FJ, Yeung DH, Kirk MD. 2014. Global incidence of human Shiga toxin–producing Escherichia coli infections and deaths: a systematic review and knowledge synthesis. Foodborne Pathog Dis 11:447–455. doi: 10.1089/fpd.2013.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batz MB, Hoffmann S, Morris JG Jr. 2012. Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. J Food Prot 75:1278–1291. doi: 10.4315/0362-028X.JFP-11-418. [DOI] [PubMed] [Google Scholar]
- 3.Gould LH, Mody RK, Ong KL, Clogher P, Cronquist AB, Garman KN, Lathrop S, Medus C, Spina NL, Webb TH. 2013. Increased recognition of non-O157 Shiga toxin-producing Escherichia coli infections in the United States during 2000−2010: epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog Dis 10:453–460. doi: 10.1089/fpd.2012.1401. [DOI] [PubMed] [Google Scholar]
- 4.Caprioli A, Tozzi A, Kaper J, O'Brien A. 1998. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. In Kaper J, O'Brien A (ed), Epidemiology of Shiga toxin-producing Escherichia coli infections in continental Europe. ASM Press, Washington, DC. [Google Scholar]
- 5.Henning PH, Tham E, Martin AA, Beare TH, Jureidini KF. 1998. Haemolytic-uraemic syndrome outbreak caused by Escherichia coli O111:H−: clinical outcomes. Med J Aust 168:552–555. [DOI] [PubMed] [Google Scholar]
- 6.Karmali MA, Petric M, Winkler M, Bielaszewska M, Brunton J, van de Kar N, Morooka T, Nair GB, Richardson SE, Arbus GS. 1994. Enzyme-linked immunosorbent assay for detection of immunoglobulin G antibodies to Escherichia coli Vero cytotoxin 1. J Clin Microbiol 32:1457–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matano S, Inamura K, Konishi M, Okumura T, Kawai H, Okamura T, Takata Y, Yamada K, Obata M, Nagata H, Muramoto Y, Sugimoto T. 2012. Encephalopathy, disseminated intravascular coagulation, and hemolytic-uremic syndrome after infection with enterohemorrhagic Escherichia coli O111. J Infect Chemother 18:558–564. doi: 10.1007/s10156-011-0336-9. [DOI] [PubMed] [Google Scholar]
- 8.Luna-Gierke R, Griffin P, Gould L, Herman K, Bopp C, Strockbine N, Mody R. 2014. Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol Infect 142:2270–2280. doi: 10.1017/S0950268813003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anonymous. 2014. Green cabbage likely caused Minnesota's recent E. coli O111 outbreak. Food Saf News http://www.foodsafetynews.com/2014/08/cabbage-likely-cause-of-e-coli-o111-outbreak-in-minnesota/#.VoRtI3lIiM8.
- 10.Hlavsa MC, Roberts VA, Kahler AM, Hilborn ED, Mecher TR, Beach MJ, Wade TJ, Yoder JS. 2015. Outbreaks of illness associated with recreational water—United States, 2011−2012. MMWR Morb Mortal Wkly Rep 64:668–672. [PMC free article] [PubMed] [Google Scholar]
- 11.Diodati ME, Bates AH, Cooley MB, Walker S, Mandrell RE, Brandl MT. 2015. High genotypic and phenotypic similarity among Shiga toxin-producing Escherichia coli O111 environmental and outbreak strains. Foodborne Pathog Dis 12:235–243. doi: 10.1089/fpd.2014.1887. [DOI] [PubMed] [Google Scholar]
- 12.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. 2013. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bielaszewska M, Mellmann A, Zhang W, Köck R, Fruth A, Bauwens A, Peters G, Karch H. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis 11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 14.Kjærgaard K, Schembri MA, Ramos C, Molin S, Klemm P. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ Microbiol 2:695–702. doi: 10.1046/j.1462-2920.2000.00152.x. [DOI] [PubMed] [Google Scholar]
- 15.Sherlock O, Vejborg RM, Klemm P. 2005. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect Immun 73:1954–1963. doi: 10.1128/IAI.73.4.1954-1963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clavijo AP, Bai J, Gómez-Duarte OG. 2010. The Longus type IV pilus of enterotoxigenic Escherichia coli (ETEC) mediates bacterial self-aggregation and protection from antimicrobial agents. Microb Pathog 48:230–238. doi: 10.1016/j.micpath.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schembri MA, Hjerrild L, Gjermansen M, Klemm P. 2003. Differential expression of the Escherichia coli autoaggregation factor antigen 43. J Bacteriol 185:2236–2242. doi: 10.1128/JB.185.7.2236-2242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol 2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- 19.Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. 2010. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology 156:2470–2483. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- 20.Olsén A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. 1993. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol 7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 21.Gerstel U, Römling U. 2003. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res Microbiol 154:659–667. doi: 10.1016/j.resmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 24.Römling U, Bian Z, Hammar M, Sierralta WD, Normark S. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol 180:722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 26.King T, Ishihama A, Kori A, Ferenci T. 2004. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J Bacteriol 186:5614–5620. doi: 10.1128/JB.186.17.5614-5620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Hammer ND, Chapman MR. 2008. The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem 283:21530–21539. doi: 10.1074/jbc.M800466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandl MT, Carter MQ, Parker CT, Chapman MR, Huynh S, Zhou Y. 2011. Salmonella biofilm formation on Aspergillus niger involves cellulose-chitin interactions. PLoS One 6:e25553. doi: 10.1371/journal.pone.0025553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter MQ, Louie JW, Huynh S, Parker CT. 2014. Natural rpoS mutations contribute to population heterogeneity in Escherichia coli O157:H7 strains linked to the 2006 US spinach-associated outbreak. Food Microbiol 44:108–118. doi: 10.1016/j.fm.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 30.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cookson AL, Cooley WA, Woodward MJ. 2002. The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int J Med Microbiol 292:195–205. doi: 10.1078/1438-4221-00203. [DOI] [PubMed] [Google Scholar]
- 32.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macarisin D, Patel J, Bauchan G, Giron JA, Sharma VK. 2012. Role of curli and cellulose expression in adherence of Escherichia coli O157:H7 to spinach leaves. Foodborne Pathog Dis 9:160–167. doi: 10.1089/fpd.2011.1020. [DOI] [PubMed] [Google Scholar]
- 34.Jeter C, Matthysse AG. 2005. Characterization of the binding of diarrheagenic strains of E. coli to plant surfaces and the role of curli in the interaction of the bacteria with alfalfa sprouts. Mol Plant Microbe Interact 18:1235–1242. doi: 10.1094/MPMI-18-1235. [DOI] [PubMed] [Google Scholar]
- 35.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol 180:2442–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida Si. 2005. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol Immunol 49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- 37.Uhlich GA, Chen CY, Cottrell BJ, Hofmann CS, Dudley EG, Strobaugh TP Jr, Nguyen LH. 2013. Phage insertion in mlrA and variations in rpoS limit curli expression and biofilm formation in Escherichia coli serotype O157:H7. Microbiology 159:1586–1596. doi: 10.1099/mic.0.066118-0. [DOI] [PubMed] [Google Scholar]
- 38.Lange R, Hengge-Aronis R. 1991. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor σS. J Bacteriol 173:4474–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okeke IN, Nataro JP. 2001. Enteroaggregative Escherichia coli. Lancet Infect Dis 1:304–313. doi: 10.1016/S1473-3099(01)00144-X. [DOI] [PubMed] [Google Scholar]
- 40.Owen P, Caffrey P, Josefsson L. 1987. Identification and partial characterization of a novel bipartite protein antigen associated with the outer membrane of Escherichia coli. J Bacteriol 169:3770–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hasman H, Chakraborty T, Klemm P. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J Bacteriol 181:4834–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerstel U, Park C, Römling U. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol Microbiol 49:639–654. doi: 10.1046/j.1365-2958.2003.03594.x. [DOI] [PubMed] [Google Scholar]
- 43.Jubelin G, Vianney A, Beloin C, Ghigo J-M, Lazzaroni J-C, Lejeune P, Dorel C. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol 187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrova OE, Sauer K. 2012. Sticky situations: key components that control bacterial surface attachment. J Bacteriol 194:2413–2425. doi: 10.1128/JB.00003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter MQ, Parker CT, Louie JW, Huynh S, Fagerquist CK, Mandrell RE. 2012. RcsB contributes to the distinct stress fitness among Escherichia coli O157:H7 curli variants of the 1993 hamburger-associated outbreak strains. Appl Environ Microbiol 78:7706–7719. doi: 10.1128/AEM.02157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishihama A. 2010. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol Rev 34:628–645. doi: 10.1111/j.1574-6976.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- 47.Uhlich GA, Keen JE, Elder RO. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl Environ Microbiol 67:2367–2370. doi: 10.1128/AEM.67.5.2367-2370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiang SM, Dong T, Edge TA, Schellhorn HE. 2011. Phenotypic diversity caused by differential RpoS activity among environmental Escherichia coli isolates. Appl Environ Microbiol 77:7915–7923. doi: 10.1128/AEM.05274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gualdi L, Tagliabue L, Bertagnoli S, Ieranò T, De Castro C, Landini P. 2008. Cellulose modulates biofilm formation by counteracting curli-mediated colonization of solid surfaces in Escherichia coli. Microbiology 154:2017–2024. doi: 10.1099/mic.0.2008/018093-0. [DOI] [PubMed] [Google Scholar]
- 50.Parker CT, Kyle JL, Huynh S, Carter MQ, Brandl MT, Mandrell RE. 2012. Distinct transcriptional profiles and phenotypes exhibited by Escherichia coli O157:H7 isolates related to the 2006 spinach-associated outbreak. Appl Environ Microbiol 78:455–463. doi: 10.1128/AEM.06251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menozzi F, Boucher P, Riveau G, Gantiez C, Locht C. 1994. Surface-associated filamentous hemagglutinin induces autoagglutination of Bordetella pertussis. Infect Immun 62:4261–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Menozzi FD, Rouse JH, Alavi M, Laude-Sharp M, Muller J, Bischoff R, Brennan MJ, Locht C. 1996. Identification of a heparin-binding hemagglutinin present in mycobacteria. J Exp Med 184:993–1001. doi: 10.1084/jem.184.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frick IM, Mörgelin M, Björck L. 2000. Virulent aggregates of Streptococcus pyogenes are generated by homophilic protein-protein interactions. Mol Microbiol 37:1232–1247. doi: 10.1046/j.1365-2958.2000.02084.x. [DOI] [PubMed] [Google Scholar]
- 54.Ochiai K, Kurita-Ochiai T, Kamino Y, Ikeda T. 1993. Effect of co-aggregation on the pathogenicity of oral bacteria. J Med Microbiol 39:183–190. doi: 10.1099/00222615-39-3-183. [DOI] [PubMed] [Google Scholar]
- 55.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 56.Carter MQ, Brandl MT, Louie JW, Kyle JL, Carychao DK, Cooley MB, Parker CT, Bates AH, Mandrell RE. 2011. Distinct acid resistance and survival fitness displayed by curli variants of enterohemorrhagic Escherichia coli O157:H7. Appl Environ Microbiol 77:3685–3695. doi: 10.1128/AEM.02315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Chapman MR. 2008. Curli provide the template for understanding controlled amyloid propagation. Prion 2:57–60. doi: 10.4161/pri.2.2.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hammer ND, Schmidt JC, Chapman MR. 2007. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci U S A 104:12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.