Abstract
In temperate and boreal forest ecosystems, nitrogen (N) limitation of tree metabolism is alleviated by ectomycorrhizal (ECM) fungi. As forest soils age, the primary source of N in soil switches from inorganic (NH4+ and NO3−) to organic (mostly proteins). It has been hypothesized that ECM fungi adapt to the most common N source in their environment, which implies that fungi growing in older forests would have greater protein degradation abilities. Moreover, recent results for a model ECM fungal species suggest that organic N uptake requires a glucose supply. To test the generality of these hypotheses, we screened 55 strains of 13 Suillus species with different ecological preferences for their in vitro protein degradation abilities. Suillus species preferentially occurring in mature forests, where soil contains more organic matter, had significantly higher protease activity than those from young forests with low-organic-matter soils or species indifferent to forest age. Within species, the protease activities of ecotypes from soils with high or low soil organic N content did not differ significantly, suggesting resource partitioning between mineral and organic soil layers. The secreted protease mixtures were strongly dominated by aspartic peptidases. Glucose addition had variable effects on secreted protease activity; in some species, it triggered activity, but in others, activity was repressed at high concentrations. Collectively, our results indicate that protease activity, a key ectomycorrhizal functional trait, is positively related to environmental N source availability but is also influenced by additional factors, such as carbon availability.
INTRODUCTION
In temperate and boreal forests, nitrogen (N) is the element that usually limits tree nutrition (1). To acquire sufficient N, trees form symbioses with microorganisms, including ectomycorrhizal (ECM) fungi (2), as well as shoot-endophytic bacteria (3). In soils, ECM fungi can take up N from both mineral and organic sources. Mineral N can be found as NH4+ or NO3− (4), while organic N can be present as part of several different organic oligomers or polymers: peptides, chitin, nucleic acids, and heterocyclic N compounds (3). Peptides are considered the dominant organic N source in forest soils (representing as much as 80% of organic N [5]), with ECM fungi typically retrieving N from this source through the use of proteases (3). Despite generally broad enzymatic capacities (6), not all ECM fungi have the ability to access peptide N, which has resulted in the classification of ECM fungi into “protein” and “nonprotein” species (7). Multiple authors have suggested that natural selection should favor traits allowing mycorrhizal fungi to utilize the most abundantly available N source in their environment (8). This would suggest that protein ECM fungal species have their ecological niche in organic-N-rich soils. Empirical support for this hypothesis has been shown by Lilleskov et al. (9), who found that ECM fungal species growing in a soil rich in mineral N had a lower ability to grow on proteins than those from poorer mineral N soils. Similarly, Tibbett et al. (10) demonstrated that strains of the ECM fungal genus Hebeloma from the arctic region (where 99% of the N is in organic form) had the ability to use seed protein as a N source, which was not the case for Hebeloma strains from temperate soils.
The ratio between organic and mineral N in forest soils is affected by many factors, with forest succession being the most prominent (11). Organic N forms become increasingly dominant as a forest ages due to the accumulation of organic matter. Hence, the organic N/inorganic N ratio increases through succession, suggesting that organic forms are increasingly important N sources in older forest soils (12). Along with shifts in the N source, changes in ECM fungal community composition have also been well documented during forest succession, though some species can be found at almost all stages (13). The stage specificity of ECM fungal community composition is more likely linked to the age of organic horizons (i.e., the litter, fragmentation, and humus layers that develop consecutively) than to the age of the tree, as experiments have shown that seedlings establishing near mature trees are generally colonized by ECM fungi typical of older forests (14). Also, experiments with litter removal and litter addition to conifer stands of various ages provided support for the greater importance of the age of the soil (including the organic layers) than that of the tree (15). Taking the data together, it appears that the later an ECM fungal species occurs in forest soil succession, the more it is likely to be in contact with organic matter, making protein its dominant N source. If the above-mentioned hypothesis about N availability is correct, one would predict ECM fungal protein degradation ability to correlate positively with forest soil age.
Although the general functioning of ECM fungi in forest N cycling has been recognized for many years, the properties of the ECM fungal enzymes involved in protein degradation are relatively poorly characterized. Research on a limited number of species showed that these enzymes are secreted proteases, which most of the time belong either to the aspartic or to the serine peptidase class (3, 16). While their secretion appears to be induced by the presence of protein (16), recent results for a model ECM species (Paxillus involutus; Boletales, Basidiomycota) also showed that uptake of N from organic matter is additionally dependent on the availability of a simple carbon (C) source (17). The latter result supports the hypothesis that mining for organic N by ECM species does not occur without an energy supply (18, 19). However, since the last two studies compared only the presence and absence of glucose, it has yet to be determined how different levels of C affect organic N degradation.
Species from the ECM genus Suillus (Boletales, Basidiomycota) offer a good model to test the relationships among protein degradation ability, forest (soil) age, and carbon availability, because (i) they are widely distributed in temperate and boreal forests and form a well-defined monophyletic group (20, 21); (ii) different Suillus species show clear preferences for soils with different organic matter contents and for trees of different ages (22) (Table 1); (iii) they associate exclusively with trees in the genus Pinaceae (20), resulting in similar environmental gradients during succession; (iv) they are easily isolated into pure culture; and (v) they have fast growth and high biomass production in vitro (22). Here, we used in vitro assays to test four hypotheses: (i) Suillus species preferentially occurring in mature forests, where soils have high organic N content, have a greater protein degradation ability than Suillus species characteristic of young forests, where soil organic N content is low; (ii) Suillus species that occur in both young and mature forests harbor protein-degrading strains (ecotypes) in organic-rich soils and non-protein-degrading strains (ecotypes) in mineral soils; (iii) protein degradation occurs through the secretion of a mixture of aspartic and serine proteases; and (iv) protease activity, when present, increases with the glucose concentration.
TABLE 1.
Ecological characteristics of the 55 Suillus strains investigated
| Species | Strain | Site | Ecotype | Host genus | Forest age preference | Soil organic matter preference | Reference | Type(s) of fungal tissue in the reference |
|---|---|---|---|---|---|---|---|---|
| Suillus americanus | 51 | Cedar Creek (USA) | NAa | Pinus | Multiple | Low organic | Personal observations | Sporocarps |
| 52 | Cloquet (USA) | |||||||
| Suillus bovinus | SboP1 | Paal (Belgium) | Young | Pinus | Multiple | Low organic | http://www.verspreidingsatlas.nl/ | Sporocarps |
| SboP3 | Paal (Belgium) | Young | ||||||
| SboP4 | Paal (Belgium) | Young | ||||||
| SboP5 | Paal (Belgium) | Young | ||||||
| SboP6 | Paal (Belgium) | Young | ||||||
| SboP7 | Paal (Belgium) | Young | ||||||
| SboZ1 | Zolder (Belgium) | Mature | ||||||
| SboZ2 | Zolder (Belgium) | Mature | ||||||
| SboZ3 | Zolder (Belgium) | Mature | ||||||
| SboZ4 | Zolder (Belgium) | Mature | ||||||
| SboZ5 | Zolder (Belgium) | Mature | ||||||
| SboZ6 | Zolder (Belgium) | Mature | ||||||
| SboZ7 | Zolder (Belgium) | Mature | ||||||
| Suillus brevipes | 13 | Yosemite (USA) | NA | Pinus | Multiple | NA | 34 | Sporocarps, mycorrhizas |
| 14 | Yosemite (USA) | |||||||
| 15 | Yosemite (USA) | |||||||
| 17 | Mendocino1 (USA) | |||||||
| 18 | Mendocino1 (USA) | |||||||
| 19 | Ocean Shores (USA) | |||||||
| 20 | Ocean Shores (USA) | |||||||
| 32 | Mendocino1 (USA) | |||||||
| 47 | Rock Creek (USA) | |||||||
| Suillus caerulescens | 38 | Mendocino2 (USA) | NA | Pseudotsuga | Multiple | NA | Personal observations | Sporocarps |
| 39 | Mendocino2 (USA) | |||||||
| Suillus cavipes | 53 | Cloquet (USA) | NA | Larix | Mature | High organic | http://www.verspreidingsatlas.nl/ | Sporocarps |
| 54 | Cloquet (USA) | |||||||
| 55 | Cloquet (USA) | |||||||
| Suillus granulatus | 56 | Cedar Creek (USA) | NA | Pinus | Young | Low organic | http://www.verspreidingsatlas.nl/ | Sporocarps |
| 57 | Cedar Creek (USA) | |||||||
| Suillus grisellus | 60 | Cedar Creek (USA) | NA | Larix | Mature | High organic | Personal observations | Sporocarps |
| 61 | Cedar Creek (USA) | |||||||
| Suillus lakei | 43 | Point Reyes (USA) | NA | Pseudotsuga | Young | NA | 35 | Mycorrhizas |
| 75 | Point Reyes (USA) | |||||||
| Suillus viscidus | 63 | Cedar Creek (USA) | NA | Larix | Mature | High organic | http://www.verspreidingsatlas.nl/ | Sporocarps |
| 64 | Cedar Creek (USA) | |||||||
| Suillus luteus | 65 | Cedar Creek (USA) | NA | Pinus | Young | Low organic | http://www.verspreidingsatlas.nl/ | Sporocarps |
| SluP1 | Paal (Belgium) | |||||||
| SluP2 | Paal (Belgium) | |||||||
| SluP3 | Paal (Belgium) | |||||||
| SluP4 | Paal (Belgium) | |||||||
| SluP8 | Paal (Belgium) | |||||||
| Suillus pungens | 27 | Berkeley Marina (USA) | NA | Pinus | Multiple | High organic | 36 | Sporocarps, mycorrhizas |
| 28 | Berkeley Marina (USA) | |||||||
| 30 | Berkeley Marina (USA) | |||||||
| Suillus tomentosus | 33 | Montana (USA) | NA | Pinus | Multiple | Low organic | 34 | Sporocarps, mycorrhizas |
| 34 | Montana (USA) | |||||||
| 35 | Montana (USA) | |||||||
| 36 | Montana (USA) | |||||||
| Suillus variegatus | SvaP1 | Paal (Belgium) | Young | Pinus | Multiple | High organic | 22 | Sporocarps |
| SvaP3 | Paal (Belgium) | Young | ||||||
| SvaZJW6 | Zolder (Belgium) | Mature | ||||||
| SvaZJW8 | Zolder (Belgium) | Mature | ||||||
| SvaZJW13 | Zolder (Belgium) | Mature |
NA, not available.
MATERIALS AND METHODS
Strains.
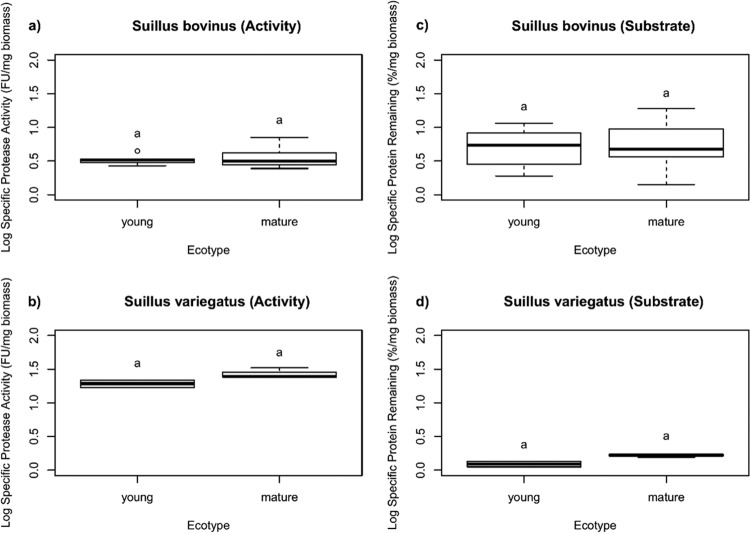
We investigated the protein degradation abilities of 55 ECM fungal strains, isolated from sporocarps, belonging to 13 species: Suillus americanus, Suillus brevipes, Suillus bovinus, Suillus cavipes, Suillus caerulescens, Suillus granulatus, Suillus grisellus, Suillus lakei, Suillus laricinus, Suillus luteus, Suillus pungens, Suillus tomentosus, and Suillus variegatus. All the species were represented by at least two strains. Thirty-two strains were isolated in North America (United States), from 9 sites (Cloquet, lat 46.704397, long −92.510528; Yosemite, lat 37.817027, long −119.712591; Mendocino1, lat 38.787030, long −123.514499; Mendocino2, lat 39.311419, long −123.760378; Cedar Creek, lat 45.407066, long −93.199801; Ocean Shores, lat 47.032500, long −124.164167; Rock Creek, lat 29.670780, long −82.371932; Point Reyes, lat 38.084533, long −122.870891; and Berkeley Marina, lat 37.859584, long −122.315999), and 23 in Europe (Belgium), from 2 sites (Paal, lat 51.058887, long 5.175981; Zolder, lat 50.995371, long 5.272788). All isolations were made from sporocarp tissue. In cases where morphological identification was not possible, species were identified by internal transcribed spacer (ITS) sequencing. The habitat preference of each species (e.g., forest age and soil type) was gathered from the primary literature and the website of the Dutch mycological society (http://www.verspreidingsatlas.nl/paddenstoelen), as well as from our direct field observations (Table 1).
Growth.
Fungi were maintained on solid minimum Melin-Norkrans (MMN) medium for 10 days and then transplanted to fresh solid MMN medium and grown again for 7 days. At this time, they were considered sufficiently active to perform the experiment. A 3- by 3-mm plug of active mycelium was then placed in a glass bead system. The system consisted of a monolayer of sterile 4-mm-diameter glass beads sitting on a 9-cm petri dish filled with 11 ml of liquid medium, which is the volume needed to cover the glass beads (16). The liquid medium consisted of standard liquid MMN medium, where the N source (ammonium chloride) was replaced by a soluble protein (bovine serum albumin [BSA]) and the elemental N concentration was kept the same (53 mg liter−1, which is 342 mg liter−1 of BSA). We chose BSA as a model protein because previous results, on the same glass bead system but with P. involutus, showed that the trends observed with the protein are the same as with organic matter (16) and are therefore ecologically relevant. The pH was adjusted to 4.5. Fungi were grown under static conditions, in the dark, at 21°C and 80% humidity for 17 days. We monitored the protease activity and the protein content of the growth medium at 0, 5, 7, 11, 12, and 17 days after inoculation. Incubation was stopped at 17 days because further sampling significantly increased the risk of contamination; the mycelium was harvested and weighed after freeze-drying.
Protease assays.
Protease assays were run using two complementary measurement procedures. The first was run on culture supernatant and therefore measured secreted protease activity. This assay was an adaptation of the fluorescein isothiocyanate (FITC)-BSA assay (23) for microplates. First, for each sample, 100 μl of medium supernatant was transferred to a 96-well microplate (flat bottom; Sarstedt). Then, we added 100 μl of 50 mM citrate buffer (pH 4.2) and 5 μl of freshly prepared 1 mg · ml−1 FITC-BSA solution (Sigma-Aldrich). A preliminary experiment with a reduced number of strains showed that the protease activity measured was highest at acidic pH values (we tested pH 3, 4, 5, and 6 [data not shown]); we chose pH 4 as a compromise between high values and ecological relevance (the soils of these conifer forests are most often between pH 4 and 5). The plate was sealed with aluminum foil to prevent evaporation and incubated overnight (24 h) at 40°C. Then, the proteins were precipitated by adding 100 μl of 10% trichloroacetic acid (TCA) (Sigma-Aldrich), incubated for 1 h at room temperature, and centrifuged for 30 min at 25°C and 2,000 rpm, and 40 μl of the supernatant was transferred to a new microplate containing 200 μl of 1 M Tris buffer (pH 9.7). The fluorescence was read with a FluoStar Omega microplate reader at 485-nm excitation and 520-nm emission. One thousand units corresponds to the fluorescence produced by 27 mg liter−1 of trypsin over 24 h at pH 8.
The second assay measured the protein left in the culture supernatant, which was the result of the activities of both secreted and cell wall-bound proteases. This measurement was done in microplates using the Bradford assay. First, 30 μl of sample was diluted with 70 μl of distilled water in a transparent 96-well microplate (flat bottom). Then, 50 μl of this solution was transferred to a new transparent microplate containing 100 μl of distilled water and 100 μl of Bradford Quick reagent. After 1 h of incubation, the absorbance of each well was read at 595 nm on a FluoStar Omega microplate reader. In order to estimate the concentration of protein from the absorbance, we also measured the absorbance of 4 standard solutions (342 mg liter−1, 224 mg liter−1, 112 mg liter−1, and 0 mg liter−1).
Determination of the protease class.
In order to determine the relative proportion of each protease class of the cocktail produced by a given strain, we measured protease activity as previously described but added 10 μl of specific protease inhibitors to the FITC-BSA mixture (as described in reference 16): E64 [trans-epoxysuccinyl-l-leucylamido(4-guanidino)butane, which inhibits cysteine proteases; final concentration, 10 μM; stock solution prepared in water], pepstatin A (which inhibits aspartic proteases; final concentration, 10 μM; in ethanol), EDTA (which inhibits metalloproteases; final concentration, 5 mM; stock solution prepared in water), and phenylmethylsulfonyl fluoride (PMSF) (which inhibits serine proteases; final concentration, 1 mM; stock solution prepared in isopropanol). All chemicals were ordered from Sigma-Aldrich. Measurements were carried out on 17-day-postincubation samples, since they contained the highest protease values for all strains.
Influence of glucose concentration on protease activity.
We measured the effect of glucose on protease activity by growing Suillus strains in BSA medium as before, but using four different glucose concentrations: 0, 1, 2.5, and 5 g liter−1. We chose to use multiple strains of 3 Suillus species with contrasting ecologies: S. luteus, which grows preferentially in early forests in organic-N-poor soils; S. variegatus, which grows preferentially in mature forests with high-organic-N-rich soils; and S. bovinus, which occurs in both young and mature forests at similar frequencies. Protease activity was measured at 1, 4, 6, 8, and 11 days after inoculation.
Soil analyses.
To more clearly assess the link between in vitro protein degradation ability and environmental N source availability, we measured soil organic and mineral N contents in two sites from which several Suillus strains used in this study were isolated: a young forest (Paal) and a mature forest (Zolder) in Belgium. At each site, 15 soil samples were taken with a soil corer (15 cm deep; 8-cm diameter) on 11 October 2014. The core locations were arranged on a 3- by 5-m grid, with the nodes separated by 4 m at Paal. Due to the topology of the Zolder site (a 500-m-long, 8-m-wide dike), we harvested 5 groups of 3 cores, with the groups separated by at least 20 m along the dike. The soil samples were then pooled by groups of three into five composite samples, which were individually passed through a 2-mm sieve. Two hundred grams of fresh composite sample was dried overnight at 60°C. Total N was measured by the Kjeldahl method (decomposition of organic N by sulfuric acid, oxidation of reduced nitrogen as ammonium sulfate, and back-titration with boric acid) and inorganic N (NO3 and NH4-N) by titrimetry after reduction by Devarda's alloy. Organic N was then deduced by calculating the difference between total N and inorganic N.
Statistics.
We tested the effects of four factors—species (13 species), forest age (young, multiple stage, and mature), soil type (low and high organic nitrogen), and host genus (Larix, Pseudotsuga, and Pinus)—on two variables, secreted protease activity and protein remaining in the medium. To account for differences in growth rates among strains, as well as the positive relationship between biomass and protease activity and protein degradation, we used specific protease activity (total protease activity divided by the dry mass of the mycelium) and specific protein remaining (percent dry mass) in the final analyses. The effects of the four factors were tested using a nested analysis of variance (ANOVA) (with strain nested within species to account for possible nonindependence) on log-transformed data (for specific protease activity) and square root of x-transformed data (for specific protein remaining). We used Tukey honestly significant difference (HSD) tests to determine significant post hoc differences among factor means. To compare the protease activities of different strains of the same species collected from different sites (ecotypes), we used the two species that had at least two strains from both a young and a mature forest site (S. bovinus and S. variegatus). Significant differences between species and ecotypes were assessed using ANOVA on 1/x-normalized data (secreted protease activity) and on untransformed data (protein remaining), followed by a Duncan post hoc test. To determine the relative contributions of different protease classes, we compared protease activities with four inhibitors with a one-way ANOVA. For that test, the data were log(x + 1) transformed to improve variance homogeneity, and a Tukey HSD test was used to determine significant differences among assay means. The effects of different glucose concentrations on protease activity were estimated using an ANOVA followed by a Tukey HSD test on the protease activities measured at the end of the experiment. Correlations between the protease activity at the end of the experiment and the percentage of protein left were evaluated by a Kendall correlation analysis. Statistics were run using R (24).
RESULTS
Relationships between protease activity and ecological traits.
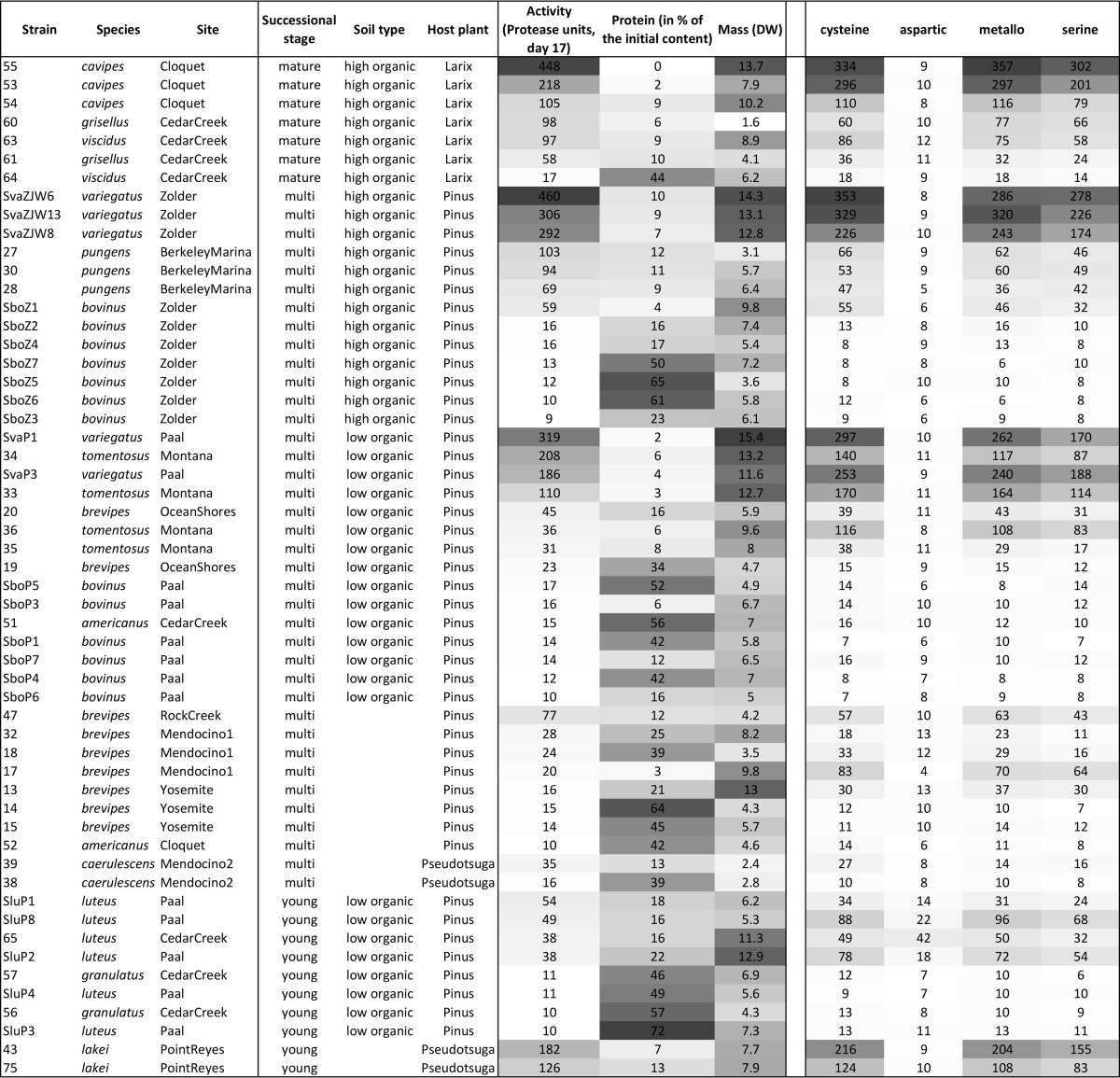
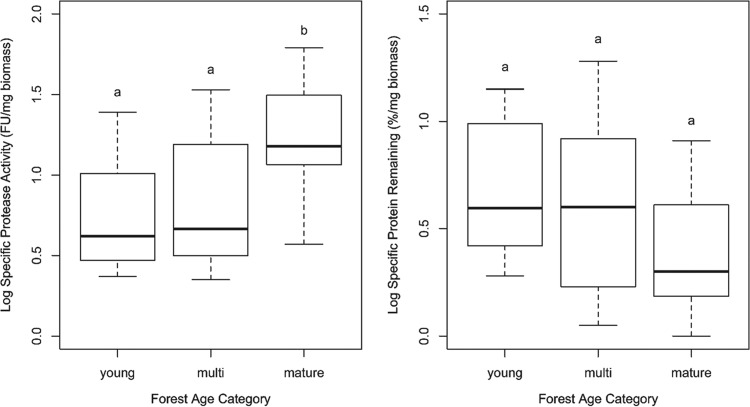
All 55 Suillus strains displayed significant (i.e., higher than the control) secreted protease activity, and the protein content decreased by at least 28% in all assays (Table 2). The protease activity and the amount of protein left at the end of the experiment were significantly and negatively correlated (Kendall's tau < 0.001) (see Fig. S1 in the supplemental material). However, there were 8 strains for which both variables were low, suggesting that in these strains, a major part of the protease activity occurred through cell wall-bound proteases (<35 fluorescence units and <25% protein left in the medium, respectively): they belonged to S. bovinus (SboP3, SboP6, SboP7, SboZ2, SboZ3, and SboZ4) and S. brevipes (strains 13 and 17). For these strains, there was no correlation between the amount of protein left in the medium and their biomass at the end of the experiment. Differences in both specific protease activity and protein remaining in the medium were significant among species (nested ANOVA; P < 0.001 and P < 0.019, respectively); there were also significant effects of soil type (P < 0.001) and forest age (P < 0.001) on specific protease activity (Table 3). Species from mature forests had significantly higher secreted protease activity and lower specific protein remaining than those from young forests and from multiple-stage forests (i.e., present in both young and mature forests) (Fig. 1). When grouped by soil type, strains of species from high-organic-N soils had significantly higher secreted protease activity than those from low-organic-N soils (Fig. 2); these strains also had smaller amounts of remaining protein in the medium, but the difference was not significant (P = 0.146) (Table 3). The nested ANOVA showed no significant effect of host tree species on protease activity and remaining protein contents; however, in pairwise comparisons, Pinus-associated species had on average significantly lower protease activities than those associated with Larix-associated hosts (see Fig. S2 in the supplemental material).
TABLE 2.
Characteristics of all 55 Suillus strains at the end of the experiment (17 days)a
Protease activity (fluorescence units), remaining protein content (percentage of the initial BSA concentration), dry biomass (milligrams), and ecological characteristics. The four columns on the right represent the protease activity in the presence of four protease inhibitors for all 55 strains: cysteine, E64 inhibitor; aspartic, pepstatin A; metallo, EDTA; serine, PMSF.
TABLE 3.
ANOVA results for factors affecting specific protease activity and protein remaining in the medium

a Significant effects are shaded.
FIG 1.
Box-and-whiskers representation of log-transformed values of specific protease activities and specific protein degradation of the 55 Suillus strains categorized by forest age. Different letters indicate significant differences among treatment forest age category means, as determined by post hoc Tukey HSD tests. The boxes represent the 2nd and 3rd interquartile ranges; the horizontal lines in the boxes represent the median; the upper and lower bars outside the boxes represent the 1st and 4th quartiles, respectively. FU, fluorescence units.
FIG 2.
Box-and-whiskers representation of log-transformed values of specific protease activities and specific protein degradation of the 55 Suillus strains categorized by soil type. Different letters indicate significant differences among treatment soil type means as determined by post hoc Tukey HSD tests. The boxes represent the 2nd and 3rd interquartile ranges; the horizontal lines in the boxes represent the median; the upper and lower bars outside the boxes represent the 1st and 4th quartiles, respectively.
Soil organic N and protein degradation abilities.
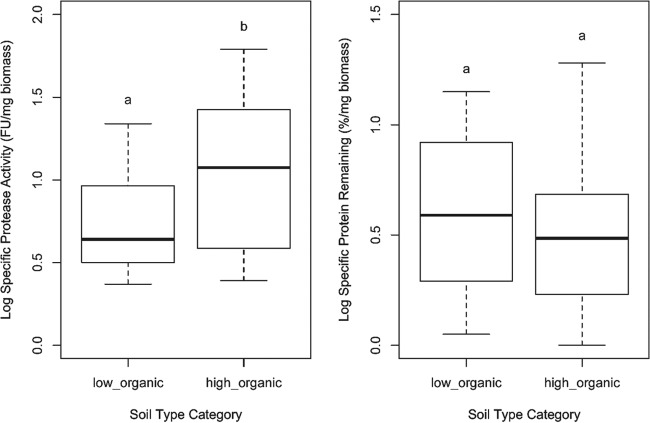
To investigate the extent to which there is local adaptation within species depending on N source availability (i.e., the presence of ecotypes), we compared the protein degradation activities of 19 strains belonging to S. bovinus and S. variegatus present at both a young (Paal; low organic N/mineral N ratio, 28 ± 9) and a mature (Zolder; high organic N/mineral N ratio, 132 ± 12) forest site. The organic N content in soil was, on average, 15 times higher in the mature forest than in the young forest (Zolder, 1,838 mg N · kg−1; Paal, 119 mg N · kg−1) (see Table S1 in the supplemental material), but there was three times more NH4+ in the mature forest soils, as well (Zolder, 13.7 mg N · kg−1; Paal, 4.2 mg N · kg−1). There was no significant difference in protease activity and protein degradation between ecotypes of the same species (Table 4). Strains of S. bovinus had low specific protease activity and protein degradation in both groups (Fig. 3a and c), while strains of S. variegatus had significantly higher protease activity and degraded significantly more protein (Fig. 3b and d).
TABLE 4.
ANOVA results for differences in specific protease activity and protein remaining in the medium between ecotypes of S. bovinus and S. variegatus at two sitesa
| Variable | Test | Result for factor: |
||
|---|---|---|---|---|
| Species × site | Species | Site | ||
| Protease activity | ANOVA | 0.89 | 1.35E−05 | 0.79 |
| Protein degradation | ANOVA | 0.80 | 3.70E−03 | 0.65 |
| df | 1 | 1 | 1 | |
Paal, with low organic matter content, and Zolder, with high organic matter content.
FIG 3.
Box-and-whiskers representation of log-transformed values of specific protease activities and specific protein degradation of strains of S. bovinus and S. variegatus isolated from two forest sites: a young forest with soil with a low organic N/mineral N ratio (Paal) and a mature forest with soil with a high organic N/mineral N ratio (Zolder). The box plots represent the variation of each parameter between species (S. bovinus, 6 strains in Paal and 7 in Zolder; S. variegatus, 2 strains in Paal and 3 in Zolder). The boxes represent the 2nd and 3rd interquartile ranges; the horizontal lines in the boxes represent the median; the upper and lower bars outside the boxes represent the 1st (Q1) and 4th quartiles, respectively; and the dots outside the bars represent the outliers (defined as values outside 1.5 times the interquartile range below Q1 and above Q3).
Identification of the secreted protease class.
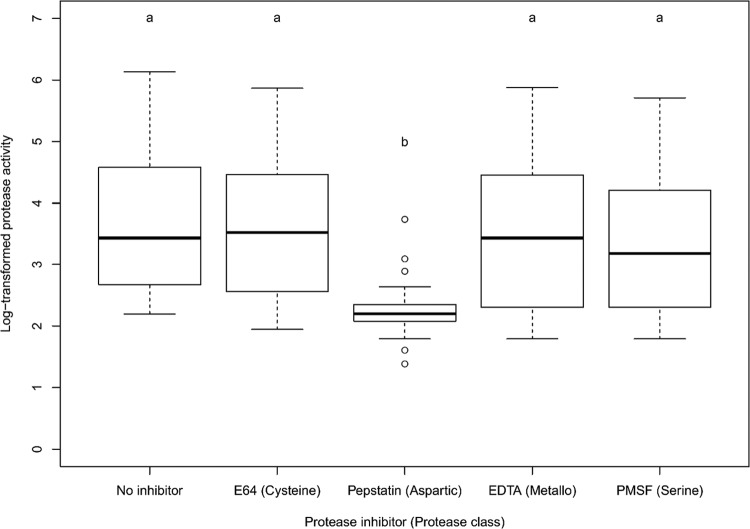
Among the four protease classes (aspartic, serine, and cysteine proteases and metalloproteases), there were no significant differences in protease activity in the presence of serine, cysteine, and metalloprotease inhibitors relative to an assay with no inhibitors present (Fig. 4). In contrast, there was a significant, ∼10-fold reduction in average secreted protease activity in the presence of pepstatin A, which inhibits aspartic proteases. This inhibition was observed in all Suillus strains in which we measured significant secreted protease activity (Table 2).
FIG 4.
Box-and-whiskers representation of log-transformed values of protease activities of the 55 Suillus strains treated with four different protease inhibitors. Different letters indicate significant differences among treatment soil type means as determined by post hoc Tukey HSD tests. The boxes represent the 2nd and 3rd interquartile ranges; the horizontal lines in the boxes represent the median; the upper and lower bars outside the boxes represent the 1st (Q1) and 4th quartiles, respectively; and the dots outside the bars represent the outliers (defined as values outside 1.5 times the interquartile range below Q1 and above Q3).
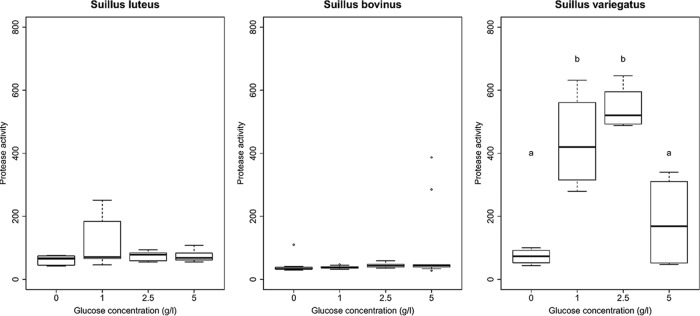
Influence of glucose concentration on protease activity.
Glucose addition had a significant effect on secreted protease activity (expressed as fluorescence units) for only one fungal species (Table 5). Strains of S. variegatus were significantly affected by the levels of glucose addition (Fig. 5). The protease activities of glucose at 1 g liter−1 and 2.5 g liter−1 were significantly higher than the protease activity without glucose (Fig. 5). However, at 5 g liter−1 of glucose addition, the protease activity was low again, without a significant difference from that with no glucose addition. For S. luteus and S. bovinus, the addition of glucose had no significant effect, and all activities were low (Fig. 5).
TABLE 5.
ANOVA results for factors affecting protease activity when fungal strains were provided with different levels of glucose
| Variable | Species | Test | Glucose concn |
|---|---|---|---|
| Protease activity | S. luteus | ANOVA | 0.21 |
| S. bovinus | ANOVA | 0.14 | |
| S. variegatus | ANOVA | 2.70E−04 | |
| df | 3 |
FIG 5.
Box-and-whiskers representation of protease activities of the Suillus strains growing in BSA medium (expressed as fluorescence units) with different glucose concentrations (0, 1, 2.5, and 5 g liter−1). Three species were investigated: S. luteus (strains P1, P3, P4, P8, and P13), S. bovinus (strains P1, P2, P4, P10, Z1, Z2, Z3, and Z4), and S. variegatus (strains Z1, ZJW3, ZJW4, ZW6, and ZJW13). For S. bovinus, strains from both Zolder and Paal sites were investigated. Different letters indicate significant differences among treatment soil type means as determined by post hoc Tukey HSD tests. The boxes represent the 2nd and 3rd interquartile ranges; the horizontal lines in the boxes represent the median; the upper and lower bars outside the boxes represent the 1st (Q1) and 4th quartiles, respectively; and the dots outside the bars represent the outliers (defined as values outside 1.5 times the interquartile range below Q1 and above Q3).
DISCUSSION
Relationships between soil N sources and protease activity.
We found that protease activities differed significantly among strains of Suillus species based on forest age and soil type. Consistent with the hypothesis about N source and protein degradation ability, strains of species restricted to mature forests and high-organic-N soils had significantly higher protease activity than those present in younger forests and low-organic-N soils. While these results indicate that protein degradation is linked to changes in forest age and soil type, the two factors are clearly not independent: mature forests are usually associated with a thick organic soil layer (13). Since many ecological factors also change significantly with forest age (e.g., host tree species composition [11], soil pH [11], and litter phenol concentration [25]), it is also possible that other factors besides soil organic N content contribute to the observed patterns. Measuring additional soil variables at each collection site was beyond the scope of this study, but future experimental work (e.g., adding organic N to young soils or removing the organic layer in mature forests [15]) will be helpful in differentiating the relative importance of changes in organic N availability from these additional environmental factors.
The strains of S. bovinus and S. variegatus from the Paal site (a young pine forest with a low organic N/mineral N ratio) and the Zolder site (a mature pine forest with a high organic N/mineral N ratio) showed no significant differences in their protease activities. Given that previous results suggested that locally adapted strains of multiple-forest-age species may have higher protease activity in high-organic-N soils (8, 9), we were surprised to find no support for this kind of variability. The overall higher protease activity of S. variegatus strains than those of S. bovinus, however, is consistent with closer observations of the ecology of the two species. Despite being a multiple-forest-age species, S. variegatus preferentially inhabits mature forests, while S. bovinus grows there only as satellite populations (22). Hence, the presence of S. variegatus may depend on the development of an organic layer in the forest soil, where its protein degradation ability would give a competitive advantage for N uptake. Moreover, S. bovinus was also stimulated by litter removal in pine stands that exposed mineral soil (26). Interestingly, S. luteus, which is classified as a species characteristic of young trees on mineral soils, does occur in older stands as well, but its root tips and mycelium are located in the mineral rather than the organic layer (27, 28). Taken together, these results suggest that the presence of S. variegatus in young forests may be attributable to local organic niches in young-forest soil, and conversely, local mineral N patches may facilitate the persistence of S. bovinus in mature forests.
For 8 of the 55 strains, the protein content of the medium significantly decreased while protease activity was low, meaning that the protease activity was very likely cell wall bound. Alternative mechanisms could involve adsorption of BSA to the mycelium (29), but this hypothesis can be partially ruled out by the fact that there was no correlation between protein left in the medium and the mycelial biomass for these strains. Therefore, we conclude that most of the protease activity of the above-mentioned strains of S. bovinus and S. brevipes was cell wall bound. Moreover, the strains preferentially inhabiting mature forests or high-organic soils were always characterized by high secreted protease activity. In the range of Suillus species tested here, secreted proteases could therefore be an adaptation to an organic-N-rich environment.
While our results suggest that ecological filtering or natural selection favors physiological capacities in ECM fungi that allow them to utilize the dominant N source in their environment, they do not imply that protein degradation is necessarily the rate-limiting step of N uptake or that ECM protein degradation controls only N availability. Litter breakdown and N mineralization depend on its lignin and polyphenolic contents (25). Lignin degradation mechanisms (e.g., lignin peroxidases, Mn peroxidases, laccases, and the Fenton reaction) hence may play a role as important as that of protein degradation in ECM-mediated plant N uptake. Moreover, proteins are not the only source of organic N in forest soils: simple amino acids, chitin (fungal or arthropod necromass), or heterocyclic N (chlorophyll and nucleic acids) can also contribute to N assimilation, and all the associated enzyme activities may contribute significantly to N mineralization.
Classes of proteases.
All protease cocktails of the strains that had significant activity were strongly repressed by pepstatin A, but the other inhibitors did not significantly decrease protease activity in the experimental assays. From this, we conclude that the protease activity was dominated by aspartic proteases. Shah et al. (16) also showed that the cocktail of proteases secreted by P. involutus was also dominated by aspartic proteases and, as a consequence, had an acidic optimum. Moreover, the authors also showed that this class of proteases accounted for most of the protease activity when the fungus was growing on BSA, but also on other N sources, such as gliadin, pollen, and dissolved soil organic matter. These findings are consistent with the ecology of these systems; where organic N accumulates, soils are acidic, as observed by Chalot and Brun (2). However, partly in contrast to our study, these authors reported that ECM fungal proteases belonged to the aspartic and serine protease classes. We therefore suggest that secreted aspartic proteases are key agents in organic N acquisition for the ECM species, at least in the order Boletales.
Effects of glucose on protease activity.
Because glucose has been previously found to trigger organic matter oxidation and N acquisition from that organic matter by the ECM fungus P. involutus (17), we measured the protease activities of strains of three species with contrasting ecologies, S. luteus (pioneer), S. bovinus (preference for young forest stages), and S. variegatus (preference for old forest stages), at different glucose concentrations. S. luteus strains did not respond to glucose input, possibly because of inherently low protease activities. For S. bovinus, we observed protease activity in only one of the strains coming from the high-organic-N site, and only at the highest glucose load (5 g liter−1). For S. variegatus, the protease activity was influenced by the glucose concentration, but not in a linear manner. Protease activity reached peak values at 1 and 2.5 g liter−1 and was relatively low at 5 g liter−1. Repression of protease activity by a high glucose concentration was reported by Colpaert and Van Laere (30) and is consistent with the use of BSA as a carbon source. Indeed, high glucose input represses genes involved in C metabolism pathways through catabolite repression (gluconeogenesis, Krebs/TCA cycle, and genes involved in metabolization of C from other sources [31]). Moreover, it is known that some ECM fungi can use the deaminated skeletons of amino acids as a C source for the TCA cycle or as a template for synthesis of new amino acids (3, 17). Therefore, we suggest that the BSA in our experiment may have also been used by Suillus species as an alternative C source, with C catabolites repressing protease activity at high glucose concentrations. This hypothesis is consistent with the fact that protease activity is not immediately induced in our assays. However, this does not explain why the tested Suillus species were not able to degrade protein without glucose, which shows that an easily available C source is needed to trigger protease activity, as already observed with P. involutus (17). One explanation could be the following: fungal protease activity is triggered by a low to average host plant C supply, while high mineral N availability in soil would result in faster uptake by the plant, a higher photosynthesis rate, and a higher C flux. High C supply rates would then be an indication of plant N sufficiency and therefore that fungal protease activity is not necessary. Alternatively, repression of protease activity by a high glucose supply could be related to the distance between the glucose concentration and the place where organic N exploitation takes place: Suillus species are long-distance exploration types, and therefore, the hyphae proliferating close to a protein-containing patch would be far away from the glucose supply, in the Hartig net of the root tip. To better understand the role of host carbon in protein degradation, more experimental work is needed in this area, for example, through the use of 13C labeling of organic N.
Conclusions and future directions.
In summary, we found that the protein degradation ability of Suillus strains (i) was highest in species adapted to high-organic soils, (ii) showed little intraspecific variability, (iii) was due primarily to aspartic peptidases, and (iv) was controlled to some extent by glucose levels. Though these data were all obtained using an in vitro experimental system, we assert they are still ecologically informative, as previous studies using pure-culture approaches have yielded results that correlate well with those observed in field settings (10, 32, 33). The results of our study imply that the ability to forage for organic N is a crucial functional trait that may have an important role in shaping ECM fungal communities, with protein-degrading species becoming more common as the soil organic matter content increases. However, this does not rule out the possibility that other important mechanisms related to N acquisition may play important roles, as well, such as chitinase activity or N storage capacity. An important next step will be to test the validity of these results in soil microcosms or field settings, particularly the role of host tree and protein carbon in vivo. Given the contrasting protein degradation abilities of cooccurring species, such as S. bovinus and S. variegatus, determining how competition for access to different N sources may mediate species interactions and vertical niche differentiation would provide a more mechanistic understanding of the drivers of ECM fungal community structure. This knowledge is particularly important in light of the strong effect of human-induced gradients on nitrogen availability in Europe and North America (9). Finally, examining the protein degradation abilities of additional Suillus species associated with these host genera will be key to determining the strength of host phylogenetic signals versus other environmental conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Branco and T. Bruns for assistance with collection of some of the North American Suillus strains. Members of the Kennedy laboratory provided constructive comments on a previous version of the manuscript. We also acknowledge constructive comments by three reviewers on an earlier version of the manuscript.
Jelle Stas and Francois Rineau are grateful to the Bijzonder Onderzoeksfonds (BOF) from Hasselt University for financing their research.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03191-15.
REFERENCES
- 1.Rees M, Condit R, Crawley M, Pacala S, Tilman D. 2001. Long-term studies of vegetation dynamics. Science 293:650–655. doi: 10.1126/science.1062586. [DOI] [PubMed] [Google Scholar]
- 2.Chalot M, Brun A. 1998. Physiology of organic nitrogen acquisition by ectomycorrhizal fungi and ectomycorrhizas. FEMS Microbiol Rev 22:21–44. doi: 10.1111/j.1574-6976.1998.tb00359.x. [DOI] [PubMed] [Google Scholar]
- 3.Carrell AA, Frank AC. 2014. Pinus flexilis and Picea engelmannii share a simple and consistent needle endophyte microbiota with a potential role in nitrogen fixation. Front Microbiol 5:333. doi: 10.3389/fmicb.2014.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attiwill PM, Adams MA. 1993. Tansley review no. 50. Nutrient cycling in forests. New Phytol 124:561–582. [DOI] [PubMed] [Google Scholar]
- 5.Clinton PW, Newman RH, Allen RB. 1995. Immobilization of 15N in forest litter studied by 15N CPMAS NMR spectroscopy. Eur J Soil Sci 46:551–556. doi: 10.1111/j.1365-2389.1995.tb01351.x. [DOI] [Google Scholar]
- 6.Lindahl BD, Tunlid A. 2015. Ectomycorrhizal fungi: potential organic matter decomposers, yet not saprotrophs. New Phytol 205:1443–1447. doi: 10.1111/nph.13201. [DOI] [PubMed] [Google Scholar]
- 7.Abuzinadah RA, Read DJ. 1986. The role of proteins in the nutrition of ectomycorrhizal plants. I. Utilization of peptides and proteins by ecotmycorrhizal fungi. New Phytol 103:481–493. [Google Scholar]
- 8.Koide R, Fernandez C, Malcolm G. 2014. Determining place and process: functional traits of ectomycorrhizal fungi that affect both community structure and ecosystem function. New Phytol 201:433–439. doi: 10.1111/nph.12538. [DOI] [PubMed] [Google Scholar]
- 9.Lilleskov EA, Fahey TJ, Horton TR, Lovett GM. 2002. Belowground ectomycorrhizal fungal community change over a nitrogen deposition gradient in Alaska. Ecology 83:104–115. doi: 10.1890/0012-9658(2002)083[0104:BEFCCO]2.0.CO;2. [DOI] [Google Scholar]
- 10.Tibbett M, Sanders FE, Cairney JWG. 1998. The effect of temperature and inorganic phosphorus supply on growth and acid phosphatase production in arctic and temperate strains of ectomycorrhizal Hebeloma spp. in axenic culture. Mycol Res 102:129–135. doi: 10.1017/S0953756297004681. [DOI] [Google Scholar]
- 11.Read DJ. 1993. Mycorrhiza in plant communities. Adv Plant Pathol 9:1–31. [Google Scholar]
- 12.LeDuc SD, Lilleskov EA, Horton TR, Rothstein DE. 2013. Ectomycorrhizal fungal succession coincides with shifts in organic nitrogen availability and canopy closure in post-wildfire jack pine forests. Oecologia 172:257–269. doi: 10.1007/s00442-012-2471-0. [DOI] [PubMed] [Google Scholar]
- 13.Dickie IA, Martinez-Garcia LB, Koele N, Grelet GA, Tylianakis JM, Pelzer DA, Richardson SJ. 2013. Mycorrhizas and mycorrhizal fungal communities, throughout ecosystem development. Plant Soil 367:11–39. doi: 10.1007/s11104-013-1609-0. [DOI] [Google Scholar]
- 14.Fleming LV. 1983. Succession of mycorrhizal fungi on birch: infection of seedlings planted around mature trees. Plant Soil 71:263–267. doi: 10.1007/BF02182661. [DOI] [Google Scholar]
- 15.Baar J, ter Braak CJF. 1996. Ectomycorrhizal sporocarp occurrence as affected by manipulation of litter and humus layers in Scots pine stands of different age. Appl Soil Ecol 4:61–73. doi: 10.1016/0929-1393(96)00097-2. [DOI] [Google Scholar]
- 16.Shah F, Rineau F, Canback B, Johansson T, Tunlid A. 2013. The molecular components of the extracellular protein-degradation pathways of the ectomycorrhizal fungus Paxillus involutus. New Phytol 200:875–887. doi: 10.1111/nph.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rineau F, Shah F, Smits MM, Persson P, Johansson T, Carleer R, Troein C, Tunlid A. 2013. Carbon availability triggers the decomposition of plant litter and assimilation of nitrogen by an ectomycorrhizal fungus. ISME J 7:2010–2022. doi: 10.1038/ismej.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talbot JM, Allison SD, Treseder KK. 2008. Decomposers in disguise: mycorrhizal fungi as regulators of soil C dynamics in ecosystems under global change. Funct Ecol 22:955–963. doi: 10.1111/j.1365-2435.2008.01402.x. [DOI] [Google Scholar]
- 19.Franklin O, Nasholm T, Hogberg P, Hogberg M. 2014. Forests trapped in nitrogen limitation: an ecological market perspective on ectomycorrhizal symbiosis. New Phytol 203:657–666. doi: 10.1111/nph.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretzer A, Li Y, Szaro TM, Bruns TD. 1996. Internal transcribed spacer sequences from 38 recognized species of Suillus sensu lato: phylogenetic and taxonomic implications. Mycologia 88:776–785. doi: 10.2307/3760972. [DOI] [Google Scholar]
- 21.Binder M, Hibbett DS. 2006. Molecular systematics and biological diversification of boletales. Mycologia 98:971–981. doi: 10.3852/mycologia.98.6.971. [DOI] [PubMed] [Google Scholar]
- 22.Dahlberg A. 1997. Population ecology of Suillus variegatus in old Swedish Scots pine forests. Mycol Res 101:47–54. doi: 10.1017/S0953756296002110. [DOI] [Google Scholar]
- 23.Twining SS. 1984. Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal Biochem 143:30–34. doi: 10.1016/0003-2697(84)90553-0. [DOI] [PubMed] [Google Scholar]
- 24.R Core Team 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org. [Google Scholar]
- 25.Northup RR, Yu Z, Dahlgren RA, Vogt KA. 1995. Polyphenol control of nitrogen release from pine litter. Nature 377:227–229. doi: 10.1038/377227a0. [DOI] [Google Scholar]
- 26.Baar J, Kuyper TW. 1998. Restoration of above-ground ectomycorrhizal flora in stands of Pinus sylvestris (Scots pine) in The Netherlands. Restoration Ecol 6:227–238. doi: 10.1046/j.1526-100X.1998.00635.x. [DOI] [Google Scholar]
- 27.Landeweert R, Leeflang P, Kuyper TW, Hoffland E, Rosling A, Wernars K, Smit E. 2003. Molecular identification of ectomycorrhizal mycelium in soil horizons. Appl Environ Microbiol 69:327–333. doi: 10.1128/AEM.69.1.327-333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosling A, Landeweert R, Lindahl BD, Larsson KH, Kuyper TW, Taylor AFS, Finlay RF. 2003. Vertical distribution of ectomycorrhizal fungal taxa in a podzol profile. New Phytol 159:775–783. doi: 10.1046/j.1469-8137.2003.00829.x. [DOI] [PubMed] [Google Scholar]
- 29.Peters T., Jr 2012. Serum albumin, p 133–175. In Putnam FW. (ed), The plasma proteins, vol 1. Structure, function, and genetic control, 2nd ed. Elsevier Science, Burlington, MA. [Google Scholar]
- 30.Colpaert JV, Van Laere A. 1996. A comparison of the extracellular enzyme activities of two ectomycorrhizal and a leaf-saprotrophic basidiomycete colonizing beech leaf litter. New Phytol 134:133–141. doi: 10.1111/j.1469-8137.1996.tb01153.x. [DOI] [Google Scholar]
- 31.Rönne H. 1995. Glucose repression in fungi. Trends Genet 11:12–17. doi: 10.1016/S0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- 32.Finlay RD, Frostegard A, Sonnerfeldt AM. 1992. Utilization of organic and inorganic nitrogen sources by ectomycorrhizal fungi in pure culture and in symbiosis with Pinus contorta Dougl. Ex Loud. New Phytol 120:105–115. doi: 10.1111/j.1469-8137.1992.tb01063.x. [DOI] [Google Scholar]
- 33.Huggins JA, Talbot J, Gardes M, Kennedy PG. 2014. Unlocking environmental keys to host specificity: differential tolerance of acidity and nitrate by Alnus-associated ectomycorrhizal fungi. Fungal Ecol 12:52–61. doi: 10.1016/j.funeco.2014.04.003. [DOI] [Google Scholar]
- 34.Visser S. 1995. Ectomycorrhizal fungal succession in jack pine stands following wildfire. New Phytol 129:389–401. doi: 10.1111/j.1469-8137.1995.tb04309.x. [DOI] [Google Scholar]
- 35.Twieg BD, Durall DM, Simard SW. 2007. Ectomycorrhizal fungal succession in mixed temperate forests. New Phytol 176:437–447. doi: 10.1111/j.1469-8137.2007.02173.x. [DOI] [PubMed] [Google Scholar]
- 36.Peay K, Bruns TD, Kennedy PG, Bergemann SE, Garbelotto M. 2007. A strong species-area relationship for eukaryotic soil microbes: island size matters for ectomycorrhizal fungi. Ecol Lett 10:470–480. doi: 10.1111/j.1461-0248.2007.01035.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.