Abstract
Enterohemorrhagic Escherichia coli (EHEC) is an important cause of diarrhea and hemolytic-uremic syndrome (HUS) worldwide. Australia's worst outbreak of HUS occurred in Adelaide in 1995 and was one of the first major HUS outbreaks attributed to a non-O157 Shiga-toxigenic E. coli (STEC) strain. Molecular analyses conducted at the time suggested that the outbreak was caused by an O111:H− clone, with strains from later in the outbreak harboring an extra copy of the genes encoding the potent Shiga toxin 2 (Stx2). Two decades later, we have used next-generation sequencing to compare two isolates from early and late in this important outbreak. We analyzed genetic content, single-nucleotide polymorphisms (SNPs), and prophage insertion sites; for the latter, we demonstrate how paired-end sequence data can be leveraged to identify such insertion sites. The two strains are genetically identical except for six SNP differences and the presence of not one but two additional Stx2-converting prophages in the later isolate. Isolates from later in the outbreak were associated with higher levels of morbidity, suggesting that the presence of the additional Stx2-converting prophages is significant in terms of the virulence of this clone.
INTRODUCTION
Enterohemorrhagic Escherichia coli (EHEC) is a subgroup of Shiga-toxigenic E. coli (STEC) that causes diarrhea and hemorrhagic colitis in humans and can lead to life-threatening sequelae such as hemolytic-uremic syndrome (HUS) (1, 2). EHEC typically possesses the genes encoding Shiga toxins (stx1 and/or stx2) and enterohemolysin (hly) as well as a large pathogenicity island named the locus of enterocyte effacement (LEE) (3). The LEE harbors genes contributing to pathogenesis, including intimin (eae), which causes the formation of characteristic attaching and effacing legions on the intestinal epithelium (4–6). Following colonization of the gut, the bacteria release Stx into the gut lumen. The toxin is absorbed systemically and targets cells expressing globotriaosylceramide receptors, which are found at high levels on renal tubular epithelial cells and at significant levels on microvascular endothelial cells of the kidney, intestine, pancreas, and brain (2, 7). Stx is directly responsible for the pathological features observed in disease, with Stx2 in particular being highly potent (8, 9). Stx is an AB5 toxin with the A and B subunit genes carried by lambda-like phages and therefore easily disseminated (10, 11). Lambda-like prophages are genetic mosaics and form an important component of the EHEC genome, with the first EHEC strain to be fully sequenced (O157:H7 isolate Sakai) being shown to contain 13 lambda-like prophages (12, 13).
Numerous EHEC/STEC serotypes have been associated with HUS. The prototypic EHEC serotype O157:H7 is undoubtedly the most notorious cause of HUS outbreaks, but in the 1990s, O111 strains emerged as another important contributor (14, 15). The worst outbreak of HUS in Australia occurred in Adelaide, South Australia, from 4 January through 20 February 1995 and was attributed to dry fermented sausage (mettwurst) contaminated with EHEC O111:H− (15, 16). The isolates were highly virulent, with the infectious dose being calculated to be as little as 1 organism per 10 g of sausage (15). Twenty-three children (age range, 4 months to 12 years) developed HUS. One child died of multiple hemorrhagic cerebral infarcts; 18 children required renal dialysis (for a median of 14 days), and 12 months after discharge, 5 still had significant impairment of renal function. Other major complications included colonic necrosis in three patients, cerebral hemorrhage/infarction in three patients, convulsions in four patients, and development of glucose intolerance in three patients (17). Pulsed-field gel electrophoresis (PFGE) analysis of the outbreak isolates suggested that they were all clones (15). These isolates were also shown by PCR to carry eae and both stx1AB and stx2AB. However, the isolates from later in the outbreak (post-25 January) appeared to be more virulent, as the patients experienced a higher proportion of complications (17), and Southern blot analysis suggested that the strains harbored an additional copy of stx2AB (15). In this paper, we further compared two representative early and late isolates using next-generation sequencing and bioinformatics tools not available at the time of this important outbreak.
MATERIALS AND METHODS
Bacterial strains.
E. coli O111:H− strains 95JB1 and 95NR1 were originally isolated from fecal samples from two patients of the South Australian outbreak, who presented with HUS on 19 and 31 January 1995, respectively (15, 18).
DNA manipulation.
Genomic DNA was extracted from cultures grown overnight in Luria-Bertani medium by using a Qiagen (Hilden, Germany) genomic DNA kit according to the manufacturer's instructions. High-throughput sequencing was performed by the Australian Genome Research Facility Ltd. (AGRF) (Brisbane, Australia), using an Illumina GAII instrument, and generated 5,533,910 and 6,259,993 paired-end 100-base reads for 95JB1 and 95NR1, respectively. Sanger sequencing of PCR products to confirm six single-nucleotide polymorphisms (SNPs) was also performed by the AGRF on the Applied Biosystems 3730xl platform using the first 12 primers listed in Table S1 in the supplemental material.
Bioinformatic analysis.
Illumina adapter sequences were trimmed from sequence reads by using Cutadapt (19). De novo contig assembly was performed by using Ray (version 2.0.0-rc5) (20). Other tools used for analysis and visualization included Artemis (version 14.0.0) (21), ACT (Artemis Comparison Tool) (version 13.0) (22), BLAST (Basic Local Alignment Search Tool) (version 2.2.24+) (23), Bowtie (24), Bowtie2 (2.0.0-beta6) (25), BWA (alignment via Burrows-Wheeler transformation) (version 0.5.9rc1) (26), Mauve (version 2.3.1) (27), RAST (Rapid Annotation Using Subsystem Technology) (28), and SAMtools (version 0.1.18) (29), as appropriate. BRIG (BLAST Ring Image Generator) was used to generate figures displaying read mapping coverage (30). To assess copy number variation, normalized mapped read depth counts were calculated for annotated features using RPKM (reads per kilobase of reference genome per million mapped reads) values in accordance with methods described previously by Mortazavi et al. (31). For end-sequence profiling, a multisequence fasta reference file was constructed by excising all of the prophage sequences from the genome of O111:H− EHEC isolate 11128 (3) and appending a set of representative phage sequences (see Table S2 in the supplemental material) as separate entries. Read alignment to the reference construct was performed by using BWA, and SAMtools view was used with the command line option −F 14 to return read pairs that were mapped but not as a proper pair. Artemis was used to search the resulting BAM files for candidate phage insertion sites. To search for prophages that were not included in the index, pairs with one mate mapped and the other mate unmapped were also retrieved by using the −f 9 option of SAMtools view.
MLST.
Multilocus sequence typing (MLST) was performed in silico by using 15 housekeeping genes described on the STEC Center website (http://www.shigatox.net/new) (ST[15]; arcA, aroE, aspC, clpX, cyaA, dnaG, fadD, grpE, icdA, lysP, mdh, mtlD, mutS, rpoS, and uidA) (32).
Phylogenetic tree.
Whole-genome alignment of the two EHEC genome assemblies and 51 additional complete E. coli reference genomes was performed by using Mugsy version 1.2.2 (33) with default parameter settings. Blocks of sequence that aligned across all 53 genomes and were larger than 10,000 bp were concatenated by using a custom python script to construct a core genome alignment. Ambiguously aligned regions of the core genome alignment were identified and trimmed by using GBLOCKS version 0.91b (34) with the following parameter settings, where n is the total number of sequences in the alignment: minimum number of sequences for a conserved position of (n/2) + 1, minimum number of sequences for a flank position of n × 0.85, maximum number of contiguous nonconserved positions of 8, minimum length of a block of 10, and no gap positions allowed.
A maximum likelihood phylogenetic tree was inferred from the trimmed core genome by using RAxML version 7.2.8 (35), using the generalized time-reversible (GTR) model with gamma correction for among-site rate variation. Support for nodes was assessed by using 1,000 bootstrap replicates. The tree visualization was manually rooted in accordance with methods described previously by Touchon et al. (36).
PCR.
PCRs were performed by using a G-Storm thermal cycler (Somerton, United Kingdom) and the Phusion Flash high-fidelity PCR system (Thermo Scientific, Victoria, Australia) according to the manufacturer's instructions. Primers were purchased through Sigma-Aldrich (New South Wales, Australia) and are listed in Table S1 in the supplemental material.
Nucleotide sequence accession numbers.
Assembled contigs for 95NR1 and 95JB1 have been deposited in the WGS division of GenBank under accession numbers AVDU00000000 and AWFJ00000000, respectively. The versions described in this paper can be found under accession numbers AVDU01000000 and AWFJ01000000, respectively.
RESULTS
95NR1 and 95JB1 are nearly identical at the core genome level.
The complete genome of another O111:H− EHEC isolate (11128), which was collected in Japan in 2001 (3), is publicly available (GenBank accession number AP010960.1), including its 5 plasmids pO111_1 to pO111_5 (accession numbers AP010961.1, AP010962.1, AP010963.1, AP010964.1, and AP010965.1, respectively). In contrast to the South Australian outbreak isolates, this strain was isolated from a sporadic case of bloody diarrhea (3, 37). To investigate the relatedness of the isolates, in silico MLST was performed. 11128 was found to belong to clonal group 14 and sequence type (ST) [15]40. Analysis of the same sequences in 95NR1 and 95JB1 identified only one SNP difference overall in comparison to 11128, which was shared by both outbreak strains in the allele of mtlD. This placed 95NR1 and 95JB1 in clonal group 14 but in ST [15]39. In addition, while 11128 is reported to be H−, it has an intact copy of fliCH8, but both outbreak strains have a deletion of T within this allele (equivalent to nucleotide [nt] 2491268 in 11128), resulting in a reading frameshift and a premature stop codon.
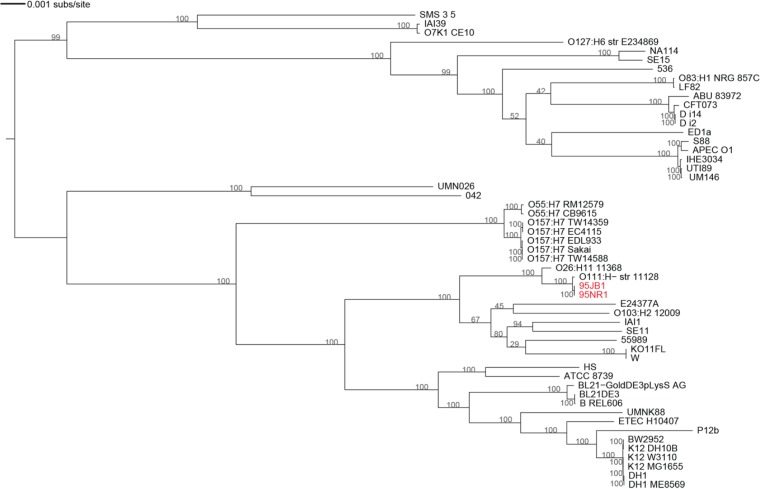
Phylogenetic analysis of the core genome shared between draft assemblies of 95NR1 and 95JB1 and 51 completely sequenced E. coli genomes currently available in public databases (3,397,484 bp, trimmed to 2,065,655 bp) showed that the two Australian outbreak strains are most closely related to each other but also group closely together with O111 strain 11128 within phylogroup B1 (Fig. 1). Whole-genome comparison of 11128, 95NR1, and 95JB1 using reads mapped to 11128 identified ∼880 SNPs that discriminate the Australian strains from 11128. Only six confirmed SNPs discriminate the two Australian strains, consistent with a clonal relationship (see Table S4 in the supplemental material).
FIG 1.
Maximum likelihood E. coli phylogenetic tree reconstructed from a trimmed nucleotide core genome alignment (2,065,655 bp). Isolates from the Adelaide HUS outbreak are indicated in red. The numbers at the nodes correspond to the bootstrap values (1,000 bootstraps). Details of the various genomes are provided in Table S3 in the supplemental material. ETEC, enterotoxigenic E. coli; APEC, avian pathogenic E. coli.
95NR1 contains additional copies of the stx2AB genes.
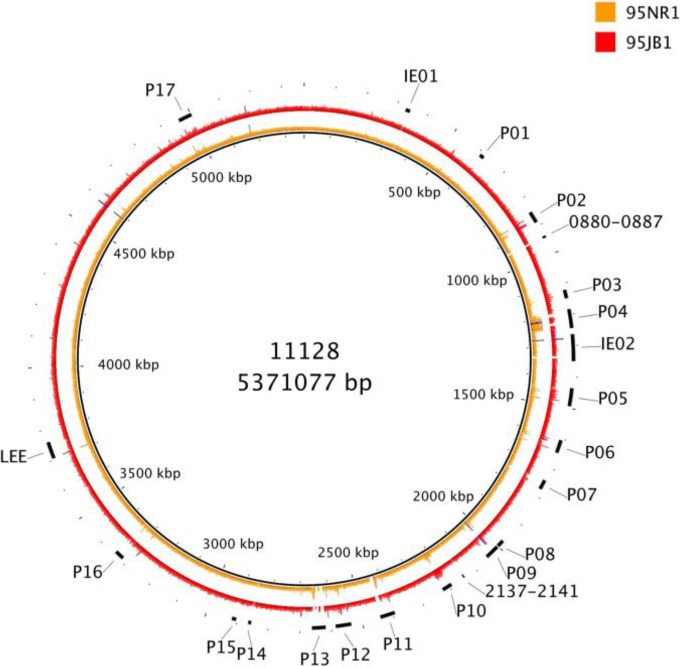
When the reads were mapped to the 11128 genome and plasmids, it was found that both 95NR1 and 95JB1 lack the large multidrug resistance plasmid pO111_1. However, both strains have a P1 plasmid, an EHEC plasmid, and two colicin plasmids, like 11128 (pO111_2 to pO111_5, respectively) (see Fig. S1 in the supplemental material). Nevertheless, 95NR1 and 95JB1 share common differences from the 11128 plasmids; in particular, >10 kb of pO111_2 as well as the immunity proteins of pO111_4 are absent from the read data for the two outbreak strains (see Fig. S1 in the supplemental material). In regard to the chromosome, there were only four regions outside prophages absent in both outbreak strains but present in strain 11128. These regions include the ferric aerobactin operon of 11128's second integrative element (IE02), which encodes a potential iron-scavenging siderophore, and part of the ydhYVWXUT oxidoreductase operon (Fig. 2; see also Table S5 in the supplemental material) (38, 39). The majority of differences were related to prophage genes, with some genes being without coverage (e.g., within P11 and P13) and other genes exhibiting more than 1× coverage (e.g., within P02 and P05). However, the modular nature of phage genomes, which can exhibit homology between one another, limited the conclusions on prophage content that could be drawn from read mapping alone (Fig. 2). Nevertheless, some prophage genes had higher read mapping coverage for 95NR1 than for 95JB1. In particular, the coverage depth of stx2AB was 3.7 times that of the stx1AB genes for 95NR1, while the coverage ratio of stx2AB and stx1AB was 1:1 for 95JB1. These results are consistent with the presence of at least two additional copies of stx2AB in 95NR1.
FIG 2.
Mapping of 95NR1 and 95JB1 reads to the 11128 genome. Reads of 95NR1 and 95JB1 were aligned to the complete 11128 genome by using Bowtie2. The inner ring represents 95NR1 coverage, and the outer ring represents 95JB1 coverage. The maximum coverage shown is 700 reads, with blue representing regions with higher coverage than this. Prophages, regions without mapping (integrative element or genes), and the LEE are indicated.
Genomic comparisons identify an additional prophage sequence in 95NR1 compared to 95JB1.
To identify the unique regions in 95NR1 and 95JB1, the draft genome assemblies of the two outbreak strains were initially compared to 11128 by using BLAST and ACT. The vast majority of sequences present in both 95NR1 and 95JB1 but absent in 11128 were found to be of prophage origin. These included sequences with similarity to those of a Mu-like prophage of O157:H7 Sakai, O103:H2 12009's prophages ECO103_P04 and ECO103_P14, O26:H11 11368's prophage ECO26_P14, as well as various lambda-like or Stx-converting prophages.
The genomes of 95NR1 and 95JB1 were also directly compared to one another. The 95NR1 draft assembly was >50 kb larger than the draft genome of 95JB1, with the additional sequence being found to have similarity to those of lambda-like prophages. Although lambda-like prophages are heterogenous, they possess a number of genes that are functionally conserved, such as repressors and replication initiators (40). By analyzing the contigs of 95NR1 containing at least some of the sequence absent in 95JB1, it was evident that 95NR1 has two additional copies of several of these genes, including cI, cII, cIII, cro (repressor), N (antiterminator), O (replication initiation), and P (replication initiation). Together with the increased depths of coverage of the stx2AB genes in 95NR1, these observations are consistent with the conclusion that 95NR1 harbored two additional Stx2-converting prophages compared with 95JB1.
95NR1 has a single additional prophage insertion site relative to 95JB1.
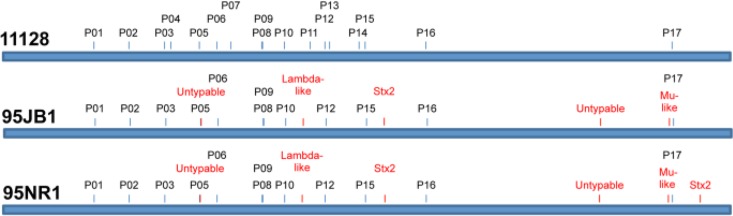
Prophage insertion sites were analyzed in the de novo assemblies. Five of the 16 prophages in the 11128 genome, ECO111_P04, ECO111_P07, ECO111_P11, ECO111_P13, and ECO111_P14, had no prophages at the corresponding insertion sites in either of the South Australian outbreak strain genomes. Although parts of these prophages could be detected by read mapping, their absence or insertion elsewhere in the genomes of 95NR1 and 95JB1 could not be determined with certainty, due to the modular nature of and high similarity between the different prophages (Fig. 2). In particular, very high levels of coverage can be seen for ECO111_P04 in 95NR1 due to sequence identity with the extra prophage sequence (Fig. 2). ECO111_P11 is the Stx2-converting prophage of 11128, and the absence of prophage sequences in the corresponding genomic location in the two HUS outbreak strains indicated that their Stx2-converting prophages were located elsewhere in the genome. Five additional prophage regions relative to 11128, inserted within or adjacent to ycfD, argW, yicC, lysC, and yebY, were identified in both of the draft genomes. Moreover, a prophage insertion site unique to 95NR1 was distinguished within the gene of a 356-amino-acid hypothetical protein (gene ECO111_5095 in 11128).
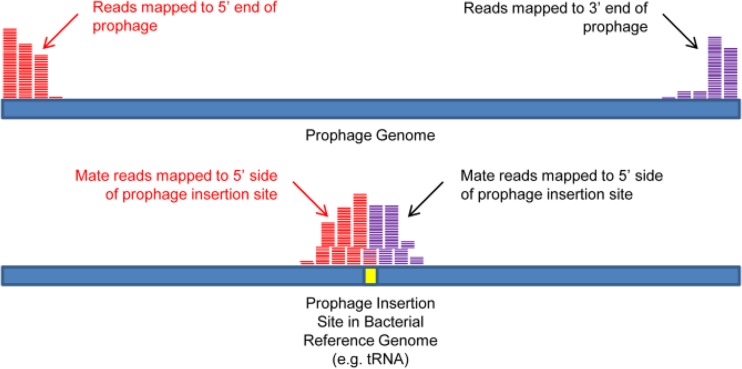
The sequencing data strongly suggested the presence of an additional prophage sequence in 95NR1 compared to 95JB1, but its insertion site could not be easily identified in the draft genome. Since read mapping can be used to detect insertions as well as deletions, we employed end-sequence profiling using a variant of the 11128 genome with all prophage sequences excised and itemized along with a range of prophages detected through BLASTn analysis of the de novo-assembled contigs (41, 42). As the sequencing data from this study were paired-end data, when sequences matching a prophage of 95NR1 or 95JB1 were represented in the index, one read of the pair would map to the end of the phage genome and the other would map adjacent to the phage insertion site, as illustrated in Fig. 3. We confirmed the above-mentioned sites of prophage insertion and predicted the type of phage present at each site using this method (summarized in Fig. 4; see also Table S6 in the supplemental material). It is likely that 95NR1 and 95JB1 possess prophages similar to those inserted at common locations in the 11128 genome, but there could be differences within the prophage genomes, which would not be detectable by end-sequence profiling. No additional unique prophage insertion sites were found, other than those already identified in the 95NR1 genome, even after filtering for phage insertion sites without a phage in the index and comparing 95NR1 directly with 95JB1 for a characteristic stack of reads present for 95NR1 but not 95JB1.
FIG 3.
End-sequence profiling for prophage insertion site detection. Paired-end reads are aligned to a multifasta reference containing a phage sequence-free bacterial genome and multiple phage genomes. When the ends of an index phage have homology to one of the sequenced strain's phages, one read of the pair will map to the index phage and the other will map to the corresponding side of the phage insertion site in the bacterial genome. Opposite ends of the phage will have mate reads mapped to different sides of the insertion site. Thus, the orientation of the phage can also be determined.
FIG 4.
Summary of end-sequence profiling results. The phage insertion sites of 95NR1 and 95JB1 are shown in relation to the 11128 genome, with the phages associated at each site being displayed (see Table S5 in the supplemental material). Phage insertion sites empty in 11128 are highlighted in red.
Two Stx prophages are arranged in tandem in 95NR1 but not 95JB1.
Analysis of contigs in 95NR1 that had no equivalent in 95JB1 identified several that contained genes with sequence identity with known Stx prophage genomes, including a 22,295-bp contig containing a >8-kb hypothetical gene also found within phage VT2-phi272 at its 3′ end. This contig also harbored five downstream genes normally found near the 5′ end of lambda-like prophages, namely, N, cro, cI, cII, and cIII, suggesting that it encompassed the boundary of two tandemly arranged prophages. A PCR product of the correct size was attainable for 95NR1 but not 95JB1 by using primers designed for the large hypothetical gene and the unique cII gene on the same contig (95NR1 contig213000000 F and 95NR1 contig213000000 R) (see Table S1 in the supplemental material), suggesting that this was not simply an assembly error (see Fig. S2 in the supplemental material). The 3′ end of the contig (harboring cII) could be linked downstream by paired-end read mapping with a contig containing the stx2AB genes. Furthermore, the 5′ end of the contig (harboring the large hypothetical gene) could similarly be linked upstream with a contig containing tail fiber genes. As tail genes are located at the 3′ end of lambda-like prophages following the head and packaging genes, this finding further supported the contention that 95NR1 contained two tandemly arranged prophages. To confirm this contig arrangement, PCR was carried out by using primers designed for a phage gene near the hypothetical gene and the tail fiber gene within the middle of the upstream contig (see Table S1 in the supplemental material). A PCR product of the expected size was attainable for 95NR1 but not 95JB1 (see Fig. S2 in the supplemental material). As a further precaution, the ends of the PCR product were sequenced, indicating that mispriming did not occur (data not shown).
There was no integrase gene identified within either of the contigs described above, strongly suggesting that the “tail”-to-“head” arrangement of the two putative prophage genomes was a feature of the genome rather than an artifact of prophage induction. Read mapping coverage suggested that there were two complete copies of an identical integrase gene within prophages harbored at argW and the unique insertion site within the 95NR1 genome, respectively. Therefore, the gene order that we have confirmed by PCR could not be explained by circular permutation of an excised prophage genome but indicated that the two prophages had inserted into the same insertion site, with the second phage subsequently losing its integrase. As the contig containing the tail fiber genes demonstrated little sequence identity with the 95JB1 genome, the two prophages could be present only in the unique prophage site of 95NR1.
The two tandemly arranged prophages were more similar to each other than to the Stx2 prophage common to both 95NR1 and 95JB1. However, some genes common to lambda-like prophages were heterogenous among all three prophages. Despite the similarity between lambda-like prophages and the draft assemblies, we were able to ascertain three unique genomic contexts for stx2AB by designing PCR primers for three unique cI genes and the common stx2A gene in order to confirm the presence of three Stx2-converting prophages in 95NR1 (see Table S1 in the supplemental material). PCR products were attainable for all three sets of primers for 95NR1 but only for the first primer set for 95JB1, as expected (see Fig. S2 in the supplemental material). Again, sequencing of the ends of the PCR products proved that the primers annealed accurately (data not shown). All copies of the stx2AB genes of 95NR1, as well as those of 95JB1 and 11128, could be subtyped as Stx2a according to the guidelines described previously by Persson et al. (43). The stx2AB genes of 95NR1 were also analyzed to identify SNP differences among the multiple copies. Only one SNP was identified in one copy of the stx2AB genes, which resulted in a conversion of thymine to cytosine in stx2A (position 2424591 in the 11128 reference genome).
Stx2 titers.
Since 95NR1 has two additional copies of stx2AB relative to 95JB1, we tested the cytotoxicity of French pressure cell lysates of cultures of the two strains grown overnight on Vero cells, as previously described (44). To neutralize the effects of Stx1, lysates were preincubated for 30 min at 37°C with polyclonal rabbit Stx1 antiserum (diluted 1:40). The residual Stx2 titers were 80 and 320 50% cytotoxic doses (CD50) per ml for the 95JB1 and 95NR1 lysates, respectively, consistent with the higher toxin gene dose in the latter strain.
DISCUSSION
In 2011, there was a major O104:H4 HUS outbreak in Germany, from which isolates were quickly sequenced and analyzed by utilizing high-throughput sequencing technologies (45–47). At the time of the 1995 O111:H− Australia outbreak, such technology was unavailable. The 1995 outbreak remains the largest in Australia to date; many victims continue to suffer chronic medical issues, including diabetes, and several have undergone kidney transplantation. PFGE analysis suggested that the strains were clonal, but Southern blot hybridization suggested that isolates from later in the outbreak carried an additional Stx2-converting phage (15). Furthermore, the patients from whom these later isolates were collected exhibited signs of more severe disease, including a fatality. Consequently, we have sequenced two isolates from different times in this outbreak and compared them to the fully sequenced and annotated O111:H− isolate 11128, which was isolated from a sporadic case (3), in contrast to our isolates.
The most notable differences between 11128 and the Australian outbreak isolates were related to phage and plasmid sequences. In particular, the Australian isolates lack a multidrug resistance plasmid and differ in their Stx2-converting prophages in both sequence and location. From analyses of the draft genomes, both 95NR1 and 95JB1 appear to have a Stx2-converting phage inserted into argW, which is a known Stx-converting phage insertion site (48, 49).
Comparison of the two Australian outbreak genomes suggests that the major difference between these two strains is the additional two Stx2-converting prophages of 95NR1. The original Southern hybridization analysis of 95NR1 genomic DNA revealed an additional stx2-reactive band, relative to 95JB1, suggestive of an additional Stx2 prophage (15). However, close inspection reveals that the hybridization signal intensity of the additional band was consistently greater than that of the other stx2-containing restriction fragment (15). This observation strongly suggests the presence of not one but two additional stx2 genes, in concordance with data from the present study. Increased toxin production was previously demonstrated for double-Stx2 phage lysogens (50) and was also demonstrated in the present study. Thus, it is likely that the additional Stx2 prophages contributed to the severity of disease seen later in the South Australian outbreak. Stx2 of all three phages was subtyped as Stx2a, which has a high association with HUS (43). Not only is the 95NR1 genome unusual in the presence of three almost identical Stx2 alleles, but the two extra Stx2-converting prophages are inserted at the same site. The second stx2 phage insertion site identified in 95NR1 was within a hypothetical gene (ECO111_5095 in 11128). The hypothetical gene has been identified in only a small number of E. coli strains to date and is not a known Stx-converting phage insertion site. Lambda-like phage integrases typically target conserved regions whose disruption will not impact host fitness (50).
The genomes of both Australian outbreak strains differed from 11128 by several hundred SNPs, but 95JB1 contained five additional nucleotide substitutions relative to both 11128 and 95NR1. Thus, the additional two tandemly arranged Stx2-converting prophages in 95NR1 may not have been acquired during the course of the outbreak but rather may have been lost from 95JB1 during the divergence of these two strains from a common ancestor. This implies that the primary outbreak source (perhaps a livestock herd) may have contained a heterogeneous population of near-identical clonal strains harboring between one and three Stx2-converting prophages and that the identification of more virulent triple-Stx2 isolates later in the outbreak was coincidental or related to epidemiological factors. A subsequent coronial inquest found that there were major processing failures at the small goods manufacturing facility responsible for the outbreak, such that multiple batches of mettwurst were contaminated. Thus, earlier batches released to the marketplace may have been produced from a carcass contaminated with a 95JB1-like strain, while later batches included meat from a carcass contaminated with a 95NR1-like strain. Further whole-genome sequencing of other isolates will help to resolve the fine details of this historical outbreak. In conclusion, the 1995 South Australian outbreak was caused by an O111:H− clone, but an isolate from later in the outbreak possessed two additional Stx2-converting prophages. This study highlights that even when a pathogen clone causes an outbreak, there can be small but highly significant genomic differences between isolates within the clone, which can have a significant impact on virulence.
Supplementary Material
ACKNOWLEDGMENTS
J.C.P. is a National Health and Medical Research Council of Australia (NHMRC) Senior Principal Research Fellow; S.A.B. is an NHMRC Career Development Fellow.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01229-15.
REFERENCES
- 1.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Paton JC, Paton AW. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev 11:450–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K, Tobe T, Hattori M, Hayashi T. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A 106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A 101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci U S A 87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A 92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindberg AA, Brown JE, Stromberg N, Westling-Ryd M, Schultz JE, Karlsson KA. 1987. Identification of the carbohydrate receptor for Shiga toxin produced by Shigella dysenteriae type 1. J Biol Chem 262:1779–1785. [PubMed] [Google Scholar]
- 8.Louise CB, Obrig TG. 1995. Specific interaction of Escherichia coli O157:H7-derived Shiga-like toxin II with human renal endothelial cells. J Infect Dis 172:1397–1401. doi: 10.1093/infdis/172.5.1397. [DOI] [PubMed] [Google Scholar]
- 9.Tesh VL, Burris JA, Owens JW, Gordon VM, Wadolkowski EA, O'Brien AD, Samuel JE. 1993. Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect Immun 61:3392–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strockbine NA, Marques LR, Newland JW, Smith HW, Holmes RK, O'Brien AD. 1986. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect Immun 53:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unkmeir A, Schmidt H. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect Immun 68:4856–4864. doi: 10.1128/IAI.68.9.4856-4864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 13.Ohnishi M, Kurokawa K, Hayashi T. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol 9:481–485. doi: 10.1016/S0966-842X(01)02173-4. [DOI] [PubMed] [Google Scholar]
- 14.Caprioli A, Luzzi I, Rosmini F, Resti C, Edefonti A, Perfumo F, Farina C, Goglio A, Gianviti A, Rizzoni G. 1994. Community-wide outbreak of hemolytic-uremic syndrome associated with non-O157 verocytotoxin-producing Escherichia coli. J Infect Dis 169:208–211. doi: 10.1093/infdis/169.1.208. [DOI] [PubMed] [Google Scholar]
- 15.Paton AW, Ratcliff RM, Doyle RM, Seymour-Murray J, Davos D, Lanser JA, Paton JC. 1996. Molecular microbiological investigation of an outbreak of hemolytic-uremic syndrome caused by dry fermented sausage contaminated with Shiga-like toxin-producing Escherichia coli. J Clin Microbiol 34:1622–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. 1995. Community outbreak of hemolytic uremic syndrome attributable to Escherichia coli O111:NM—South Australia 1995. MMWR Morb Mortal Wkly Rep 44:550–551, 557–558. [PubMed] [Google Scholar]
- 17.Henning PH, Tham EB, Martin AA, Beare TH, Jureidini KF. 1998. Haemolytic-uraemic syndrome outbreak caused by Escherichia coli O111:H−: clinical outcomes. Med J Aust 168:552–555. [DOI] [PubMed] [Google Scholar]
- 18.Paton AW, Voss E, Manning PA, Paton JC. 1997. Shiga toxin-producing Escherichia coli isolates from cases of human disease show enhanced adherence to intestinal epithelial (Henle 407) cells. Infect Immun 65:3799–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17:10–12. [Google Scholar]
- 20.Boisvert S, Laviolette F, Corbeil J. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comp Biol 17:1519–1533. doi: 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 22.Smith DL, Rooks DJ, Fogg PC, Darby AC, Thomson NR, McCarthy AJ, Allison HE. 2012. Comparative genomics of Shiga toxin encoding bacteriophages. BMC Genomics 13:311. doi: 10.1186/1471-2164-13-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darling AE, Mau B, Perna NT. 2010. progresiveMauve: multiple genome alignment with gene gain, loss, and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 32.Qi W, Lacher DW, Bumbaugh AC, Hyma KE, Ouellette LM, Large TM, Tarr CL, Whittam TS. 2004. EcMLST: an online database for multi locus sequence typing of pathogenic Escherichia coli, p 520–521. In Proceedings of the 2004 IEEEE Computational Systems Bioinformatics Conference, Los Alamitos, CA. Institute of Electrical and Electronics Engineers, Washington, DC. [Google Scholar]
- 33.Angiuoli SV, Salzberg SL. 2011. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27:334–342. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 35.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 36.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguenec C, Lescat M, Mangenot S, Martinez-Jehanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Medigue C, Rocha EP, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogura Y, Ooka T, Asadulghani Terajima J, Nougayrede JP, Kurokawa K, Tashiro K, Tobe T, Nakayama K, Kuhara S, Oswald E, Watanabe H, Hayashi T. 2007. Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol 8:R138. doi: 10.1186/gb-2007-8-7-r138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Lorenzo V, Bindereif A, Paw BH, Neilands JB. 1986. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J Bacteriol 165:570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Partridge JD, Browning DF, Xu M, Newnham LJ, Scott C, Roberts RE, Poole RK, Green J. 2008. Characterization of the Escherichia coli K-12 ydhYVWXUT operon: regulation by FNR, NarL and NarP. Microbiology 154:608–618. doi: 10.1099/mic.0.2007/012146-0. [DOI] [PubMed] [Google Scholar]
- 40.Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol 299:27-51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 41.Raphael BJ, Volik S, Collins C, Pevzner PA. 2003. Reconstructing tumor genome architectures. Bioinformatics 19(Suppl 2):ii162–ii171. [DOI] [PubMed] [Google Scholar]
- 42.Volik S, Zhao S, Chin K, Brebner JH, Herndon DR, Tao Q, Kowbel D, Huang G, Lapuk A, Kuo WL, Magrane G, De Jong P, Gray JW, Collins C. 2003. End-sequence profiling: sequence-based analysis of aberrant genomes. Proc Natl Acad Sci U S A 100:7696–7701. doi: 10.1073/pnas.1232418100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persson S, Olsen KE, Ethelberg S, Scheutz F. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J Clin Microbiol 45:2020–2024. doi: 10.1128/JCM.02591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paton AW, Bourne AJ, Manning PA, Paton JC. 1995. Comparative toxicity and virulence of Escherichia coli clones expressing variant and chimeric Shiga-like toxin type II operons. Infect Immun 63:2450–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin C-S, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Moller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med 365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohde H, Qin J, Cui Y, Li D, Loman NJ, Hentschke M, Chen W, Pu F, Peng Y, Li J, Xi F, Li S, Li Y, Zhang Z, Yang X, Zhao M, Wang P, Guan Y, Cen Z, Zhao X, Christner M, Kobbe R, Loos S, Oh J, Yang L, Danchin A, Gao GF, Song Y, Li Y, Yang H, Wang J, Xu J, Pallen MJ, Wang J, Aepfelbacher M, Yang R, E. coli O104:H4 Genome Analysis Crowd-Sourcing Consortium. 2011. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med 365:718–724. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- 48.Kotewicz ML, Mammel MK, LeClerc JE, Cebula TA. 2008. Optical mapping and 454 sequencing of Escherichia coli O157:H7 isolates linked to the US 2006 spinach-associated outbreak. Microbiology 154:3518–3528. doi: 10.1099/mic.0.2008/019026-0. [DOI] [PubMed] [Google Scholar]
- 49.Ohnishi M, Terajima J, Kurokawa K, Nakayama K, Murata T, Tamura K, Ogura Y, Watanabe H, Hayashi T. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc Natl Acad Sci U S A 99:17043–17048. doi: 10.1073/pnas.262441699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bobay L-M, Rocha EPC, Touchon M. 2013. The adaptation of temperate bacteriophages to their host genomes. Mol Biol Evol 30:737–751. doi: 10.1093/molbev/mss279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.