Abstract
Comamonas plasmids play important roles in shaping the phenotypes of their hosts and the adaptation of these hosts to changing environments, and understanding the evolutionary strategy of these plasmids is thus of great concern. In this study, the sequence of the 119-kb 3,5-dibromo-4-hydroxybenzonitrile-catabolizing plasmid pBHB from Comamonas sp. strain 7D-2 was studied and compared with those of three other Comamonas haloaromatic catabolic plasmids. Incompatibility group determination based on a phylogenetic analysis of 24 backbone gene proteins, as well as TrfA, revealed that these four plasmids all belong to the IncP-1β subgroup. Comparison of the four plasmids revealed a conserved backbone region and diverse genetic-load regions. The four plasmids share a core genome consisting of 40 genes (>50% similarities) and contain 12 to 50 unique genes each, most of which are xenobiotic-catabolic genes. Two functional reductive dehalogenase gene clusters are specifically located on pBHB, showing distinctive evolution of pBHB for haloaromatics. The higher catabolic ability of the bhbA2B2 cluster than the bhbAB cluster may be due to the transcription levels and the character of the dehalogenase gene itself rather than that of its extracytoplasmic binding receptor gene. The plasmid pBHB is riddled with transposons and insertion sequence (IS) elements, and ISs play important roles in the evolution of pBHB. The analysis of the origin of the bhb genes on pBHB suggested that these accessory genes evolved independently. Our work provides insights into the evolutionary strategies of Comamonas plasmids, especially into the adaptation mechanism employed by pBHB for haloaromatics.
INTRODUCTION
Comamonas species are ubiquitous in the environment and have been isolated from soil, mud, water, activated sludge, clinical samples, and organic pollutant- and heavy metal-contaminated environments (1). Comamonas species play an important role in matter cycling in nature and have been noted for their capabilities to degrade various aromatics (1–5). Interestingly, many Comamonas strains have been found to harbor plasmids, most of which have been confirmed to be related to the catabolism of xenobiotic aromatics (4, 6–8). For example, the plasmid pI2, responsible for the degradation of aniline/3-chloroaniline, was isolated from Comamonas testosteroni I2 (4, 9, 10). Another transferable plasmid, pTB30, which is involved in the mineralization of 3-chloroaniline, was isolated from C. testosteroni TB30 (9, 11). In addition, the plasmid pCNB1, encoding the catabolic pathway to convert 4-chloronitrobenzene to 5-chloro-4-hydroxy-2-oxovalerate, was sequenced in Comamonas sp. strain CNB-1 (12, 13). Recently, the plasmid pBHB was characterized in Comamonas sp. strain 7D-2. This plasmid is involved in the mineralization of the herbicide bromoxynil (3,5-dibromo-4-hydroxybenzonitrile), which includes the reductive dehalogenation of the bromoxynil nitrile hydrolyte 3,5-dibromo-4-hydroxybenzoate (DBHB) (14).
Plasmids are among the most important elements in the evolution of prokaryotes and their adaptation to changing environments (15). In addition to the conserved backbone, which is important for DNA replication and mobilization, plasmids usually carry highly variable regions of accessory genes that are responsible for the phenotypic diversity of their hosts, such as the capability to catabolize organic pollutants and to resist heavy metals (13, 16). Furthermore, the plasmid-meditated horizontal transfer of catabolic genes contributes to the evolution of catabolic pathways in recipients (9, 16). Given the important role of plasmids in shaping microbial phenotypes and adaptation to changing environments, understanding their biology is central to understanding microbial evolution and ecology (17). However, the evolution of plasmids has been poorly investigated, mainly due to the lack of extensive similarities among plasmids, which hampers classical phylogenetic analyses based on gene genealogy and synteny (18, 19). To understand the evolution of the plasmids and their adaptation to toxic compounds in the environment, complete plasmid sequences are needed. Fortunately, cutting-edge DNA-sequencing technologies and rapidly growing data sets have facilitated comparisons of the genomes of plasmids, enhancing our understanding of the evolutionary adaptation of plasmids.
The evolutionary dynamics of the Comamonas plasmid genome (plasmidome) are a particularly attractive subject of study because of their xenobiotic-catabolic characteristics. Therefore, in this work, the catabolic plasmid pBHB from Comamonas sp. 7D-2 was compared in detail with three other Comamonas catabolic plasmids, pI2, pCNB1, and pTB30, to (i) present a possible evolutionary scenario of IncP-1β plasmids within the genus Comamonas and (ii) reveal the adaptation mechanism of pBHB to haloaromatics. The two clusters of reductive dehalogenase genes on pBHB were also compared and investigated at the biochemical level to further reveal the evolutionary adaptation of pBHB.
MATERIALS AND METHODS
Chemicals.
The standard compounds DBHB and 3-bromo-4-hydroxybenzoate (BHB) were purchased from Sigma-Aldrich (St. Louis, MO, USA), and bromoxynil, 3,5-dichloro-4-hydroxybenzoate (DCHB), 3-chloro-4-hydroxybenzoate (CHB), 2,6-dibromophenol, and 2-bromophenol were purchased from Energy Chemical Co., Ltd. (Shanghai, China), or J&K Chemical Co., Ltd. (Shanghai, China). The purities of all standard compounds exceeded 98%. High-performance liquid chromatography (HPLC)-grade methanol, ethyl acetate, and acetonitrile were purchased from the Shanghai Chemical Reagent Co., Ltd., Shanghai, China. All other reagents used in this study were of analytical grade.
Bacterial strains, plasmids, primers, media, and culture conditions.
The bacterial strains and plasmids used for biological experiments are described in Table 1, and the primers are described in Table S1 in the supplemental material. Comamonas strains were grown at 30°C in lysogeny broth (LB) or minimal salts medium (MM) (1.0 g NH4NO3, 1.6 g K2HPO4, 0.5 g KH2PO4, 0.2 g MgSO4, and 1.0 g NaCl per liter of water, pH 7.0) supplemented with substrates (0.2 to 0.3 mM bromoxynil, DBHB, or BHB) as described previously (14). Solid-medium plates were prepared by adding 2.0% (wt/vol) agar to the medium. Antibiotics were added as necessary at the following concentrations: ampicillin (Amp), 100 mg liter−1; kanamycin (Km), 50 mg liter−1; and gentamicin (Gm), 80 mg liter−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Comamonas sp. strain 7D-2 | Bromoxynil-degrading strain with plasmid pBHB; bxn2+ bhb+ Gmr | 14 |
| Comamonas sp. strain 2B | Plasmid pBHB-cured mutant of Comamonas sp. strain 7D-2; bxn2− bhb− Gmr | 14 |
| Comamonas sp. strain 2B-bhbAB | Complementary strain of Comamonas sp. strain 2B containing pMCSQ2-bhbAB | 14 |
| Comamonas sp. strain 2B-bhbA2B2 | Complementary strain of Comamonas sp. strain 2B containing pMCSQ2-bhbA2B2 | This study |
| Comamonas sp. strain 2B-bhbA2B | Complementary strain of Comamonas sp. strain 2B containing pMCSQ2-bhbA2B | This study |
| Comamonas sp. strain 2B-bhbAB2 | Complementary strain of Comamonas sp. strain 2B containing pMCSQ2-bhbAB2 | This study |
| E. coli DH5α | Host strain for cloning vectors | TaKaRa |
| E. coli HB101(pRK2013) | Conjugation helper strain; Kmr | Laboratory stores |
| Plasmids | ||
| pBBR1MCS-2 | Broad-host-range vector | 44 |
| pMCSQ2 | 527-bp fragment containing promoter sequence upstream of bhbF inserted into the HindIII-BamHI sites of pBBR1MCS2 | 14 |
| pMCSQ2-bhbAB | 4,293-bp fragment containing bhbAB inserted into the BamHI-XbaI sites of pMCSQ2 | 14 |
| pMCSQ2-bhbA2B2 | 4,252-bp fragment containing bhbA2B2 inserted into the BamHI-XbaI sites of pMCSQ2 | This study |
| pMCSQ2-bhbB | 1,046-bp fragment of bhbB inserted into the HindIII-XbaI sites of pMCSQ2 | This study |
| pMCSQ2-bhbB2 | 1,023-bp fragment of bhbB2 inserted into the HindIII-XbaI sites of pMCSQ2 | This study |
| pMCSQ2-bhbA2B | 323-bp fragment of bhbA2 with BamHI-XbaI sites inserted into the BglII-XbaI sites of pMCSQ2-bhbB | This study |
| pMCSQ2-bhbAB2 | 3,254-p fragment of bhbA with BamHI-XbaI sites inserted into the BglII-XbaI sites of pMCSQ2-bhbB2 | This study |
454 sequencing, ORF annotation, and sequence analysis.
Approximately 100 μg of DNA of the plasmid pBHB isolated from Comamonas sp. 7D-2 (14) was delivered to Shanghai Majorbio Bio-Pharm Biotechnology Co., Ltd. (Shanghai, China), for complete sequencing. One-plate whole-run sequencing was performed on the 454 GsFLX Illumina GAIIx platform (Roche Diagnostics, Indianapolis, IN, USA), following the manufacturer's protocol. Redundant sequences were removed, and the high-quality reads were assembled using the software GS De Novo Assembler (Newbler 2.6; Roche). Gaps were closed using PCR. The sequence of pBHB was automatically annotated by the J. Craig Venter Institute Annotation Service (http://www.jcvi.org/annotation/service/) and further manually annotated using Manatee (http://manatee.sourceforge.net). Open reading frames (ORFs) predicted by the GeneMark software were manually analyzed with BLASTp and the NCBI database. The G+C contents were determined with DNAMAN (version 5.1; Lynnon Biosoft). Transposons and insertion sequences (ISs) were identified using ISfinder (20).
Incompatibility group assignment.
The complete sequences of 16 representative IncP-1 plasmids, including all six plasmids from the genus Comamonas, were downloaded from GenBank. The assignment of plasmids to an incompatibility group was carried out by a phylogenetic analysis of the sequences of 24 “backbone” gene proteins (9) and the replication initiator TrfA. The 24 backbone gene protein sequences were aligned, manually adjusted, and concatenated into a single alignment using MUSCLE (21). The conserved regions of the alignments were selected with Gblocks v0.91b (22) using the default parameters. The best amino acid substitution model (the WAG+I+G model) was selected using ProtTest v3.2 (23). The maximum-likelihood (ML) phylogenetic analyses were performed using RAxML (24) via the RAxML-HPC Black Box Web server at CIPRES (http://www.phylo.org/) with the best-fit model. Maximum-parsimony (MP) and neighbor-joining (NJ) trees were generated using PAUP v4.0b10 and MEGA v 5.05, respectively.
Orthology reconstruction.
OrthoMCL version 2.0 (25) was used to delineate orthologous protein sequences among the four Comamonas plasmidomes. The first step in the OrthoMCL procedure was to perform an all-against-all BLASTp search within and between all genome pairs. Proteins that shared over 50% sequence similarity were grouped into clusters. A P value cutoff of 1E−5 was selected for putative orthologs. Putative orthologous relationships of pBHB to the other three Comamonas plasmids were then converted into a circular graph using CIRCOS software, in which weighted edges in different colors represent these relationships.
Specificity of the extracytoplasmic binding receptor (BhbB/BhbB2) to reductive dehalogenase (BhbA/BhBA2).
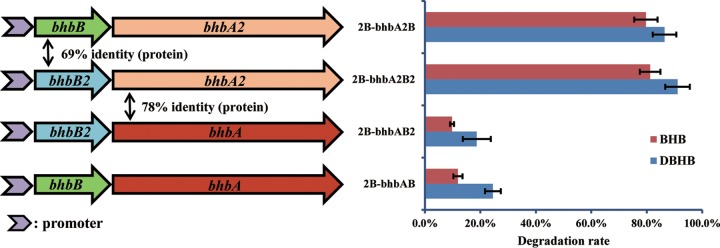
To study the specificity of the extracytoplasmic binding receptor (BhbB/BhbB2) and its effect on the degradation activity of reductive dehalogenase (BhbA/BhbA2), different combinations of the extracytoplasmic binding receptor gene and the reductive dehalogenase gene (specifically, bhbAB, bhbA2B2, bhbA2B, and bhbAB2) were constructed and ligated into pMCSQ2 (see Fig. 6) as described previously (14). These constructs were then transformed into strain 2B to generate strains 2B-bhbAB, 2B-bhbA2B2, 2B-bhbA2B, and 2B-bhbAB2, respectively. The four strains were precultured in LB containing 0.2 mM DBHB for 14 h, harvested by centrifugation at 6,000 × g for 5 min, and washed twice with ice-cold MM. The four strains were then individually inoculated into 100 ml MM (containing 0.2 mM DBHB or BHB) to a final optical density at 600 nm (OD600) of 0.2. The inocula were incubated at 30°C on a shaker at 150 rpm. Samples were collected at 8 h, and the concentration of DBHB or BHB was detected by HPLC as described previously (14).
FIG 6.
Diagram of the combinations of the extracytoplasmic binding receptor gene (bhbB/B2) and the reductive dehalogenase gene (bhbA/bhbA2) (left) and the degradation rates of DBHB and BHB by strain 2B carrying pMCSQ2-bhbAB, pMCSQ2-bhbAB2, pMCSQ2-bhbA2B, or pMCSQ2-bhbA2B2 (right). The data are presented as the means ± standard deviations for triplicate incubations. When the error bar is not visible, it is within the data point.
The crude enzymatic activities of BhbA and BhbA2, which were coexpressed with their native extracytoplasmic binding receptors, were also assayed. Strains 2B-bhbAB and 2B-bhbA2B2 were harvested in late exponential phase at the same OD600 and disrupted by sonication as described previously (14). The activity of the crude dehalogenase was assayed in a standard system (26). One unit of reductive dehalogenase activity was defined as the amount of crude protein that catalyzes the release of 1 nmol of halide ions from the substrate per minute. All assays were performed independently three times, and the means and standard deviations were calculated.
Transcription of the bhbAB and bhbA2B2 gene cluster in response to different substrates.
Comamonas sp. 7D-2 harboring the plasmid pBHB was incubated in MM with different substrates (0.2 mM glucose, bromoxynil, DBHB, DCHB, BHB, or DBP) at 30°C and 150 rpm on a rotary shaker for 8 h. The total RNA was isolated with an RNAprep pure kit for bacteria (Tiangen Biotech, Beijing, China) and reverse transcribed into cDNA using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Dalian, China). Quantitative PCR (qPCR) was performed with a 7500 real-time PCR system (Applied Biosystems, USA) with SYBR Premix Ex Taq II (Tli RNase H Plus; TaKaRa, Dalian, China). The primers used to amplify the partial fragments of the bhbA, bhbA2, bhbB, and bhbB2 genes are listed in Table S1 in the supplemental material. The 175-bp fragment of the trfA gene of pBHB was used as the reference to evaluate the relative differences in integrity between individual RNA samples. The 2−ΔΔCT threshold cycle method was used to calculate relative changes in gene expression (27).
RESULTS
Sequence features of pBHB.
To sequence the plasmid pBHB, 14,560 reads with a total length of 5,345,347 bp were obtained after the removal of the redundant sequences. The sequencing coverage was 44.8-fold. The gaps were filled using PCR by considering the relationships between contigs. The complete sequence of pBHB is a circular molecule of 119,225 bp harboring 122 predicted ORFs (see Fig. S1 in the supplemental material). The average G+C content was 63.57% (Table 2). The total length of the coding sequence (CDS) is 101,190 bp and accounts for a coding percentage of 84.87%. The 122 ORFs appear to be involved in (i) plasmid core functions, such as the initiation of vegetative replication, maintenance/stability, and conjugal transfer; (ii) resistance to mercury; and (iii) the catabolism of haloaromatics. The backbone region of pBHB contains 46 genes in clusters responsible for the core functions. Twelve tra genes (traO, traN, traM, traL, traK, traJ, traI, traG, traE, traE, traD, and traC) and 16 trb genes (trbP, trbO, trbN, trbM, trbL, trbK, trbJ, trbI, trbH, trbG, trbF, trbE, trbD, trbC, trbB, and trbA) are included among these 46 genes, making pBHB transferrable. The genetic-load region of pBHB is inserted between trfA and the plasmid stabilization gene (parE) and contains 53 accessory genes, including the mercury resistance elements and the haloaromatic catabolic genes.
TABLE 2.
Features of four catabolic plasmids from Comamonas species compared in this study
| Plasmid | Origina | Host | Length (bp) | ORF | G+C% | Coding % | Degrading substrate | Accession no. | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|
| pCNB1 | Industrial WWTP in China | C. testosteroni CNB-1 | 91,181 | 91 | 63.4 | 81.6 | 4-Chloronitrobenzene | NC_010935.1 | 13 |
| pI2/pNB2 | Activated sludge in Belgium | C. testosteroni I2 | 84,204 | 85 | 64.7 | 90.4 | Aniline, 3-chloroaniline | NC_016978.1 | 4, 9, 10 |
| pTB30 | 3-Chloroaniline-treated agricultural soil in Belgium | C. testosteroni TB30 | 79,291 | 76 | 63.9 | 89.3 | 3-Chloroaniline | NC_016968.1 | 9, 11 |
| pBHB | Bromoxynil-contaminated soil in China | Comamonas sp. 7D-2 | 119,225 | 122 | 63.57 | 84.9 | Bromoxynil (3,5-dibromo-4-hydroxybenzonitrile) | KC771559 | This study |
WWTP, wastewater treatment plant.
Incompatibility group assignment.
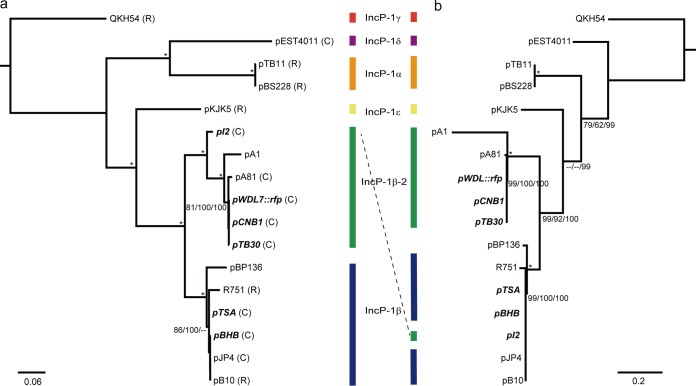
A phylogenetic analysis using 24 concatenated protein sequences encoded by the IncP-1 plasmids showed that pBHB belonged to the IncP-1β subgroup and clustered together with pJP4, pB10, R75, pTSA, and pBP136 with 100% support (Fig. 1). Other catabolic plasmids from the genus Comamonas (pI2, pTB30, pCNB1, and pWDL7::rfp) clustered tightly with 100% support and belonged to the IncP-1β-2 subgroup, which was derived from the IncP-1β subgroup and renamed by Norberg et al. (28) (Fig. 1). The TrfA protein of pBHB exhibited 100%, 87%, and 87% similarity to the corresponding sequences in pI2, pTB30, and pCNB1, respectively. The TrfA-based phylogenetic tree showed a consistent topological structure and showed only slight discrepancy for pI2 (Fig. 1), which may reflect the unique evolutionary status of pI2. Nevertheless, the consistency suggests the reliability of both methods, although phylogeny estimation based on backbone genes is expected to produce conflicting gene tree topologies (29). Based on these results, all six Comamonas catabolic plasmids belong to the IncP-1β subgroup of plasmids (Fig. 1, boldface italic).
FIG 1.
Phylogenetic trees based on 24 backbone gene proteins (a) and TrfA (b) showing the relationship of the plasmids pBHB, pTB30, pCNB1, and pI2 to five IncP-1 subgroup plasmids. The ML/MP/NJ bootstrap values are sequentially indicated at the branches, and the asterisks indicate that all three values are 100. The α, β, β-2, δ, and ε taxonomies of IncP-1 subgroup plasmids are marked by vertical lines with different colors. The 24 backbone gene protein-based and TrfA-based phylogenetic trees are in accordance, except for a slight discrepancy in the plasmid pI2. The plasmids from the genus Comamonas are in boldface italics. R, resistance plasmid; C, catabolic plasmid. The substrates of catabolic plasmids are as follows: pEST4011, 2,4-dichlorophenoxyacetic acid; pI2, 3-chloroaniline; pA81, chlorobenzoate; pWDL7::rfp, 3-chloroaniline; pCNB1, 4-chloronitrobenzene; pTB30, 3-chloroaniline; pTSA, p-toluenesulfonate; pBHB, 3,5-dibromo-4-hydroxybenzonitrile; pJP4, 2,4-dichlorophenoxyacetic acid.
Core genome of the four Comamonas catabolic plasmids.
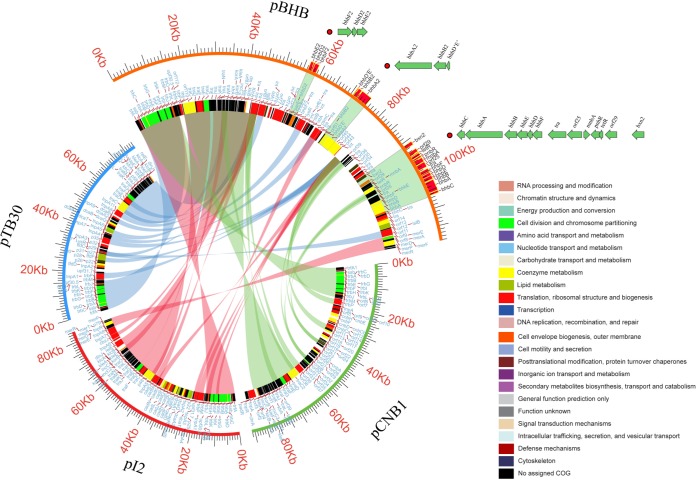
Due to the common hosts, close phylogenetic relationships, and capacity for haloaromatic catabolism, the genomes of three other catabolic plasmids—pCNB1, pI2, and pTB30—were compared with that of pBHB. The plasmid pWDL7::rfp was not included in this analysis because it has been genetically modified (9). The basic information on the four plasmids is listed in Table 2. The four plasmids are all circular and have similar G+C% contents (63.4 to 64.7%). The complete sequence of plasmid pBHB shows 39.6%, 33.6%, and 30.2% similarity to the sequences of pI2, pCNB1, and pTB30, respectively. The complete sequence similarity was defined as the product of the percentage of the conserved sequence among the complete sequence and the similarity between the conserved sequences (see Table S2 in the supplemental material). Plasmid pBHB encoded 44 (36.1% of the total proteins encoded by pBHB), 42 (34.4%), and 35 (28.7%) proteins with homology values higher than 0.81 compared with pI2, pCNB1, and pTB30, respectively (Fig. 2). According to the OrthoMCL analysis, these four Comamonas plasmids shared a core genome consisting of 40 genes (32.8% of the ORFs of pBHB), which encode proteins sharing at least 50% similarity among all four plasmids (Fig. 3; see Fig. S2 in the supplemental material). The core genome somewhat resembles the backbone genes of pBHB and is responsible for the core functions. The conjugal transfer proteins (Tra and Trb); plasmid replication, central control, stable inheritance, and partitioning proteins (TrfA, Klc, Kor, Kle, and KfrA); single-stranded DNA binding protein (Ssb); plasmid stabilization system protein (Xf2080); damage-inducible-like protein (Din); and a putative outer membrane protein (Upf30.5) are highly conserved. Din and KlcA are 100% identical among all four plasmids.
FIG 2.
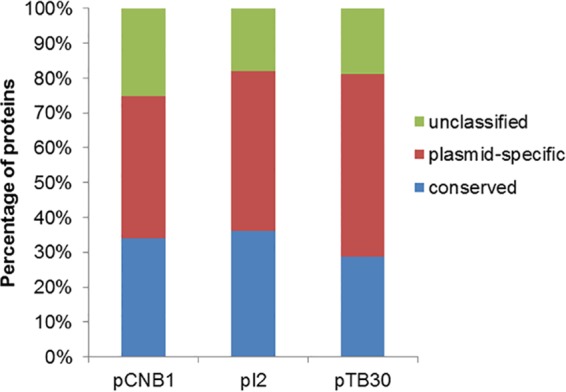
Percentages of conserved and plasmid-specific proteins encoded by pBHB compared with the other three Comamonas catabolic plasmids. Proteins with a BLASTp-derived homology value higher than 0.81 were classified as conserved, whereas those with a homology value lower than 0.42 were considered plasmid specific. The remaining genes were designated unclassified.
FIG 3.
Alignments of the plasmidome of pBHB with pTB30, pI2, and pCNB1. The putative genes are represented by 24 colors, based on cluster of orthologous groups (COG) assignments. The putative orthologous relationships of pBHB to the other three plasmids were then converted into a circular graph in which weighted edges in different colors (light blue, pBHB to pTB30; pink, pBHB to pI2; pale green, pBHB to pCNB1) represent their relationships. Bromoxynil catabolic genes, including two reductive dehalogenase gene clusters on plasmid pBHB, are marked.
Genetic-load regions of the four Comamonas catabolic plasmids.
In addition to the commonly shared core genome that encodes plasmid-associated core functions, the four Comamonas plasmids include a genetic-load region that contains a number of accessory genes. Plasmids pBHB, pCNB1, pI2, and pTB30 contain 50, 33, 12, and 19 unique genes (plasmid-specific genes among all four plasmids), respectively, most of which are located in the genetic-load region. In pBHB, orf11, orf12, orf13, orf14, bhbC, mnbA, pnbR, tetR, orf29, bxn2, tra1-4, orf34, orf35, orf36, orf39, orf43, orf44, orf45, orf51, osmC, int, orf58, istA, orf61, lysR, parE, orf73, orf74, orf77, orf78, istB, orf81, orf110, res, orf112, bhbA, bhbA2, bhbB, bhbB2, bhbC, bhbD, bhbD2, bhbE, bhbE2, bhbF, bhbF2, and int are unique, and most of them are transposase genes or genes involved in haloaromatic catabolism. In addition, the dca and tfd 3-chloroaniline catabolic gene clusters on pTB30, the dca (halo)aniline catabolic gene cluster on pI2, and the cnb and cat 4-chloronitrobenzene and catechol catabolic gene clusters on pCNB1 are all located in the genetic-load region, showing that the genetic-load region plays critical roles in haloaromatic catabolism.
Although the other three Comamonas catabolic plasmids—pCNB1, pI2, and pTB30—reportedly catabolize haloaromatics (3-chloroaniline and 4-chloronitrobenzene), dehalogenase genes were not identified. Plasmid pBHB encodes 56 (45.9% of the proteins encoded by pBHB), 50 (41.0%), and 64 (52.5%) proteins with homology values lower than 0.42 compared with pI2, pCNB1, and pTB30, respectively (Fig. 2). A functional reductive dehalogenase gene cluster, bhbAB, has been previously identified on pBHB (14). Notably, pBHB was isolated from the aerobic strain of Comamonas sp. 7D-2, and aerobes were originally thought to rarely exhibit reductive dehalogenation activity (30, 31). Moreover, in addition to the previously identified bhbAB cluster, pBHB contains another reductive dehalogenase gene cluster, bhbA2B2. These data indicate that pBHB has specifically evolved to catabolize haloaromatics. In addition to the reductive dehalogense gene clusters, pBHB contains the complete set of genes for bromoxynil catabolism, including the nitrilase gene bxn2, the 4-hydroxybenzoate 3-monooxygenase gene bhbF-bhbF2, and the protocatechuate 4,5-dioxygenase genes bhbDE-bhbD2E2-bhbD′E′, which are located between the mer cluster and the praE gene. In addition, a putative 3-nitrobenzoate oxygenase gene, mnbA, was also found adjacent to the bhb cluster.
The role of mobile genetic elements in the evolution of pBHB.
Interestingly, many of the accessory genes in pBHB were found to be associated with mobile genetic elements, including transposons and IS elements (Table 3 and Fig. 4; see Fig. S3 in the supplemental material). The 2.6-kb IS1071-like element on pBHB contains an intact 50-bp right end, whereas a left end was not identified. However, one 106-bp sequence containing an intact left end (100% similarity with that of IS1071 on pBRC60) was found 5,500 bp upstream of the right IS1071-like element (see Fig. S3 in the supplemental material). An ISThsp19-like element (32) was found to be inserted between the above-mentioned left and right ends, and an ISPsy30-like element (33) was found 180 bp upstream of the 106-bp sequence, suggesting that the truncated IS1071-like element was caused by the insertion of other ISs. Two identical ISPps1 (34) elements oriented in the same direction were found on pBHB. The right ISPps1 element is only 550 bp upstream of the IS1071-like element. This result is consistent with the location of ISPps1, which often resides near other mobile elements, most of which are IS1071 (35).
TABLE 3.
Features of putative insertion sequences on pBHB
| Insertion sequence | Length (bp) | IS family | IS databaseb | Coverage (%) | Identity (%) | E value | IR count | Copy no. |
|---|---|---|---|---|---|---|---|---|
| IS1071-like | 2,635 | Tn3 | IS1071 | 100 | 99 | 0 | 1 | 2 |
| ISPps1 | 1,864 | IS91 | ISPps1 | 100 | 100 | 0 | 0 | 2 |
| ISAav1-like | 2,500 | IS21 | ISAav1 | 55 | 83 | 5.00E−67 | 2 | 2 |
| ISbhb1a | 1,662 | IS3 | IS1240, ISSba13 | 100, 100 | 68, 47 | 5E−97, 2E−19 | 2 | 2 |
| ISThsp19-like | 2,356 | IS21 | ISThsp19 | 50 | 89 | 1.00E−101 | 2 | 1 |
| TnAs2-like | 4,677 | Tn3 | TnAs2 | 94 | 89 | 0 | 1 | 1 |
| ISPsy30-like | 3,716 | Tn3 | ISPsy30 | 98 | 84 | 0 | 2 | 1 |
The two amino acid sequences encoded by ORF15 and ORF16 in ISbhb1 were used to perform the BLASTp analysis.
The best hit(s) in the IS database.
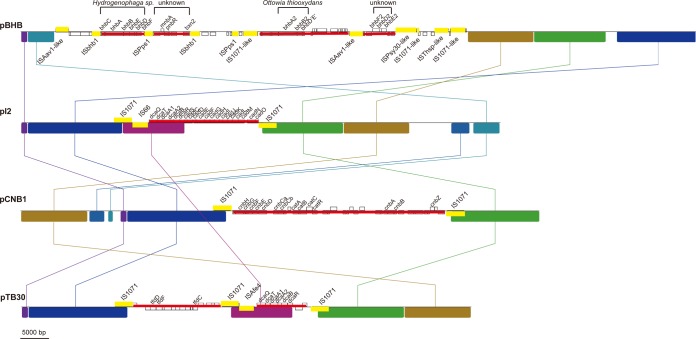
FIG 4.
Supposed insertions and rearrangements occurred on the four Comamonas catabolic plasmids. The colored boxes represent syntenic regions, and the white areas refer to the unique regions. The lines link blocks with homology between genomes. The gene distributions in the white areas are shown in small open boxes, and catabolic genes are marked with names. The thick yellow lines represent transposons and ISs, while the thin red lines represent the possible cargos of the transposons. The details of the transposons and ISs are shown in Fig. S3 in the supplemental material. The four catabolic gene clusters that evolved independently on pBHB are shown with square brackets. The potential source (ancestor) of the two bhb gene clusters (bbhABCDEF and bhbA2B2D′E′) on pBHB is indicated, while no clear evolutionary patterns were detected for the other two clusters.
Two identical 1.6-kb sequences oriented in the same direction were detected on pBHB. Each sequence encoded a transposase, i.e., ORF15/ORF16 and ORF30/ORF31. The transposase encoded by ORF15/ORF30 showed 68% similarity to the transposase of IS1240 from Pseudomonas syringae (34), whereas the transposase encoded by ORF16/ORF31 showed only 47% similarity to the transposase of ISSba13 from Shewanella baltica OS155 (34). Moreover, an imperfect 21-bp inverted repeat (IR) was found at the boundaries of both sequences. Based on these results, the 1.6-kb sequence was identified as a novel IS, which was designated ISbhb1.
Two identical ISAav1-like elements (20) were also detected on pBHB. The two identical ISAav1-like elements form a 54-kb composite transposon. Several other IS elements were also found to constitute the composite transposon, including two ISPps1 elements, one IS1071-like element, and two ISbhb1 elements. Intriguingly, the 54-kb ISAav1-like composite transposon and the 39-kb IS1071-like composite transposon overlapped. Moreover, the 19-kb ISbhb1 composite transposon and 17-kb ISPps1 composite transposon also overlapped (see Fig. S3 in the supplemental material). These results showed that a series of insertions may have occurred inside each other. Unfortunately, since no direct repeats of these ISs were found, it was difficult to determine the relationships of the insertions.
The origin of the catabolic genes in pBHB.
The bhbFDE and bhbAB genes are clustered together, although the bhbFDE genes are not functional (14). The phylogenetic analysis showed that all bbhABCDEF genes clustered (showing the highest similarity) with the corresponding homologous genes from Hydrogenophaga (Fig. 5), suggesting that the bbhABCDEF genes evolved from a common ancestor of Comamonas and Hydrogenophaga (considering the close phylogenetic relationship between them) or originated from horizontal gene transfer (HGT), because these genes are included in the ISbhb1 or ISAav1 composite transposon. However, similar patterns were not detected for bxn2 and the other genes between ORF24 and bxn2, indicating that the cluster ORF24-bxn2 originated independently of the bbhABCDEF genes.
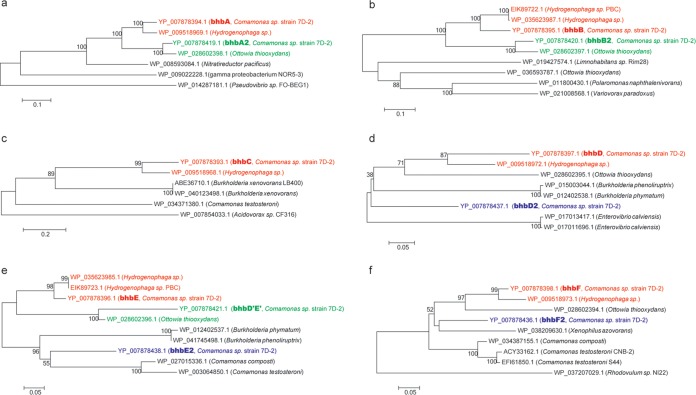
FIG 5.
Phylogenetic relationships of the bromoxynil catabolic genes with other related genes based on the highest identities of their amino acid sequences. Unrooted phylogenetic NJ trees of amino acid sequences from BhbA and BhbA2 (a); BhbB and BhbB2 (b); BhbC (c); BhbD and BhbD2 (d); BhbE, BhbE2, and BhbD′E′ (e); and BhbF and BhbF2 (f) were constructed. The hosts harboring these genes are shown in parentheses. The bootstrap values are indicated above the branches. Clades in red, green, and blue indicate genes that form the bhbABCDEF cluster, the bhbA2B2D′E′ cluster, and the bhbE2D2F2 cluster, respectively.
The bhbA2B2D′E′ genes are located 15 kb upstream of the bhbABCDEF cluster (Fig. 3 and 4). Other than clustering with bhbABDE in the phylogenetic tree, the bhbA2B2D′E′ genes all clustered with corresponding genes from Ottowia thiooxydans (Fig. 5). These results suggested that the bhbA2B2D′E′ cluster was not a simple duplication of bhbABDE but, rather, evolved independently. The functional bhbF2D2E2 cluster was found 31 kb upstream of the bhbFDE cluster. Although the phylogenetic analysis did not identify a clear evolutionary pattern (Fig. 5), the bhbF2D2E2 cluster was found to also have originated independently instead of being a duplication of the bhbFDE cluster.
The activity and substrate range of BhbA2B2.
The previously characterized bhbAB gene cluster has been functionally confirmed to have reductive dehalogenase activity (14), and the function of the bhbA2B2 cluster was investigated in the present study. BhbA2 and BhbB2 were 78% and 69% identical to BhbA and BhbB, respectively. Strain 2B-bhbA2B2 was able to reductively dehalogenate 81.2% and 91.1% of 0.2 mM BHB and DBHB, respectively, within 8 h, which indicated that its degradation rate was significantly higher than that of strain 2B-bhbAB (Fig. 6). The crude enzymatic activities of BhbA and BhbA2 coexpressed with their native extracytoplasmic binding receptors were also compared (Table 4). Consistent with the degradation rate, the activity of BhbA2 was 3.5- to 30.9-fold higher than that of BhbA for various substrates. Although the enzymatic activity of BhbA2 was much higher, the substrate range of BhbA2 was the same as that of BhbA. BhbA2 dehalogenated ortho-halogens with respect to the hydroxyl group in brominated and chlorinated hydroxybenzoate and, overall, preferred DBHB as a substrate (Table 4).
TABLE 4.
Crude reductive dehalogenation activities of BhbA and BhbA2 extracted from strains 2B-bhbA2B2 and 2B-bhbAB, respectively
| Substrate | Activity (nmol min−1 mg−1) |
|
|---|---|---|
| BhbA | BhbA2 | |
| Bromoxynil | 0.19 ± 0.01 | 1.24 ± 0.13 |
| DBHB | 0.82 ± 0.09 | 13.79 ± 0.47 |
| BHB | 0.22 ± 0.01 | 3.05 ± 0.11 |
| DCHB | 0.19 ± 0.03 | 6.02 ± 0.42 |
| CHB | 0.05 ± 0.01 | 0.16 ± 0.03 |
| DBP | 0.20 ± 0.05 | 3.47 ± 0.38 |
Specificity of the extracytoplasmic binding receptor (BhbB/BhbB2) for the reductive dehalogenase (BhbA/A2).
The extracytoplasmic binding receptor BhbB has been confirmed to anchor the reductive dehalogenase BhbA to the cytoplasmic membrane and to greatly enhance the dehalogenation activity of BhbA on the membrane (14). To investigate the specificity of the extracytoplasmic binding receptor, the effects of different combinations of reductive dehalogenase genes and the extracytoplasmic binding receptor gene in strain 2B on the degradation rates were investigated. The specificity of the extracytoplasmic binding receptor was not strict. Specifically, the exchange of the extracytoplasmic binding receptor did not significantly affect the degradation rate (Fig. 6). For example, the degradation rates of strains 2B-bhbAB and 2B-bhbAB2 were nearly the same, and a similar phenomenon was also observed in strains 2B-bhbA2B and 2B-bhbA2B2 (Fig. 6). Therefore, the higher degradation rates of strains containing bhbA2 than the rates of strains containing bhbA occurred because of the reductive dehalogenase gene itself instead of the extracytoplasmic binding receptor gene.
Transcription of the bhbAB and bhbA2B2 gene cluster in response to different substrates.
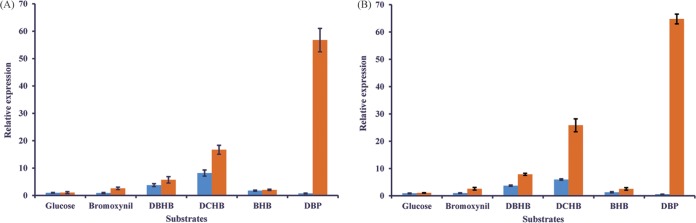
The transcriptions of the bhbAB and bhbA2B2 clusters on the plasmid pBHB in response to the dehalogenated substrates or glucose were determined using reverse transcription (RT)-qPCR. As shown in Fig. 7, the transcriptional level of bhbA was consistent with that of bhbB, and the same status was also found for bhbA2 and bhbB2. These data suggest that the bhbAB gene cluster (as well as bhbA2B2) was located in one operon. Moreover, the transcript level of bhbA2B2 was generally higher than that of bhbAB for various dehalogenated substrates. The transcript levels of bhbA2 and bhbB2 were more than 76- and 108-fold higher than those of bhbA and bhbB, respectively, when DBP was used as the substrate.
FIG 7.
Transcriptional analysis of bhbA (A, blue), bhbA2 (A, orange), bhbB (B, blue), and bhbB2 (B, orange) in Comamonas sp. strain 7D-2 in response to different substrates. The levels of gene expression in each sample were calculated as the fold expression ratio after normalization to the trfA gene transcriptional levels. The error bars indicate standard deviations.
DISCUSSION
The plasmids in the ubiquitous and versatile Comamonas species are of great interest because Comamonas plasmids are highly abundant in the bacterial populations of many habitats and often provide exchangeable niche-specific fitness factors (4, 6–11, 13, 14, 36). To gain insight into the evolutionary adaptation of Comamonas plasmids and their roles in the catabolism of xenobiotics, the complete sequences of the four catabolic plasmids pI2, pCNB1, pTB30, and pBHB were investigated and compared.
Incompatibility group assignment by phylogenetic analyses of 24 backbone gene proteins, as well as TrfA, showed that all six Comamonas catabolic plasmids belonged to the IncP-1β subgroup (Fig. 1, boldface). Most IncP-1 plasmids described to date carry genes for xenobiotic catabolism, mercury resistance, or multidrug resistance (13, 37). Except for pBP136 and pA1, all the representative plasmids used are catabolic (pA81, pCNB1, pTB30, pWDL7::rfp, pI2, pJP4, pEST4011, pTSA, and pBHB) or resistance (pB10, R751, pTB11, pBS228, and pKJK5) plasmids. Interestingly, our data indicate that IncP-1β subgroup plasmids are likely haloaromatic catabolic plasmids. All haloaromatic catabolic plasmids, including pBHB, pCNB1, pI2, pTB30, pJP4 (the chloroaromatic pollutant catabolic plasmid from Ralstonia eutropha), pA81 (the haloaromatic acid catabolic plasmid from Achromobacter xylosoxidans), and pWDL7::rfp (the chloroaniline catabolic plasmid from Comamonas), are located in the IncP-1β subgroup.
Comparison of the four Comamonas plasmids revealed the conservation of the backbone region and the diversity of the genetic-load region. All four plasmids share 40 orthologous genes (>50% similarities) that encode the core functions (Fig. 3; see Fig. S2 in the supplemental material). These results reinforced the fact that plasmids from different, often geographically separated sites may nonetheless share core genomes (38, 39). The number of orthologous genes is relatively high, suggesting that these plasmids evolved from a common ancestor. Among the four plasmids, pBHB and pI2 shared 17 proteins with 100% identity and the highest complete sequence similarity. Notably, only pBHB and pI2 contained mer (mercury resistance) operons, although pCNB1 contained arsenate and chromate resistance genes. Antibiotic resistance genes were not found in any of the four plasmids, which corroborates previous reports indicating that rare plasmids carry both antibiotic resistance and degradative genes (40).
In addition to the conserved backbone region, these plasmids contained a diverse genetic-load region (Fig. 3). Not only the hosts, but also plasmids, are under differential selective pressures, and consequently, they gain and lose genes to form a dynamic organization. IncP-1β plasmids can reportedly undergo recombination in the environment, enhancing plasmid diversity (40). The genetic-load regions of the four plasmids contained relatively high numbers of unique genes, most of which are catabolic genes or heavy-metal resistance genes. The conservation of the backbone region and the diversity of the genetic-load region might be the outcomes of an evolutionary molecular history starting from ancestral plasmid backbones, which very likely underwent different evolutionary paths in different hosts, gaining and/or losing genes from their plasmidomes.
The genetic-load region seemed to have been subject to rearrangements and HGT events (Fig. 3). In addition, all four plasmids carry ISs or composite transposons (Fig. 4). For example, a 44.5-kb large transposon, TnCNB1, containing 4-chloronitrobenzene-degrading genes and arsenate resistance genes was found on pCNB1 (13). pBHB is riddled with numerous relics of mobile elements, including transposons and IS elements (Fig. 4 and Table 3). IS1071 seemed to play an important role in the evolution of plasmids (41, 42). IS1071 was found on all four of these plasmids (4, 13). However, the IS1071-like element on pBHB showed only 76% similarity with IS1071 on pCNB1 (13). Furthermore, no orthologs shared by these two plasmids were found to be inserted in the IS1071-like element, which suggests that the IS1071-like element on pBHB evolved independently from that on pCNB1.
ISPps1, similar to other IS91 family elements, often resides on plasmids and is frequently associated with genes responsible for the degradation of halo- or nitroaromatics (35, 43). The nonfunctional bhbF on pBHB is approximately 350 bp shorter than previously reported HB-3-monooxygenase genes. The functional loss of bhbFDE may be due to ISPps1, because ISPps1 was found only 253 bp upstream of the bhbF gene. Similarly, the loss of the approximately 450-bp sequence of bhbC compared with other, homologous genes might be caused by ISbhb1, which is located adjacent to bhbC. In addition, bbhABCDEF genes were included in the ISbhb1 or ISAav1 composite transposon. These results revealed that ISs and transposons play pivotal roles in the modular evolution of pBHB.
The analysis of the origin of the catabolic genes on pBHB suggested that the bbhABCDEF cluster derived from a common ancestor of Comamonas and Hydrogenophaga or via HGT, whereas the ORF24-bxn2 cluster originated independently (Fig. 5). However, the bhbA2B2D′E′ genes all clustered (showing the highest similarity) with the corresponding genes from O. thiooxydans instead of Hydrogenophaga (Fig. 5), which indicated that the bhbA2B2D′E′ cluster was not a simple duplication of bhbABDE but also evolved independently. Both the IS1071 and ISAav1 composite transposons might be involved in the acquisition of the bbhA2B2D′E′ cluster. The functional bhbF2D2E2 cluster also originated independently instead of by duplication of the bhbFDE cluster (Fig. 5). The bhbF2D2E2 cluster was in the IS1071 composite transposon, and the ISPsy30-like element, which was located 180 bp downstream, might be involved in the evolution of the bhbF2D2E2 cluster. The independent origin of catabolic gene clusters and the occurrence of many truncated genes strongly suggested that the effects of different mobile genetic elements on the evolution of pBHB were crossed and that these accessory genes in the genetic-load region were obtained multiple times.
Plasmid pBHB contains an approximately 50-kb region of accessory genes, is larger (119 kb) than the other three plasmids (79 to 91 kb), and encodes 21 to 36 more ORFs (Table 2). Although the other three Comamonas plasmids were capable of catabolizing haloaromatics (3-chloroaniline or 4-chloronitrobenzene), no dehalogenase genes have been identified. The identification of functionally reductive dehalogenase genes on pBHB suggests that pBHB contains recently acquired genetic materials that are specific for haloaromatic catabolism. The presence of two reductive dehalogenase gene clusters on pBHB is interesting (Fig. 3 and 4). These two copies of dehalogenase gene clusters may confer on the host additional advantages in the utilization of specific haloaromatic substrates and confer fitness in unusual ecological niches (i.e., in niches contaminated with different types or concentrations of haloaromatics).
The degradation rates and the crude enzymatic activities of the reductive dehalogenase extracted from strain 2B-bhbA2B2 were higher than those of strain 2B-bhbAB, which is surprising (Fig. 6 and Table 4). Although the extracytoplasmic binding receptor is important for the activity of the reductive dehalogenase, the extracytoplasmic binding receptor was not strictly specific for the reductive dehalogenase. The RT-qPCR results revealed that the transcript levels of the bhbA2B2 cluster in the wild-type strain were higher than those of bhbAB, especially when the substrates DCHB and DBP were used. The increase in bhbA2B2 transcription compared with bhbAB might be explained by their different regulatory mechanisms. Transcription regulatory genes (lysR or marR) were found to be closely associated with bhb homologous clusters in many strains, including Ruegeria conchae DSS-3, R. conchae TW15, Jannaschia sp. strain CCS1, Roseibium sp. strain TrichSKD4, Rhodobacteraceae bacterium HIMB11, Nitratireductor pacificus pht-3B, Phaeobacter caeruleus DSM 24564, and O. thiooxydans, indicating that reductive dehalogenase genes are regulated. A candidate bhbA2B2 regulator gene, lysR (ORF66), whose amino acid sequence was 55% identical to that of the above-mentioned LysR in O. thiooxydans, was found to be closely related to bhbD2E2F2. In addition, a tetR regulator gene was found adjacent to the bhbABCDEF cluster on pBHB. However, the regulation of the two bhb clusters requires further investigation. In general, the transcription level and the character of the dehalogenase itself, instead of the extracytoplasmic binding receptor, might explain the differences between the activities of BhbA2 and BhbA. Overall, our work provides insights into the evolutionary strategies of Comamonas plasmids, particularly the adaptation mechanism employed by pBHB for the degradation of haloaromatics.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Outstanding Youth Foundation of Jiangsu Province (BK20130029), the NSFC-ISF joint program (31461143009), the Chinese National Science Foundation for Excellent Young Scholars (31222003), the Program for New Century Excellent Talents in University (NCET-12-0892), the Fundamental Research Funds for the Central Universities (KYZ201422 and KJQN201529), the National Natural Science Foundation of China (31400105), and the China Postdoctoral Science Foundation (2014M561666).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02930-15.
REFERENCES
- 1.Willems A, De Vos P. 2006. Comamonas, p 723–736. In Prokaryotes. Springer-Verlag, New York, NY. [Google Scholar]
- 2.Arai H, Akahira S, Ohishi T, Kudo T. 1999. Adaptation of Comamonas testosteroni TA441 to utilization of phenol by spontaneous mutation of the gene for a trans-acting factor. Mol Microbiol 33:1132–1140. [DOI] [PubMed] [Google Scholar]
- 3.Boon N, Goris J, De Vos P, Verstraete W, Top EM. 2000. Bioaugmentation of activated sludge by an indigenous 3-chloroaniline-degrading Comamonas testosteroni strain, I2gfp. Appl Environ Microbiol 66:2906–2913. doi: 10.1128/AEM.66.7.2906-2913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boon N, Goris J, De Vos P, Verstraete W, Top EM. 2001. Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl Environ Microbiol 67:1107–1115. doi: 10.1128/AEM.67.3.1107-1115.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hein P, Powlowski J, Barriault D, Hurtubise Y, Ahmad D, Sylvestre M. 1998. Biphenyl-associated meta-cleavage dioxygenases from Comamonas testosteroni B-356. Can J Microbiol 44:42–49. [PubMed] [Google Scholar]
- 6.De Rore H, Demolder K, De Wilde K, Top E, Houwen F, Verstraete W. 1994. Transfer of the catabolic plasmid RP4::Tn4371 to indigenous soil bacteria and its effect on respiration and biphenyl breakdown. FEMS Microbiol Ecol 15:71–77. [Google Scholar]
- 7.Hrywna Y, Tsoi TV, Maltseva OV, Quensen JF III, Tiedje JM. 1999. Construction and characterization of two recombinant bacteria that grow on ortho- and para-substituted chlorobiphenyls. Appl Environ Microbiol 65:2163–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Junker F, Cook AM. 1997. Conjugative plasmids and the degradation of arylsulfonates in Comamonas testosteroni. Appl Environ Microbiol 63:2403–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Król JE, Penrod JT, McCaslin H, Rogers LM, Yano H, Stancik AD, Dejonghe W, Brown CJ, Parales RE, Wuertz S, Top EM. 2012. Role of IncP-1β plasmids pWDL7::rfp and pNB8c in chloroaniline catabolism as determined by genomic and functional analyses. Appl Environ Microbiol 78:828–838. doi: 10.1128/AEM.07480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y, Arumugam K, Tay MQ, Seshan H, Mohanty A, Cao B. 2015. Comparative genome analysis reveals genetic adaptation to versatile environmental conditions and importance of biofilm lifestyle in Comamonas testosteroni. Appl Microbiol Biotechnol 99:3519–3532. doi: 10.1007/s00253-015-6519-z. [DOI] [PubMed] [Google Scholar]
- 11.Dejonghe W, Goris J, Dierickx A, De Dobbeleer V, Crul K, De Vos P, Verstraete W, Top EM. 2002. Diversity of 3-chloroaniline and 3,4-dichloroaniline degrading bacteria isolated from three different soils and involvement of their plasmids in chloroaniline degradation. FEMS Microbiol Ecol 42:315–325. doi: 10.1111/j.1574-6941.2002.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 12.Wu JF, Jiang CY, Wang BJ, Ma YF, Liu ZP, Liu SJ. 2006. Novel partial reductive pathway for 4-chloronitrobenzene and nitrobenzene degradation in Comamonas sp. strain CNB-1. Appl Environ Microbiol 72:1759–1765. doi: 10.1128/AEM.72.3.1759-1765.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma YF, Wu JF, Wang SY, Jiang CY, Zhang Y, Qi SW, Liu L, Zhao GP, Liu SJ. 2007. Nucleotide sequence of plasmid pCNB1 from Comamonas strain CNB-1 reveals novel genetic organization and evolution for 4-chloronitrobenzene degradation. Appl Environ Microbiol 73:4477–4483. doi: 10.1128/AEM.00616-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K, Huang L, Xu C, Liu X, He J, Zinder SH, Li S, Jiang J. 2013. Molecular characterization of the enzymes involved in the degradation of a brominated aromatic herbicide. Mol Microbiol 89:1121–1139. doi: 10.1111/mmi.12332. [DOI] [PubMed] [Google Scholar]
- 15.Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 16.Brown Kav A, Sasson G, Jami E, Doron-Faigenboim A, Benhar I, Mizrahi I. 2012. Insights into the bovine rumen plasmidome. Proc Natl Acad Sci U S A 109:5452–5457. doi: 10.1073/pnas.1116410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall JPJ, Harrison E, Lilley AK, Paterson S, Spiers AJ, Brockhurst MA. 13 May 2015. Environmentally co-occurring mercury resistance plasmids are genetically and phenotypically diverse and confer variable context-dependent fitness effects. Environ Microbiol. doi: 10.1111/1462-2920.12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernández-López R, Garcillán-Barcia MP, Revilla C, Lázaro M, Vielva L, de la Cruz F. 2006. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol Rev 30:942–966. doi: 10.1111/j.1574-6976.2006.00042.x. [DOI] [PubMed] [Google Scholar]
- 19.Bentley SD, Parkhill J. 2004. Comparative genomic structure of prokaryotes. Annu Rev Genet 38:771–792. doi: 10.1146/annurev.genet.38.072902.094318. [DOI] [PubMed] [Google Scholar]
- 20.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 23.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Stoeckert CJ Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen K, Jiang JD. 2014. The BhbA enzyme assay. Bio-Protocol 4:e1111. [Google Scholar]
- 27.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Norberg P, Bergström M, Jethava V, Dubhashi D, Hermansson M. 2011. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat Commun 2:268. doi: 10.1038/ncomms1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sen D, Brown CJ, Top EM, Sullivan J. 2013. Inferring the evolutionary history of IncP-1 plasmids despite incongruence among backbone gene trees. Mol Biol Evol 30:154–166. doi: 10.1093/molbev/mss210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janssen DB, Oppentocht JE, Poelarends GJ. 2001. Microbial dehalogenation. Curr Opin Biotechnol 12:254–258. doi: 10.1016/S0958-1669(00)00208-1. [DOI] [PubMed] [Google Scholar]
- 31.van Pée KH, Unversucht S. 2003. Biological dehalogenation and halogenation reactions. Chemosphere 52:299–312. doi: 10.1016/S0045-6535(03)00204-2. [DOI] [PubMed] [Google Scholar]
- 32.Duquesne K, Lieutaud A, Ratouchniak J, Muller D, Lett MC, Bonnefoy V. 2008. Arsenite oxidation by a chemoautotrophic moderately acidophilic Thiomonas sp.: from the strain isolation to the gene study. Environ Microbiol 10:228–237. [DOI] [PubMed] [Google Scholar]
- 33.Bardaji L, Pérez-Martínez I, Rodríguez-Moreno L, Rodríguez-Palenzuela P, Sundin GW, Ramos C, Murillo J. 2011. Sequence and role in virulence of the three plasmid complement of the model tumor-inducing bacterium Pseudomonas savastanoi pv. savastanoi NCPPB 3335. PLoS One 6:e25705. doi: 10.1371/journal.pone.0025705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schleinitz KM, Vallaeys T, Kleinsteuber S. 2010. Structural characterization of ISCR8, ISCR22, and ISCR23, subgroups of IS91-like insertion elements. Antimicrob Agents Chemother 54:4321–4328. doi: 10.1128/AAC.00006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalhoefer D, Thole S, Voget S, Lehmann R, Liesegang H, Wollher A, Daniel R, Simon M, Brinkhoff T. 2011. Comparative genome analysis and genome-guided physiological analysis of Roseobacter litoralis. BMC Genomics 12:324. doi: 10.1186/1471-2164-12-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Top EM, Springael D, Boon N. 2002. Catabolic mobile genetic elements and their potential use in bioaugmentation of polluted soils and waters. FEMS Microbiol Ecol 42:199–208. doi: 10.1111/j.1574-6941.2002.tb01009.x. [DOI] [PubMed] [Google Scholar]
- 38.Heuer H, Szczepanowski R, Schneiker S, Pühler A, Top EM, Schlüter A. 2004. The complete sequences of plasmids pB2 and pB3 provide evidence for a recent ancestor of the IncP-1β group without any accessory genes. Microbiology 150:3591–3599. doi: 10.1099/mic.0.27304-0. [DOI] [PubMed] [Google Scholar]
- 39.Jerke K, Nakatsu CH, Beasley F, Konopka A. 2008. Comparative analysis of eight Arthrobacter plasmids. Plasmid 59:73–85. doi: 10.1016/j.plasmid.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Schlüter A, Heuer H, Szczepanowski R, Forney LJ, Thomas CM, Pühler A, Top EM. 2003. The 64,508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1β group. Microbiology 149:3139–3153. doi: 10.1099/mic.0.26570-0. [DOI] [PubMed] [Google Scholar]
- 41.Nakatsu C, Ng J, Singh R, Straus N, Wyndham C. 1991. Chlorobenzoate catabolic transposon Tn5271 is a composite class I element with flanking class II insertion sequences. Proc Natl Acad Sci U S A 88:8312–8316. doi: 10.1073/pnas.88.19.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Providenti MA, Shaye RE, Lynes KD, McKenna NT, O'Brien JM, Rosolen S, Wyndham RC, Lambert IB. 2006. The locus coding for the 3-nitrobenzoate dioxygenase of Comamonas sp. strain JS46 is flanked by IS1071 elements and is subject to deletion and inversion events. Appl Environ Microbiol 72:2651–2660. doi: 10.1128/AEM.72.4.2651-2660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcillán-Barcia MP, de la Cruz F. 2002. Distribution of IS91 family insertion sequences in bacterial genomes: evolutionary implications. FEMS Microbiol Ecol 42:303–313. doi: 10.1111/j.1574-6941.2002.tb01020.x. [DOI] [PubMed] [Google Scholar]
- 44.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.