Abstract
Geochemical data indicate that protons released during pyrite (FeS2) oxidation are important drivers of mineral weathering in oxic and anoxic zones of many aquatic environments, including those beneath glaciers. Oxidation of FeS2 under oxic, circumneutral conditions proceeds through the metastable intermediate thiosulfate (S2O32−), which represents an electron donor capable of supporting microbial metabolism. Subglacial meltwaters sampled from Robertson Glacier (RG), Canada, over a seasonal melt cycle revealed concentrations of S2O32− that were typically below the limit of detection, despite the presence of available pyrite and concentrations of the FeS2 oxidation product sulfate (SO42−) several orders of magnitude higher than those of S2O32−. Here we report on the physiological and genomic characterization of the chemolithoautotrophic facultative anaerobe Thiobacillus sp. strain RG5 isolated from the subglacial environment at RG. The RG5 genome encodes genes involved with pathways for the complete oxidation of S2O32−, CO2 fixation, and aerobic and anaerobic respiration with nitrite or nitrate. Growth experiments indicated that the energy required to synthesize a cell under oxygen- or nitrate-reducing conditions with S2O32− as the electron donor was lower at 5.1°C than 14.4°C, indicating that this organism is cold adapted. RG sediment-associated transcripts of soxB, which encodes a component of the S2O32−-oxidizing complex, were closely affiliated with soxB from RG5. Collectively, these results suggest an active sulfur cycle in the subglacial environment at RG mediated in part by populations closely affiliated with RG5. The consumption of S2O32− by RG5-like populations may accelerate abiotic FeS2 oxidation, thereby enhancing mineral weathering in the subglacial environment.
INTRODUCTION
The comminution of bedrock in subglacial systems promotes weathering processes by exposing fresh minerals with a high surface area (1–5). Subglacial water chemical profiles (e.g., see references 5 and 6), field- and laboratory-based microcosm experiments (e.g., see references 7–9), and molecular analyses (e.g., see references 6, 10, and 11) indicate the presence of an active and diverse subglacial microbiome founded on chemical energy that functions to enhance rates of mineral weathering (8). Given that ice covers approximately 10% of the present-day continental landmass, the subglacial environment is a widespread habitat for microbial life and for mineral weathering.
Aqueous geochemical data collected from the meltwaters of numerous glaciers suggest that pyrite (FeS2) oxidation and the concomitant production of hydrogen ions are key drivers of subglacial bedrock weathering (5, 6, 9, 12, 13). It has also been inferred that FeS2 weathering in the subglacial environment may be microbially mediated (3, 6, 9). This inference is supported by DNA-based molecular data that show the presence in subglacial systems of a number of taxa closely related to organisms capable of Fe and S oxidation (6, 7, 9, 11, 14, 15). Moreover, Mitchell et al. (7) showed that microbial communities colonizing FeS2 incubated in situ in a subglacial meltwater stream at Robertson Glacier (RG), Canada, were phylogenetically more similar at the level of 16S rRNA genes to communities associated with native subglacial sediments and suspended sediments than communities colonizing other iron-bearing minerals (i.e., magnetite, hematite, and olivine) and carbonate minerals (i.e., calcite) (7). These data suggest a relationship between microbial community structure and bedrock mineralogy and imply that bedrock minerals serve as a source of energy for subglacial microbial communities. Further evidence for the role of FeS2 in supporting subglacial microbial communities comes from the recovery of 16S rRNA gene transcripts from RG sediments that exhibit a close affiliation with known Fe- and S-oxidizing taxa (11). However, FeS2 oxidation pathways in the subglacial system and the role of microbes in these geochemical transformations, especially those FeS2 oxidation processes that occur under hypoxic or anoxic conditions thought to characterize significant sectors of subglacial drainage networks, are poorly understood (16).
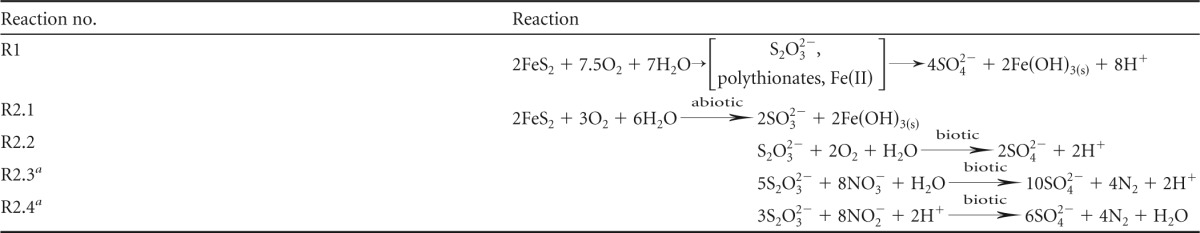
In acidic (pH < 4) environments, abiotic FeS2 oxidation is known to occur through both oxic and anoxic processes (17–20). Anoxic FeS2 oxidation in these systems is achieved through surface oxidation with aqueous Fe3+ (e.g., see references 18 and 20). Geochemical and isotopic measurements of subglacial waters suggest that FeS2 oxidation under anoxic conditions occurs in several subglacial environments (5, 21). However, abiotic, anoxic FeS2 oxidation via aqueous Fe3+ is not possible at the circumneutral to alkaline pHs that characterize many subglacial outflow waters (22, 23) due to the rapid precipitation of ferric iron as iron hydroxide [Fe(OH)3], which does not promote FeS2 oxidation (24). Consequently, FeS2 oxidation in systems with circumneutral to alkaline pHs is thought to be driven primarily by oxidation with O2. Unlike in acidic systems, however, the reduced sulfur intermediates released during oxic FeS2 oxidation (Table 1, reaction R1), such as thiosulfate (S2O32−), are metastable in O2-rich waters at circumneutral to alkaline pHs (20). The metastable S2O32− produced in oxic, circumneutral subglacial waters may be oxidized by aerobic microorganisms, resulting in the production of SO42− (Table 1, reaction R2.2), or may be transported to anoxic zones where anaerobic microbial processes may drive its oxidation to SO42− (25). Previous work has demonstrated the importance of nitrate as an oxidant capable of supporting microbial metabolism in a variety of subglacial environments (26–28), including RG (29).
TABLE 1.
Proposed mechanisms of FeS2 and S2O32− oxidation in oxic and anoxic environments at circumneutral to alkaline pH observed in meltwaters emanating from RG
a Potential reduced, nitrogenous intermediates are not shown.
Here we describe the isolation of Thiobacillus sp. strain RG5 from RG subglacial sediments using S2O32− as an electron donor and NO3− as an electron acceptor. Strain RG5 is a chemoautotrophic facultative anaerobe. Genomic data indicate that it is closely affiliated with Thiobacillus denitrificans. For the remainder of this communication, this strain is referred to as Thiobacillus sp. RG5 or as strain RG5. We demonstrate that Thiobacillus sp. RG5 is (i) capable of catalyzing the oxidation of S2O32− with O2, NO2−, or NO3− as the electron acceptor at circumneutral pH and (ii) adapted to the low temperatures that characterize subglacial habitats, on the basis of physiological and energetic data. Consistent with these findings, transcripts of soxB, which encodes a component of the S2O32−-oxidizing complex, were recovered from RG subglacial sediments. They exhibited a close affiliation with the soxB in the genome of Thiobacillus sp. RG5. Considering the widespread distribution of thiobacilli in contemporary subglacial environments (e.g., see references 6, 11, and 30 to 32), these results suggest a key role for autotrophic organisms like Thiobacillus sp. RG5 in driving oxic and anoxic subglacial mineral weathering on a global scale.
MATERIALS AND METHODS
Site description.
Robertson Glacier (RG) is located in Peter Lougheed Provincial Park, Alberta, Canada. The RG catchment bedrock is sedimentary, part of a Late Devonian marine carbonate sequence with minor clastic input, and primarily composed of impure carbonates, including limestones and dolostones, with silicic interbeds of siltstone, shale, and sandstone (33, 34). Trace amounts of FeS2 (1 to 2 wt%) form nodules within glacial till and bedrock and provide the only substantive source of reduced sulfur (S) to the subglacial environment (33). Sediments contain mineral abundances similar to those in bedrock and small amounts (<1 to 2 wt%) of FeS2 (35). Gypsum and other sulfate-bearing minerals are not observed in RG bedrock (33). Bassanite (2CaSO4·H2O) has been identified to be a minor component in a single RG till sample; however, its absence from the bedrock suggests that it may be a secondary precipitate (33). One, predominant subglacial outflow stream drains from the western edge of the glacier terminus. The subglacial outflow waters, whose temperatures are <1°C, form an ice cave-like incision beneath the glacier, exposing basal sediments. The SO42− concentrations in supraglacial waters that have only ice melt as their source are 1 to 2 μM (9). These waters that are input into the subglacial system have sulfate concentrations that are 1 to 2 orders of magnitude lower than those measured in the subglacial outflow stream, where they are 23 to 571 μM, indicating that there is a sulfate source in the subglacial environment (9).
Sample collection.
Fine-grained basal sediments were collected, using aseptic techniques, on 10 October 2010 from within an ice cave that formed at the terminus of RG where the subglacial melt stream discharges. The samples were collected for RNA-based analysis and enrichment culturing as previously described (11). Briefly, triplicate samples of sediment submerged beneath the melt stream were collected from a 0.5-m2 area of the sediment surface within the ice cave, and these were pooled in order to minimize the influence of spatial heterogeneity on culture- and molecular-based analyses. Sediments were collected from this 0.5-m2 area using a flame-sterilized spatula or spoon. Sediment aliquots (∼1 g) for RNA-based analysis were collected in sterile 2-ml tubes containing 0.5 ml RNAlater (Qiagen, Valencia, CA, USA) and flash-frozen in a dry ice-ethanol slurry. Samples were stored on dry ice during transport to the field station and back to Montana State University, where they were stored at −80°C until further processing. Sediments for enrichment and culture isolations (described below) were collected, using a flame-sterilized spoon, from the same location that the sediment samples for RNA analyses were collected. These sediments for enrichment and culture isolations were placed in a sterile 500-ml screw-cap container and transported and maintained at <4°C until used for enrichment and isolation. Water samples were collected from the subglacial stream outlet every 6 h over diurnal cycles on 17 and 18 July, 5 and 6 August, and 26 and 27 September 2014 and filtered using a 0.4-μm-pore-size Nuclepore membrane for analysis of major anion and cation concentrations as previously described (30).
Enrichment and isolation.
A basal medium containing 5.1 mM NaCl, 10 mM K2HPO4, 5 mM NH4Cl, 0.25 mM MgCl2·6H2O, and 6 mM Na2S2O3 was prepared as previously described (36). Thirty milliliters of medium was dispensed into 70-ml serum bottles, their contents were degassed with N2 passed over H2-reduced and heated (210°C) copper shavings, and the bottles were sealed with butyl rubber stoppers and autoclaved at 121°C for 20 min. All media were subsequently amended with anoxic, filter-sterilized solutions to achieve final concentrations of 20 mM NaHCO3, 0.1% (vol/vol) SL 10 trace metal solution (37), and 0.1% (vol/vol) Wolfe's vitamin solution. The anoxic medium was then amended with KNO3 as the sole electron acceptor to a final concentration of 5 mM.
The inoculum was prepared by mixing 5 g of fine-grained basal sediment with 10 ml of sterile, anoxic medium to create a slurry. Three milliliters of slurry was added to 70-ml serum bottles containing 30 ml of prepared medium. The enrichment was immediately subjected to a series of 10-fold dilutions (6 in total). Following a month of incubation at 5°C, a 2-ml aliquot of each dilution culture was removed aseptically, filter sterilized, and assayed for sulfate (SO42−), nitrate (NO3−), and nitrite (NO2−) using ion chromatography (described below). The analyte concentrations in the inoculated medium were compared to those in uninoculated medium to quantify S2O32− oxidation and NO3− reduction activities. The most dilute culture from the first dilution-to-extinction series exhibiting an increase in the SO42− concentration (as a proxy for S2O32− oxidation) and a decrease in the NO3− concentration relative to the concentrations for the uninoculated controls was used as the inoculum for a second additional dilution-to-extinction sequence following the same procedure described above. The same process was repeated for a third and final dilution-to-extinction series. The most dilute culture of the third sequence was used for further physiological experimentation as well as DNA extraction, quantification, and genome sequencing, as described below.
DNA extraction, quantification, and sequencing.
Two milliliters of the most dilute culture from the third dilution-to-extinction series was concentrated via centrifugation (14,000 × g, 15 min, 5°C). Genomic DNA was extracted, purified, and quantified as previously described (11). Total genomic DNA was sequenced at the Genomics Core Facility at the University of Wisconsin—Madison using a paired-end Illumina MiSeq platform (Illumina, San Diego, CA). DNA fragments were prepared according to the manufacturer's protocol. The quality of the reads was checked with the FastQC program (www.bioinformatics.babraham.ac.uk/projects/fastqc). The reads were quality trimmed from both ends using Trimmomatic (38). Reads containing more than 3 nucleotides were also removed, and reads with an average quality score of less than the Q20 standard or a sequence length of less than 50 bp were removed. The trimmed paired-end reads were assembled using the Velvet algorithm (39) as previously described (40), with a kmer length of 49 base pairs being specified. Contigs were annotated using the RAST server (41).
RNA extraction, quantification, and sequencing.
RNA extraction and initial purification were performed using a FastRNA Pro soil-direct kit (MP Biomedical, Solon, OH). RNA was extracted in triplicate from approximately 400 mg (wet weight) of sediment. After initial purification, RNA was subjected to DNase I digestion (Roche, Indianapolis, IN) for 1 h at room temperature (∼22°C). Following digestion, RNA was further purified using a High Pure RNA isolation kit (Roche, Indianapolis, IN) and was stored at −80°C in a solution of 100% ethanol and 0.3 M sodium acetate until it was further processed. The subglacial sediment dry mass was determined by drying (80°C, 24 h) and weighing the residue after RNA extraction.
The concentration of RNA was determined using a Qubit RNA assay kit (Molecular Probes, Eugene, OR) and a Qubit (version 2.0) fluorometer (Invitrogen, Carlsbad, CA). RNA extracts were screened for the presence of contaminating genomic DNA by performing a PCR using ∼1 ng of RNA as the template and archaeal and bacterial 16S rRNA gene-specific primers, as described previously (11). Equal volumes of each DNA-free RNA extract were pooled and subjected to cDNA synthesis. cDNA was synthesized from 20 ng of purified RNA using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) and the following reaction cycling conditions: 5 min at 25°C, 30 min at 42°C, and 5 min at 85°C. Following the synthesis of cDNA, samples were purified by ethanol precipitation and resuspended in nuclease-free water.
The amplification of soxB transcript fragments from cDNA was performed using primers soxB693F (5′-ATCGGNCARGCNTTYCCNTA-3′) and soxB1446R (5′-CATGTCNCCNCCRTGYTG-3′) (42). Approximately 10 ng of purified cDNA was subjected to PCR in triplicate using the reaction and cycling conditions described previously (42), with the following exceptions: an initial 10 cycles of PCR was conducted at an annealing temperature of 55°C followed by 25 cycles of PCR at an annealing temperature of 47°C. Equal volumes of each replicate reaction mixture were combined, purified, cloned, and sequenced as described previously (43).
Phylogenetic analysis.
soxB transcript sequences obtained from RG, the soxB gene sequence from strain RG5, as well as soxB sequences from related strains were compiled and then translated using the ExPASy server (http://web.expasy.org/translate/). SoxB sequences were aligned with the program ClustalW (version 1.8.1) (44), specifying default alignment parameters. A maximum likelihood phylogenetic reconstruction was inferred using the PhyML (version 3.0) program (45), with the Le and Gascuel (LG) substitution matrix and a four-category gamma substitution model being specified. The phylogeny was projected from 100 bootstrap replicates using the program FigTree (version 1.4.2; http://tree.bio.ed.ac.uk/software/figtree/). SoxB sequences from Thermus thermophilus strains HB8 and HB27 were used to root the tree.
Characterization of Thiobacillus sp. RG5 growth.
The growth of strain RG5 was characterized under oxic (O2-dependent) and anoxic (NO3−- and NO2−-dependent) conditions at 5.1 and 14.4°C. Sixty-milliliter cultures were prepared in 120-ml serum bottles as described above (see “Enrichment and isolation” above). Medium was inoculated to an initial cell concentration of 4 × 106 ± 3 × 106 (1σ) cells ml−1. Triplicate biotic and abiotic (uninoculated) cultures were incubated at 5.1 ± 0.8°C and 14.4 ± 0.7°C (1σ). Temperatures below 5°C were not tested to avoid freeze-thaw cycles that could occur due to the observed ±1°C fluctuation in the incubator temperature. The average pH of the medium for all assays at the start of incubation was 8.3 ± 0.1 (1σ). Samples (1 ml) were removed from the cultures every 24 to 72 h. The culture bottle headspace was periodically pressurized with filter-sterilized air or N2 gas for aerobic and anaerobic cultures, respectively, to prevent contamination and oxidation via the development of a vacuum as samples were removed over time. This additional headspace pressure also allowed the removal of 10-ml gas samples from the culture bottles without pulling a vacuum. Filter-sterilized, concentrated formaldehyde was added to aliquots for cell counting to achieve a final formaldehyde concentration of 5% (vol/vol). Samples were refrigerated at 5°C until they were used for analyses. Aliquots (2 to 100 μl, contingent on the predicted cell concentration) of formaldehyde-fixed samples were vortexed lightly and adjusted to 1 ml with filter-sterilized H2O. Cell suspensions were amended with 0.01% vol/vol SYTO 9 (Thermo Fisher, Waltham, MA) and incubated in the dark for 1 h prior to filtration onto 0.2-μm-pore-size black polycarbonate filters (Millipore, Billerica, MA). Cells were counted using fluorescence microscopy on an Axioskop 2 Plus microscope with a Zeiss 100× oil immersion objective. Sample aliquots for major anion and cation analysis were filtered immediately through 0.2-μm-pore-size nylon filters and diluted gravimetrically (i.e., by mass) with 18.2 MΩ cm−1 H2O prior to analysis via ion chromatography. Ten-milliliter gas samples were taken from the headspace of each anaerobic culture following the plateau of the growth curves and stored under a positive pressure in 20- or 50-ml vials containing a saturated NaCl solution for subsequent N2O analysis.
Geochemical analyses.
The concentrations of major anions and cations (i.e., NO3−, NO2−, SO42−, S2O32−, and NH4+) in field and laboratory samples were determined using a Metrohm ion chromatograph (Riverview, FL) equipped with 25- mm Metrosep A Supp 5 and C4 anion and cation columns, respectively, with 13- and 20-μl sample injection volumes, respectively. Total run times were 50 min. Linear calibration curves (R2 = 0.99 or better) were fitted to standard data spanning from 1.2, 1.6, 0.8, 0.7, and 4.2 μM, the lower detection limit, to 484, 652, 312, 265, and 1,109 μM, the upper detection limit, for NO3−, NO2−, SO42−, S2O32−, and NH4+, respectively. Corresponding anion and cation concentration detection limits for samples from RG5 cultures were 0.06, 0.08, 0.04, 0.03, and 0.21 mM to 24.2, 32.6, 15.6, 13.3, and 55.4 mM for NO3−, NO2−, SO42−, S2O32−, and NH4+, respectively, because of the 50× sample dilutions. A lower range of standards was used when analyzing field samples with the lowest standard for S2O32− at 0.1 μM. The nitrous oxide (N2O) concentrations in the headspace of anaerobic cultures provided NO3− as an oxidant were measured on an HP 5890 series II gas chromatograph outfitted with an electron capture detector (ECD) and a 1-cm3 sample loop. Two analytical columns (both 183 cm by 0.32 cm [outer diameter]) packed with Chromosorb 102 80/100 mesh and Porapak-Q 80/100 mesh, respectively, were used in series for gas separation. Certified standard mixtures of N2O in N2 (Scott Specialty Gases, Houston, TX) were used for instrument calibration (either directly or following volumetric dilution into carrier N2). The limit of quantification for N2O was approximately 0.003 ppm by volume. pH was monitored during the course of the incubation. Despite the strong buffering capacity of the medium (20 mM HCO3−, 10 mM PO43−), the pH in biotic incubations dropped by 1.1 to 1.4 units in aerobic cultures incubated at 5.1 and 14.4 °C but did not change substantially in anaerobic incubations or abiotic assays (data not shown).
Geochemical and thermodynamic modeling.
SO42− was chosen for comparative geochemical modeling to enable more accurate detection of the reaction progress, because S2O32− concentrations approach the analytical detection limit during growth of Thiobacillus sp. RG5 (described below). Best-fit logistic curves for the average SO42− and cell concentrations from triplicate assays as a function of time (t) were developed for each temperature and geochemical condition (equation 1):
| (1) |
where C is the concentration of SO42− or cells at time t, Cmax is the predicted maximum concentration of SO42− or cells produced, α is the maximum growth rate or the slope of the curve at the point of inflection, and thalf is the time at which the sigmoid midpoint occurs.
The amount of energy released as a function of time for aerobic S2O32− oxidation was determined on the basis of logistic curve models describing SO42− production and with the assumption of the stoichiometric conversion of S2O32− to SO42− in a ratio of 1:2. The energy resulting from S2O32− oxidation coupled to NO3− reduction was determined on the basis of a nitrogen (N) mass balance for time points during which NO3− was consumed, assuming the formation of NO2− and then N2. The activities of redox-active analytes [α, i.e., S2O32−, SO42−, O2(aq), NO3−, NO2−, N2(aq)] were determined with the PHREEQC program and the Lawrence Livermore National Laboratory thermodynamics database (http://wwwbrr.cr.usgs.gov/projects/GWC_coupled/phreeqc/). The standard-state Gibbs free energy of each reaction () and the log of the reaction quotient at each time step (logQr,t) were calculated from the activities of the reactants and products using the CHNOSZ thermodynamics database (http://www.chnosz.net/) and subcrt function. The Gibbs free energy of the reaction at each time point (ΔGr,t) was calculated according to equation 2
| (2) |
where R is the gas constant, T is temperature (in Kelvin), and Qr,t is the reaction quotient at time t.
Stepwise energy release (Et; equation 3) was calculated as follows on the basis of the reaction progress, where α(t) and α(t − 1) are the activities of sulfate at time t and t − 1, respectively, and nα is the stoichiometric coefficient of SO42− in each respective metabolic reaction:
| (3) |
Energy requirements per cell produced (Ec) during exponential growth phase for S2O32− oxidation coupled to O2 and NO3− reduction were calculated on the basis of the average of the model-derived and average cell concentrations (Ccell), respectively, according to equation 4.
| (4) |
Nucleotide sequence accession numbers.
Raw sequencing reads from this research have been deposited at DDBJ/EMBL/GenBank under accession number LDUG00000000. The version described in this paper is LDUG01000000.
RESULTS AND DISCUSSION
Enrichment, isolation, and genomic characterization of Thiobacillus sp. RG5.
Three rounds of dilution to extinction in cultures incubated at 5.1°C with thiosulfate and NO3− resulted in a culture comprising a single morphotype (RG5) that exhibited a close affiliation with Thiobacillus denitrificans, on the basis of genomic characterization. The Thiobacillus sp. RG5 draft genome, which consists of 3.29 Mbp, is predicted to be 95% complete based on the presence of conserved phylogenetic marker genes (Table 2; see also Table S1 in the supplemental material). The size of this genome is slightly larger than that of the genome of T. denitrificans, which consists of 2.90 Mbp (46). The average GC content of the RG isolate draft genome, 62.7%, is less than that of the T. denitrificans genome, which has an average GC content of 66.0%.
TABLE 2.
Statistics associated with the assembly of the Thiobacillus sp. RG5 draft genome
| Genome feature | Value |
|---|---|
| Total no. of base pairs | 3,293,652 |
| No. of contigs | 86 |
| N50 (bp) | 146,048 |
| % GC content | 62.71 |
| Longest contig (bp) | 312,422 |
| No. of tRNAs | 44 |
| No. of protein-coding genes | 3,259 |
Thiobacillus denitrificans is a facultative anaerobe and chemolithoautotroph capable of coupling the complete oxidation of inorganic sulfur compounds, such as S2O32−, to the reduction of oxidized nitrogen compounds (e.g., NO3−, NO2−) or O2 (46). Similar to the T. denitrificans genome (46), the Thiobacillus sp. RG5 draft genome encodes the genes for a number of enzymes necessary for aerobic and anaerobic respiration. The genome also encodes the genes for proteins involved in the oxidation of reduced sulfur compounds, including the Sox enzyme complex (SoxB, SoxAX, and SoxYZ), which is a component of a widely distributed pathway for the oxidation of S2O32− to elemental sulfur (S80) (25). The genome also encodes the genes for the reverse dissimilatory sulfite reductase (rDsr) protein complex. The gene cluster (dsrABEFHCMKLJOPNR) is similar to that in other lithotrophic sulfur-oxidizing organisms including T. denitrificans (46, 47). The rDsr enzyme complex is involved in the oxidation of intracellular S80 to SO42−. The presence of genes encoding both Sox and rDsr suggests that Thiobacillus sp. RG5 is capable of catalyzing the complete oxidation of S2O32− to SO42−. Other genes encoding proteins with putative roles in sulfur oxidation are also present in the genome. These include genes that encode a thiosulfate sulfurtransferase (rhodanese), sulfate adenylyltransferase, adenylylsulfate reductase, tetrathionate reductase, dimethyl sulfoxide reductase, and a sulfide:quinone oxidoreductase.
The Thiobacillus sp. RG5 genome encodes genes for the enzymatic machinery for dissimilatory nitrate reduction, including nitrate reductase (NarGHIJ) and nitrite reductase (NirBD), as well as the enzymes necessary for the complete denitrification of nitrate to nitrogen, which includes nitrate reductase (NarGHIJ), nitrite reductase (NirS), nitric oxide reductase (NorBC), and nitrous oxide reductase (NosZ). The genome also encodes genes for the enzymatic machinery necessary for aerobic respiration. Similar to T. denitrificans, the genome encodes an NADH:ubiquinone oxidoreductase, a succinate dehydrogenase, and a cytochrome b/c1 ubiquinol oxidoreductase complex. The genome also encodes aa3- and cbb3-type cytochrome c oxidases and an F-type ATPase.
Characterized members of the Thiobacillus genus are chemoautotrophic and capable of fixing CO2 using the Calvin-Benson-Bassham cycle (46). A cluster of genes encoding homologs of the ribulose bisphosphate carboxylase (cbbRLQO) is encoded in the draft genome of Thiobacillus sp. RG5 in the same orientation as in the genome of T. denitrificans (46), providing a putative mechanism allowing for autotrophic growth in this organism.
Molecular and chemical evidence for S2O32− oxidation at RG: a putative role for Thiobacillus sp. RG5.
SoxB from Thiobacillus sp. RG5 exhibited 90% amino acid sequence identity with SoxB of Thiobacillus thioparus and 89% amino acid sequence identity with SoxB of T. denitrificans strain 25259. Intriguingly, SoxAYXZ, all of which are located on the same contig in an apparent operon (organized as soxBAZYX) in the partial genome of Thiobacillus sp. RG5, were more closely affiliated with those proteins encoded by the T. denitrificans genome (81 to 90% amino acid sequence identities) than with proteins encoded by the T. thioparus genome (76 to 86% amino acid sequence identities). Thus, although SoxB from Thiobacillus sp. RG5 is more closely related to SoxB from T. thioparus, the rest of the proteins forming the Sox complex exhibited closer phylogenetic affiliations with Sox complex proteins of T. denitrificans.
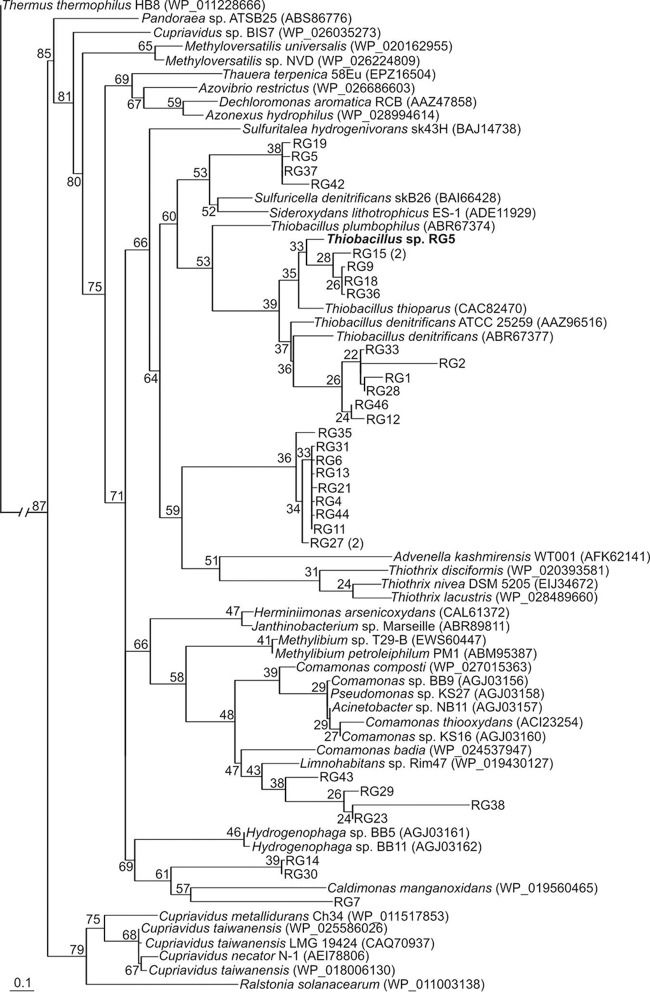
We extracted RNA from RG sediments and amplified soxB from cDNA in an effort to determine the potential for S2O32− oxidation by Thiobacillus sp. RG5 in the subglacial environment. A total of 31 soxB transcripts were sequenced from RG subglacial sediments sampled in October 2010. Phylogenetic reconstruction of the translated soxB transcripts revealed five distinct sequence clusters (Fig. 1). One of the five clusters was most closely related to T. thioparus (∼90% amino acid sequence identities). SoxB from T. denitrificans formed a sister group to this cluster. This cluster also included the SoxB sequence from Thiobacillus sp. RG5, suggesting that the strain isolated in the present study is representative of several Thiobacillus-like populations that may be involved in S2O32− oxidation in the RG subglacial environment. Despite not being collected on the same dates, evidence for a diverse community of putative S2O32− oxidizers in the RG subglacial environment in 2010 is consistent with geochemical data collected in the outflowing stream at RG over a summer melt season in 2014. Only one sample, collected at 5:00 a.m. on 27 September 2014, contained S2O32− at a concentration of 0.3 μM; the concentrations in all other samples were below the detection limit (data not shown). This early-morning sample should have contained a high proportion of subglacially derived water because the supraglacial input from surficial melt would have been negligible, given that it was late in the melt season, it was dark (i.e., no insolation to drive surface melt), and the air temperature was close to freezing (48). This sample also had an SO42− concentration of 332 μM, the second highest measured in 2014 (data not shown).
FIG 1.
Maximum likelihood phylogenetic reconstruction of translated SoxB protein sequences recovered from RG sediment cDNA, the genome of Thiobacillus sp. RG5, and reference sequences obtained from the GenBank database (in parentheses). Single-digit numbers in parentheses indicate the number of times that a given operational taxonomic unit (defined at a level of 97% amino acid sequence identity) was detected in RG sediment cDNA.
The recovery of soxB transcripts affiliated with Thiobacillus in RG subglacial sediments is consistent with previous molecular data indicating the presence of members of the genus in this environment. For example, RubisCO form II (cbbL) transcripts recovered from the same aliquot of subglacial sediments analyzed here included a sequence closely affiliated (92% amino acid sequence identity) with CbbL from T. denitrificans (9). CbbL from Thiobacillus sp. RG5 exhibited 92% amino acid sequence identity to CbbL from T. denitrificans. This provides further support that the strain isolated in the present study is likely representative of thiobacilli in the subglacial environment at RG. Mitchell et al. (7) also recovered 16S rRNA genes that exhibited a close affiliation with Thiobacillus-like populations (>94% sequence identities) from RG subglacial sediments. On the basis of the 16S rRNA gene sequence similarity, thiobacilli have also been detected in a wide range of subglacial systems, including Bench Glacier, AK (6); John Evans Glacier, Ellesmere Island (6); and the Kamb (31) and Whillans (30) Ice Streams in West Antarctica. Sattley and Madigan (49) isolated a sulfur-oxidizing Thiobacillus strain that was most closely related to the mesophile Thiobacillus thioparus, from the cold (<2°C) water column of ice-covered Lake Fryxell, Antarctica. This organism was shown to grow optimally at 18°C, consistent with it being psychrotolerant (49). Collectively, these data reveal a widespread occurrence of thiobacilli in both polar and alpine subglacial systems and indicate a role for these cold-adapted populations in the aerobic and anaerobic oxidation of sulfur compounds in these systems.
Influence of oxidants on growth of Thiobacillus sp. RG5.
We generated growth curves for cultures grown at 5.1 and 14.4°C with S2O32− as the sole electron donor and O2, NO3−, or NO2− as the electron acceptor (Fig. 2 to 4). The composition of bulk medium was monitored in each culture until S2O32− oxidation and SO42− production ceased (i.e., the concentrations reached an asymptote), indicating that the culture had reached stationary phase. Cell production was observed in cultures of Thiobacillus sp. RG5 under all growth conditions except anaerobic growth at 14.4°C with NO2− as the sole oxidant. The level of SO42− production in abiotic controls was less than 0.1 mM for all oxidants and temperatures studied, suggesting that abiotic S2O32− oxidation was minimal or absent over the duration of the incubation.
FIG 2.
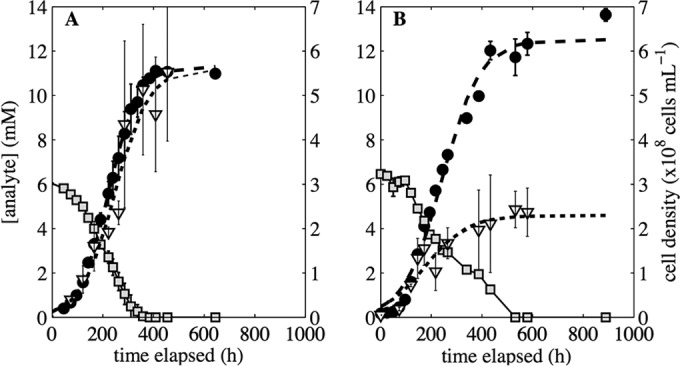
Concentrations of SO42− (●) and S2O32− ( ) and cell density (▽) during the growth of Thiobacillus sp. RG5 when it was incubated at 5.1°C (A) or 14.4°C (B) with S2O32− provided as the electron donor and O2 provided as the electron acceptor. Values represent the averages from triplicate biotic assays. Error bars are 1σ. Best-fit logistic models describe average SO42− concentration (– –) and cell count (···) data.
) and cell density (▽) during the growth of Thiobacillus sp. RG5 when it was incubated at 5.1°C (A) or 14.4°C (B) with S2O32− provided as the electron donor and O2 provided as the electron acceptor. Values represent the averages from triplicate biotic assays. Error bars are 1σ. Best-fit logistic models describe average SO42− concentration (– –) and cell count (···) data.
FIG 3.

Concentrations of SO42− (●), S2O32− ( ), NO3− (
), NO3− ( ), and NO2− (▲) and cell density (▽) during the growth of Thiobacillus sp. RG5 incubated at 5.1°C (A and B) and 14.4°C (C and D) with S2O32− provided as the sole electron donor and NO3− provided as the sole electron acceptor. Values represent the averages from triplicate biotic assays, with error bars showing 1σ. NO3− reduction results in the intermediate production of aqueous NO2−, which is subsequently utilized as an electron acceptor (B and D). The dashed line shows the best-fit logistic models describing SO42− production.
), and NO2− (▲) and cell density (▽) during the growth of Thiobacillus sp. RG5 incubated at 5.1°C (A and B) and 14.4°C (C and D) with S2O32− provided as the sole electron donor and NO3− provided as the sole electron acceptor. Values represent the averages from triplicate biotic assays, with error bars showing 1σ. NO3− reduction results in the intermediate production of aqueous NO2−, which is subsequently utilized as an electron acceptor (B and D). The dashed line shows the best-fit logistic models describing SO42− production.
FIG 4.
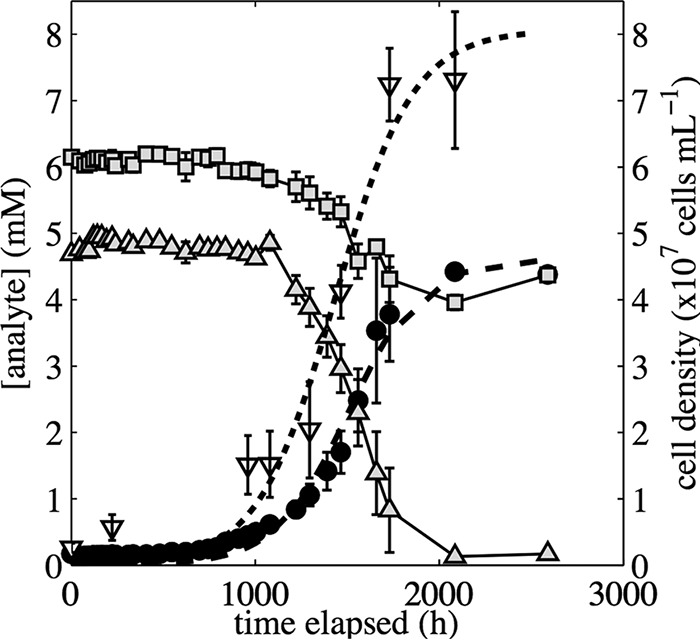
Concentrations of SO42− (●), S2O32− (■), and NO2− (▲) and cell density (▽) during the growth of Thiobacillus sp. RG5 when it was incubated at 5.1°C with S2O32− provided as the electron donor and NO2− provided as the electron acceptor. Values represent the average from triplicate biotic assays. Error bars are 1σ. Dashed line and dotted lines show the best-fit logistic model describing SO42− production and cell formation data, respectively.
Cultures provided S2O32− and O2 (Table 1, reaction R2.2) offer the most available energy, with the initial ΔGr being −871 and −837 kJ mol−1 in cultures incubated at 5.1 and 14.4°C, respectively. The nearly complete oxidation of S2O32− to SO42− (Fig. 2; Table 3) was observed in cultures exposed to atmospheric levels of O2 (21%). The rates of SO42− production and, by inference, S2O32− oxidation were more rapid when these processes were coupled to O2 as an electron acceptor than when they were coupled to NO2− or NO3− in cultures incubated at 5.1 or 14.4°C (see Table S2 in the supplemental material). It is important to note that cells in anoxic cultures provided with NO2− or NO3− as the sole electron acceptor were visibly smaller than cells in oxic cultures. It is possible that numerous, interconnected factors, including, but not limited to, cell size, cell concentration, and cell growth efficiency, contribute to the more rapid S2O32− oxidation in oxic cultures than in anoxic cultures.
TABLE 3.
Net production and consumption of oxidized and reduced chemical species during growth of Thiobacillus sp. RG5 on S2O32− with O2 or NO3− at 5 and 15°C
| Temp and oxidant | Change in concna (mM) |
Reaction stoichiometryb |
|||||
|---|---|---|---|---|---|---|---|
| NO2− | NO3− | SO42− | S2O32− | S2O32− | Oxidant | SO42− | |
| 5°C | |||||||
| O2 | 10.9 | −6.0 | 1.0 | 1.8 | |||
| NO3− | −4.0 | 7.0 | −3.0 | 1.0 | 1.3 | 2.3 | |
| NO2− | −4.5 | 4.2 | −1.8 | 1.0 | 2.5 | 2.4 | |
| 15°C | |||||||
| O2 | 13.5 | −6.5 | 1.0 | 2.1 | |||
| NO3− | −4.6 | 7.0 | −3.9 | 1.0 | 1.2 | 1.8 | |
| NO2− | NAc | NA | NA | NA | NA | NA | |
The data presented represent the difference in the average of triplicate measurements taken from triplicate biological controls.
The stoichiometry of the production or consumption of a given chemical species is based on measured values and was normalized to net S2O32− consumption.
NA, not available.
NO3− and NO2− supported the nearly stoichiometric oxidation of S2O32− to SO42− (Fig. 3 and 4; Table 3). S2O32− oxidation coupled to NO3− reduction resulted in the release of NO2− into bulk solution. Once the concentration of NO3− dropped below the analytical detection limit (0.04 mM) in the cultivation medium, Thiobacillus sp. RG5 utilized the NO3− reduction product NO2− to further oxidize S2O32− to SO42−. The intermediate production of NO2− within Thiobacillus sp. RG5 cultures was not, however, stoichiometric with respect to the level of NO3− reduction (Fig. 3). Claus and Kutzner (50) observed the same phenomenon of aqueous NO2− accumulation and subsequent reduction during the anaerobic growth of a T. denitrificans isolate in batch culture with S2O32− oxidation coupled to NO3− reduction at 30°C.
After a plateau in SO42− production was reached, neither ammonium (NH4+) nor nitrous oxide (N2O) was detected at a level above the initial background concentration (data not shown) in the anaerobic cultures incubated at 5.1 and 14.4°C and supplied with NO3− as the sole oxidant (Fig. 3). This suggests that NO3− and NO2− were likely reduced to N2 in these cultures (Table 1, reactions R2.3 and R2.4). These findings are consistent with previous results from other T. denitrificans strains in which neither NH4+ nor N2O was detected during NO3−-dependent growth (50). The nonstoichiometric production of NO2− in cultures supplied with NO3− as the sole oxidant and the lack of N2O and NH4+ accumulation suggest a range of possible explanations, including the following: (i) some NO3− may have been completely reduced to N2 without the formation of intermediates; (ii) NO3− and NO2− reduction steps may have occurred simultaneously with the observed, subsequent NO2− reduction step; (iii) an unmeasured, reduced nitrogen intermediate, such as NO, may have been produced and was potentially utilized as an oxidant; and (iv) a portion of the NO3− or NO2− may have been assimilated into biomass.
Incomplete, abiotic FeS2 oxidation (Table 1, reaction R1) can produce metastable reduced S intermediates, such as S2O32−, in aquatic environments with circumneutral pHs (20), such as the subglacial environment at RG (e.g., see reference 33). Characterization of strain RG5 (Fig. 2 to 4) suggests that aerobic S2O32− oxidation and anaerobic S2O32− oxidation coupled to NO3− and NO2− reduction are plausible metabolic strategies supporting chemoautotrophic life at low temperatures in the RG subglacial system. Additional work is required, however, to determine if populations such as the Thiobacillus sp. RG5 population are capable of enhancing the rate or extent of FeS2 oxidation by oxidizing S2O32− under such conditions (Table 1, reaction R2.1).
Effect of temperature on growth and metabolic efficiency of Thiobacillus sp. RG5.
Cell concentration was chosen as a metric for evaluating cold adaptation since the differences in the efficiency of cell synthesis and cell concentration integrate the cumulative enzymatic and physiological factors that enhance or inhibit growth at low temperatures. There was no visual difference in cell size between cultures containing the same electron donor but incubated at different temperatures. We therefore restricted cell concentration and energy-per-cell comparisons to cultures with the same electron donor but different incubation temperatures. This limitation takes into account large differences in cell biomass due to differences in cell size between oxic and anoxic cultures.
The maximum cell concentrations in aerobic cultures incubated at 5.1°C were higher than those in aerobic cultures incubated at 14.4°C (Fig. 2), suggesting that the coupling of energy generation to cellular synthesis is more efficient at low temperatures. Indeed, normalization of best-fit model-derived maximum cell concentrations to the maximum concentrations of oxidized S2O32− in aerobic cultures incubated at 5.1 and 14.4°C revealed that 9.9 × 107 ± 1.0 × 107 (1σ) and 3.7 × 107 ± 0.3 × 107 (1σ) cells were produced per mM S2O32− oxidized, respectively (see Table S3 in the supplemental material). Moreover, the doubling times for O2-dependent RG5 growth when it was incubated at 5.1 and 14.4°C were 140 and 294 h, respectively (see Table S3 in the supplemental material). While these metrics strongly suggest that this organism is cold adapted, it is possible that the increased solubility of O2(g) and, therefore, the higher activity of O2(aq) at 5.1°C are partially responsible for these observations.
We calculated the energy requirements for aerobic S2O32−-dependent growth (Ec) to account for the influence of solution chemistry and temperature variation on cellular synthesis in cultures of RG5 incubated at 5.1 versus 14.4°C. This thermodynamic assessment determines the Gibbs free energy of the reaction at each sampling time point during exponential growth (ΔGr,t) on the basis of the incubation temperature and the calculated activities of model-derived reactants and products, including deviations in O2(aq) activity as a function of incubation temperature. The results of this analysis indicate that the cellular synthesis of Thiobacillus sp. RG5 by aerobic S2O32− oxidation at 5.1 and 14.4°C requires an Ec of 3.2 ± 0.6 and 7.7 ± 2.4 μJ cell−1 (1σ), respectively, (see Table S3 in the supplemental material) over the duration of incubation. Comparison of the Ec values via the t test showed that the energy requirements for the synthesis of a cell at 5.1 and 14.4°C are significantly different (P < 0.01). These findings indicate that Thiobacillus sp. RG5 is more efficient at coupling energy generation to cell production at 5.1 than 14.4°C and are evidence that the organism is cold adapted.
A similar pattern in cellular growth, including higher cell concentrations (Fig. 2) and shorter generation times (see Table S3 in the supplemental material) for cultures incubated at 5.1°C than for cultures incubated at 14.4°C, was observed in anaerobic cultures provided NO3− as the sole oxidant. The Ec for synthesis of a cell under NO3−-dependent conditions was calculated from data obtained prior to the onset of NO2− consumption and the assumed production of N2. The resulting Ec values for synthesis of a cell at 5.1 and 14.4°C were 2.3 ± 1.7 and 7.6 ± 3.4 μJ cell−1 (1σ), respectively (see Table S2 in the supplemental material). Despite a t-test analysis suggesting that the Ec values for the NO3−-dependent synthesis of a cell at 5.1 and 14.4°C are not significantly different (P = 0.07), these data and the temperature-dependent differences in cell concentration during NO3− reduction (Fig. 3) are consistent with cell synthesis data from aerobic cultures and together suggest that Thiobacillus sp. RG5 is cold adapted. It should be noted that Ec values for the synthesis of a cell under O2- and NO3−-reducing conditions cannot be directly compared, because of the noticeably smaller size of cells when cells are grown with NO3− than when cells are grown with O2, regardless of the incubation temperature (data not shown).
Growth was not detected at 14.4°C with NO2− as the sole oxidant. Cultures of the isolate Thiobacillus sp. RG5 provided with NO2− as an oxidant and incubated at 14.4°C produced only 0.06 mM SO42− after 1,345 h of incubation (data not shown); cultures of the isolate produced approximately 4 mM SO42− when it was incubated at 5.1°C (Fig. 4). As a result, we did not calculate the energetics associated with cell production under the NO2−-reducing growth condition.
Numerous adaptations can contribute to more energy-efficient cellular synthesis at low temperatures. These include enzyme variations that result in increased substrate conversion efficiencies or lower activation energies for reactions at low temperatures (e.g., see references 51 and 52). The same adaptations can, however, impart a high degree of temperature sensitivity to an organism. Exposing cold-adapted microorganisms to elevated temperatures can hinder growth due to enzyme denaturation, morphological and membrane modifications, and inhibition of RNA synthesis and cell division (51). It is possible that multiple factors, including both adaptations to low temperature and sensitivity at high temperature, are responsible for the enhanced growth rates and more efficient production of cells observed at 5.1°C than at 14.4°C in cultures of Thiobacillus sp. RG5. The use of cell concentration, generation time, and the energetics of cellular production as metrics to describe cold adaptation, as opposed to the use of the more widely used approach of normalization of the growth rate to protein synthesis, may provide a more robust and integrated assessment of cold adaptation by taking into account growth efficiency at the level of the entire cell.
Conclusions.
Thiobacillus sp. RG5, isolated from RG subglacial sediments and closely related to T. denitrificans, is a relevant member of the subglacial community on the basis of soxB transcript analysis. An evaluation of its genome reveals two enzyme complexes associated with S2O32− oxidation to S80 (Sox) and, ultimately, SO42− (rDsr). Additional genes that encode enzyme complexes capable of complete dissimilatory denitrification from NO3− to N2 and the Calvin-Benson-Bassham cycle, which provides a putative mechanism for NO3−-dependent autotrophic growth, were identified.
Microbially mediated anaerobic S2O32− oxidation has never been demonstrated, to our knowledge, under conditions relevant to subglacial environments, such as the environment found in RG, which has a temperature of <1°C (10). The suite of putative enzymes whose genes are encoded by the Thiobacillus sp. RG5 genome is consistent with the activity and cellular production data presented here, which indicate that Thiobacillus sp. RG5 is capable of aerobic (O2) and anaerobic (NO3− and NO2−) S2O32− oxidation at low temperatures (5.1°C) and under solution pH conditions relevant to the RG subglacial system. In cultures grown under O2- or NO3−-dependent conditions, the shorter generation times and the more energy-efficient cellular production at 5.1°C than at 14.4°C indicate that this Thiobacillus isolate is cold adapted.
The recovery of soxB transcripts from RG subglacial sediments closely affiliated with strain RG5 and the physiological characterization of the isolate reported here, coupled with previously reported cbbL transcript data from RG that also exhibit a close affiliation with the data for RG5 (9), suggest that aerobic and anaerobic S2O32− oxidation by the autotrophic Thiobacillus sp. RG5 likely contributes to primary production within the subglacial system at RG. Given that the most probable source of SO42− in this system is FeS2 (7, 35), these results indicate a role for Thiobacillus sp. RG5 in catalyzing the oxidation of S2O32− in situ, thereby maintaining its low concentration (<0.3 μM) in RG meltwaters. The widespread distribution of Thiobacillus-like populations in subglacial environments beneath both valley glaciers (6) and continental ice sheets, where sediments have been isolated from surficial input for tens of thousands of years (30, 31), is suggestive of a key role for autotrophic organisms such as Thiobacillus sp. RG5 in driving mineral weathering in subglacial systems both today and in Earth's past. The activity of chemolithoautotrophs, such as Thiobacillus sp. RG5, would contribute to the local microbial food web by generating chemosynthate and biomass (9). This biomass would then be available to support numerous heterotrophic taxa, including bacterivorous eukaryotes, that have previously been detected in subglacial environments (11).
Supplementary Material
ACKNOWLEDGMENTS
T.L.H. acknowledges support for this work from the NASA postdoctoral program.
We thank the Biogeosciences Institute at the University of Calgary's Kananaskis Field Station for the use of field and laboratory facilities. We thank Chris Allen for assistance with ion chromatography analyses and John Dore for assistance with N2O measurements.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03398-15.
REFERENCES
- 1.Sharp M, Parkes J, Cragg B, Fairchild IJ, Lamb H, Tranter M. 1999. Widespread bacterial populations at glacier beds and their relationship to rock weathering and carbon cycling. Geology 27:107–110. doi:. [DOI] [Google Scholar]
- 2.Hodson A, Anesio AM, Tranter M, Fountain A, Osborn M, Priscu J, Laybourn-Parry J, Sattler B. 2008. Glacial ecosystems. Ecol Monogr 78:41–67. doi: 10.1890/07-0187.1. [DOI] [Google Scholar]
- 3.Wadham J, Tranter M, Skidmore M, Hodson A, Priscu J, Lyons W, Sharp M, Wynn P, Jackson M. 2010. Biogeochemical weathering under ice: size matters. Global Biogeochem Cycles 24:GB3025. [Google Scholar]
- 4.Statham PJ, Skidmore M, Tranter M. 2008. Inputs of glacially derived dissolved and colloidal iron to the coastal ocean and implications for primary productivity. Global Biogeochem Cycles 22:GB3013. [Google Scholar]
- 5.Tranter M, Sharp M, Lamb H, Brown G, Hubbard B, Willis I. 2002. Geochemical weathering at the bed of Haut Glacier d'Arolla, Switzerland, a new model. Hydrol Processes 16:959–993. doi: 10.1002/hyp.309. [DOI] [Google Scholar]
- 6.Skidmore M, Anderson SP, Sharp M, Foght J, Lanoil BD. 2005. Comparison of microbial community compositions of two subglacial environments reveals a possible role for microbes in chemical weathering processes. Appl Environ Microbiol 71:6986–6997. doi: 10.1128/AEM.71.11.6986-6997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell AC, Lafreniere MJ, Skidmore ML, Boyd ES. 2013. Influence of bedrock mineral composition on microbial diversity in a subglacial environment. Geology 41:855–858. doi: 10.1130/G34194.1. [DOI] [Google Scholar]
- 8.Montross SN, Skidmore M, Tranter M, Kivimäki A-L, Parkes RJ. 2013. A microbial driver of chemical weathering in glaciated systems. Geology 41:215–218. doi: 10.1130/G33572.1. [DOI] [Google Scholar]
- 9.Boyd ES, Hamilton TL, Havig JR, Skidmore ML, Shock EL. 2014. Chemolithotrophic primary production in a subglacial ecosystem. Appl Environ Microbiol 80:6146–6153. doi: 10.1128/AEM.01956-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd ES, Skidmore M, Mitchell AC, Bakermans C, Peters JW. 2010. Methanogenesis in subglacial sediments. Environ Microbiol Rep 2:685–692. doi: 10.1111/j.1758-2229.2010.00162.x. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton TL, Peters JW, Skidmore ML, Boyd ES. 2013. Molecular evidence for an active endogenous microbiome beneath glacial ice. ISME J 7:1402–1412. doi: 10.1038/ismej.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tranter M, Raiswell R. 1991. The composition of the englacial and subglacial component in bulk meltwaters draining the Gornergletscher, Switzerland. J Glaciol 37:59–66. [Google Scholar]
- 13.Tranter M, Huybrechts P, Munhoven G, Sharp MJ, Brown GH, Jones IW, Hodson AJ, Hodgkins R, Wadham JL. 2002. Direct effect of ice sheets on terrestrial bicarbonate, sulphate and base cation fluxes during the last glacial cycle: minimal impact on atmospheric CO2 concentrations. Chem Geol 190:33–44. doi: 10.1016/S0009-2541(02)00109-2. [DOI] [Google Scholar]
- 14.Skidmore M. 2011. Microbial communities in Antarctic subglacial aquatic environments. Geophys Monogr Ser 192:61–81. [Google Scholar]
- 15.Christner BC, Skidmore ML, Priscu JC, Tranter M, Foreman CM. 2008. Bacteria in subglacial environments, p 51–71. In Psychrophiles: from biodiversity to biotechnology. Springer, New York, NY. [Google Scholar]
- 16.Tranter M, Skidmore M, Wadham J. 2005. Hydrological controls on microbial communities in subglacial environments. Hydrol Processes 19:995–998. doi: 10.1002/hyp.5854. [DOI] [Google Scholar]
- 17.Edwards KJ, Goebel BM, Rodgers TM, Schrenk MO, Gihring TM, Cardona MM, McGuire MM, Hamers RJ, Pace NR, Banfield JF. 1999. Geomicrobiology of pyrite (FeS2) dissolution: case study at Iron Mountain, California. Geomicrobiol J 16:155–179. doi: 10.1080/014904599270668. [DOI] [Google Scholar]
- 18.Moses CO, Nordstrom DK, Herman JS, Mills AL. 1987. Aqueous pyrite oxidation by dissolved oxygen and by ferric iron. Geochim Cosmochim Acta 51:1561–1571. doi: 10.1016/0016-7037(87)90337-1. [DOI] [Google Scholar]
- 19.Rimstidt JD, Vaughan DJ. 2003. Pyrite oxidation: a state-of-the-art assessment of the reaction mechanism. Geochim Cosmochim Acta 67:873–880. doi: 10.1016/S0016-7037(02)01165-1. [DOI] [Google Scholar]
- 20.Chandra AP, Gerson AR. 2010. The mechanisms of pyrite oxidation and leaching: a fundamental perspective. Surface Sci Rep 65:293–315. doi: 10.1016/j.surfrep.2010.08.003. [DOI] [Google Scholar]
- 21.Bottrell SH, Tranter M. 2002. Sulphide oxidation under partially anoxic conditions at the bed of the Haut Glacier d'Arolla, Switzerland. Hydrol Processes 16:2363–2368. doi: 10.1002/hyp.1012. [DOI] [Google Scholar]
- 22.Moses CO, Herman JS. 1991. Pyrite oxidation at circumneutral pH. Geochim Cosmochim Acta 55:471–482. doi: 10.1016/0016-7037(91)90005-P. [DOI] [Google Scholar]
- 23.Jorgensen C, Jacobsen OS, Elberling B, Aamand J. 2009. Microbial oxidation of pyrite coupled to nitrate reduction in anoxic groundwater sediment. Environ Sci Technol 43:4851–4857. doi: 10.1021/es803417s. [DOI] [PubMed] [Google Scholar]
- 24.Schippers A, Jorgensen BB. 2002. Biogeochemistry of pyrite and iron sulfide oxidation in marine sediments. Geochim Cosmochim Acta 66:85–92. doi: 10.1016/S0016-7037(01)00745-1. [DOI] [Google Scholar]
- 25.Friedrich CG, Rother D, Bardischewsky F, Quentmeier A, Fischer JR. 2001. Oxidation of reduced inorganic sulfur compounds by Bacteria: emergence of a common mechanism? Appl Environ Microbiol 67:2873–2882. doi: 10.1128/AEM.67.7.2873-2882.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skidmore ML, Foght JM, Sharp MJ. 2000. Microbial life beneath a high arctic glacier. Appl Environ Microbiol 66:3214–3220. doi: 10.1128/AEM.66.8.3214-3220.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wynn PM, Hodson AJ, Heaton THE, Chenery SR. 2007. Nitrate production beneath a high Arctic glacier, Svalbard. Chem Geol 244:88–102. [Google Scholar]
- 28.Hodson A, Roberts TJ, Engvall A-C, Holmén K, Mumford P. 2010. Glacier ecosystem response to episodic nitrogen enrichment in Svalbard, European High Arctic. Biogeochemistry 98:171–184. doi: 10.1007/s10533-009-9384-y. [DOI] [Google Scholar]
- 29.Boyd ES, Lange RK, Mitchell AC, Havig JR, Hamilton TL, Lafrenière MJ, Shock EL, Peters JW, Skidmore M. 2011. Diversity, abundance, and potential activity of nitrifying and nitrate-reducing microbial assemblages in a subglacial ecosystem. Appl Environ Microbiol 77:4778–4787. doi: 10.1128/AEM.00376-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christner BC, Priscu JC, Achberger AM, Barbante C, Carter SP, Christianson K, Michaud AB, Mikucki JA, Mitchell AC, Skidmore ML, Vick-Majors TJ, WISSARD Science Team . 2014. A microbial ecosystem beneath the West Antarctic ice sheet. Nature 512:310–313. doi: 10.1038/nature13667. [DOI] [PubMed] [Google Scholar]
- 31.Lanoil B, Skidmore M, Priscu JC, Han S, Foo W, Vogel SW, Tulaczyk S, Engelhardt H. 2009. Bacteria beneath the West Antarctic ice sheet. Environ Microbiol 11:609–615. doi: 10.1111/j.1462-2920.2008.01831.x. [DOI] [PubMed] [Google Scholar]
- 32.Skidmore M, Tranter M, Tulaczyk S, Lanoil B. 2010. Hydrochemistry of ice stream beds—evaporitic or microbial effects? Hydrol Processes 24:517–523. [Google Scholar]
- 33.Sharp M, Creaser RA, Skidmore M. 2002. Strontium isotope composition of runoff from a glaciated carbonate terrain. Geochim Cosmochim Acta 66:595–614. doi: 10.1016/S0016-7037(01)00798-0. [DOI] [Google Scholar]
- 34.McMechan M. 1989. Geology of Peter Lougheed Provincial Park, Rocky Mountain Frontier Ranges, Alberta. Open File Report 2057. Geological Survey of Canada, Calgary, Alberta, Canada. [Google Scholar]
- 35.Griggs R. 2013. Characterization of subglacial till from Robertson Glacier, Alberta, Canada: implications for biogeochemical weathering. M.S. thesis, Department of Earth Sciences, Montana State University Bozeman, MT. [Google Scholar]
- 36.Sorokin DY, Tourova TP, Braker G, Muyzer G. 2007. Thiohalomonas denitrificans gen. nov., sp. nov. and Thiohalomonas nitratireducens sp. nov., novel obligately chemolithoautotrophic, moderately halophilic, thiodenitrifying Gammaproteobacteria from hypersaline habitats. Int J Syst Evol Microbiol 57:1582–1589. doi: 10.1099/ijs.0.65112-0. [DOI] [PubMed] [Google Scholar]
- 37.Widdel F, Kohring G, Mayer F. 1983. Studies on the dissimilatory sulfate that decomposes fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov., sp. nov., and Desulfonema magnum sp. nov. Arch Microbiol 134:286–294. [Google Scholar]
- 38.Lohse M, Bolger A, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. 2012. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, Frischkorn KR, Williams KH, Tringe SG, Banfield JF. 2013. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1:22. doi: 10.1186/2049-2618-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petri R, Podgorsek L, Imhoff JF. 2001. Phylogeny and distribution of the soxB gene among thiosulfate-oxidizing bacteria. FEMS Microbiol Lett 197:171–178. doi: 10.1111/j.1574-6968.2001.tb10600.x. [DOI] [PubMed] [Google Scholar]
- 43.Boyd ES, Jackson RA, Encarnacion G, Zahn JA, Beard T, Leavitt WD, Pi Y, Zhang CL, Pearson A, Geesey GG. 2007. Isolation, characterization, and ecology of sulfur-respiring Crenarchaea inhabiting acid-sulfate-chloride-containing geothermal springs in Yellowstone National Park. Appl Environ Microbiol 73:6669–6677. doi: 10.1128/AEM.01321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 45.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 46.Beller HR, Chain PSG, Letain TE, Chakicherla A, Larimer FW, Richardson PM, Coleman MA, Wood AP, Kelly DP. 2006. The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans. J Bacteriol 188:1473–1488. doi: 10.1128/JB.188.4.1473-1488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dahl C, Engels S, Pott-Sperling AS, Schulte A, Sander J, Lübbe Y, Deuster O, Brune DC. 2005. Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J Bacteriol 187:1392–1404. doi: 10.1128/JB.187.4.1392-1404.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gurnell AM, Clark MJ. 1987. Glacio-fluvial sediment transfer: an alpine perspective. John Wiley & Sons Ltd, Chichester, United Kingdom. [Google Scholar]
- 49.Sattley WM, Madigan MT. 2006. Isolation, characterization, and ecology of cold-active, chemolithotrophic, sulfur-oxidizing bacteria from perennially ice-covered Lake Fryxell, Antarctica. Appl Environ Microbiol 72:5562–5568. doi: 10.1128/AEM.00702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Claus G, Kutzner HJ. 1985. Autotrophic denitrification by Thiobacillus denitrificans in a packed bed reactor. Appl Microbiol Biotechnol 22:289–296. [Google Scholar]
- 51.Margesin R, Schinner F. 1994. Properties of cold-adapted microorganisms and their potential role in biotechnology. J Biotechnol 33:1–14. doi: 10.1016/0168-1656(94)90093-0. [DOI] [Google Scholar]
- 52.Gerday C, Aittaleb M, Bentahir M, Chessa J-P, Claverie P, Collins T, D'Amico S, Dumont J, Garsoux G, Georlette D, Hoyoux A, Lonhienne T, Meuwis M-A, Feller G. 2000. Cold-adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol 18:103–107. doi: 10.1016/S0167-7799(99)01413-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.