Abstract
Cellulose and xylan are two major components of lignocellulosic biomass, which represents a potentially important energy source, as it is abundant and can be converted to methane by microbial action. However, it is recalcitrant to hydrolysis, and the establishment of a complete anaerobic digestion system requires a specific repertoire of microbial functions. In this study, we maintained 2-year enrichment cultures of anaerobic digestion sludge amended with cellulose or xylan to investigate whether a cellulose- or xylan-digesting microbial system could be assembled from sludge previously used to treat neither of them. While efficient methane-producing communities developed under mesophilic (35°C) incubation, they did not under thermophilic (55°C) conditions. Illumina amplicon sequencing results of the archaeal and bacterial 16S rRNA genes revealed that the mature cultures were much lower in richness than the inocula and were dominated by single archaeal (genus Methanobacterium) and bacterial (order Clostridiales) groups, although at finer taxonomic levels the bacteria were differentiated by substrates. Methanogenesis was primarily via the hydrogenotrophic pathway under all conditions, although the identity and growth requirements of syntrophic acetate-oxidizing bacteria were unclear. Incubation conditions (substrate and temperature) had a much greater effect than inoculum source in shaping the mature microbial community, although analysis based on unweighted UniFrac distance found that the inoculum still determined the pool from which microbes could be enriched. Overall, this study confirmed that anaerobic digestion sludge treating nonlignocellulosic material is a potential source of microbial cellulose- and xylan-digesting functions given appropriate enrichment conditions.
INTRODUCTION
Cellulose and xylan, two major structural components of plant cell walls, represent important resources for renewable energy (1–3). Cellulose, hemicellulose (the major component is xylan), and lignin make up 35 to 50%, 20 to 35%, and 10 to 25% of total lignocellulose by dry weight, respectively (4). Cellulose and xylan from lignocellulosic biomass not only represent new energy sources but also could reduce carbon dioxide emissions, as lignocellulosic biofuels are considered carbon neutral. However, digesting recalcitrant lignocellulosic components to fermentable sugars is a rate-limiting step (5), making lignocellulosic biofuels currently challenging to produce while being price competitive with fossil fuels (6, 7). Microbes have evolved strategies (8, 9) to digest lignocellulosic components concurrent with the evolution of plant cell walls, making the investigation of microbial cellulose and xylan digestion potentially important to this emerging energy source.
Metagenomic and microbial diversity studies of ruminant animals (e.g., cow [10], sheep [11], and deer [12]) have revealed rich taxonomic and functional microbial diversity, including mechanisms for cellulose and xylan digestion. Microbial cellulose and xylan digestion systems have been identified in insects, e.g., termite gut (13–15), forest soils (16, 17), and marsh sediments (18). As well as these natural systems, artificial cellulose- and xylan-digesting systems have been developed, e.g., composters (19, 20) and laboratory enrichment cultures seeded with various animal fecal matters as inoculum (21, 22). Many of these studies represent mature cellulose- and/or xylan-digesting systems where digestion is active and which have been maintained for a long time. However, we know relatively little about the potential for cellulose- and xylan-digesting functions in microbial communities where cellulosic biomass digestion is not initially prominent.
In this study, a 2-year enrichment culture experiment was set up in the laboratory with microbial communities from wastewater anaerobic digesters that were not treating cellulose or xylan waste as the seed inocula. The inocula were from two different anaerobic digesters located at separate treatment facilities that treated different waste streams under different operating conditions. We hypothesized that since digester microbial communities contain a broad repertoire of microbes with diverse metabolic functions (23–25), cellulose- and xylan-digesting microbes should be present and able to be enriched under appropriate conditions. We further aimed to investigate whether culturing under similar conditions would cause the taxonomic compositions of the different seed inocula to converge. Cultures from each seed inoculum were incubated at different temperatures (35°C or 55°C) and given different substrates (cellulose or xylan), and the similarities between the enriched cultures were evaluated with high-throughput sequencing of the 16S rRNA gene.
MATERIALS AND METHODS
Collection of inocula.
The inocula were collected from an industrial anaerobic digester treating high-strength wastewater from a beverage manufacturing facility located in Guangzhou (GZ), China, and from a municipal anaerobic digester treating waste activated sludge located in Shek Wu Hui (SWH), Hong Kong (details of the digesters can be found in reference 26). Three separate samples from the midsection of the anaerobic digester were collected and pooled as described previously (27). Sample collection was conducted in late 2011 or early 2012. The collected samples were separated into two sets; one set (seed samples) was immediately stored at −80°C for genomic DNA (gDNA) extraction, while the other set was used for culture enrichment.
Establishment of enrichment cultures.
Fifty-milliliter aliquots of the original samples were centrifuged, the supernatant was removed, and the pellet was inoculated into 100 ml of defined minimal salts anaerobic medium (28) with 0.5% (wt/vol) cellulose (Sigmacell cellulose type 101; Sigma-Aldrich, Germany) or xylan from beechwood (Sigma-Aldrich, Germany) as the sole carbon source. All cultures with xylan as the carbon source were cultivated at 35°C, and cultures amended with cellulose were cultivated at both 35°C and 55°C in order to cultivate thermophilic microbes (29, 30). All cultures were maintained under static conditions and were transferred when the amended substrates were consumed. Five milliliters of culture at 35°C or 10 ml of culture at 55°C was inoculated into 100 ml of fresh medium with the corresponding carbon source. A culture typically required 3 to 4 weeks to consume the substrate, which was determined as the point where decreasing substrate concentration plateaued. Six enrichments were set up initially, but after several transfers, the culture inoculated from GZ failed to consume cellulose at 55°C and was terminated. The remaining five enrichments were continuously subcultured by dilution at 1:20 (35°C) or 1:10 (55°C) for 2 years (2012 to 2014). The cultures were designated SWH-C-35, SWH-C-55, SWH-X-35, GZ-C-35, and GZ-X-35 according to the scheme “inoculum-substrate-temperature,” where C denotes cellulose and X denotes xylan. The time point (Y1 or Y2 for 1- or 2-year enrichment) and replicate number also were added to name samples collected for sequencing during enrichment. For example, SWH-C-35-Y2-1 represents the first replicate sample of the enrichment culture inoculated from SWH, using cellulose as a substrate, and incubated at 35°C for 2 years. Inoculum (seed) samples were named by the scheme “inoculum-Seed-replicate” e.g., SWH-Seed-1.
Culture profiles and analytical analyses.
After 2 years of enrichment, triplicate subcultures were established from each of the five long-term enrichments for a 4-week time course experiment, during which physical parameters and metabolite concentrations were closely monitored. The concentration of hydrogen (H2), pH of the culture medium, and concentrations of the volatile fatty acids (VFAs), cellulose, and xylan were analyzed as previously described (31, 32). The methane (CH4) concentration was measured using a gas chromatograph (GC-2010; Shimadzu, Japan) equipped with a flame ionization detector. The column (30 m by 0.53 mm inner diameter) was a 5A molecular sieve (Restek, PA) with helium as a carrier gas. The injector temperature was 120°C, the column oven temperature was 35°C, and the detector temperature was 200°C.
DNA extraction and 16S rRNA gene sequencing.
gDNA was extracted using the PowerSoil DNA isolation kit (MO BIO Laboratories, Carlsbad, CA) with minor modifications as described previously (33). Inoculum gDNA was extracted in triplicate. For samples taken at the year one time point, triplicate cultures were prepared for each condition and the gDNA from each culture was pooled for sequencing after the amended substrate had been consumed. For year two samples, a gDNA sample was sequenced from each of the triplicate cultures at the end of the time course experiment described above. gDNA concentration and quality were determined by NanoDrop (NanoDrop 2000; Thermo Scientific). The 515F/806R primer pair (34) was used to amplify the V4 region of the 16S rRNA gene in triplicate for each sample, and the amplicons were pooled and purified as described previously (33). The Illumina TruSeq DNA sample preparation kit v2 was used to attach adapters to the amplicons, and paired-end 150-bp reads were sequenced on the Illumina MiSeq platform by Health GeneTech Corporation (Taoyuan City, Taiwan).
Sequencing read analysis.
The Quantitative Insights Into Microbial Ecology (QIIME) software package (release 1.8.0) (35) and the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) were used to process raw reads. Chimeras were removed using ChimeraSlayer (36) via the QIIME script “parallel_identify_chimeric_seqs.py.” Quality filtering and operational taxonomic unit (OTU) formation by clustering against the Greengenes rRNA gene sequence database (version 13_8) were performed as previously described (37). All data processing and statistical analyses were performed using custom R (version 3.1.1) and Perl (version 5.18.2) scripts. Global singletons were removed from all downstream analyses. Since the forward and reverse reads gave similar results, only the forward reads were used.
For OTU relative abundances, rarefaction curves, and α-diversity analyses, reads from Bacteria and Archaea were considered separately. Rarefaction curves for the three α-diversity metrics (observed OTUs, Faith's phylogenetic diversity [FPD], and Chao1 richness estimator) were constructed using the QIIME script “alpha_rarefaction.py.” Each metric was independently calculated 10 times at 10 even intervals between 10 and the maximum read count of all samples, and the values were averaged. Normalized α-diversity metrics were calculated from OTU tables randomly subsampled to 22,333 reads for Bacteria and 1,394 for Archaea. SWH-C-55 samples contained too few archaeal reads and were excluded. Normalized bacterial and archaeal α-diversity metrics were compared between the seed and year two enrichment samples using the nonparametric Mann-Whitney test with the P value calculated from 1,000 permutations. Box plots were drawn with the R package ggplot2 (38).
For the β-diversity analyses, bacterial and archaeal OTUs were not separated. The year one samples were omitted as metabolite profiles were not available. Both the abundance-weighted and unweighted UniFrac distances (39) were computed. Principal coordinate analysis (PCoA) ordinations of the UniFrac distances were drawn with the R package vegan (2.0-9). Analysis of similarities (ANOSIM) and nonparametric permutational multivariate analysis of variance (MANOVA) (“adonis”) analyses were performed with vegan to determine the major factors contributing to the differences between sample groups. The formula “UniFrac ∼ Origin * Condition” was used for adonis; temperature and substrate were combined in the variable “Condition,” as there was only one 55°C culture. Similarity percentage (SIMPER) analyses based on the Bray-Curtis dissimilarities between samples were performed with PRIMER (version 6; Plymouth, United Kingdom) to determine the OTUs contributing most to the differences between sample groups. Venn diagrams of OTUs shared between samples were drawn with the R package VennDiagram (40) based on total OTUs observed in each sample with triplicate cultures pooled.
Nucleotide sequence accession number.
Illumina amplicon sequencing reads generated for this study have been deposited in the NCBI Sequence Read Archive under project PRJNA281520.
RESULTS
Profiles of the year two enrichment cultures.
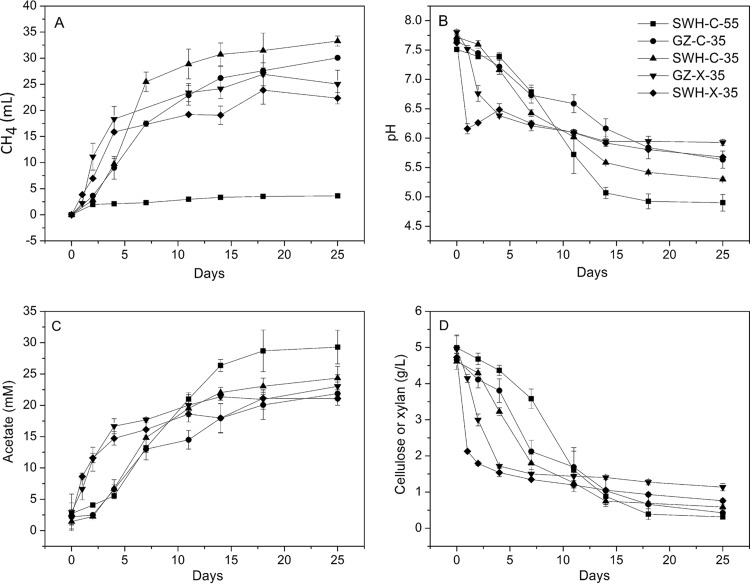
Each culture underwent at least 22 transfers during the 2-year period of enrichment in defined medium (the inoculum from GZ was unable to consume cellulose at 55°C). At transfer dilutions of 1:20 for cultures at 35°C and 1:10 for cultures at 55°C, the original inocula were diluted at least 1022-fold over this period. After 2 years of enrichment, the microbial communities were considered stable. To determine the metabolic characteristics of the cultures, CH4 and H2 volumes, VFA and substrate concentrations, and pH were measured during a 4-week time course (Fig. 1). Except for the SWH-C-55 culture (<5 ml), accumulated CH4 ranged from 22 to 33 ml (Fig. 1A), while almost no H2 was detected (data not shown). The initial pH decrease, indicative of anaerobic fermentation, was much faster for the cultures amended with xylan than with cellulose (Fig. 1B). Simultaneous with the pH decrease was VFA accumulation (Fig. 1C) and substrate consumption (Fig. 1D). The major accumulated VFA was acetate, reaching a final concentration of 20 to 30 mM in all cultures (Fig. 1C), while concentrations of formate, propionate, and butyrate were negligible (data not shown). Seventy-eight to 94% of the substrate (cellulose or xylan) was digested after 25 days (Fig. 1D), with xylan being consumed rapidly within the first few days and cellulose taking more than 2 weeks. The slower consumption of cellulose was accompanied by a slower pH drop and acetate accumulation (Fig. 1). In the thermophilic (55°C) culture, cellulose digestion was slower than that in the cultures at 35°C, with significant consumption after about 1 week. Overall, a relatively similar trend was obtained between the enrichment cultures amended with the same substrate and incubated under the same conditions regardless of the inoculum.
FIG 1.
Characteristics of the five enrichment cultures after 2 years of enrichment. (A) Production of CH4; (B) pH of the culture medium; (C) concentration of acetate; (D) concentration of carbon substrate. Error bars represent the standard deviations from biological triplicate measurements.
Overview of the year two microbial communities.
A total of 1,066,869 high-quality bacterial reads (22,333 to 82,962 reads per sample) were clustered into 15,248 unique OTUs that were classified into 65 phyla (including candidate divisions) (see Table S1 in the supplemental material). Taking together all seed, year 1, and year 2 samples, the most abundant phyla were Firmicutes (62%), Chloroflexi (8.5%), Proteobacteria (7.3%), Bacteroidetes (6.9%), Spirochaetes (4.3%), Actinobacteria (2.4%), Armatimonadetes (1.7%), Thermotogae (1.3%), and OP1 (1.2%), with the remaining bacterial phyla all detected at <1%. A dramatic increase in Clostridiales was observed between the seed and year 2 samples, from an average of 6.8% of the seed bacterial communities to 80% in the 2-year enriched samples (Fig. 2B). An overall comparison of the seed, year one, and year two data showed that although most change occurred during the first year of enrichment, the second year of enrichment further simplified the microbial communities (see Table S1).
FIG 2.
Microbial community structure of each sample at the OTU level for Archaea (A) and taxonomic rank of order (B) for Bacteria. The relative abundance was calculated based on the total number of reads in a sample (i.e., the relative abundances of Archaea and Bacteria sum to 100%).
At the genus level, cultures incubated with the same substrate at the same temperature had relatively similar microbial compositions regardless of the inoculum source. For the two cellulose cultures at 35°C after 2 years of enrichment, OTUs classified to the genus Ruminococcus were high in relative abundance (41 to 57%), while for the two xylan cultures, Coprococcus (26 to 45%) and Ruminococcus (27 to 30%) OTUs together dominated the communities. Thermophilic genera such as Thermoanaerobacterium (38 to 50%) were dominant in the SWH-C-55 cultures.
For Archaea, a total of 151,023 high-quality reads (1,394 to 15,296 reads per sample, excluding SWH-C-55) were clustered into 710 unique OTUs within three phyla (see Table S1 in the supplemental material). Euryarchaeota was the most abundant phylum, comprising 99.7% of the total archaeal reads. The archaeal communities differed from the inocula following enrichment. In the seed samples, the dominant archaeal order was the methanogenic Methanosarcinales, while in the enriched samples a small number of OTUs classified to the methanogenic Methanobacteriales comprised 98.3% of Archaea, almost all of which were from the genus Methanobacterium (Fig. 2A). For the SWH-C-55 culture, very few archaeal or methanogenic OTUs were present (<0.4% of total reads) (Fig. 2A; also see Table S1 in the supplemental material), consistent with the low methane generation (Fig. 1A).
α-Diversity of the seed and enriched samples.
When triplicate samples for each culture were pooled, many OTUs were found in both the GZ (9,753) and SWH (10,102) seed samples, while substantially fewer were detected in the enrichments (<2,000). The rarefaction curves indicated that the sequencing depth was sufficient to detect most community members (see Fig. S1 in the supplemental material), with Good's coverage of Bacteria across all samples of >94% and Archaea of >98% (excluding SWH-C-55). Three α-diversity metrics (taxonomy-based observed OTUs, Chao1, and phylogeny-based FPD) were calculated to further investigate the effect of enrichment on OTU diversity, and we consistently found both the bacterial and archaeal community diversity decreased significantly (P < 0.05 by Mann-Whitney test) after 2 years of enrichment across all samples (Fig. 3; also see Table S1 and Fig. S1). The decrease occurred mainly during the first year but continued during the second year of enrichment to a smaller extent (see Table S1).
FIG 3.
Box-and-whisker plot of alpha-diversity metrics for the bacterial (A) and archaeal (B) communities. The alpha-diversity of each sample was computed from biological triplicate samples. The P value was calculated by Mann-Whitney test between the seed and enrichment groups. Sample SWH-C-55-Y2 contained too few archaeal reads and was excluded.
Comparison and clustering of the seed and year two enrichment communities.
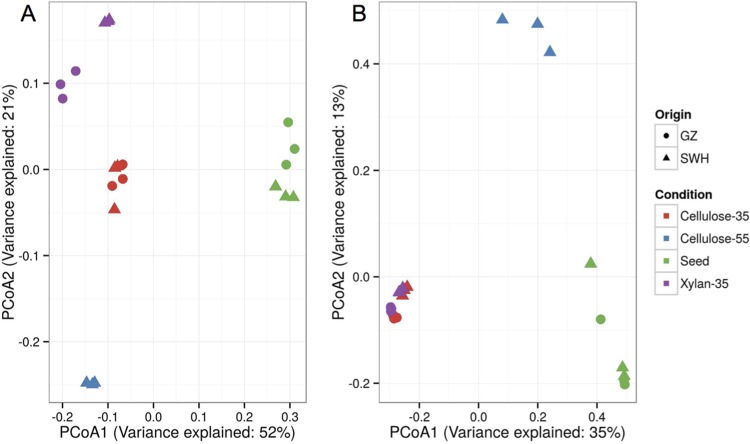
In order to study the factors shaping the mature (2-year enrichment) microbial communities, the abundance-weighted and unweighted UniFrac distances between samples (β-diversity) were calculated. PCoA ordination of the weighted UniFrac distances showed microbial communities in the enrichment cultures differed significantly from their inocula (ANOSIM R = 1, P < 0.05 for both inocula) (Fig. 4A; also see Table S2 in the supplemental material). Communities enriched under different conditions (i.e., combinations of substrate and temperature) were very different (ANOSIM R = 0.997, P = 0.001), while the inoculum source had little influence on the final community structure (ANOSIM P = 0.667) (Fig. 4A). Adonis analysis similarly found enrichment conditions (i.e., combination of temperature and substrate; R2 = 0.80, P = 0.001) played a much greater role than inoculum source (R2 = 0.089, P = 0.001) in determining the weighted UniFrac distances between samples. The communities cultured at 35°C differed strongly and significantly by substrate (ANOSIM R = 0.993, P = 0.003), and the community cultured with cellulose at 55°C also was significantly different from the cellulose cultures at 35°C (ANOSIM R = 1, P = 0.014).
FIG 4.
PCoA ordinations of the abundance-weighted (A) and unweighted (B) UniFrac distances between samples. Cultures are from 2-year enrichments. Symbols represent the origin of the cultures, while colors indicate the different conditions.
The unweighted UniFrac distance gave slightly different results than abundance-weighted UniFrac (Fig. 4B). Using unweighted UniFrac, the five enrichment cultures clustered significantly by both inoculum source (ANOSIM R = 0.423, P = 0.002) and enrichment condition (ANOSIM R = 0.746, P = 0.001), although condition was still the major factor. Adonis analysis confirmed that the enrichment condition (R2 = 0.40, P = 0.001) played a greater role than inoculum source (R2 = 0.20, P = 0.001). Substrate (ANOSIM R = 0.459, P = 0.012) and temperature (cellulose cultures only; ANOSIM R = 1.0, P = 0.011) also significantly grouped the enrichment cultures.
OTUs enriched from the seed communities.
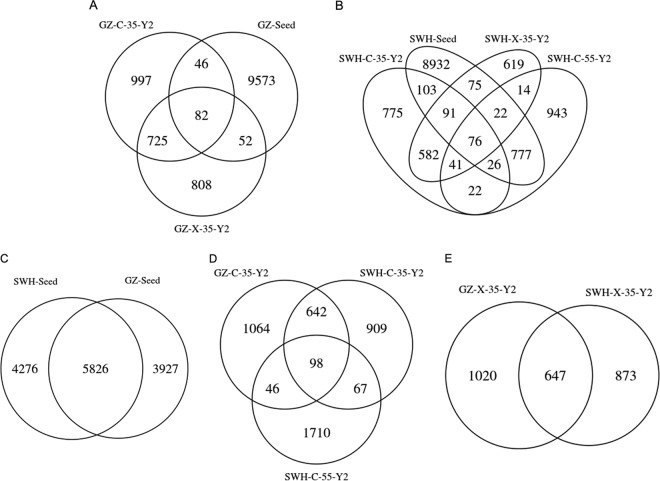
Venn diagrams were constructed to determine the extent of OTU overlap between samples (Fig. 5). Eighty to 90% of the OTUs in each enrichment culture were not observed in their respective seed samples, indicating most enriched OTUs were scarce in the source inocula (Fig. 5A and B). The two mature enrichments from the GZ inoculum at 35°C, GZ-C-35-Y2 and GZ-X-35-Y2, shared <50% of their OTUs (Fig. 5A). Similarly, 35°C enrichments from the SWH inoculum shared ∼50% of OTUs (Fig. 5B). However, the thermophilic sample SWH-C-55-Y2 shared ∼5% of OTUs with SWH-Seed and <10% with the two 35°C cultures from the same inoculum (Fig. 5B), confirming growth temperature strongly affected community composition. While the two inocula shared <60% of their OTUs (Fig. 5C), mature cultures enriched from different inocula but under the same conditions shared ∼40% of OTUs (Fig. 5D and E). SWH-C-55-Y2 still only shared <10% of OTUs with GZ-C-35-Y2 (Fig. 5D).
FIG 5.
Venn diagrams comparing the overlap of OTUs between the seeds (inocula) and the five enrichment cultures. Numbers indicate the number of OTUs shared between samples. Panels A and B are comparisons within the same seed, while panels C to E show interseed comparisons.
SIMPER analysis was used to identify the OTUs that drove the differences between the inocula and mature cultures (see Tables S3 to S7 in the supplemental material). In the GZ-C-35-Y2 culture (see Table S3), two OTUs classified to the genus Ruminococcus (760263 and 114730) increased from 0% in the inoculum to average abundances of 35% and 2.5%, respectively, and contributed a total of 19% to the dissimilarity between the GZ-C-35-Y2 and GZ-Seed sample. Other enriched OTUs in GZ-C-35-Y2 included those classified to the family Porphyromonadaceae (1122596; enriched from 0% to 7.8%), genus Treponema (New.ReferenceOTU49; 0% to 7.2%), and genus Methanobacterium (New.ReferenceOTU237 and 101553; 0% to 4.8% and 0.1% to 2.9%, respectively). In the GZ-X-35-Y2 culture (see Table S4), the two OTUs contributing most (12%) to the dissimilarity from the inoculum were from the genus Coprococcus (New.ReferenceOTU19 and 1139736; enriched from 0% to 14% and 10%, respectively). OTUs classified to the genera Ruminococcus (564026, 114730, and New.ReferenceOTU67; enriched from 0% to 8.6%, 7.2%, and 2.6%, respectively), Clostridium (4380971; 0% to 8.6%), Methanobacterium (New.ReferenceOTU237 and 101553; 0% to 3.8% and 0.1% to 3.0%, respectively), and Oscillospira (520720; 0% to 3.6%) cumulatively contributed 19% of the dissimilarity between GZ-X-35-Y2 and the seed. Most of the OTUs diluted to extinction and contributing >1% to the dissimilarity between the GZ seed and enrichments belonged to the methanogenic genera Methanolinea and Methanosaeta (see Tables S3 and S4).
OTUs of the genus Ruminococcus also were major contributors to the shift from the SWH seed sample to SWH-C-35-Y2 (see Table S5 in the supplemental material), increasing from 0% to 21% (OTU 760263), 15% (1089636), and 2.9% (4307829) and contributing 19% of the total dissimilarity. Also highly enriched were OTUs classified to the families Porphyromonadaceae (1122596; increasing from 0% to 16%) and Veillonellaceae (4373061; 0% to 9.0%) and the genera Methanobacterium (249915; 0% to 4.7%) and Treponema (New.ReferenceOTU49; 0% to 3.4%). In the SWH-C-55-Y2 culture (see Table S6), a single OTU (13760) classified to the thermophilic species Thermoanaerobacterium saccharolyticum was enriched from 0% to 42%, contributing 21% of the dissimilarity. Only two other OTUs were enriched to an average abundance of >1% in SWH-C-55-Y2, from the genus Ethanoligenens (548689; enriched from 0% to 27%) and the family Ruminococcaceae (53642; enriched from 0% to 13%). The most enriched OTUs in the SWH-X-35-Y2 culture (see Table S7) were of the genera Methanobacterium (249915 and New.ReferenceOTU237; enriched from 0% to 20% and 8.7%, respectively), Coprococcus (1139736 and New.ReferenceOTU19; 0% to 17% and 11%, respectively), Clostridium (4380971; 0% to 7.2%), and Ruminococcus (114730, 760263, and 4307829; from 0% to 6.0%, 4.3%, and 2.4%, respectively). OTUs diluted to extinction in the SWH cultures and contributing highly to the dissimilarity from the SWH seed were taxonomically similar to those in the GZ cultures (see Tables S5, S6, and S7) and were dominated by an OTU classified to the genus Methanosaeta (149173).
DISCUSSION
Microbial communities that can digest cellulose and/or xylan into soluble sugars are important to the development of sustainable biofuels. In this study, high-throughput sequencing was used to monitor the development of microbial communities enriched from two different inocula on cellulose or xylan for 2 years. This culture-dependent approach was used to test the hypothesis that cellulose- or xylan-degrading bacteria could be enriched from communities that were not initially consuming lignocellulosic components. Furthermore, we were interested in whether the taxonomic profiles of the communities would converge after cultivation under the same conditions, as the inocula originated from different systems (the GZ digester was treating high-strength wastewater and the SWH digester was treating waste sludge). Community convergence indicates that the effect of identical culturing conditions was strong enough to overcome any initial numerical advantage in the inocula.
After 2 years of enrichment, all cultures had a high cellulose- or xylan-digesting capability, with CH4 and acetate as the main accumulated metabolic products (Fig. 1). ANOSIM and adonis tests of the abundance-weighted UniFrac distances consistently supported a much greater role for growth condition (substrate and temperature) than inoculum origin in determining the taxonomic compositions of the enriched communities. In contrast, ANOSIM tests of the unweighted UniFrac distances found that both origin and growth condition had strong and significant effects. While a slightly larger proportion of OTUs were shared between the inocula, some were unique to one (Fig. 5). Thus, the disagreement between the two UniFrac distances on the effect of origin reflects the role of the inoculum as defining the initial pool from which microbes are selected, rather than influencing their relative abundances after multiple dilutions and years of growth. However, ANOSIM R values for enrichment conditions (substrate and temperature) and substrate alone were higher with the weighted than unweighted UniFrac distance, confirming similar enrichment conditions caused both taxonomic composition and abundance to converge.
Most OTUs (82% to 93%) in the enrichment cultures were not observed in the seed samples, suggesting that initially they were present at very low abundances. For example, the most enriched bacterial OTU in the SWH-C-55 culture (13760; species Thermoanaerobacterium saccharolyticum; relative abundance, 42%) was undetected in SWH-Seed. This confirms that under appropriate conditions, species comprising a complete cellulose- or xylan-digesting and methanogenic system can be assembled from very small initial populations. These initially undetectable OTUs likely were sustained in their source digesters by low concentrations of lignocellulosic substrates introduced through the feeding streams. Similarly, many OTUs abundant in the seed inocula were absent following enrichment (Fig. 2; also see Tables S3 to S7 in the supplemental material), and the overall diversity was substantially lower (see Table S1). This is likely due to the selective pressure of a single carbon source combined with the long time period (2 years) available for the communities to stabilize. Previous reports of enrichments with more complex substrates have found richer microbial communities than our enriched communities (e.g., see references 21 and 22), although the same bacterial (Clostridiales) and archaeal (Euryarchaeota) groups are consistently among the most abundant. However, our previous study of short-term incubation (without enrichment) of GZ and SWH sludge, with sole carbon sources including cellulose and xylan, also found a more diverse set of dominant archaeal (most samples dominated by an even mix of Methanomicrobiales and Methanosarcinales) and bacterial (Clostridiales dominant in only some samples) groups than the present study (41). While this could be attributable to the sequencing of transcribed 16S rRNA rather than the 16S rRNA gene and, thus, differences between the active (i.e., rRNA-transcribing) and resident populations, the shorter incubation time of the previous study and high relative proportion of inoculum likely supported a more complex anaerobic digestion consortium. In particular, the higher abundances of the bacterial phyla Proteobacteria and Chloroflexi in the RNA-based study indicated that multiple bacterial groups were contributing to hydrolysis, fermentation, and acetogenesis rather than the Clostridiales-dominated communities observed in this study. Further, in the previous study there was no statistically significant clustering of culture communities by either inoculum source or substrate, although there was a significant tendency for culture communities to resemble their inocula more than other seed samples. This suggests that the convergent pressure of culture conditions on community structure acts only over longer time scales and/or multiple subcultures.
Although both inocula were from systems operated at <35°C, the culture inoculated from SWH could digest cellulose at 55°C, while the one from GZ failed to digest cellulose after a few transfers at 55°C and was discontinued. The thermophilic culture SWH-C-55 had a higher accumulation of acetate and lower CH4 accumulation than any mesophilic culture (Fig. 1) and a very low abundance of Archaea (Fig. 2). Another work (22) similarly reported a decrease in CH4 production of 70 to 80% by volume and increase in acetate accumulation in biogas reactors following a shift from 39°C to 55°C, and it noted that in at least one reactor this was linked to a community shift toward hydrogenotrophic methanogens, particularly the genus Methanoculleus, and possibly bacterial partners responsible for syntrophic acetate oxidation. A similar shift was reported for incubations of rice field soil slurries (42). In our study, however, methane production in the 55°C SWH culture was low (Fig. 1) and the GZ culture failed to yield any methane. While the year 1 SWH 55°C cellulose culture contained 2.8% by relative abundance of the hydrogenotrophic methanogenic genus Methanobacterium (Fig. 2), in year 2 the cultures contained negligible (<0.5%) archaeal populations consistent with the poor methane yield. In our previous short-term incubation experiment with inocula from GZ and SWH (41), the transcribed 16S rRNA profiles of cultures supplemented with cellulose and incubated at 55°C indicated a large population of the hydrogenotrophic methanogen genera Methanolinea (GZ) and Methanoculleus (SWH), along with putative bacterial syntrophic acetate-oxidizing partners, e.g., members of the phyla Chloroflexi (GZ) and Synergistetes (SWH). These thermophilic cultures produced methane at rates comparable to those of mesophilic incubations, although with slightly elevated acetate concentrations. Given that these archaeal methanogen and putative bacterial syntrophic acetate-oxidizing taxa also were present in the GZ and SWH seed samples and the SWH thermophilic cellulose culture in the present study, it is unclear why a complete anaerobic digestion pathway failed to become established in the GZ cultures and yielded little methane in the SWH cultures. Another previous study reported that an enrichment culture inoculated with SWH sludge and incubated with cellulose at 55°C initially was able to convert cellulose to methane, albeit at low efficiency, but following subculturing it failed to consume cellulose or yield methane and instead accumulated H2 (43). This was attributed to the lack of a monosaccharide cosubstrate which can increase cellulose utilization, although cultures supplemented with glucose and xylose similarly failed following subculturing. Notably, the authors found the abundance of the monosaccharide-utilizing bacterial genus Thermoanaerobacterium was strongly negatively correlated with cellulose conversion and associated with acidic conditions; an OTU classified to Thermoanaerobacterium saccharolyticum (13760) was the most abundant in the SWH-C-55 culture, which also attained the lowest pH of all cultures (∼5) at the end of the time course experiment (Fig. 1). Experimentally maintaining a pH of 6.0 and glucose supplementation allowed the continuous thermophilic digestion of cellulose with an SWH inoculum for over a year (44). These results suggest that the success of thermophilic cellulose digestions with or without a cosubstrate is sensitive to both pH and inoculum composition. Additionally, most of the bacteria responsible for syntrophic acetate oxidation in thermophilic conditions are poorly described or unknown (22, 42), although some parameters, e.g., ammonia concentration (45, 46), have been shown to influence syntrophic acetate-oxidizing activity and, therefore, methanogenesis. Further research on the identity and growth requirements of syntrophic acetate-oxidizing bacteria is needed to optimize the operation of anaerobic digestion systems at thermophilic temperatures.
For both substrates and inoculum sources, the dominant Archaea genera shifted from Methanosaeta and (in GZ) Methanolinea in the seed samples to Methanobacterium in the enriched cultures. A high abundance of the acetoclastic Methanosaeta typically indicates lower acetate concentration and/or retention time (47, 48) and previously has been observed in GZ and SWH sludge (26). Given the high acetate concentrations observed toward the end of the time course experiment (Fig. 1) and, thus, presumably toward the end of each subculture during the long-term enrichment, it is unsurprising this genus was suppressed. In the enriched cultures, the near-complete dominance of Methanobacterium OTUs suggests methanogenesis was proceeding almost entirely via the hydrogenotrophic pathway. This differs from our previous short-term incubation of GZ and SWH sludge with the same substrates, which found a mix of hydrogenotrophic Methanobacteriales and acetoclastic/hydrogenotrophic Methanosarcinales (41). The hydrogenotrophic route may be favored over long time scales under the acetate-accumulating, acidic pH conditions that developed in our mesophilic cultures (Fig. 1) (49). It is also noteworthy that, unlike in most natural or artificial anaerobic digestion systems, the cultures in this study were hermetically sealed for long periods (i.e., several weeks between transfers or collection of headspace gas by syringe), during which the partial pressure of H2 was determined entirely by microbial activity, with H2 unable to leave the system by diffusion. Bacterial acetogenesis from fermentation products is thermodynamically favored only under low H2 concentrations (50), and this may have contributed to the development of a hydrogenotrophy-dominated system over time.
The most enriched bacteria in all cultures were associated with the catabolism of the respective amended substrates. Ruminococcus (class Clostridium) was the most enriched genus in the cellulose-digesting cultures (see Tables S3 and S5 in the supplemental material). Many Ruminococcus species have been found in herbivore rumen or gut and are believed to be important in assisting the hosts to digest cellulosic biomass (51), as their ability to hydrolyze and ferment cellulose is well described (52, 53). The second most enriched bacterial OTU (OTU 1122596) in the cellulose cultures was classified to the family Porphyromonadaceae (order Bacteroidales), which currently has no isolated representatives, although it has been found in enrichment cultures amended with cow feces and complex polysaccharides (54). As other Bacteroidales taxa are able to digest cellulose or hemicellulose (55), this OTU likely is involved in cellulose hydrolysis.
The genus Treponema also was enriched in the cellulose-amended cultures. Some Treponema species have been shown to interact with cellulolytic bacteria and increase the degradation rate of cellulose (56), and Treponema spp. also are involved in degradation of soluble fibers (57). Recent studies also have identified abundant Treponema in a termite gut metagenome (58) and enriched in the gut microbiomes of humans with highly fibrous hunter-gatherer diets (59). Bacteria from this genus likely are able to metabolize exopolysaccharides produced by the primary cellulolytic bacteria, functioning as fermenters (15). The bacterial family Veillonellaceae (phylum Firmicutes) also was highly enriched. Although the role of this family in cellulose digestion is poorly understood, they have been identified in various anaerobic cellulose or hemicellulose digesting systems, for example, in ruminants (60), termite gut (61), and other anaerobic cellulose enrichment cultures (54).
Coprococcus was the most enriched genus in the xylan-amended cultures. Coprococcus spp. are the major bacterial taxa in the rumen microbiota of some ruminants (62, 63). Although bacteria from this family might be involved in the digestion of some cellulosic materials (64), they were not enriched in cellulose cultures in this study (<1%), suggesting that they cannot degrade crystalline cellulose. As with the cellulose-amended cultures, Ruminococcus spp. were enriched in the xylan cultures, although the most enriched OTUs differed. OTUs 760263 and 1089636 were abundant in the cellulose cultures, while OTUs 114730 and 564026 were dominant in xylan cultures (see Tables S3 to S5 and S7 in the supplemental material). This suggests that within the same genus, different species and/or strains possess different substrate-digesting abilities or are adapted to work with different syntrophic partners selected for by the amended substrate.
The most abundant bacterial OTU in the 55°C cellulose culture was the thermophilic anaerobe Thermoanaerobacterium saccharolyticum (13760) (see Table S6 in the supplemental material). The growth temperature of Thermoanaerobacterium species is typically 45 to 72°C (65), and many Thermoanaerobacterium saccharolyticum strains are able to digest cellulose or hemicellulose (66, 67). The other abundant OTU specific to the 55°C cellulose culture was of the genus Ethanoligenens (548689) from the family Ruminococcaceae. Two strains from this genus isolated from an anaerobic activated sludge of molasses wastewater were found to hydrolyze various mono-, di-, and oligosaccharides (68). Members of this genus also have been reported to produce hydrogen (69), but cellulolytic ability has yet to be demonstrated.
As hydrogenotrophic Methanobacterium organisms were dominant in the archaeal populations of all mesophilic cultures (Fig. 2), it is likely that the enriched communities included bacterial syntrophic acetate-oxidizing partners. The identities of syntrophic acetate-oxidizing bacteria remain mostly unknown, although candidates include species of the genus Clostridium (70), which was enriched in both xylan cultures (see Tables S4 and S7 in the supplemental material). Given the important role syntrophic acetate-oxidizing bacteria play in anaerobic digestion (71), a better understanding of these species is one of the most pressing needs in anaerobic digestion research.
This study aimed to test the hypothesis that a complete, methanogenic cellulose- or xylan-digesting community could be enriched from anaerobic digester sludge with no lignocellulose specificity, i.e., wastewater treatment systems. While this was confirmed at mesophilic temperatures with both substrate degradation and methane production, only one thermophilic culture was able to degrade cellulose, and it produced substantially less methane than comparable mesophilic cultures. An improved understanding of the bacteria responsible for syntrophic oxidation of acetate is likely to shed light on this. This study confirmed that given a long culturing time, a minimal (i.e., low-diversity) but functional and complete cellulose- or xylan-digesting biogas system could be assembled. It also showed that the inoculum composition had little effect relative to the growth conditions, with the communities converging toward a consortium of substrate-specific bacterial hydrolyzers and fermenters and hydrogenotrophic methanogens under all mesophilic conditions. Our results demonstrate the wealth of functional potential available in complex substrate anaerobic digestion sludge and confirm that microbially mediated cellulose and xylan digestion is a practical means for biogas production.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Research Grants Council of Hong Kong through projects 116111 and 11206514.
We thank the plant operators for their assistance with sampling and Xiaoying Lu during the initial phase of the project.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03360-15.
REFERENCES
- 1.Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. 2006. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci U S A 103:11206–11210. doi: 10.1073/pnas.0604600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamelinck CN, Hooijdonk GV, Faaij APC. 2005. Ethanol from lignocellulosic biomass: techno-economic performance in short-, middle- and long-term. Biomass Bioenergy 28:384–410. doi: 10.1016/j.biombioe.2004.09.002. [DOI] [Google Scholar]
- 3.Metzger JO, Hüttermann A. 2009. Sustainable global energy supply based on lignocellulosic biomass from afforestation of degraded areas. Naturwissenschaften 96:279–288. doi: 10.1007/s00114-008-0479-4. [DOI] [PubMed] [Google Scholar]
- 4.Saha BC. 2004. Lignocellulose biodegradation and applications in biotechnology. American Chemical Society, Oxford University Press, New York, NY. [Google Scholar]
- 5.Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 6.Sendich EN, Laser M, Kim S, Alizadeh H, Laureano-Perez L, Dale B, Lynd L. 2008. Recent process improvements for the ammonia fiber expansion (AFEX) process and resulting reductions in minimum ethanol selling price. Bioresour Technol 99:8429–8435. doi: 10.1016/j.biortech.2008.02.059. [DOI] [PubMed] [Google Scholar]
- 7.Wyman CE. 2007. What is (and is not) vital to advancing cellulosic ethanol. Trends Biotechnol 25:153–157. doi: 10.1016/j.tibtech.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Wilson DB. 2008. Three microbial strategies for plant cell wall degradation. Ann N Y Acad Sci 1125:289–297. doi: 10.1196/annals.1419.026. [DOI] [PubMed] [Google Scholar]
- 9.Li LL, McCorkle SR, Monchy S, Taghavi S, van der Lelie D. 2009. Bioprospecting metagenomes: glycosyl hydrolases for converting biomass. Biotechnol Biofuels 2:10. doi: 10.1186/1754-6834-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 11.Toyoda A, Iio W, Mitsumori M, Minato H. 2009. Isolation and identification of cellulose-binding proteins from sheep rumen contents. Appl Environ Microbiol 75:1667–1673. doi: 10.1128/AEM.01838-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pope PB, Mackenzie AK, Gregor I, Smith W, Sundset MA, McHardy AC, Morrison M, Eijsink VGH. 2012. Metagenomics of the svalbard reindeer rumen microbiome reveals abundance of polysaccharide utilization loci. PLoS One 7:e38571. doi: 10.1371/journal.pone.0038571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brune A. 2014. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol 12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 14.Ni J, Tokuda G. 2013. Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol Adv 31:838–850. doi: 10.1016/j.biotechadv.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, McHardy AC, Djordjevic G, Aboushadi N, Sorek R, Tringe SG, Podar M, Martin HG, Kunin V, Dalevi D, Madejska J, Kirton E, Platt D, Szeto E, Salamov A, Barry K, Mikhailova N, Kyrpides NC, Matson EG, Ottesen EA, Zhang X, Hernández M, Murillo C, Acosta LG, Rigoutsos I, Tamayo G, Green BD, Chang C, Rubin EM, Mathur EJ, Robertson DE, Hugenholtz P, Leadbetter JR. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 16.Valášková V, Šnajdr J, Bittner B, Cajthaml T, Merhautová V, Hofrichter M, Baldrian P. 2007. Production of lignocellulose-degrading enzymes and degradation of leaf litter by saprotrophic basidiomycetes isolated from a Quercus petraea forest. Soil Biol Biochem 39:2651–2660. doi: 10.1016/j.soilbio.2007.05.023. [DOI] [Google Scholar]
- 17.Baldrian P, Voříšková J, Dobiášová P, Merhautová V, Lisá L, Valášková V. 2010. Production of extracellular enzymes and degradation of biopolymers by saprotrophic microfungi from the upper layers of forest soil. Plant Soil 338:111–125. doi: 10.1007/s11104-010-0324-3. [DOI] [Google Scholar]
- 18.Benner R, Hodson RE. 1985. Microbial degradation of the leachable and lignocellulosic components of leaves and wood from Rhizophora mangle in a tropical mangrove swamp. Mar Ecol Prog Ser 23:221–230. doi: 10.3354/meps023221. [DOI] [Google Scholar]
- 19.Allgaier M, Reddy A, Park JI, Ivanova N, D'haeseleer P, Lowry S, Sapra R, Hazen TC, Simmons BA, Vander Gheynst JS, Hugenholtz P. 2010. Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. PLoS One 5:e8812. doi: 10.1371/journal.pone.0008812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sizova MV, Izquierdo JA, Panikov NS, Lynd LR. 2011. Cellulose- and xylan-degrading thermophilic anaerobic bacteria from biocompost. Appl Environ Microbiol 77:2282–2291. doi: 10.1128/AEM.01219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Lelie D, Taghavi S, McCorkle SM, Li LL, Malfatti SA, Monteleone D, Donohoe BS, Ding SY, Adney WS, Himmel ME, Tringe SG. 2012. The metagenome of an anaerobic microbial community decomposing poplar wood chips. PLoS One 7:e36740. doi: 10.1371/journal.pone.0036740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziganshin AM, Liebetrau J, Pröter J, Kleinsteuber S. 2013. Microbial community structure and dynamics during anaerobic digestion of various agricultural waste materials. Appl Microbiol Biotechnol 97:5161–5174. doi: 10.1007/s00253-013-4867-0. [DOI] [PubMed] [Google Scholar]
- 23.Nelson MC, Morrison M, Yu Z. 2011. A meta-analysis of the microbial diversity observed in anaerobic digesters. Bioresour Technol 102:3730–3739. doi: 10.1016/j.biortech.2010.11.119. [DOI] [PubMed] [Google Scholar]
- 24.Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, Sørensen SJ, Karlsson A. 2013. 454 Pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol 85:612–626. doi: 10.1111/1574-6941.12148. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Yu K, Xia Y, Lau FTK, Tang DTW, Fung WC, Fang HHP, Zhang T. 2014. Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl Microbiol Biotechnol 98:5709–5718. doi: 10.1007/s00253-014-5648-0. [DOI] [PubMed] [Google Scholar]
- 26.Wilkins D, Lu XY, Shen Z, Chen J, Lee PKH. 2015. Pyrosequencing of mcrA and archaeal 16S rRNA genes reveals diversity and substrate preferences of methanogen communities in anaerobic digesters. Appl Environ Microbiol 81:604–613. doi: 10.1128/AEM.02566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu X, Rao S, Shen Z, Lee PKH. 2013. Substrate induced emergence of different active bacterial and archaeal assemblages during biomethane production. Bioresour Technol 148:517–524. doi: 10.1016/j.biortech.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 28.He J, Holmes VF, Lee PKH, Alvarez-Cohen L. 2007. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl Environ Microbiol 73:2847–2853. doi: 10.1128/AEM.02574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wongwilaiwalin S, Rattanachomsri U, Laothanachareon T, Eurwilaichitr L, Igarashi Y, Champreda V. 2010. Analysis of a thermophilic lignocellulose degrading microbial consortium and multi-species lignocellulolytic enzyme system. Enzyme Microb Technol 47:283–290. doi: 10.1016/j.enzmictec.2010.07.013. [DOI] [Google Scholar]
- 30.Yang SJ, Kataeva I, Hamilton-Brehm SD, Engle NL, Tschaplinski TJ, Doeppke C, Davis M, Westpheling J, Adams MWW. 2009. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl Environ Microbiol 75:4762–4769. doi: 10.1128/AEM.00236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu H, Lee PKH. 2015. Effects of cellulose concentrations on the syntrophic interactions between Clostridium cellulovorans 743B and Rhodopseudomonas palustris CGA009 in coculture fermentation for biohydrogen production. Int J Hydrogen Energy 40:11800–11808. doi: 10.1016/j.ijhydene.2015.05.135. [DOI] [Google Scholar]
- 32.Dubois M, Gilles KA, Ton JKH, Rebers PA, Smith F. 1956. Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- 33.Leung MHY, Wilkins D, Li EKT, Kong FKF, Lee PKH. 2014. Indoor-air microbiome in an urban subway network: diversity and dynamics. Appl Environ Microbiol 80:6760–6770. doi: 10.1128/AEM.02244-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leung MHY, Wilkins D, Lee PKH. 2015. Insights into the pan-microbiome: skin microbial communities of Chinese individuals differ from other racial groups. Sci Rep 5:11845. doi: 10.1038/srep11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wickham H. 2009. ggplot2: elegant graphics for data analysis, 1st ed Springer-Verlag, New York, NY. [Google Scholar]
- 39.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. 2011. UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H. 2014. VennDiagram: generate high-resolution Venn and Euler plots. R package version 1.6.7. [Google Scholar]
- 41.Wilkins D, Rao S, Lu X, Lee PKH. 2015. Effects of sludge inoculum and organic feedstock on active microbial communities and methane yield during anaerobic digestion. Front Microbiol 6:1114. doi: 10.3389/fmicb.2015.01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noll M, Klose M, Conrad R. 2010. Effect of temperature change on the composition of the bacterial and archaeal community potentially involved in the turnover of acetate and propionate in methanogenic rice field soil. FEMS Microbiol Ecol 73:215–225. doi: 10.1111/j.1574-6941.2010.00883.x. [DOI] [PubMed] [Google Scholar]
- 43.Xia Y, Cai L, Zhang T, Fang HHP. 2012. Effects of substrate loading and co-substrates on thermophilic anaerobic conversion of microcrystalline cellulose and microbial communities revealed using high-throughput sequencing. Int J Hydrogen Energy 37:13652–13659. doi: 10.1016/j.ijhydene.2012.02.079. [DOI] [Google Scholar]
- 44.Xia Y, Wang Y, Fang HHP, Jin T, Zhong H, Zhang T. 2014. Thermophilic microbial cellulose decomposition and methanogenesis pathways recharacterized by metatranscriptomic and metagenomic analysis. Sci Rep 4:6708. doi: 10.1038/srep06708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun L, Müller B, Westerholm M, Schnürer A. 2014. Syntrophic acetate oxidation in industrial CSTR biogas digesters. J Biotechnol 171:39–44. doi: 10.1016/j.jbiotec.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 46.Schnürer A, Nordberg Å. 2008. Ammonia, a selective agent for methane production by syntrophic acetate oxidation at mesophilic temperature. Water Sci Technol 57:735–740. doi: 10.2166/wst.2008.097. [DOI] [PubMed] [Google Scholar]
- 47.Ma J, Zhao B, Frear C, Zhao Q, Yu L, Li X, Chen S. 2013. Methanosarcina domination in anaerobic sequencing batch reactor at short hydraulic retention time. Bioresour Technol 137:41–50. doi: 10.1016/j.biortech.2013.03.101. [DOI] [PubMed] [Google Scholar]
- 48.Conklin A, Stensel HD, Ferguson J. 2006. Growth kinetics and competition between Methanosarcina and Methanosaeta in mesophilic anaerobic digestion. Water Environ Res 78:486–496. doi: 10.2175/106143006X95393. [DOI] [PubMed] [Google Scholar]
- 49.Kotsyurbenko OR, Friedrich MW, Simankova MV, Nozhevnikova AN, Golyshin PN, Timmis KN, Conrad R. 2007. Shift from acetoclastic to H2-dependent methanogenesis in a West Siberian peat bog at low pH values and isolation of an acidophilic Methanobacterium strain. Appl Environ Microbiol 73:2344–2348. doi: 10.1128/AEM.02413-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schink B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61:262–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dassa B, Borovok I, Ruimy-Israeli V, Lamed R, Flint HJ, Duncan SH, Henrissat B, Coutinho P, Morrison M, Mosoni P, Yeoman CJ, White BA, Bayer EA. 2014. Rumen cellulosomics: divergent fiber-degrading strategies revealed by comparative genome-wide analysis of six ruminococcal strains. PLoS One 9:e99221. doi: 10.1371/journal.pone.0099221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moon YH, Iakiviak M, Bauer S, Mackie RI, Cann IKO. 2011. Biochemical analyses of multiple endoxylanases from the rumen bacterium Ruminococcus albus 8 and their synergistic activities with accessory hemicellulose-degrading enzymes. Appl Environ Microbiol 77:5157–5169. doi: 10.1128/AEM.00353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brulc JM, Yeoman CJ, Wilson MK, Miller ME, Jeraldo P, Jindou S, Goldenfeld N, Flint HJ, Lamed R, Borovok I, Vodovnik M, Nelson KE, Bayer EA, White BA. 2011. Cellulosomics, a gene-centric approach to investigating the intraspecific diversity and adaptation of Ruminococcus flavefaciens within the rumen. PLoS One 6:e25329. doi: 10.1371/journal.pone.0025329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ziemer CJ. 2014. Newly cultured bacteria with broad diversity isolated from eight-week continuous culture enrichments of cow feces on complex polysaccharides. Appl Environ Microbiol 80:574–585. doi: 10.1128/AEM.03016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanokratana P, Mhuantong W, Laothanachareon T, Tangphatsornruang S, Eurwilaichitr L, Pootanakit K, Champreda V. 2013. Phylogenetic analysis and metabolic potential of microbial communities in an industrial bagasse collection site. Microb Ecol 66:322–334. doi: 10.1007/s00248-013-0209-0. [DOI] [PubMed] [Google Scholar]
- 56.Kudo H, Cheng KJ, Costerton JW. 1987. Interactions between Treponema bryantii and cellulolytic bacteria in the in vitro degradation of straw cellulose. Can J Microbiol 33:244–248. doi: 10.1139/m87-041. [DOI] [PubMed] [Google Scholar]
- 57.Bekele AZ, Koike S, Kobayashi Y. 2011. Phylogenetic diversity and dietary association of rumen Treponema revealed using group-specific 16S rRNA gene-based analysis. FEMS Microbiol Lett 316:51–60. doi: 10.1111/j.1574-6968.2010.02191.x. [DOI] [PubMed] [Google Scholar]
- 58.Do TH, Nguyen TT, Nguyen TN, Le QG, Nguyen C, Kimura K, Truong NH. 2014. Mining biomass-degrading genes through Illumina-based de novo sequencing and metagenomic analysis of free-living bacteria in the gut of the lower termite Coptotermes gestroi harvested in Vietnam. J Biosci Bioeng 118:665–671. doi: 10.1016/j.jbiosc.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 59.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, Fiori J, Gotti R, De Bellis G, Luiselli D, Brigidi P, Mabulla A, Marlowe F, Henry AG, Crittenden AN. 2014. Gut microbiome of the Hadza hunter-gatherers. Nat Commun 5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu S, Baldwin RL, Li W, Li C, Connor EE, Li RW. 2012. The bacterial community composition of the bovine rumen detected using pyrosequencing of 16S rRNA genes. Metagenomics 1:1–11. doi: 10.4303/mg/235571. [DOI] [Google Scholar]
- 61.Schauer C, Thompson CL, Brune A. 2012. The bacterial community in the gut of the cockroach Shelfordella lateralis reflects the close evolutionary relatedness of cockroaches and termites. Appl Environ Microbiol 78:2758–2767. doi: 10.1128/AEM.07788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kong Y, Xia Y, Seviour R, He M, Mcallister T, Forster R. 2012. In situ identification of carboxymethyl cellulose-digesting bacteria in the rumen of cattle fed alfalfa or triticale. FEMS Microbiol Ecol 80:159–167. doi: 10.1111/j.1574-6941.2011.01295.x. [DOI] [PubMed] [Google Scholar]
- 63.Omoniyi LA, Jewell KA, Isah OA, Neumann AP, Onwuka CFI, Onagbesan OM, Suen G. 2014. An analysis of the ruminal bacterial microbiota in West African Dwarf sheep fed grass- and tree-based diets. J Appl Microbiol 116:1094–1105. doi: 10.1111/jam.12450. [DOI] [PubMed] [Google Scholar]
- 64.Szczepańska AM. 2011. Functional metagenomic analysis of carbohydrate degrading enzymes from the human gut microbiota. Ph.D. thesis University of East Anglia, Norwich, United Kingdom. [Google Scholar]
- 65.Cann IKO, Stroot PG, Mackie KR, White BA, Mackie RI. 2001. Characterization of two novel saccharolytic, anaerobic thermophiles, Thermoanaerobacterium polysaccharolyticum sp. nov. and Thermoanaerobacterium zeae sp. nov., and emendation of the genus Thermoanaerobacterium. Int J Syst Evol Microbiol 51:293–302. doi: 10.1099/00207713-51-2-293. [DOI] [PubMed] [Google Scholar]
- 66.Joe Shaw A, Jenney FE, Adams MWW, Lynd LR. 2008. End-product pathways in the xylose fermenting bacterium, Thermoanaerobacterium saccharolyticum. Enzyme Microb Technol 42:453–458. doi: 10.1016/j.enzmictec.2008.01.005. [DOI] [Google Scholar]
- 67.Hung KS, Liu SM, Tzou WS, Lin FP, Pan CL, Fang TY, Sun KH, Tang SJ. 2011. Characterization of a novel GH10 thermostable, halophilic xylanase from the marine bacterium Thermoanaerobacterium saccharolyticum NTOU1. Process Biochem 46:1257–1263. doi: 10.1016/j.procbio.2011.02.009. [DOI] [Google Scholar]
- 68.Xing D, Ren N, Li Q, Lin M, Wang A, Zhao L. 2006. Ethanoligenens harbinense gen. nov., sp. nov., isolated from molasses wastewater. Int J Syst Evol Microbiol 56:755–760. doi: 10.1099/ijs.0.63926-0. [DOI] [PubMed] [Google Scholar]
- 69.Ren N, Wang A, Cao G, Xu J, Gao L. 2009. Bioconversion of lignocellulosic biomass to hydrogen: potential and challenges. Biotechnol Adv 27:1051–1060. doi: 10.1016/j.biotechadv.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 70.Müller B, Sun L, Schnürer A. 2013. First insights into the syntrophic acetate-oxidizing bacteria–a genetic study. Microbiologyopen 2:35–53. doi: 10.1002/mbo3.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stams AJM, Plugge CM. 2009. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol 7:568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.