Abstract
We present here the findings from a study of the microbiome of the southern basin of the Caspian Sea, the largest water body on Earth disconnected from any ocean and a brackish inland sea. By high-throughput metagenomics, we were able to reconstruct the genomes of representative microbes. The gross community structure (at the phylum level) was different from the structure of typical marine and freshwater communities in temperate open oceans, with the Caspian Sea having freshwater-like amounts of Actinobacteria and Alphaproteobacteria, while Gammaproteobacteria and Betaproteobacteria were present at intermediate levels. We assembled the genomes of several groups and provide detailed descriptions of partial genomes from Actinobacteria, Thaumarchaea, and Alphaproteobacteria. Most belonged to hitherto unknown groups, although they were related to either marine or freshwater groups. The phylogenetic placement of the Caspian genomes indicates that the organisms have multiple and separate phylogenetic origins and that they are related to organisms with both freshwater and marine lineages. Comparative recruitment from global aquatic metagenomes indicated that most Caspian microbes are endemic. However, some Caspian genomes were recruited significantly from either marine water (a member of the Alphaproteobacteria) or freshwater (a member of the Actinobacteria). Reciprocally, some genomes of other origins, such as the marine thaumarchaeon “ Candidatus Nitrosopelagicus” or the actinobacterium “Candidatus Actinomarina,” were recruited from the Caspian Sea, indicating some degree of overlap with the microbiota of other water bodies. Some of these microbes seem to have a remarkably widespread geographic and environmental distribution.
INTRODUCTION
Salinity is a major factor determining the microbiota of an aquatic environment (1–3). The microbial communities of freshwater and marine habitats that have similar characteristics (for example, oligotrophic and euphotic habitats) are characterized by the presence of different microbes (for a review, see reference 4). In particular, differences in salt concentration can affect microbial energetic costs and metabolic pathways (5, 6). However, despite a consistent correlation, there is no definite proof that salinity is indeed the main reason for the differences in community structure. There are other parameters which covary with salinity; for example, due to their larger volume and depth, marine water bodies tend to be more stable and less affected by climatic or seasonal fluctuations. Along similar lines, terrestrial water bodies, such as rivers or lakes, are much more strongly influenced by the surrounding terrain and contain much more organic matter of terrestrial origin (4). One way to overcome these difficulties with the assessment of the effects of salinity on the microbiome is the study of salinity gradients, such as those in estuaries, in which salinity changes over relatively short spatial and time ranges. This allows comparison of water masses that are similar in all other environmental parameters. However, estuaries are also interfaces in which different microbial communities get mixed, and this mixing makes it very difficult to distinguish autochthonous from allochthonous microbes (7–10). Brackish seas, such as the Black Sea or the Baltic Sea, are also connected to the global ocean through straits.
The southern basin of the Caspian Sea, on the other hand, is permanently brackish and has been so for 2 million to 3 million years (11). It has no significant mixing with other water bodies (the main rivers feeding the Caspian Sea, e.g., the Volga River, enter through the northern basin hundreds of kilometers away) and is deep, like marine offshore waters. The Caspian Sea itself not only is the largest enclosed body of water on Earth by area but also is considered to be the only ancient lake with an oceanic origin (12), as it is a remnant of the ancient Tethys Ocean with an estimated age of 2 million to 3 million years. It can be broadly separated into three different sections: the northern shallow part, which can be considered an almost freshwater lake; the middle part, which has a maximum depth of 790 m; and the deep southern part, which has a maximum depth of 1,025 m. The last two sections contain brackish water with a salinity of about one-third of that of the open ocean. The north-south salinity gradient of the Caspian Sea is especially steep in the northern section, while the southern section has a very stable salinity with only minor changes, mostly due to the rainfall-evaporation balance, occurring during the year (13). Therefore, inhabitants of these waters are exposed to permanent brackish conditions rather than to a salinity gradient. The synergy of salinity and temperature sets up vertical currents in the Caspian Sea, resulting in the oxygenation of its deep waters, a characteristic which makes it unlike other large brackish water bodies, like the Black Sea and the Baltic Sea, and more similar to the global ocean. A strong stratification based on temperature during the summer is another one of the features of the Caspian Sea that makes it similar to temperate open oceans (11). These characteristics make the Caspian Sea a model of the ocean, but with less than one-third of its typical salinity.
In the work described here, we studied a depth profile of the Caspian Sea by direct high-throughput, deep sequencing and reconstructed several genomes of the local microbiota. We focused on the assembled reconstructed genomes because they provide more reliable information than short individual metagenomic reads. We compared the results with those for marine water and freshwater bodies at equivalent latitudes and show that the brackish water microbiota of the Caspian Sea shares commonalities with the microbiota of both marine water and freshwater but retains its own individual characteristics that we suggest might be shared by brackish water microbes at large.
MATERIALS AND METHODS
Sampling and sequencing.
A single depth profile was obtained on 1 October 2013 from the southern part of the Caspian Sea near Babolsar, Iran (52°36′22.7″E, 36°51′7.6″N) (see Fig. S1 in the supplemental material). The sampling site was 13 km from the coast and had a bottom depth of 230 m. Samples were taken using a Rosette Niskin bottle sampler (multi-water sampler MWS 12; Hydro-Bios, Germany). Twelve bottles closing at 1-m intervals were taken from depths in the ranges of 14 to 25, 39 to 50, and 149 to 160 m (50 liters of water was collected at each depth). For simplicity, the samples from the three depths are identified here as Caspian15, Caspian40, and Caspian150, respectively. The salinity, temperature, and conductivity profile of the water column was determined with a conductivity-temperature-depth (CTD) sensor in the Rosette Niskin bottle sampler (Ocean Seven 316 Plus CTD for oceanography; Idronaut, Italy). To retrieve the biomass, samples were sequentially prefiltered through a 20-μm-pore-size prefilter (Albet DP5891; Hahnemuehle, Germany) and a 5-μm-pore-size prefilter (Albet DP5895; Hahnemuehle, Germany) and were finally concentrated on a 0.22-μm-pore-size filter (catalog number 11107-142G; Sartorius, Germany) using a peristaltic pump system. The filters were stored at −20°C until DNA extraction. DNA was extracted by a standard phenol-chloroform protocol (14) and sequenced by use of an Illumina HiSeq 2000 PE 101 sequencer (Beijing Genomics Institute, Hong Kong). For each sample, 1 library with an insert size of 350 bp was constructed, and one lane of paired-end sequences for each library provided 510 million, 639 million, and 675 million reads for the Caspian15, Caspian40, and Caspian150 samples, respectively.
16S rRNA gene classification.
A nonredundant version of the RDP database (15) was created by clustering its ca. 2.3 million 16S rRNA gene sequences into approximately 800,000 sequences at the 90% nucleotide sequence identity level using the UCLUST algorithm (16). All reads from the Illumina data sets were compared to this reduced set, and an E-value cutoff of 1e−5 was used to identify candidate 16S rRNA gene sequences. The candidate sequences were further examined using the ssu-align software package to separate them into archaeal, bacterial, and eukaryal 16S/18S rRNA or non-16S rRNA gene sequences (17).
Only these bona fide sequences were finally compared to the sequences from the complete RDP database and classified into a high-level taxon if the sequence identity was ≥80% and the alignment length was ≥90 bp. Sequences failing these thresholds were discarded.
Assembly and annotation.
All three data sets were assembled together using the IDBA assembler (18). The Prodigal algorithm (in the metagenomic mode) was used for predicting protein-coding genes in the assembled contigs (19). tRNA prediction was performed using the tRNAscan-SE server (20), and rRNA genes were identified with meta_rna software (21). Multiple methods were used to annotate the predicted proteins in the assembled genomes. The sequences of all proteins were compared to those in a local NCBI NR database using the BLASTP program, and functions were assigned if the query shared >80% similarity and >80% alignment coverage with the hit in the database. Additional annotations were made using the Clusters of Orthologous Groups (COG) (22) and TIGRfam (23) databases, which provide high-quality annotations. Functions for these were also assigned if the protein had >80% coverage both on the gene model and on the query protein and an E value of 1e−3. The RAST server (24) provides automated protein functional annotations, and pathway analysis for complete genomes, performed using FIGfams sequences, was also used to annotate all the assembled genomes.
Identification of bona fide contigs in each phylum and genome reconstruction.
Only contigs that were longer than 10 kb were used in the genome reconstructions. A contig was considered to belong to a phylum if a majority of its genes gave best BLAST hits to that phylum. Within each phylum, contigs were grouped using multiple parameters of taxonomy, principal component analysis of tetranucleotide frequencies, percent GC content, and coverage in three metagenomes, as described previously (25–27). Tetranucleotide frequencies were computed using the wordfreq program in the EMBOSS package (28). Principal component analysis was performed using the FactoMineR package in R (29).
Metagenomic recruitment.
To avoid bias in recruitment results owing to the presence of highly related rRNA sequences, as a first step we masked the rRNA sequences from both the genomes and the metagenomes. After masking, recruitments were performed using the BLASTN program (30), and a hit was considered only when it was at least 50 nucleotides long, the sequence identity was >95%, and the E value was ≤1e−5. These hits were used to compute RPKG values (the number of reads recruited per kilobase of genome per gigabase of metagenome), which provide a normalized value that is comparable across different metagenomes.
ANI calculation, genome size estimation, and genomic phylogenetic tree construction.
The average nucleotide identity (ANI) was calculated as it is defined elsewhere (31). Two sets of previously described genes, one with 35 single-copy orthologous genes (32) and another with 112 essential genes (25) found in bacteria, were used to estimate genome completeness for the bacterial genomes. For the Archaea, a set of 53 highly conserved core gene functions which are universally present in all archaeal genomes on the basis of COG annotations were used (33). To create whole-genome phylogenies, conserved proteins in the reconstructed genomes and the reference genomes were identified using the COG database (22), the sequences of these proteins were concatenated and aligned using Kalign software (34), and the alignment was trimmed using the trimAL tool (35). A maximum likelihood tree was constructed with the FastTree2 program (36), using a JTT+CAT model, a gamma approximation, and 100 bootstrap replicates.
Accession numbers.
The metagenomic data sets have been submitted to NCBI SRA and are accessible under BioProject accession number PRJNA279271. The assembled genome sequences have been deposited in the DDBJ/EMBL/GenBank database and can be accessed using the accession numbers LFEM00000000, LFEN00000000, LFEO00000000, LFEP00000000, LFEQ00000000, LFER00000000, LFES00000000, LFET00000000, LFEU00000000, and LFEV00000000 for Alphaproteobacteria genomes Caspian-Alpha1 to Caspian-Alpha10, respectively; LFEW00000000, LFEX00000000, LFEY00000000, and LFEZ00000000 for Thaumarchaeota genomes Caspian-Thauma1 to Caspian-Thauma4, respectively; and LFFA00000000, LFFB00000000, LFFC00000000, LFFD00000000, LFFE00000000, LFFF00000000, LFFG00000000, LFFH00000000, LFFI00000000, LFFJ00000000, LFFK00000000, and LFFL00000000 for Actinobacteria genomes Caspian-Actino1 to Caspian-Actino12, respectively.
RESULTS
Community structure based on rRNA reads.
We took an offshore sample (13 km from the coast) to minimize anthropogenic impacts. The water column there is relatively rich in biomass and has a Secchi disk reading of 6.5 m. The euphotic zone typically extends to 2 to 2.5 times this depth, what would give an estimated depth of 15 to 20 m for this specific location in the Caspian Sea (see Fig. S1 and Table S1 in the supplemental material for the CTD profile and physicochemical features). According to the analysis of water parameters and biological monitoring of these coastal waters of the southern Caspian Sea, it has been considered a moderately productive water body with a mesotrophic status (37), similar to coastal marine waters.
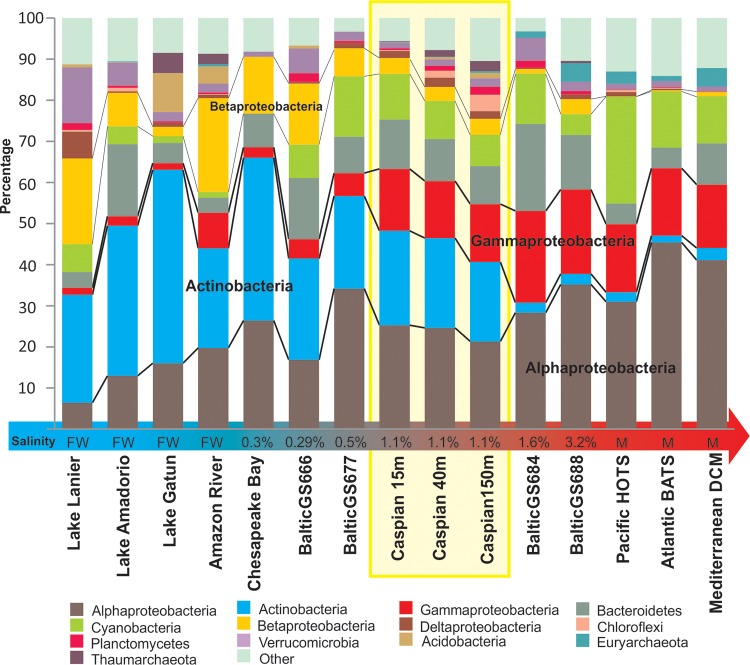
A broad overview of the community structure could be obtained from rRNA reads retrieved from the unassembled data. A total of ca. 44,000, 63,000, and 72,000 reads of 16S rRNA gene fragments were obtained for the Caspian15, Caspian40, and Caspian150 samples, respectively. Alphaproteobacteria, Actinobacteria, Gammaproteobacteria, Bacteroidetes, and Cyanobacteria were the most dominant groups (Fig. 1; see Table S2 in the supplemental material for the numerical values). Overall, the samples from the three depths of the Caspian Sea showed similar profiles at the level of the main phyla, although the numbers of sequences belonging to the Planctomycetes, Verrucomicrobia, Chloroflexi, and Archaea were found to increase with depth. This increase was particularly dramatic for archaeal sequences (nearly all reads belonged to the Thaumarchaea, although a few reads for members of the marine group II [MGII] clade were also present), which comprised barely 1% of all rRNA sequences in the Caspian15 sample but more than 6% of all rRNA sequences in the Caspian150 sample. Remarkably, there was not a major change in the fraction of Cyanobacteria in the three samples, although the depth at which the Caspian150 sample was obtained should be well below the euphotic zone. We also compared the proportions of major phylogenetic groups detected in our samples with those found in selected aquatic environments studied by similar means and covering the entire salinity gradient from freshwater to marine (photic zone only) (Fig. 1). Interestingly, the Caspian Sea data sets showed a combination of phylogenetic groups similar to the combinations found in marine and freshwater communities. The previously described variation from the dominance of Actinobacteria (7) in freshwater to Alphaproteobacteria in marine water (38) is apparent in Fig. 1. The known decrease in Betaproteobacteria from freshwater toward marine environments (7, 39) is also evident. There seemed to be a sharp shift in the microbial community composition over a salinity range of 1 to 1.6%, particularly regarding the presence of Actinobacteria, which makes the southern Caspian Sea, which has a stable 1.1 to 1.2% salinity, a good model for this major transition between marine and freshwater environments. In addition to this gross change in the proportions of the major phyla, it is also known that there are even more dramatic changes at the level of fine-grained diversity, i.e., genera and species that are found only in freshwater or seawater habitats. A paradigmatic example is the SAR11 group of Alphaproteobacteria, which has marine water representatives (Pelagibacter) and freshwater ones (the LD12 clade) (40). To study the microbes present in the Caspian Sea at a finer level, we used assembly to generate fragments of some of the predominant genomes and to get a more refined classification of the microbes present.
FIG 1.
Prokaryotic community structure based on 16S rRNA gene reads from unassembled data sets from the Caspian Sea depth profile in comparison to that based on 16S rRNA gene reads from freshwater, marine water, and brackish water data sets. Marine photic zone data sets include HOTS (Hawaii Ocean Time-series; depth, 25 m), BATS (Bermuda Atlantic Time-series; depth, 20 m), and the Mediterranean DCM (deep chlorophyll maximum; depth, 55 m) (68, 83, 99), and freshwater data sets include those for Lake Lanier, Lake Amadorio, and Lake Gatun together with the Amazon River (81, 100–102). For intermediate salinities, Chesapeake Bay (101) and Baltic Sea (5) salinity gradient data sets were used. The salinity of brackish habitats is indicated at the base of each column (FW, freshwater [0% salinity]; M, marine water [ca. 3.5% salinity]).
Assembled genomes as a first glimpse at Caspian Sea microbes.
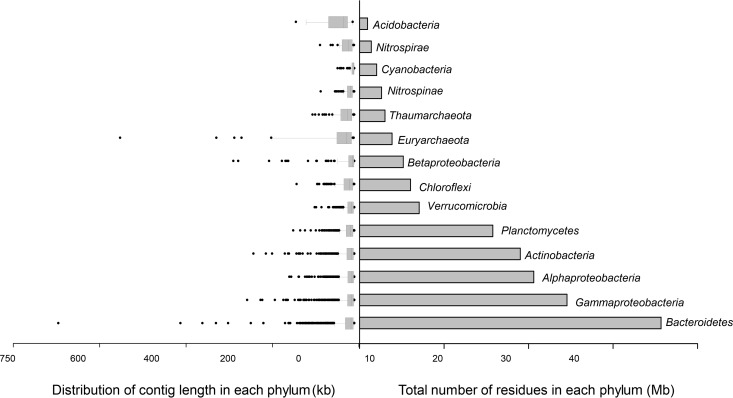
In order to obtain larger contigs, all three data sets were assembled together. Thus, approximately 9,000 contigs longer than 10 kb were obtained and classified after annotation. Contigs of similar genomic parameters were binned together (see Materials and Methods). The contigs fell into 12 different phyla of bacteria and archaea (Fig. 2). In our own experience, assembly favors taxa with a lower level of complexity at the level of fine-grained diversity, and therefore, the presence of long contigs for a group is not directly related to its abundance. However, although the relative proportions (as depicted in Fig. 1) changed, the phyla found by assembly were the same as those found by 16S rRNA gene fragment classification, which (together with metagenomic recruitment; see below) supports the environmental relevance of the microbes represented by the scaffolds described here. The highest total number of mega-base pairs assembled and the longest contigs were for the Bacteroidetes (Data Set S1 in the supplemental material provides the annotation of the largest contig). Bacteroidetes are the most abundant group of bacteria in the ocean after Proteobacteria and Cyanobacteria (41–43), and almost 16 different Bacteroidetes genomes could be separated from the contigs assembled from the Caspian Sea. They belonged to the families Flavobacteriaceae, Cryomorphaceae, Sphingobacteriaceae, Chitinophagaceae, and Rhodothermaceae. The genome fragments retrieved belonged to representatives of novel taxa with low values for the average nucleotide identity (ANI) to the closest relatives (less than 70%), which suggests that they belong to completely new genera within these families. The next most abundant group by the number of assembled contigs was the Gammaproteobacteria, for which a total of almost 25 Mb of assembled sequence fell into six different genomes according to our binning approach. Among them, the highest recruitment value was observed for genomes related to the SAR86 clade, which is considered a marine gammaproteobacterial group (44). However, a marine SAR86 isolate (45) did not recruit from the Caspian raw reads (see Fig. S2 in the supplemental material). The sequences of other genome fragments were more similar to those of different clades of oligotrophic marine Gammaproteobacteria (OMG), like SAR92 and OM60. OMG includes physiologically diverse heterotrophs (46). Members of this group could be detected globally along the ocean euphotic zone and comprise a significant portion of the community in some marine locations (47, 48). They seemed to be abundant in the Caspian Sea samples examined, as deduced from the 16S rRNA gene-related sequences as well.
FIG 2.
Different phyla in the assembled contigs of the Caspian Sea data sets. (Left) Distribution of the contig lengths (in kilobases) for each phylum; (right) the total length of the assembled contigs (in megabases). The data are sorted in increasing order from top to bottom (the minimum total length of the assembled sequences to the maximum total length). The data for the phylum Proteobacteria were further separated at the class level.
Betaproteobacteria are among the highly dominant microbial groups in freshwater environments (1). Genomes belonging to the Methylophilaceae and Hydrogenophilaceae betaproteobacterial families were assembled from the Caspian reads. The family Methylophilaceae includes clade OM43, which comprises uncultured aerobic and obligate methylotrophic Betaproteobacteria (49). The genome fragments of the Methylophilaceae assembled from the Caspian Sea belonged to clade OM43.
Cyanobacteria are the main primary producers in both marine and freshwater environments (50, 51). According to our 16S rRNA gene analysis, the Cyanobacteria were one of the most abundant microbial groups in the Caspian microbial community. However, only 2 Mb of assembled sequences belonged to this phylum, and they fell into three separate bins (data not shown). The sequences of two of these genomes showed the highest similarity to sequences of the genus Cyanobium, which is a member of family Synechococcaceae and which has a mixed marine water and freshwater distribution (52, 53). This genus has also been found to be the predominant picocyanobacterium in the Baltic Sea (54). The other genome was similar to that of the genus Leptolyngbya, which is a filamentous cyanobacterium with a broad distribution as well (55–57). There were very few large contigs related to Synechococcus among our assembled contigs of Caspian Cyanobacteria, although on the basis of recruitment analysis and the sequences of the 16S rRNA gene fragments, representatives of this genus were present in our Caspian metagenomes. Specifically, Synechococcus sp. strain CB0205 from picocyanobacteria subcluster 5.2 (58) showed a very high (100%) fragment recruitment from the Caspian metagenomes. On the other hand, Prochlorococcus, the main component of the picocyanobacterial populations in temperate and tropical oligotrophic marine waters, was absent from our samples (see Fig. S3 in the supplemental material). For a more in-depth analysis, we focus on the genomes of the Alphaproteobacteria, Actinobacteria, and Thaumarchaeota retrieved from the Caspian Sea. These are groups extensively studied in both marine and freshwater ecosystems and contain clades that are characteristic of both.
Alphaproteobacteria.
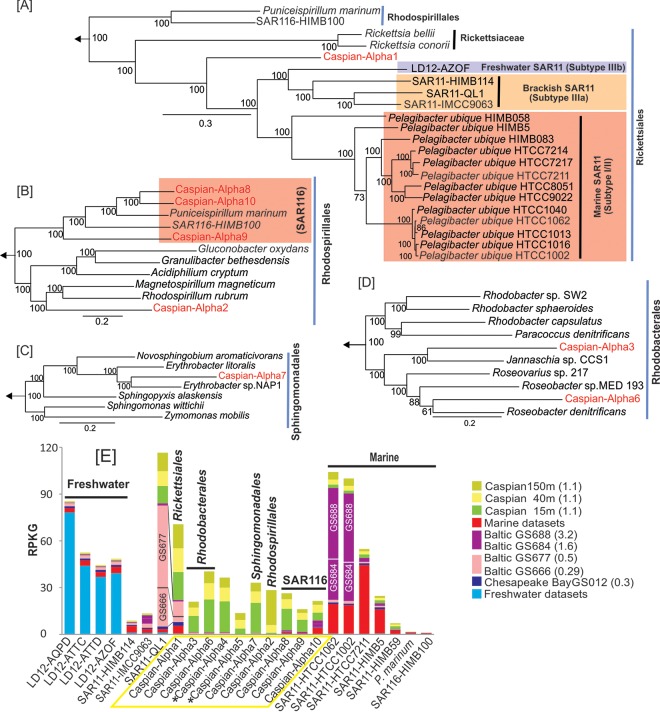
The phylum Alphaproteobacteria contains the SAR11 clade, the members of which are the most abundant microbes in the ocean (59). Within the SAR11 clade there are marine and freshwater representatives. The freshwater clade, referred to as LD12, corresponds to subtype IIIb of SAR11 (40), while SAR11 I and SAR11 II are considered to be offshore marine microbes and SAR11 IIIa comprises coastal water (SAR11-HIMB114) and mesohaline water (SAR11-IMCC9063) representatives (60, 61). A partial genome was recently reconstructed from a metagenome from brackish Lake Qinghai in China (62). This draft genome, which was identified as SAR11-QL1, belongs to rRNA group SAR11 IIIa. We performed metagenomic recruitments with all these representative genomes (see Fig. S4 in the supplemental material) and found a significant recruitment at the same species similarity level (above 95%) only for the brackish water SAR11-QL1 genome. Neither freshwater nor marine water representatives had any significant recruitment. On the other hand, the SAR11 clade made up the largest portion of the bacterial community in all three Caspian Sea data sets, as quantified by the numbers of 16S rRNA gene fragments recovered, which is similar to the findings for pristine marine environments (63). In addition, almost 20 Mb of assembled contigs belonged to the Alphaproteobacteria. They could be assigned to 10 different genomic bins (see Materials and Methods) (Table 1). Although these contig clusters could originate from multiple, closely related organisms (as in any metagenomic assembly), we refer to them as “genomes” for the sake of simplicity. They were classified into the orders Rickettsiales (like SAR11 or LD12), Rhodobacterales, Rhodospirillales, Sphingomonadales, and the SAR116 clade on the grounds of a phylogenetic tree of concatenated proteins (Fig. 3). The trees were built separately to maximize the numbers of genes used in the alignment. Caspian-Alpha1, the closest to the SAR11 clade, was the most abundant group of Alphaproteobacteria in the Caspian Sea on the basis of overall coverage estimates. To establish the phylogenomic placement of this genome, 190 concatenated genes of 24 genomes of related isolates and a single-cell amplified genome (SAG) of the freshwater LD12 clade (64) were included in the tree. Although additional LD12 clade SAGs were available at the time of the analysis, they were too small for retrieval of the 190 genes used for the alignment. In the tree shown in Fig. 3A, aquatic Rickettsiales microbes appeared to have a pattern of adaptation to salinity, with the marine SAR11 clade in subtype I/II, the freshwater LD12 clade in subtype IIIb, and the brackish genomes in subtype IIIa forming separate clusters. Caspian-Alpha1 belongs to the order Rickettsiales, but it branched a long distance from the marine “Candidatus Pelagibacter ubique” representatives and from the genomes from brackish water or freshwater as well. Caspian-Alpha1 contained 213 contigs with a total size of 3.3 Mb and a GC content of 31.3%. This genome showed low values of ANI (less than 76%) to any other available genomes in the order Rickettsiales and clade SAR116. This set of contigs appears to be comprised of 3 closely related organisms which could not be separated from each other by the binning approach used. Genome size estimates suggested a size of ca. 1.14 Mb for Caspian-Alpha1. The median intergenic spacer size of 11 bp (see Fig. S5 in the supplemental material) was also consistent with a small highly streamlined genome. We also found three proteorhodopsin genes in these contigs, what indicates a likely photoheterotrophic metabolism. Like in all the other aquatic Rickettsiales, there was no indication of the presence of motility or chemotaxis genes, indicating a planktonic, free-living lifestyle (65, 66).
TABLE 1.
Statistics for reconstructed Alphaproteobacteria, Actinobacteria, and Thaumarchaeota genomesa
| Genome | No. of contigs | Total bin size (Mb) | % GC content | No. of CDSs | Completeness (%) | No. of genomes | Estimated genome size (Mb) | Protein(s) encoded by or function of featured gene(s) |
|---|---|---|---|---|---|---|---|---|
| Rickettsiales, Alpha1 | 213 | 3.3 | 31.3 | 3,496 | 97.2 | 3 | 1.14 | Proteorhodopsin |
| Rhodobacterales | ||||||||
| Alpha3 | 81 | 1.2 | 61.8 | 1,229 | 1 | |||
| Alpha6 | 68 | 3 | 49.7 | 2,916 | 88.6 | 1 | 3.4 | CO2 fixation in Calvin cycle |
| Sphingomonadales, Alpha7 | 50 | 1.1 | 56.7 | 1,121 | 1 | Proteorhodopsin | ||
| Rhodospirillales | ||||||||
| Alpha2 | 84 | 1.8 | 58.2 | 1,819 | 88.6 | 1 | 2 | Inorganic sulfur assimilation |
| SAR116 | ||||||||
| Alpha8 | 39 | 1.7 | 52.1 | 1,684 | 80 | 1 | 2.2 | Proteorhodopsin, CODH |
| Alpha9 | 85 | 2.2 | 47.6 | 2,165 | 95 | 2 | 1.16 | Proteorhodopsin, CODH |
| Alpha10 | 85 | 3.3 | 50.8 | 3,206 | 2 | Proteorhodopsin, CODH | ||
| acIB lineage | ||||||||
| Actino1 | 94 | 2.5 | 44.3 | 2,532 | 74.8 | 2 | 1.7 | |
| Acidimicrobidae | ||||||||
| Actino2 | 82 | 1.6 | 56.2 | 1,610 | 57 | 1 | 2.8 | |
| Actino5 | 32 | 1.9 | 55.9 | 1,897 | 73 | 1 | 2.6 | Acidirhodopsin |
| Actino8 | 51 | 1 | 63 | 1,029 | 50.5 | 1 | 2 | |
| Actino9 | 155 | 5.7 | 63.4 | 5,665 | 74.8 | 3 | 2.5 | Acidirhodopsin |
| Mycobacteriaceae, Actino6 | 119 | 2.05 | 67.6 | 2,014 | 50 | 1 | 4.1 | |
| Actinomarinales, Actino12 | 82 | 2.1 | 31.3 | 2,376 | 68.5 | 3 | 1.03 | MACrhodopsin |
| Nitrosopumilus | ||||||||
| Thauma1 | 29 | 1.04 | 30.3 | 1,321 | 90.5 | 1 | 1.15 | NH4+ transporter, AmoBC, urease, UreEFGD, NorQ |
| Thauma3 | 31 | 1.2 | 32.8 | 1,501 | 92.5 | 1 | 1.32 | NH4+ transporter, AmoBC, urease, UreEFGD, NorQ |
| Nitrosopelagicus, Thauma4 | 25 | 0.66 | 32.5 | 832 | 77.4 | 1 | 0.85 | AmoBC |
Data for groups smaller than 500 kb are not shown. Genome completeness values are shown for genomes with greater than 50% completeness. Genomes size estimation was performed for genomes with more than 50% completeness. All the genomes described here originated from the Caspian Sea, and the prefix Caspian has been omitted from the names. CDS, coding sequence; CODH, carbon monoxide dehydrogenase; AmoBC, ammonia monooxygenase subunits B and C; UreEFGD, urease accessory proteins; NorQ, nitric oxide reductase activation protein.
FIG 3.
Phylogeny of the genomes belonging to the phylum Alphaproteobacteria in the Caspian Sea and their metagenomic recruitments across diverse aquatic habitats. (A to D) Maximum likelihood phylogenies, obtained using a concatenation of conserved proteins, of Caspian Sea genomes and their close relatives in the orders Rickettsiales (190 proteins) (A), Rhodospirillales (377 proteins) (B), Sphingomonadales (167 proteins) (C), and Rhodobacterales (D). Genomes from the class Gammaproteobacteria were used to root these trees. The Caspian Sea genomes are shown in red. Bootstrap values (in percent) are indicated at each node. (E) Metagenomic recruitment of Caspian Sea alphaproteobacterial genomes and related reference genomes in different data sets. The data sets used are described in Fig. 1. (Left) Genomes of organisms of freshwater origin; (middle) genomes of organisms of brackish water origin; (right) genomes of organisms of marine water origin. The salinity of the samples is indicated in parentheses. An asterisk next to a Caspian Sea genome indicates a small genome (<500 kb) that could be classified only as belonging to the Alphaproteobacteria and no further.
A phylogenetic tree of 377 concatenated genes placed the Caspian-Alpha2, -8, -9, and -10 reconstructed genomes in the order Rhodospirillales, which contains the marine clade SAR116 (Fig. 3B). SAR116 is widespread in marine surface samples on the basis of 16S rRNA gene amplification (67) and metagenomic (68) studies. Caspian-Alpha8, -9, and -10 formed a monophyletic clade with the reference genomes of SAR116, HIMB100, and the isolate Puniceispirillum marinum IMCC1322. The Caspian SAR116 clade representatives had genes encoding proteins that are characteristic of those encoded by SAR116 reference genomes and that are of putative biogeochemical importance in the ocean surface, like proteorhodopsin, carbon monoxide dehydrogenase, and proteins involved in C1 compound metabolism (69). However, no genes encoding dimethylsulfoniopropionate (DMSP) demethylase could be found in these genomes, although the gene for DMSP demethylase has been present in all reference SAR116 genomes described until now (69, 70). Caspian-Alpha2, the other member of the order Rhodospirillales detected, was more closely related to freshwater aquatic bacteria, such as Rhodospirillum and Magnetospirillum (71–76).
The Roseobacter clade is abundant in marine environments and could comprise up to 25% of the microbial community in some of them (77). Caspian-Alpha3 and -6 belonged to this clade (Fig. 3D). The Caspian-Alpha6 genome carried genes for aerobic anoxygenic photosynthesis, which is a common feature of these organisms (78). Caspian-Alpha7 could be assigned to the order Sphingomonadales (Fig. 3C), clustering with marine photoheterotrophic Erythrobacter isolates (79).
To assess the presence of reference genomes in the Caspian Sea and the Caspian Sea contigs in aquatic environments of different salinities (including the Caspian Sea itself), we selected metagenomic data sets for environments with salinities ranging from those in freshwater to those in marine water and some reference data sets for brackish water (the Chesapeake Bay estuary and the Baltic Sea salinity gradient) to carry out recruitment analysis (Fig. 3E). The Caspian Sea contigs were mostly recruited from samples from the Caspian Sea, while LD12 SAGs recruited well from the freshwater data sets but not from the marine or brackish water data sets, and the marine water SAR11 genomes of marine subtype I/II recruited well from marine environments and the Baltic Sea salinity gradient with salinities of greater than 1.6%. Caspian-Alpha1 and Caspian-Alpha10 were the only Caspian Sea contigs that recruited from outside the Caspian Sea data sets. On the other hand, SAR11-QL1, the SAR11 IIIa subtype assembled from a brackish lake in China (62), showed a cosmopolitan distribution in brackish waters, including Baltic Sea samples of less than 1.6% salinity and the Caspian Sea samples, but it did not recruit from freshwater or marine environments. The Caspian-Alpha1 genome, a very distant relative judging from the concatenated tree, showed a recruitment pattern similar to that of SAR11-QL1.
Actinobacteria.
Actinobacteria are considered to be the most abundant freshwater bacteria (80–82), they decrease in prevalence as salinity increases, and the members of this phylum comprise a small portion of the community in marine environments (66, 83). In addition to this overall trend at the phylum level, the lower-level actinobacterial clades change dramatically from freshwater to marine habitats. Thus, in freshwater environments, the most abundant lineages are acI (a member of the Actinomycetales) and acIV (a member of the Acidimicrobiales) (84), while in marine environments, only representatives of the orders “Candidatus Actinomarinales” (66) and Acidimicrobiales (83) have been found. The proportion of Actinobacteria analyzed in the Caspian Sea samples was similar to that analyzed in samples from freshwater environments (Fig. 1).
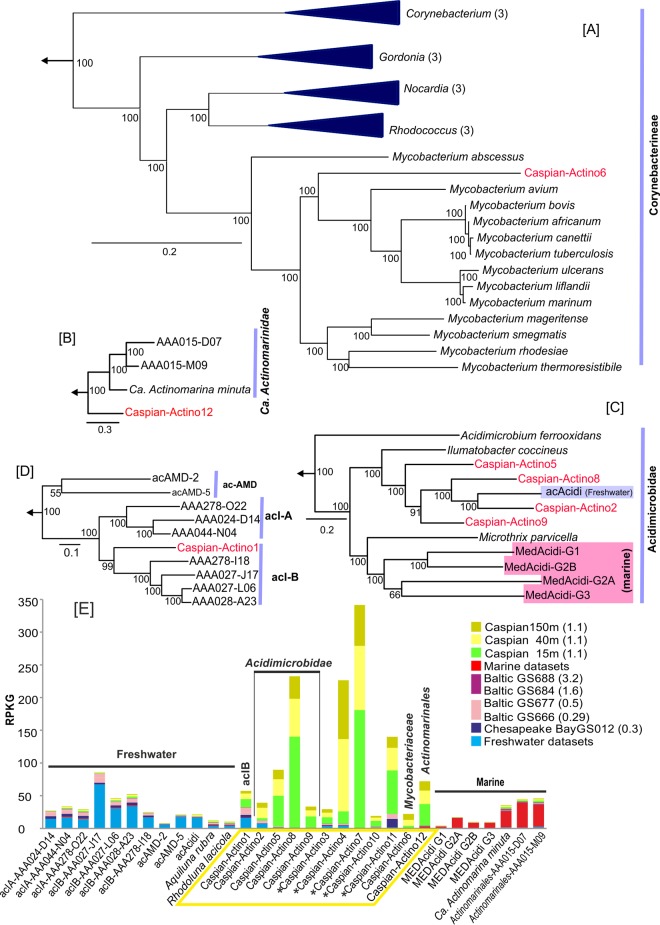
The assembled contigs of Caspian Actinobacteria could be separated into 12 different genomes (Table 1). We found representatives of the ac lineages (freshwater) and the “Candidatus Actinomarinidae” (marine) and Acidimicrobidae subclasses. The Caspian-Actino2, -5, -8, and -9 genomes belong to the Acidimicrobidae subclass, which comprises both freshwater (81) and marine (83) representatives. On the basis of a phylogenetic tree of 77 concatenated proteins, the Caspian Sea genomes clustered with freshwater representatives of this subclass (Fig. 4C). The Caspian-Actino5 and -9 genomes contained genes for rhodopsins that could be classified as acidirhodopsins similar to those encoded by the genomes of marine Acidimicrobidae (83). Caspian-Actino1 clustered near the acIB lineage on the grounds of 165 concatenated genes (Fig. 4D). This genome showed values of ANI to freshwater members of the acI lineage of less than 70%, so the similarity was at the level of a different genus or even family.
FIG 4.
Phylogeny of the genomes belonging to the phylum Actinobacteria in the Caspian Sea and their metagenomic recruitment across diverse aquatic habitats. (A to D) Maximum likelihood phylogenies, obtained using a concatenation of conserved proteins, of Caspian Sea genomes and their close relatives in the suborder Corynebacterineae (226 proteins) (A), the class “Ca. Actinomarinidae” (100 proteins) (B), the subclass Acidimicrobidae (77 proteins) (C), and the acI lineage in the order Actinomycetales (165 proteins) (D). Some nodes have been collapsed for simplicity, and the number of collapsed genomes at the node is indicated in parentheses. Genomes from the Caspian Sea are shown in red, and genomes from marine water and freshwater are highlighted in blue and pink boxes, respectively. Genomes from the order Actinomycetales were used to root these trees. Bootstrap values (in percent) are indicated at each node. (E) Metagenomic recruitment of Caspian actinobacterial genomes and related reference genomes against different data sets. The data sets used are described in Fig. 1. (Left) Genomes of organisms of freshwater origin; (middle) genomes of organisms of brackish water origin; (right) genomes of organisms of marine water origin. The salinity of the samples is indicated in parentheses. An asterisk next to a Caspian Sea genome indicates a small genome (<500 kb) that could be classified only as belonging to the Alphaproteobacteria and no further.
The subclass “Ca. Actinomarinidae” has been described to be a group that is widespread in marine environments (66), and Caspian-Actino12 could be assigned to this group (Fig. 4B). The 82 contigs of this genome had a GC content of 31.3%, and it was calculated to be 68% complete. The values of the ANI to the few “Ca. Actinomarinidae” genomes available, one metagenome assembly and two SAGs (66, 85), were rather low (less than 72%). The estimated genome size of Caspian-Actino12 was about 1 Mb, and it contained a rhodopsin gene with ∼70% similarity to the “Candidatus Actinomarina minuta” MACrhodopsin gene (66). We report here a brackish water representative of this recently described class, whose best-known representative, “Ca. Actinomarina minuta,” is considered to be the smallest free-living microbe (according to both its cell size and its genome size) described so far (66).
Caspian-Actino6 belongs to the family Mycobacteriaceae in the Corynebacterineae subclass (Fig. 4A). It is unique in that it is the first assembled genome of this family to be derived from environmental shotgun sequences. The estimated complete genome size of this organism was 4.1 Mb, and the contigs provided a genome that was almost 50% complete and that had low values (less than 73%) of ANI to all other members of the family.
Selected freshwater and marine genomes together with our contigs were used for recruitment from the selected freshwater, marine water, and brackish water data sets (Fig. 4E). The freshwater genomes in the ac group recruited well from freshwater environments and showed some recruitment from environments with salinities of less than 0.5%. The same pattern of recruitment could be observed among the Caspian actinobacterial genomes. The genomes reconstructed from the Caspian Sea, like those of the Alphaproteobacteria (see above), recruited very little outside this water body. However, Caspian-Actino12 recruited low but significant numbers of reads from marine metagenomes, and reciprocally, the marine “Candidatus Actinomarinales” recruited small but significant amounts from the Caspian Sea. Caspian-Actino3, -4, -7, -10, and -11 could not be classified because of their small size in the binned contigs, but they were used for recruitment (Fig. 4E). Caspian-Actino11 recruited well from the Chesapeake Bay, Baltic Sea (with 0.5% salinity), and Caspian Sea metagenomes and could be representative of a cosmopolitan brackish water actinobacterial genome. Unfortunately, the size of the assemblies for this group (ca. 300 kb) prevented a precise phylogenetic affiliation from being made. Genomes Caspian-Actino5 to -10 are all Caspian Sea specific, with the different recruitments from different depths representing their vertical adaptation in the water column (see below).
Thaumarchaeota.
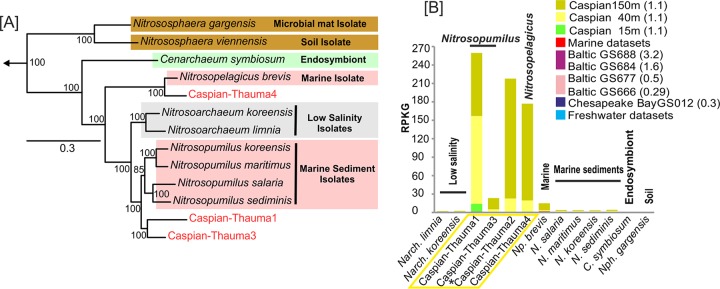
Thaumarchaeota are among the most abundant Archaea in aquatic and terrestrial environments (86). All characterized members of this phylum are chemolithotrophs oxidizing ammonia aerobically to nitrite. Accordingly, they are also known as ammonia-oxidizing archaea (AOA) (87). So far there are three alternative pathways speculated for ammonia oxidation in AOA on the basis of research with Nitrosopumilus maritimus (88). In two of the suggested pathways, ammonia oxidation by archaeal ammonia monooxygenase would result in hydroxylamine, and the two pathways differ in the origin of electrons required to initiate ammonia oxidation by the monooxygenase. The third pathway considers nitroxyl to be the immediate product of the archaeal ammonia monooxygenase. Owing to the fact that the immediate product of archaeal ammonia monooxygenase has not yet been demonstrated, the archaeal ammonia oxidation pathway could not be considered to be fully resolved (88). The assembled contigs affiliated with Thaumarchaeota in the Caspian Sea were separated into four different genomes (Fig. 5A). Caspian-Thauma1 and -3 had low values (less than 82%) of ANI to different species of the genus Nitrosopumilus, consistent with them belonging to new species within this genus. Both of these genomes contained ammonia monooxygenase (AmoA), an AmoB-like protein, and ammonium transporter genes, confirming that these genomes are also AOA. Both genomes were estimated to be almost 90% complete on the basis of the presence of 53 core archaeal genes (33). Caspian-Thauma1 and -3 have GC contents of 30.3 and 32.8%, respectively, and predicted sizes of 1 and 1.2 Mb, respectively. The other two sets of contigs were smaller, with sizes of only 84.8 kb and 660 kb being found for Caspian-Thauma2 and -4, respectively, and GC contents of about 32.5% being detected. They both had values of ANI to the single available representative of “Candidatus Nitrosopelagicus brevis” CN25 of about 86% (89). The phylogenetic tree of 164 concatenated genes confirmed that Caspian-Thauma4 clearly belongs to the genus Nitrosopelagicus (Fig. 5A). This microbe has been recovered in pure culture from oligotrophic marine waters and is highly streamlined, with a genome size of only 1.23 Mb (89). The Caspian-Thauma4 genome appeared to be 75 to 80% complete on the basis of the presence of 53 core archaeal genes, which would indicate a very small and streamlined genome as well (see Fig. S6 in the supplemental material). The genes required for ammonia oxidation were also found in Caspian-Thauma4.
FIG 5.
Phylogeny of the genomes belonging to the phylum Thaumarchaeota in the Caspian Sea and their metagenomic recruitment across aquatic data sets. (A) Maximum likelihood phylogeny obtained using a concatenation of 164 conserved proteins of Caspian Sea genomes in the phylum Thaumarchaeota and related genomes. Sequences from the Euryarchaeota were used to root the tree. The Caspian Sea groups are highlighted in red. Bootstrap values (in percent) are indicated at each node. (B) Metagenomic recruitment of Caspian Sea thaumarchaeal genomes and related reference genomes against different data sets. The data sets used are described in Fig. 1. The genomes of organisms from various habitats are shown. The salinity of the samples is indicated in parentheses. An asterisk next to a Caspian Sea genome indicates a small genome (<500 kb) that could be classified only as belonging to the thaumarchaea and no further.
There is now an abundant representation of AOA from multiple environments. Nitrosoarchaeum limnia BD20 is an AOA that was obtained from low-salinity samples of the San Francisco Bay estuary (90, 91), and two different strains of this species have been sequenced by single-cell genomics and metagenomic assembly of enrichment cultures. Nitrosoarchaeum koreensis is another isolate also from a low-salinity rhizosphere. Nitrososphaera gargensis is a microbial mat isolate, and other genomes are considered to be those of marine sediment isolates, except for the genome of “Ca. Nitrosopelagicus brevis,” which is a truly pelagic marine isolate (89, 92, 93). All these genomes were used for recruitment with the data sets, and the results are shown in Fig. 5B. Of the reference genomes, only “Ca. Nitrosopelagicus brevis” showed some recruitment from the deep-water data set of the Caspian Sea (see Fig. S7 in the supplemental material). The other genomes did not recruit from any depth. Caspian thaumarchaeal genomes showed remarkably higher levels of recruitment from the deeper-water data sets of the Caspian Sea metagenomes, indicating a preference for deeper waters. Caspian thaumarchaeal assemblies did not recruit from other environments and could be considered Caspian Sea-specific AOAs that might have an important role in the nitrogen cycle of this endorheic basin.
Depth-variable recruitment of the reconstructed genomes.
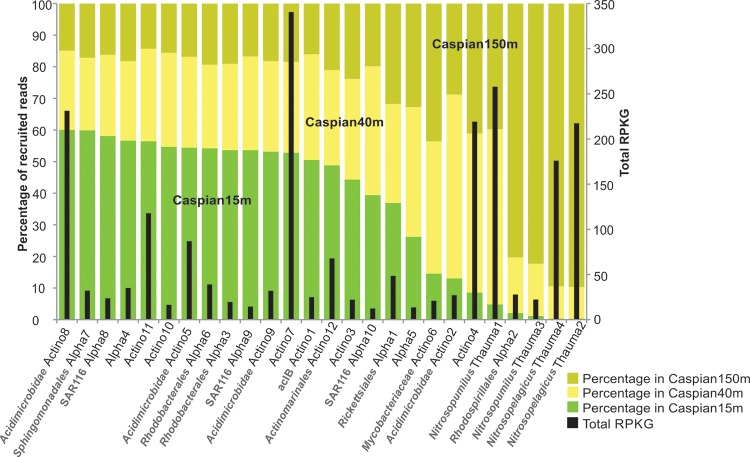
As it was mentioned above, on the basis of the 16S rRNA gene fragment profiles of the three Caspian Sea data sets, the community composition did not change significantly along the depth profile. However, when the assembled genomes of the Alphaproteobacteria, Actinobacteria, and Thaumarchaeota were recruited against the data sets for the three depths, some clear trends were apparent, with the actinobacterial contigs recruiting preferentially from the surface and the thaumarchaeal ones recruiting from the deep (Fig. 6). Among the actinobacterial genomes, Caspian-Actino2, -4, and -6 recruited more often from the deep. Caspian-Actino4 is a small set of contigs (473 kb) which could not be taxonomically affiliated further. The Alphaproteobacteria appeared at all depths, although Caspian-Alpha2 (Rhodospirillales) appeared preferentially in the deeper sample and Caspian-Alpha7 (Sphingomonadales) appeared preferentially at the surface.
FIG 6.
Abundance of genomes of the Alphaproteobacteria, Actinobacteria, and Thaumarchaeota at different depths in the Caspian Sea. Black bars, the total RPKG (number of reads recruited per kilobase of genome per gigabase of metagenome) value for each genome in all three data sets combined. The percentage of reads recruited at each depth is shown separately for each genome. All the genomes for which results are shown originated from the Caspian Sea, and the prefix Caspian has been omitted from the names.
DISCUSSION
As mentioned above, the Caspian Sea is the largest water body on Earth not connected to the ocean. Actually, the long distance to the nearest shore (500 km to the also brackish Black Sea or 700 km to the Persian Gulf) makes the transfer of marine microbes unlikely (although small inoculations might take place sporadically, for example, by microbes carried in fine water particles that travel with weather systems or water birds). The Caspian Sea has inputs from rivers, but they are mostly located at the northern end, very far from the location where our samples were obtained. Therefore, we can consider the microbes described here to be true inhabitants of a brackish water body with minimal terrestrial influence (certainly much less terrestrial influence than the lakes in other studies [94, 95]). The main difference between our Caspian Sea samples and coastal marine waters at a similar latitude is salinity. The animals are also significantly different between those associated with the Caspian Sea and those associated with coastal marine waters (96), but it is doubtful that they would be a major factor determining the microbial community structure. Our results indicate that the brackish waters with salinity in the salinity range of the Caspian Sea actually represent a different habitat with a specific microbiota adjusted to live at this salinity. Actually, the microbes found in the Caspian Sea are found in similar brackish waters and nowhere else. Reciprocally, marine or freshwater microbes are found in the Caspian Sea but always as very minor members of the community. There were a few exceptions, such as Caspian-Actino1, which recruited in freshwater lakes, or marine “Ca. Nitrosopelagicus,” which recruited in the Caspian Sea.
Have the microbes described here evolved specifically in the isolation of the Caspian Sea? The Caspian water body has remained isolated for at least 2 million years, which is a very long time but which is not enough reason to justify the sequence differences found in our reconstructed genomes. In addition, there are clear cases of microbes from distant places, like SAR11-QL1, that appear in the Caspian Sea, recruiting even more than some local assemblies. Besides, the phylogenomic trees indicate that the Caspian taxa are distributed within clades that are also found in the ocean or other aquatic environments. We do not expect that the Caspian microbes described here are really endemic and predict that similar microbes will be found in other medium- to high-salinity brackish water environments worldwide. The apparent exclusivity derives from the still scarce representation of the sequences of microbes from these environments in sequence databases. One important novelty found in this work is the bona fide mycobacterial genome represented by Caspian-Actino6. This seems to be the first case of a well-represented mycobacterial genome in an aquatic environment. There are mycobacterial isolates from marine waters (97, 98), but they are not significantly represented in marine metagenomes. Actually, this genome was among those that recruited only from the Caspian metagenomes.
From a phylogenetic point of view, the Caspian genomes associated with characteristic freshwater groups (as in the case of the Acidimicrobidae) or acIB similarly to the way in which they associated with marine ones, such as the Rhodospirillales or the Rhodobacterales. In the case of the alphaproteobacterium represented by Caspian-Alpha1, it seems to be a separate Caspian branch, which was confirmed by recruiting only from brackish water metagenomes in the salinity range of the Caspian Sea. Still, the main question remains: why is salinity so relevant for the selection of the microbes that comprise aquatic communities (3)? The physiology of transport or respiration can be severely affected by the presence of salts (6), and from this point of view, brackish water environments should be much more similar to marine than freshwater environments, but this does not seem to be the case. More extensive screening of brackish lakes and habitats and in-depth studies of the genomes retrieved, together with the study of isolates of these microbes that we now know to be predominant, will help to clarify this conundrum of biology.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the help of the Research Council of the University of Tehran and also acknowledge the help of the Iranian National Institute for Oceanography and Atmospheric Science (Nowshahr Branch) for the sampling equipment.
This work was supported by a grant from the Iranian Biological Resource Centre (IBRC) (MI-1391-20) (to M. A. Amoozegar) and project MEDIMAX BFPU2013-48007-P of the Spanish Ministerio de Economia y Competitividad, MaCuMBA project 311975 of the European Commission FP7 (FEDER funds supported this project), and PROMETEO II2014/012 project AQUAMET of the Generalitat Valenciana (to F. Rodriguez-Valera).
Funding Statement
This work was supported by a grant from the Iranian Biological Resource Centre (IBRC) (MI-1391-20) (to M. A. Amoozegar) and project MEDIMAX BFPU2013-48007-P of the Spanish Ministerio de Economia y Competitividad, MaCuMBA project 311975 of the European Commission FP7 (FEDER funds supported this project), and PROMETEO II2014/012 project AQUAMET of the Generalitat Valenciana (to F. Rodriguez-Valera).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03381-15.
REFERENCES
- 1.Zwart G, Crump B, Kamst-van Agterveld M, Hagen F, Han S. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol 28:141–155. doi: 10.3354/ame028141. [DOI] [Google Scholar]
- 2.Macleod RA. 1965. The question of the existence of specific marine bacteria. Bacteriol Rev 29:9–24. [PMC free article] [PubMed] [Google Scholar]
- 3.Lozupone CA, Knight R. 2007. Global patterns in bacterial diversity. Proc Natl Acad Sci U S A 104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logares R, Bråte J, Bertilsson S, Clasen JL, Shalchian-Tabrizi K, Rengefors K. 2009. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol 17:414–422. doi: 10.1016/j.tim.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Dupont CL, Larsson J, Yooseph S, Ininbergs K, Goll J, Asplund-Samuelsson J, McCrow JP, Celepli N, Allen LZ, Ekman M, Lucas AJ, Hagström Å Thiagarajan M, Brindefalk B, Richter AR, Andersson AF, Tenney A, Lundin D, Tovchigrechko A, Nylander JAA, Brami D, Badger JH, Allen AE, Rusch DB, Hoffman J, Norrby E, Friedman R, Pinhassi J, Venter JC, Bergman B. 2014. Functional tradeoffs underpin salinity-driven divergence in microbial community composition. PLoS One 9:e89549. doi: 10.1371/journal.pone.0089549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penn K, Jensen PR. 2012. Comparative genomics reveals evidence of marine adaptation in Salinispora species. BMC Genomics 13:86. doi: 10.1186/1471-2164-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirchman DL, Dittel AI, Malmstrom RR, Cottrell MT. 2005. Biogeography of major bacterial groups in the Delaware estuary. Limnol Oceanogr 50:1697–1706. doi: 10.4319/lo.2005.50.5.1697. [DOI] [Google Scholar]
- 8.Campbell BJ, Kirchman DL. 2013. Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J 7:210–220. doi: 10.1038/ismej.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortunato CS, Herfort L, Zuber P, Baptista AM, Crump BC. 2012. Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. ISME J 6:554–563. doi: 10.1038/ismej.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikenaga M, Guevara R, Dean AL, Pisani C, Boyer JN. 2010. Changes in community structure of sediment bacteria along the Florida coastal Everglades marsh-mangrove-seagrass salinity gradient. Microb Ecol 59:284–295. doi: 10.1007/s00248-009-9572-2. [DOI] [PubMed] [Google Scholar]
- 11.Dumont HJ. 1998. The Caspian Lake: history, biota, structure, and function. Limnol Oceanogr 43:44–52. doi: 10.4319/lo.1998.43.1.0044. [DOI] [Google Scholar]
- 12.Cristescu ME, Adamowicz SJ, Vaillant JJ, Haffner DG. 2010. Ancient lakes revisited: from the ecology to the genetics of speciation. Mol Ecol 19:4837–4851. doi: 10.1111/j.1365-294X.2010.04832.x. [DOI] [PubMed] [Google Scholar]
- 13.Jamshidi S, Abu Bakar NB. 2012. Seasonal variations in temperature, salinity and density in the southern coastal waters of the Caspian Sea. Oceanology 52:380–396. doi: 10.1134/S0001437012030034. [DOI] [Google Scholar]
- 14.Martín-Cuadrado A-B, López-García P, Alba J-C, Moreira D, Monticelli L, Strittmatter A, Gottschalk G, Rodríguez-Valera F. 2007. Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS One 2:e914. doi: 10.1371/journal.pone.0000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 17.Nawrocki E. 2009. Structural RNA homology search and alignment using covariance models. Ph.D. dissertation Washington University in St. Louis, St. Louis, MO. [Google Scholar]
- 18.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 19.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Gilna P, Li W. 2009. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25:1338–1340. doi: 10.1093/bioinformatics/btp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, White O. 2001. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res 29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 26.Ghai R, Pašić L, Fernández AB, Martin-Cuadrado A-B, Mizuno CM, McMahon KD, Papke RT, Stepanauskas R, Rodriguez-Brito B, Rohwer F, Sánchez-Porro C, Ventosa A, Rodríguez-Valera F. 2011. New abundant microbial groups in aquatic hypersaline environments. Sci Rep 1:135. doi: 10.1038/srep00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu W-T, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 28.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 29.Le S, Josse J, Husson F. 2008. FactoMineR: an R package for multivariate analysis. J Stat Soft 25:1–18. doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- 30.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P. 2007. Prediction of effective genome size in metagenomic samples. Genome Biol 8:R10. doi: 10.1186/gb-2007-8-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puigbò P, Wolf YI, Koonin EV. 2009. Search for a “tree of life” in the thicket of the phylogenetic forest. J Biol 8:59. doi: 10.1186/jbiol159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lassmann T, Sonnhammer ELL. 2005. Kalign—an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nasrollahzadeh HS, Din ZB, Foong SY, Makhlough A. 2008. Trophic status of the Iranian Caspian Sea based on water quality parameters and phytoplankton diversity. Cont Shelf Res 28:1153–1165. doi: 10.1016/j.csr.2008.02.015. [DOI] [Google Scholar]
- 38.Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I, Somerfield P, Fuhrman JA. Field D. 2012. Defining seasonal marine microbial community dynamics. ISME J 6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouvier TC, del Giorgio PA. 2002. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol Oceanogr 47:453–470. doi: 10.4319/lo.2002.47.2.0453. [DOI] [Google Scholar]
- 40.Salcher MM, Pernthaler J, Posch T. 2011. Seasonal bloom dynamics and ecophysiology of the freshwater sister clade of SAR11 bacteria “that rule the waves” (LD12). ISME J 5:1242–1252. doi: 10.1038/ismej.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bauer M, Kube M, Teeling H, Richter M, Lombardot T, Allers E, Würdemann CA, Quast C, Kuhl H, Knaust F, Woebken D, Bischof K, Mussmann M, Choudhuri JV, Meyer F, Reinhardt R, Amann RI, Glöckner FO. 2006. Whole genome analysis of the marine Bacteroidetes ‘Gramella forsetii’ reveals adaptations to degradation of polymeric organic matter. Environ Microbiol 8:2201–2213. doi: 10.1111/j.1462-2920.2006.01152.x. [DOI] [PubMed] [Google Scholar]
- 42.Fernández-Gómez B, Richter M, Schüler M, Pinhassi J, Acinas SG, González JM, Pedrós-Alió C. 2013. Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J 7:1026–1037. doi: 10.1038/ismej.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirchman D. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol 39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 44.Dupont CL, Rusch DB, Yooseph S, Lombardo M-J, Richter RA, Valas R, Novotny M, Yee-Greenbaum J, Selengut JD, Haft DH, Halpern AL, Lasken RS, Nealson K, Friedman R, Venter JC. 2012. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J 6:1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rusch DB, Novotny M, Brinkac LM, Lasken RS, Dupont CL. 2013. Draft genome sequence of a single cell of SAR86 clade subgroup. Genome Announc 1(1):e00030–12. doi: 10.1128/genomeA.00030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho JC, Giovannoni SJ. 2004. Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl Environ Microbiol 70:432–440. doi: 10.1128/AEM.70.1.432-440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thrash JC, Cho J-C, Ferriera S, Johnson J, Vergin KL, Giovannoni SJ. 2010. Genome sequences of strains HTCC2148 and HTCC2080, belonging to the OM60/NOR5 clade of the Gammaproteobacteria. J Bacteriol 192:3842–3843. doi: 10.1128/JB.00511-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spring S, Riedel T, Spröer C, Yan S, Harder J, Fuchs BM. 2013. Taxonomy and evolution of bacteriochlorophyll a-containing members of the OM60/NOR5 clade of marine Gammaproteobacteria: description of Luminiphilus syltensis gen. nov., sp. nov., reclassification of Haliea rubra as Pseudohaliea rubra gen. nov., comb. nov. BMC Microbiol 13:118. doi: 10.1186/1471-2180-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chistoserdova L, Lidstrom M. 2013. Aerobic methylotrophic prokaryotes, p 267–285. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes. Springer, Berlin, Germany. [Google Scholar]
- 50.Stockner J, Callieri C, Cronberg G. 2002. Picoplankton and other non-bloom-forming cyanobacteria in lakes, p 195–231. In Whitton BS, Potts M (ed), The ecology of cyanobacteria: their diversity in time and space. Springer, Dordrecht, Netherlands. [Google Scholar]
- 51.Scanlan DJ, West NJ. 2002. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol Ecol 40:1–12. doi: 10.1111/j.1574-6941.2002.tb00930.x. [DOI] [PubMed] [Google Scholar]
- 52.Komárek J, Kling H, Komárková J. 2003. Filamentous cyanobacteria, p 117–196. In Wehr JD, Sheath RG (ed), Freshwater algae of North America: ecology and classification (aquatic ecology), 1st ed. Academic Press, Burlington, MA. [Google Scholar]
- 53.Alquezar R, Anastasi A. 2013. The use of the cyanobacteria, Cyanobium sp., as a suitable organism for toxicity testing by flow cytometry. Bull Environ Contam Toxicol 90:684–690. doi: 10.1007/s00128-013-0977-8. [DOI] [PubMed] [Google Scholar]
- 54.Ininbergs K, Bergman B, Larsson J, Ekman M. 2015. Microbial metagenomics in the Baltic Sea: recent advancements and prospects for environmental monitoring. Ambio 44(Suppl 3):S439–S450. doi: 10.1007/s13280-015-0663-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steffen MM, Li Z, Effler TC, Hauser LJ, Boyer GL, Wilhelm SW. 2012. Comparative metagenomics of toxic freshwater cyanobacteria bloom communities on two continents. PLoS One 7:e44002. doi: 10.1371/journal.pone.0044002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Komárek J. 2007. Phenotype diversity of the cyanobacterial genus Leptolyngbya in the maritime Antarctic. Polish Polar Res 28:211–231. [Google Scholar]
- 57.Cai H, Wang K, Huang S, Jiao N, Chen F. 2010. Distinct patterns of picocyanobacterial communities in winter and summer in the Chesapeake Bay. Appl Environ Microbiol 76:2955–2960. doi: 10.1128/AEM.02868-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larsson J, Celepli N, Ininbergs K, Dupont CL, Yooseph S, Bergman B, Ekman M. 2014. Picocyanobacteria containing a novel pigment gene cluster dominate the brackish water Baltic Sea. ISME J 8:1892–1903. doi: 10.1038/ismej.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 60.Oh H-M, Kang I, Lee K, Jang Y, Lim S-I, Cho J-C. 2011. Complete genome sequence of strain IMCC9063, belonging to SAR11 subgroup 3, isolated from the Arctic Ocean. J Bacteriol 193:3379–3380. doi: 10.1128/JB.05033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herlemann DPR, Woelk J, Labrenz M, Jürgens K. 2014. Diversity and abundance of “Pelagibacterales” (SAR11) in the Baltic Sea salinity gradient. Syst Appl Microbiol 37:601–604. doi: 10.1016/j.syapm.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 62.Oh S, Zhang R, Wu QL, Liu W. 2014. Draft genome sequence of a novel SAR11 clade species abundant in a Tibetan lake. Genome Announc 2(6):e01137–14. doi: 10.1128/genomeA.01137-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown MV, Lauro FM, DeMaere MZ, Muir L, Wilkins D, Thomas T, Riddle MJ, Fuhrman JA, Andrews-Pfannkoch C, Hoffman JM, McQuaid JB, Allen A, Rintoul SR, Cavicchioli R. 2012. Global biogeography of SAR11 marine bacteria. Mol Syst Biol 8:595. doi: 10.1038/msb.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaremba-Niedzwiedzka K, Viklund J, Zhao W, Ast J, Sczyrba A, Woyke T, McMahon K, Bertilsson S, Stepanauskas R, Andersson SGE. 2013. Single-cell genomics reveal low recombination frequencies in freshwater bacteria of the SAR11 clade. Genome Biol 14:R130. doi: 10.1186/gb-2013-14-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin-Cuadrado A-B, Garcia-Heredia I, Molto AG, Lopez-Ubeda R, Kimes N, Lopez-Garcia P, Moreira D, Rodriguez-Valera F. 2015. A new class of marine Euryarchaeota group II from the Mediterranean deep chlorophyll maximum. ISME J 9:1619–1634. doi: 10.1038/ismej.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghai R, Mizuno CM, Picazo A, Camacho A, Rodriguez-Valera F. 2013. Metagenomics uncovers a new group of low GC and ultra-small marine Actinobacteria. Sci Rep 3:2471. doi: 10.1038/srep02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rappé M. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol Ecol 33:219–232. doi: 10.1111/j.1574-6941.2000.tb00744.x. [DOI] [PubMed] [Google Scholar]
- 68.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 69.Oh H-M, Kwon KK, Kang I, Kang SG, Lee J-H, Kim S-J, Cho J-C. 2010. Complete genome sequence of “Candidatus Puniceispirillum marinum” IMCC1322, a representative of the SAR116 clade in the Alphaproteobacteria. J Bacteriol 192:3240–3241. doi: 10.1128/JB.00347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grote J, Bayindirli C, Bergauer K, Carpintero de Moraes P, Chen H, D'Ambrosio L, Edwards B, Fernández-Gómez B, Hamisi M, Logares R, Nguyen D, Rii YM, Saeck E, Schutte C, Widner B, Church MJ, Steward GF, Karl DM, Delong EF, Eppley JM, Schuster SC, Kyrpides NC, Rappé MS. 2011. Draft genome sequence of strain HIMB100, a cultured representative of the SAR116 clade of marine Alphaproteobacteria. Stand Genomic Sci 5:269–278. doi: 10.4056/sigs.1854551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reslewic S, Zhou S, Place M, Zhang Y, Briska A, Goldstein S, Churas C, Runnheim R, Forrest D, Lim A, Lapidus A, Han CS, Roberts GP, Schwartz DC. 2005. Whole-genome shotgun optical mapping of Rhodospirillum rubrum. Appl Environ Microbiol 71:5511–5522. doi: 10.1128/AEM.71.9.5511-5522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anil Kumar P, Aparna P, Srinivas TNR, Sasikala C, Ramana CV. 2008. Rhodospirillum sulfurexigens sp. nov., a phototrophic alphaproteobacterium requiring a reduced sulfur source for growth. Int J Syst Evol Microbiol 58:2917–2920. doi: 10.1099/ijs.0.65689-0. [DOI] [PubMed] [Google Scholar]
- 73.Matsunaga T, Okamura Y, Fukuda Y, Wahyudi AT, Murase Y, Takeyama H. 2005. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res 12:157–166. doi: 10.1093/dnares/dsi002. [DOI] [PubMed] [Google Scholar]
- 74.Geelhoed JS, Sorokin DY, Epping E, Tourova TP, Banciu HL, Muyzer G, Stams AJM, van Loosdrecht MCM. 2009. Microbial sulfide oxidation in the oxic-anoxic transition zone of freshwater sediment: involvement of lithoautotrophic Magnetospirillum strain J10. FEMS Microbiol Ecol 70:54–65. doi: 10.1111/j.1574-6941.2009.00739.x. [DOI] [PubMed] [Google Scholar]
- 75.Schleifer KH, Schüler D, Spring S, Weizenegger M, Amann R, Ludwig W, Köhler M. 1991. The genus Magnetospirillum gen. nov. Description of Magnetospirillum gryphiswaldense sp. nov and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb nov. Syst Appl Microbiol 14:379–385. [Google Scholar]
- 76.Favinger J, Stadtwald R, Gest H. 1989. Rhodospirillum centenum, sp. nov., a thermotolerant cyst-forming anoxygenic photosynthetic bacterium. Antonie Van Leeuwenhoek 55:291–296. doi: 10.1007/BF00393857. [DOI] [PubMed] [Google Scholar]
- 77.Wagner-Döbler I, Biebl H. 2006. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol 60:255–280. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- 78.Brinkhoff T, Giebel H-A, Simon M. 2008. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol 189:531–539. doi: 10.1007/s00203-008-0353-y. [DOI] [PubMed] [Google Scholar]
- 79.Koblízek M, Janouskovec J, Oborník M, Johnson JH, Ferriera S, Falkowski PG. 2011. Genome sequence of the marine photoheterotrophic bacterium Erythrobacter sp. strain NAP1. J Bacteriol 193:5881–5882. doi: 10.1128/JB.05845-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ghai R, McMahon KD, Rodriguez-Valera F. 2012. Breaking a paradigm: cosmopolitan and abundant freshwater actinobacteria are low GC. Environ Microbiol Rep 4:29–35. doi: 10.1111/j.1758-2229.2011.00274.x. [DOI] [PubMed] [Google Scholar]
- 81.Ghai R, Mizuno CM, Picazo A, Camacho A, Rodriguez-Valera F. 2014. Key roles for freshwater Actinobacteria revealed by deep metagenomic sequencing. Mol Ecol 23:6073–6090. doi: 10.1111/mec.12985. [DOI] [PubMed] [Google Scholar]
- 82.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. 2011. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mizuno CM, Rodriguez-Valera F, Ghai R. 2015. Genomes of planktonic Acidimicrobiales: widening horizons for marine Actinobacteria by metagenomics. mBio 6:e02083–14. doi: 10.1128/mBio.02083-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Warnecke F, Amann R, Pernthaler J. 2004. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ Microbiol 6:242–253. doi: 10.1111/j.1462-2920.2004.00561.x. [DOI] [PubMed] [Google Scholar]
- 85.Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, González JM, Luo H, Wright JJ, Landry ZC, Hanson NW, Thompson BP, Poulton NJ, Schwientek P, Acinas SG, Giovannoni SJ, Moran MA, Hallam SJ, Cavicchioli R, Woyke T, Stepanauskas R. 2013. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci U S A 110:11463–11468. doi: 10.1073/pnas.1304246110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pester M, Schleper C, Wagner M. 2011. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14:300–306. doi: 10.1016/j.mib.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schleper C, Nicol GW. 2010. Ammonia-oxidising archaea—physiology, ecology and evolution. Adv Microb Physiol 57:1–41. doi: 10.1016/B978-0-12-381045-8.00001-1. [DOI] [PubMed] [Google Scholar]
- 88.Stahl DA, de la Torre JR. 2012. Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol 66:83–101. doi: 10.1146/annurev-micro-092611-150128. [DOI] [PubMed] [Google Scholar]
- 89.Santoro AE, Dupont CL, Richter RA, Craig MT, Carini P, McIlvin MR, Yang Y, Orsi WD, Moran DM, Saito MA. 2015. Genomic and proteomic characterization of “Candidatus Nitrosopelagicus brevis”: an ammonia-oxidizing archaeon from the open ocean. Proc Natl Acad Sci U S A 112:1173–1178. doi: 10.1073/pnas.1416223112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA. 2012. Genome sequence of “Candidatus Nitrosoarchaeum limnia” BG20, a low-salinity ammonia-oxidizing archaeon from the San Francisco Bay estuary. J Bacteriol 194:2119–2120. doi: 10.1128/JB.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. 2011. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 93.Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA. 2012. Genome sequence of “Candidatus Nitrosopumilus salaria” BD31, an ammonia-oxidizing archaeon from the San Francisco Bay estuary. J Bacteriol 194:2121–2122. doi: 10.1128/JB.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu SK, Xie P, Liang GD, Wang SB, Liang XM. 2006. Relationships between microcystins and environmental parameters in 30 subtropical shallow lakes along the Yangtze River, China. Freshwater Biol 51:2309–2319. doi: 10.1111/j.1365-2427.2006.01652.x. [DOI] [Google Scholar]
- 95.Comte J, Lindstrom ES, Eiler A, Langenheder S. 2014. Can marine bacteria be recruited from freshwater sources and the air? ISME J 8:2423–2430. doi: 10.1038/ismej.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mordukhai-Boltovskoi. 1964. Caspian fauna beyond the Caspian Sea. Int Rev Gesamte Hydrobiol Hydrogr 49:139–176. doi: 10.1002/iroh.19640490105. [DOI] [Google Scholar]
- 97.Padgitt P, Moshier S. 1987. Mycobacterium poriferae sp. nov., a scotochromogenic, rapidly growing species isolated from a marine sponge. Int J Syst Bacteriol 37:186–191. doi: 10.1099/00207713-37-3-186. [DOI] [Google Scholar]
- 98.Jacobs J, Rhodes M, Sturgis B, Wood B. 2009. Influence of environmental gradients on the abundance and distribution of Mycobacterium spp. in a coastal lagoon estuary. Appl Environ Microbiol 75:7378–7384. doi: 10.1128/AEM.01900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Coleman ML, Chisholm SW. 2010. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci U S A 107:18634–18639. doi: 10.1073/pnas.1009480107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ghai R, Rodriguez-Valera F, McMahon KD, Toyama D, Rinke R, Cristina Souza de Oliveira T, Wagner Garcia J, Pellon de Miranda F, Henrique-Silva F. 2011. Metagenomics of the water column in the pristine upper course of the Amazon River. PLoS One 6:e23785. doi: 10.1371/journal.pone.0023785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers Y-H, Falcón LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. 2007. The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oh S, Caro-Quintero A, Tsementzi D, DeLeon-Rodriguez N, Luo C, Poretsky R, Konstantinidis KT. 2011. Metagenomic insights into the evolution, function, and complexity of the planktonic microbial community of Lake Lanier, a temperate freshwater ecosystem. Appl Environ Microbiol 77:6000–6011. doi: 10.1128/AEM.00107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.