Abstract
In different bacteria, primarily cytosolic and metabolic proteins are characterized as surface localized and interacting with different host factors. These moonlighting proteins include glycolytic enzymes, and it has been hypothesized that they influence the virulence of pathogenic species. The presence of surface-displayed glycolytic enzymes and their interaction with human plasminogen as an important host factor were investigated in the genome-reduced and cell wall-less microorganism Mycoplasma pneumoniae, a common agent of respiratory tract infections of humans. After successful expression of 19 glycolytic enzymes and production of polyclonal antisera, the localization of proteins in the mycoplasma cell was characterized using fractionation of total proteins, colony blot, mild proteolysis and immunofluorescence of M. pneumoniae cells. Eight glycolytic enzymes, pyruvate dehydrogenases A to C (PdhA-C), glyceraldehyde-3-phosphate dehydrogenase (GapA), lactate dehydrogenase (Ldh), phosphoglycerate mutase (Pgm), pyruvate kinase (Pyk), and transketolase (Tkt), were confirmed as surface expressed and all are able to interact with plasminogen. Plasminogen bound to recombinant proteins PdhB, GapA, and Pyk was converted to plasmin in the presence of urokinase plasminogen activator and plasmin-specific substrate d-valyl-leucyl-lysine-p-nitroanilide dihydrochloride. Furthermore, human fibrinogen was degraded by the complex of plasminogen and recombinant protein PdhB or Pgm. In addition, surface-displayed proteins (except PdhC) bind to human lung epithelial cells, and the interaction was reduced significantly by preincubation of cells with antiplasminogen. Our results suggest that plasminogen binding and activation by different surface-localized glycolytic enzymes of M. pneumoniae may play a role in successful and long-term colonization of the human respiratory tract.
INTRODUCTION
Members of the class Mollicutes are known to be the smallest and simplest self-replicating prokaryotes. Among them, Mycoplasma pneumoniae is a common pathogen in humans. The cell wall-less bacterium causes infections of the respiratory tract, including severe interstitial pneumonia (1), and usually accounts for 5 to 10% of all cases of community-acquired pneumonia among pediatric (2) and adult (3) patients. Because of the time-dependent incidence of infections, >25% of pneumonia cases in epidemic periods are attributed to this agent (4). In addition, a number of extrapulmonary manifestations and complications are described, affecting mainly the skin and the central nervous system (5).
Due to the greatly reduced genome of M. pneumoniae (816 kbp, 689 open reading frames [ORFs]), the metabolic pathways and the repertoire of virulence factors are limited (6). However, M. pneumoniae has developed effective strategies to infect humans and allow long-term survival in the respiratory mucosa. These include the targeted adherence of the bacteria to epithelial cells via a specialized complex of adhesins and adhesion-related proteins (7) as the first step in colonization, followed by the release of cell-damaging substances such as community-acquired respiratory distress syndrome toxin (8) and hydroxide radicals (9).
Glycolytic enzymes are another class of proteins that have been regarded as virulence factors in a broader sense (10, 11). First described in streptococci (12), glycolytic enzymes are not only involved in cytosol-located metabolic processes of glycolysis but can also be transported by an unknown mechanism to the surfaces of microorganisms (13). There, proteins are able to interact with host factors, mainly with components of the human extracellular matrix (ECM) and fibrinolysis system. Different studies have confirmed this dual role of enzymes that function primarily in the metabolism of different bacterial, fungal, and parasitic species, suggesting a general mechanism of host-pathogen interaction. Furthermore, the number of host factors shown to interact with these bacterial proteins is increasing (14). Among them, plasminogen (Plg) plays a crucial role in fibrinolysis and has been described as the binding partner of many microbial surface molecules, including glycolytic enzymes. As a result of this interaction, Plg can be converted to the serine protease plasmin; it has been suggested that plasmin's cleavage activity promotes the growth and dissemination of bacteria within infected host tissue (15, 16).
Surface-displayed glycolytic enzymes have also been demonstrated in mycoplasmas such as the important veterinary species Mycoplasma bovis (17), Mycoplasma gallisepticum (18), Mycoplasma suis (19, 20), and Mycoplasma synoviae (21), as well in the human pathogens Mycoplasma fermentans (22) and Mycoplasma genitalium (23). As described for other bacteria species, enolase (Eno) and glutaraldehyde-6-phosphate dehydrogenase (GapA) were confirmed as the main surface-located glycolytic enzymes. In M. pneumoniae, GapA was reported as a surface-associated protein that can bind human fibrinogen (24). Additionally, pyruvate dehydrogenase (Pdh) subunits A to C were found on the cell surface interacting with human Plg (25, 26). In the case of PdhB, the protein complex activates Plg, resulting in the degradation of human fibrinogen (26). Furthermore, PdhB, one of the proteins that interact with various host factors (11), is also able to bind human fibronectin (27). Interestingly, PdhD and Eno of M. pneumoniae were also reported as binding partners of human Plg, but they do not occur on the surfaces of the bacteria (25, 26). In addition to PdhA, PdhB, and GapA, other glycolytic enzymes such as lactate dehydrogenase (Ldh) are detected in the Triton X-100-insoluble fraction that represents the membrane-associated proteins of M. pneumoniae (28). Finally, additional glycolytic enzymes are described in other bacterial species but have not hitherto been characterized in mycoplasmas as surface displayed, indicating a role for other proteins as potential interaction partners with host factors (29–32). In summary, the present information regarding surface-localized glycolytic enzymes suggests a complex network of interactions with different host factors, which may be important in understanding the pathogenesis of M. pneumoniae infections.
Therefore, we performed the first systematic study of the complete set of glycolytic enzymes of M. pneumoniae to determine their location on the cell surface. Furthermore, the comparative interaction of surface-displayed enzymes with human Plg as an example of an important host factor was analyzed. Collectively, 8 of 19 glycolytic proteins investigated can be assumed to be surface displayed, and all of them are able to interact with Plg.
MATERIALS AND METHODS
Bacteria, cell lines, and growth conditions.
M. pneumoniae strain M129 (ATCC 29342) and Escherichia coli strains NovaBlue (Novagen, Darmstadt, Germany), BL21(DE3) (Novagen), and E. cloni (Lucigen, Middleton, WI), as well as A549 cells (human lung carcinoma cell line, ATCC CCL-185), were grown as described previously (25). The protein concentrations were measured using a BCA protein assay kit (Pierce, Rockford, IL).
Expression and purification of recombinant proteins and production of guinea pig polyclonal antibodies.
DNA of M. pneumoniae was extracted using the QIAamp DNA minikit (Qiagen, Hilden, Germany) as described by the manufacturer. Overall, 19 glycolytic enzymes were included in the study (Table 1; see also Fig. S1 in the supplemental material). PCR using primers summarized in Table S1 in the supplemental material amplified the complete genes coding for glycolytic enzymes, and PCR products were purified with a QIAquick PCR purification kit (Qiagen). If TGA codons occurred in the gene, amplificates were used as the template for the multiple mutation reaction (36). The products were cloned into the pET-30/LIC vector (Novagen) to transform E. coli cells as described by the manufacturer. The inserts were checked by sequencing. N-terminal His6-tagged recombinant proteins were expressed, purified, and concentrated as described recently (26).
TABLE 1.
Characteristics of glycolytic enzymes of M. pneumoniae
| Enzyme (abbreviation) | Genea | No. of TGA codon(s) | Putative mol mass (kDa) | Phosphorylationb | Remarksc |
|---|---|---|---|---|---|
| Acetate kinase (AckA) | mpn533 | 0 | 43.8 | Yes | |
| ATP-dependent 6-phosphofructokinase (Pfk) | mpn302 | 0 | 36.0 | Yes | |
| Fructose-bisphosphate aldolase (Fba) | mpn025 | 1 | 31.1 | Yes | |
| Enolase (Eno) | mpn606 | 1 | 49.2 | Yes | Not surface displayed, binds human Plg (25) |
| Glucose-6-phosphate isomerase (Pgi) | mpn250 | 2 | 48.8 | No | Internal helix* |
| Glyceraldehyde-3-phosphate dehydrogenase (GapA) | mpn430 | 1 | 36.8 | Yes | Surface displayed, binds human fibrinogen (24) |
| Lactate dehydrogenase (Ldh) | mpn674 | 0 | 33.9 | Yes | Internal helix* |
| Phosphate acetyltransferase (Pta) | mpn428 | 1 | 35.2 | Yes | |
| Phosphofructokinase (FruK) | mpn079 | 1 | 33.6 | No | |
| Phosphoglycerate kinase (Pgk) | mpn429 | 2 | 44.2 | Yes | |
| Phosphoglycerate mutase (Pgm) | mpn628 | 0 | 56.4 | Yes | |
| Phosphomannomutase (ManB) | mpn066 | 3 | 62.7 | Yes | |
| Pyruvate dehydrogenase complex component E1 subunit alpha (PdhA) | mpn393 | 4 | 40.6 | Yes | Surface displayed, binds human Plg (26) |
| Pyruvate dehydrogenase complex component E1 subunit beta (PdhB) | mpn392 | 1 | 35.9 | Yes | Surface displayed, binds human Plg and fibronectin (25–27) |
| Pyruvate dehydrogenase complex component E2 (PdhC) | mpn391 | 0 | 42.4 | Yes | Surface displayed, binds human Plg (26) |
| Pyruvate dehydrogenase complex component E3 (PdhD) | mpn390 | 1 | 49.4 | Yes | Membrane associated but not surface displayed, binds human Plg (26) |
| Pyruvate kinase (Pyk) | mpn303 | 3 | 57.3 | Yes | |
| Transketolase (Tkt) | mpn082 | 3 | 72.4 | Yes | |
| Triose-phosphate isomerase (TpiA) | mpn629 | 2 | 27.0 | Yes |
M. pneumoniae strain M129 (NCBI RefSeq no. NC_000912.1) (34).
Schmidl et al. (33).
*, http://www.psort.org (PSORTb 3.0.2 [35]).
For the production of polyclonal antisera, guinea pigs (Charles River, Sulzfeld, Germany) were immunized. The animal experiments were approved by the ethical board of Landesdirektion Sachsen, Dresden, Germany (permit 24-9168.25-1/2011-1). Intranasal infection of guinea pigs with freshly harvested mycoplasmas, subcutaneous immunization with total proteins of M. pneumoniae and with recombinant proteins, booster immunization, and serum collection were performed as described previously (25).
SDS-PAGE, Western blotting, and enzyme-linked immunosorbent assay (ELISA).
The results of expression were investigated after separation of recombinant proteins by SDS-PAGE, Coomassie blue staining, and immunoblotting by standard procedures as reported recently (26). To test the specificity of polyclonal antisera to recombinant proteins, separated proteins of M. pneumoniae whole cells (100 μg) were incubated with corresponding antisera (1:500 each), and detection was performed with horseradish peroxidase (HRP)-conjugated secondary antibody specific for guinea pig IgG (1:2,000; Dako, Hamburg, Germany). The chemiluminescence signal was recorded using a LAS-3000 imager (Fujifilm, Düsseldorf, Germany).
The quantitative reactivity of the polyclonal sera was tested using ELISAs. Cavities of a 96-well plate (Greiner, Frickenhausen, Germany) were coated with M. pneumoniae total proteins (10 μg/ml carbonate buffer), washed, and blocked with 10% fetal calf serum (Gibco, Darmstadt, Germany). Wells were incubated with guinea pig sera to total proteins of M. pneumoniae (obtained after three intranasal infections of an animal) and to recombinant glycolytic enzymes (1:250 each). For detection, HRP-conjugated secondary anti-guinea pig IgG antibody (1:750) was used. The substrate reaction (TMB Super Slow; Sigma, St. Louis, MO) was stopped with 1 M HCl, and the absorbance was measured at 450 nm.
To analyze the presence of specific antibodies in the serum of a host infected with M. pneumoniae, total proteins of M. pneumoniae (positive control) and surface-displayed glycolytic enzymes were used to coat wells of an ELISA plate as described previously. The wells were incubated with guinea pig serum (1:250) obtained after three intranasal infections and anti-guinea pig whole immunoglobulin (1:750; Dako).
Fractionation of M. pneumoniae total proteins.
To analyze the localization of glycolytic enzymes, freshly grown M. pneumoniae cells were harvested, and total proteins were separated into membrane and cytosolic fractions by ultracentrifugation, as described previously (37). The presence of glycolytic enzymes in both fractions was detected by ELISA. Wells of 96-well plates were coated with the membrane and cytosolic protein fractions (15 μg/ml) as described previously, incubated with the corresponding antisera against the glycolytic enzymes (1:250 each), and detected with peroxidase-conjugated secondary antibody (1:1,000). Sera to Eno of M. pneumoniae as cytosolic protein (25, 26) and to C-terminal part (rP12) of the main P1 adhesin as membrane-associated protein (38) acted as controls. Additionally, guinea pig serum to total proteins of M. pneumoniae was included.
Surface location of glycolytic enzymes in M. pneumoniae.
Surface association is an important precondition for interaction of bacterial proteins with human ECM components. To determine the presence of glycolytic enzymes on mycoplasma cells, different immunological techniques were used. Fluorescence microscopy, colony blotting, and proteolysis of M. pneumoniae cells with chymotrypsin were carried out as described (25, 26; details can be found in the supplemental material).
Binding of human Plg to recombinant glycolytic enzymes of M. pneumoniae and influence of antisera on binding.
The interaction of glycolytic enzymes of M. pneumoniae with human Plg was investigated as described previously (26). Briefly, binding of Plg and the influence of guinea pig antisera to surface-displayed glycolytic enzymes on binding to Plg was tested in ELISAs. Wells of a 96-well plate were coated with recombinant proteins (15 μg/ml). In parallel, Plg (2.5 μg/ml) was preincubated with the corresponding sera (1:50 each) for 90 min at room temperature in an overhead shaker. Guinea pig preimmune serum (1:50) acted as a control. After blocking and washing, the serum-Plg mixtures were added to the coated wells, followed by incubation for 90 min at 37°C. Bound Plg was detected as described above.
The importance of ionic interactions for binding of Plg to recombinant proteins was analyzed by ELISA. Wells were coated with the surface-associated glycolytic enzymes (10 μg/ml). Mixtures of Plg (5 μg/ml) and increasing concentrations (62.5, 125, 250, 350, and 650 mM) of NaCl (Carl Roth, Karlsruhe, Germany) were added to the proteins, followed by incubation for 1.5 h at 37°C. Bound Plg was detected as described previously.
Furthermore, the influence of lysine residues on Plg binding was investigated using the lysine analogue ε-aminocaproic acid (ACA; Sigma). Wells of ELISA plates were coated with recombinant proteins. After washing and blocking, Plg (5 μg/ml) with or without ACA (1 mM) was added to the coated wells, followed by incubation for 1.5 h at 37°C. Detection of bound Plg was carried out as described above.
Binding of recombinant proteins to A549 cells.
A549 cells were harvested, and the protein concentration was adjusted to 100 μg/ml. The presence of Plg in the cell preparation was confirmed by ELISA using cells to coat the wells of a plate. Detection was performed by incubating wells with antibodies against human Plg produced in rabbits (Sigma; 1:2,000) and peroxidase-conjugated anti-rabbit IgG serum (1:1,000; Dako). Using the same primary antibody and TRITC (tetramethyl rhodamine isothiocyanate)-labeled anti-rabbit IgG, localization of Plg on the surfaces of fixed cells was confirmed as described above.
The binding of glycolytic enzymes to the cells was investigated as reported recently (26). Briefly, the cell suspension was used to coat wells of ELISA plates. In parallel, surface-exposed recombinant glycolytic enzymes (10 μg/ml) were immobilized in further wells. Recombinant proteins rP12 (involved in cytoadherence) and rP8 (part of the central region of the P1 protein which is not able to interact with cells [38]) were used as controls. After blocking, recombinant proteins (10 μg/ml PBS) were added to the cell-coated wells for 2 h at 37°C. In parallel, wells coated with recombinant proteins were incubated with PBS under the same experimental conditions. Detection was carried out in all wells with antisera specific to the corresponding proteins (1:1,000) and HRP-conjugated secondary antibody (1:1,000).
To test the influence of guinea pig antisera on binding of recombinant proteins to cells, A549 cells (100 μg/ml) were immobilized in the wells of ELISA plates. In parallel, surface-exposed recombinant proteins were preincubated with the corresponding antisera or with preimmune serum (1:50) in PBS for 2 h at room temperature in an overhead shaker. The mixture of serum and recombinant protein was added to the cell-coated wells, followed by incubation for 2 h at 37°C. Detection of bound proteins was performed as described above.
The importance of Plg binding for the interaction of surface-localized proteins with A549 cells was estimated by preincubation of cells (100 μg) with anti-Plg (1:50) at room temperature in an overhead shaker for 1.5 h. After centrifugation, the cells were used to coat wells of an ELISA plate and incubated with recombinant proteins (15 μg/ml) at 37°C for 1.5 h. Cells preincubated with PBS were used as a control. Bound proteins were detected as described previously.
Plg activity assay.
The ability of recombinant proteins to activate Plg to plasmin was tested by a quantitative assay as reported elsewhere (39). The cavities of 96-well plates were coated with surface-exposed recombinant proteins, M. pneumoniae cells, or membrane-associated proteins of M. pneumoniae, respectively. Bovine serum albumin (BSA) and the recombinant protein PdhB-P3 (part of PdhB of M. pneumoniae) as an example of a protein with Plg binding based not on lysine residues (25) were used as controls. After washing and blocking, human Plg (10 μg/ml) was added and incubated for 2 h at 37°C. Next, urokinase plasminogen activator (uPA; 4 ng/well; Millipore, Billerica, MA) and the plasmin-specific substrate d-valyl-leucyl-lysine-p-nitroanilide dihydrochloride (0.5 mM; Sigma) were added, followed by incubation at 37°C overnight. Controls without uPA, substrate, and Plg were prepared in parallel. Signals were measured at 405 nm.
Fibrinogen degradation assay.
Plasmin is a proteolytic enzyme which degrades human fibrinogen. To test which combinations of recombinant proteins with Plg resulted in degradation of fibrinogen, we performed an assay as reported previously (40). Surface-exposed glycolytic enzymes of M. pneumoniae or BSA were immobilized on wells of a 96-well plate. After washing and blocking, human Plg (10 μg/ml) was added (2 h, 37°C). Unbound Plg was removed by washing, and uPA (75 ng/ml), as well as human fibrinogen (15 μg/ml, plasminogen depleted; Millipore), was incubated at 37°C. After different time points, aliquots were taken, and sample buffer was added. The samples were boiled for 10 min, separated by SDS-PAGE, and blotted to nitrocellulose membrane. Fibrinogen was detected with goat antibody specific to human fibrinogen (1:1,500; Sigma) and HRP-conjugated secondary antibody (1:2,000; Dako). Signals were digitally recorded using a LAS-3000 imager.
Statistics.
Statistical analysis was carried out using a Student t test. A P value of <0.05 was considered statistically significant.
RESULTS
Generation of recombinant glycolytic proteins of M. pneumoniae and polyclonal antisera.
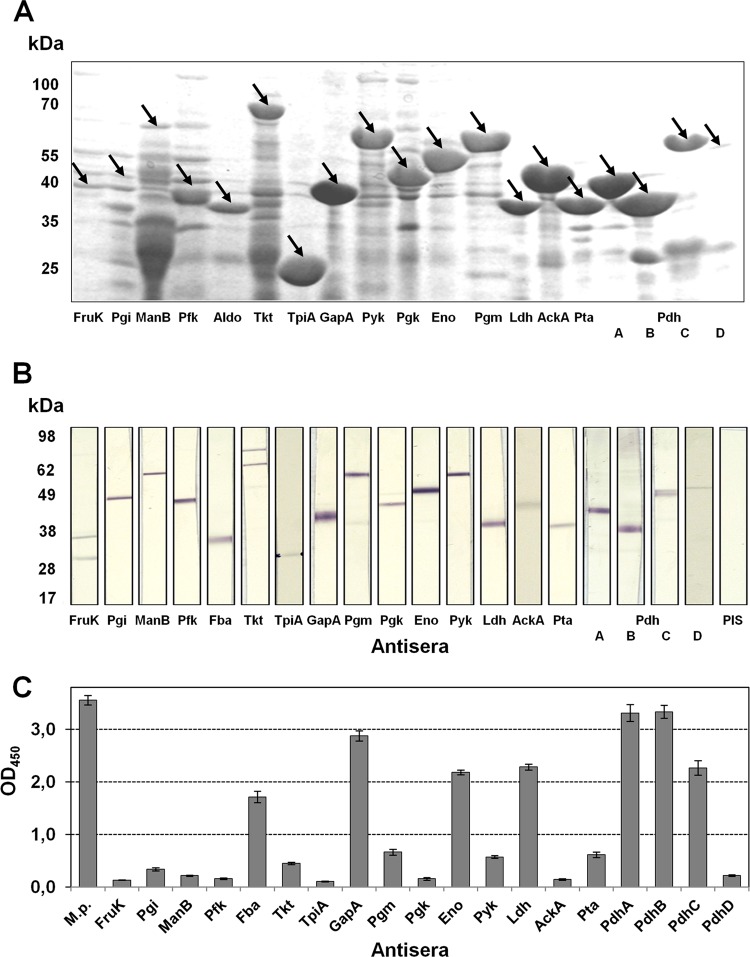
In a first step, we successfully exchanged the TGAs occurring in 14 of 19 genes that code for glycolytic enzymes. Complete genes were cloned into the pET vector and expressed in E. coli. After purification and concentration, the expression products were separated using SDS-PAGE and stained with Coomassie blue. Staining resulted in most cases in prominent proteins with molecular masses as expected for the glycolytic enzyme of interest (Fig. 1A and Table 1). In addition, detection of blotted proteins with monoclonal antibody against the His tag confirmed these results (data not shown).
FIG 1.
Expression of glycolytic enzymes of M. pneumoniae in E. coli and reactivity of guinea pig antisera to recombinant proteins. (A) The expression of recombinant proteins was analyzed by SDS-PAGE and Coomassie blue staining. Arrow, expected molecular mass of glycolytic enzyme. (B) Reaction of guinea pig antisera to recombinant proteins against total proteins of M. pneumoniae in immunoblotting. PIS, preimmune serum. (C) Reactivity of antisera with total proteins of M. pneumoniae in ELISA. The experiment was repeated twice, with eight replicates each time. The results from one experiment are shown, and bars represent means and standard deviations.
The recombinant proteins were used to produce guinea pig polyclonal antibodies. Specificity was investigated by incubating blotted total proteins of M. pneumoniae with antisera, and defined signals with the expected molecular masses of corresponding proteins were demonstrated (Fig. 1B). Obviously, only antisera to FruK and Tkt showed a reaction with a further component of total proteins of M. pneumoniae. To interpret the data from other experiments, the comparative reactivity of all antibodies was quantified by ELISA with immobilized M. pneumoniae total proteins; this resulted in a great variability of optical density (OD) values (Fig. 1C). Polyclonal antibodies specific for recombinant proteins GapA, PdhA, and PdhB reached values (>2.8) that are comparable to the result obtained with serum against total proteins of M. pneumoniae. Sera to Fba, Eno, Ldh, and PdhC demonstrated reactivity of 2.0. All other antisera (anti-FruK, -Pgi, -ManB, -Pfk, -Tkt, -TpiA, -Pgm, -Pgk, -Pyk, -AckA, -Pta, and -PdhD) showed an absorbance of <0.7, indicating limited affinity of antibodies, a low concentration of antigens among total proteins of M. pneumoniae, and/or a low density of protein epitopes.
Localization of glycolytic enzymes in M. pneumoniae cells.
The first indication of the location of glycolytic enzymes in the mycoplasma cell was the quantitative reaction of antibodies against the recombinant proteins with the membrane and cytosolic protein fraction of M. pneumoniae whole cells using ELISA (Fig. 2). As expected, the antiserum to total proteins resulted in strong signals with both fractions. Acting as a control for a surface-exposed protein, antibodies to the C-terminal part of the main P1 adhesin showed low reactivity with the cytosolic protein fraction but a high OD value after incubation with the membrane fraction. In contrast, antiserum to the cytosolic protein Eno (25, 26) yielded a low OD value (0.19) with the membrane protein fraction and high absorbance after incubation with the cytosolic protein fraction. As expected from results with unfractionated total proteins, the data demonstrated a low reactivity of antibodies to different recombinant proteins with both fractions (Fruk and TpiA). The presence of other glycolytic enzymes (Pfk, Fba, Tkt, and AckA) in the cytosolic protein fraction was confirmed (OD > 0.30), but reactivity with membrane-associated proteins was low (<0.20). The sera to the remaining glycolytic enzymes (n = 12) demonstrated a measurable reaction (OD > 0.25) with the membrane protein fraction of M. pneumoniae whole cells, indicating that these enzymes are membrane associated.
FIG 2.
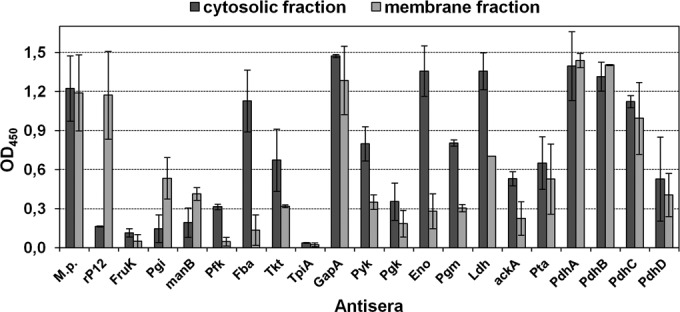
Reactivity of guinea pig antisera to glycolytic enzymes against cytosolic and membrane protein fractions of M. pneumoniae by ELISA. M.p., antiserum to whole proteins of M. pneumoniae. rP12, antiserum to C-terminal part of main P1 adhesin of M. pneumoniae (control for a confirmed membrane-associated protein region [38]). The means and standard deviations of two independent experiments with eight replicates each are shown.
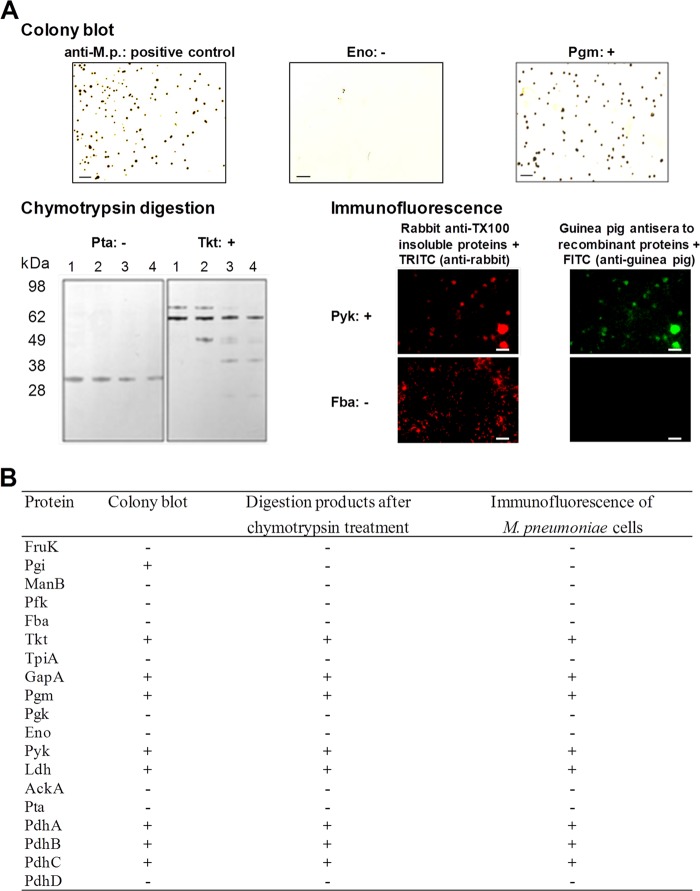
The presence of glycolytic enzymes on the surface of the cells was analyzed by colony blot, mild proteolysis, and immunofluorescence analyses of M. pneumoniae cells. The positive reaction of Pgm in the colony blot, the degradation of Tkt after treatment with chymotrypsin in contrast to stable Pta, and the immunofluorescence of mycoplasma cells after incubation with anti-Pyk are illustrated in Fig. 3 as examples. Experiments with preimmune serum remained negative in all approaches, excluding an influence of unspecific cross-reactions on the results. In contrast, the C-terminal part of P1 adhesin (positive control) was degradable, and positive colonies, as well strong immunofluorescence, were found after use of corresponding antiserum, confirming the surface localization of this protein region (data not shown). To verify degradability, recombinant proteins were treated with chymotrypsin and yielded degradation products (data not shown). Regarding the glycolytic enzymes tested, the results of the three approaches are consistent. The suitability of Eno as a control for a cytosolic protein confirmed the results of previous studies (25, 26). Overall, exposure of glycolytic enzymes on the surface of M. pneumoniae cells can be assumed for PdhA to -C, GapA, Ldh, Tkt, Pgm, and Pyk, respectively. As reported above, this result is in agreement with the presence of proteins PdhA to -C, GapA, and Ldh in the membrane-associated protein fraction, whereas Pgm and Pyk were detected in a lower but measurable proportion. Tkt was found in low concentration in both fractions.
FIG 3.
Localization of glycolytic enzymes on the surfaces of M. pneumoniae cells. (A) Examples of results. Colony blot using antiserum to whole proteins of M. pneumoniae and to recombinant proteins (scale bar, 200 μm). Chymotrypsin treatment of M. pneumoniae cells and incubation of blotted total proteins with antiserum to recombinant proteins were performed (lane 1, 0 μg; lane 2, 5 μg; lane 3, 10 μg; and lane 4, 40 μg of chymotrypsin). Immunofluorescence of fixed M. pneumoniae cells after incubation with antisera to TX100-insoluble proteins (control) and to recombinant proteins (scale bars, 10 μm), respectively, was assessed. –, Negative reaction; +, positive reaction. (B) Summary of results.
To detect the presence of specific antibodies against these surface-displayed glycolytic enzymes in an infected host, immobilized recombinant proteins were incubated with antiserum of an animal treated intranasally with viable M. pneumoniae cells (Fig. 4). Compared to total mycoplasma proteins as antigen, high OD values were measured for PdhA and PdhC only. The absorbance after incubation of serum with six other recombinant proteins remained below an OD of 0.08.
FIG 4.
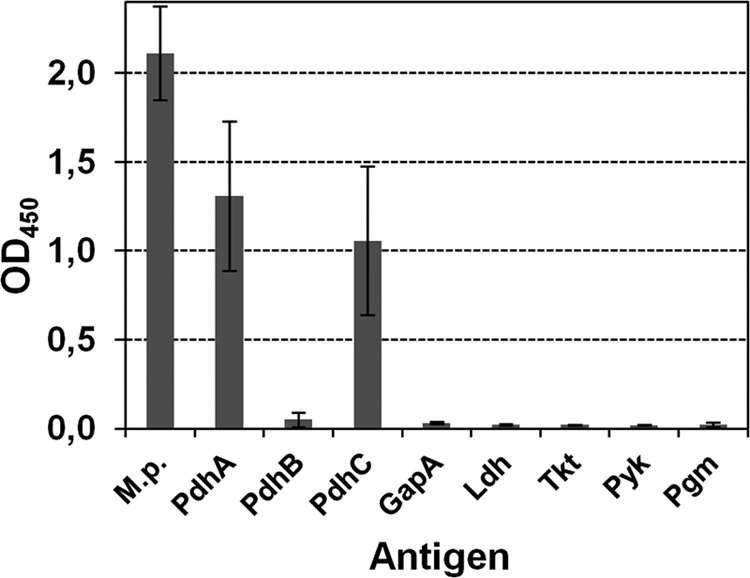
ELISA-reactivity of immobilized surface-located glycolytic enzymes and whole proteins of M. pneumoniae (M.p.) with an antiserum obtained after three intranasal infections of guinea pig with M. pneumoniae. Bars represent the means and standard deviations of two independent experiments, with eight replicates each time.
Binding of recombinant glycolytic enzymes to human Plg.
Using ELISAs, eight recombinant glycolytic enzymes of M. pneumoniae with confirmed surface localization were investigated for an interaction with Plg. All recombinant proteins are able to bind Plg in a concentration-dependent manner (see Fig. S2 in the supplemental material). Furthermore, preincubation of 2.5 μg of Plg/ml with guinea pig antibodies to the glycolytic enzymes reduces the binding of Plg to recombinant proteins significantly compared to incubation with preimmune serum (Fig. 5A), confirming the specificity of protein-protein interaction. In the presence of preimmune serum, absorbance varied between 0.78 (Tkt) and 1.31 (Ldh). The OD values of preparations with antigen-specific antisera decreased by between 60% (PdhC and Ldh) and 93% (Pgm) compared to parallel controls using preimmune serum.
FIG 5.
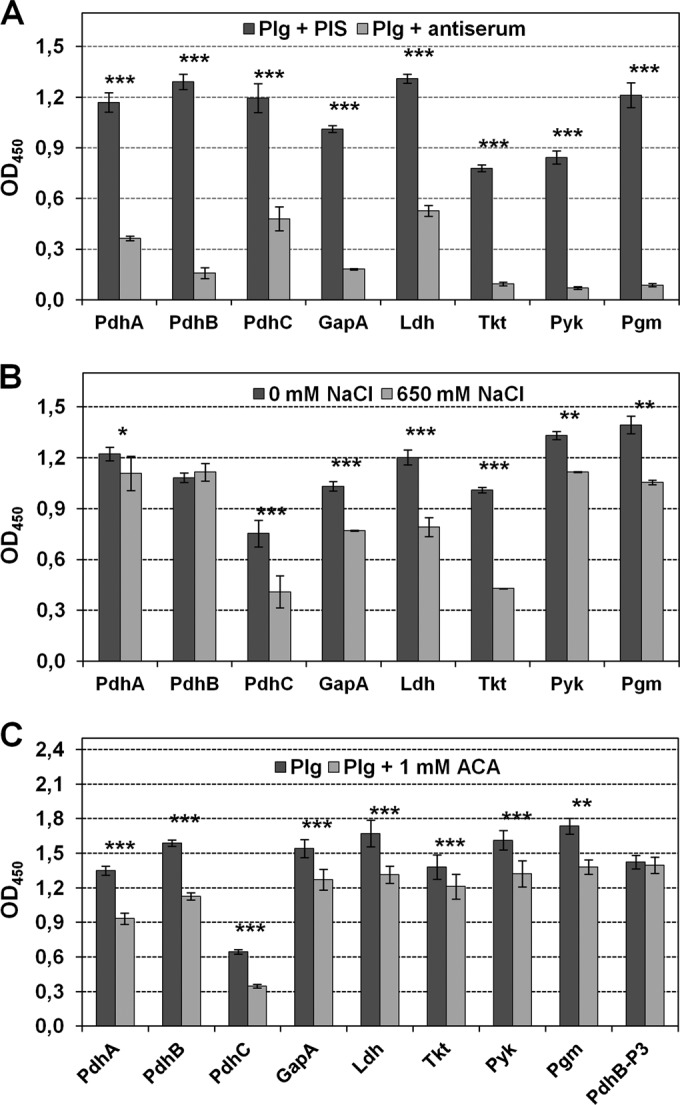
Binding of human Plg to surface-displayed glycolytic enzymes of M. pneumoniae analyzed by ELISA. (A) Influence of preincubation of Plg with corresponding guinea pig antisera (1:50) and preimmune serum (PIS) on binding to immobilized recombinant proteins. ***, P < 0.001. (B) Binding of Plg to recombinant proteins in the presence and absence of 650 mM NaCl. **, P < 0.01; *, P < 0.05. (C) Binding of Plg in the presence and absence of 1 mM ε-aminocaproic acid (ACA). Recombinant protein PdhB-P3 was used as control for a Plg-binding protein without ACA influence on this interaction (25, 26). All experiments were carried out twice, with eight wells each. The means and standard deviations of one experiment are shown.
The importance of ionic interactions for the binding of recombinant proteins with Plg was analyzed by adding increasing concentrations of NaCl to diluted Plg. Using 650 mM NaCl, Plg binding to seven of eight recombinant proteins was reduced significantly, confirming the influence of ionic forces on interactions (Fig. 5B). In contrast, the binding of Plg to recombinant protein PdhB is not dependent on the presence of NaCl up to a concentration of 650 mM. Similar experiments using NaBr and CaCl2 in increasing concentrations yielded comparable results (data not shown).
Lysine residues of microbial proteins play an important role in binding to Plg. Surface-located glycolytic enzymes of M. pneumoniae have between 5.5% (PdhB) and 8.9% (Ldh) lysine residues of total amino acids (34). Using an ELISA, addition of the lysine analog ε-aminocaproic acid (ACA) to Plg reduced binding to the eight recombinant proteins significantly (Fig. 5C), confirming the importance of lysine in all interactions. The absence of an influence on Plg binding to negative-control protein PdhB-P3 (25) was confirmed.
Binding of recombinant proteins to human A549 cells.
The lung epithelial cell line A549 was chosen to test for an interaction of recombinant proteins with human respiratory tract cells. In addition, the possible participation of Plg in protein-cell binding was analyzed. The presence of Plg resulted in strong ELISA reactivity (Fig. 6A) and fluorescence of A549 cells (Fig. 6B) after incubation with anti-Plg. In further ELISAs, immobilized A549 cells were incubated with recombinant proteins derived from the genes coding for the eight glycolytic enzymes confirmed as surface-exposed (Fig. 6C). Bound proteins were detected by using the corresponding antisera, and reactivity was compared to that of recombinant proteins without cells immobilized in the same concentration in parallel. The nonadherent central part of the P1 protein (recombinant protein rP8, negative control) demonstrated poor binding to A549 cells. This lack of reactivity was not due to the inability of the antisera to recognize bound protein as evidenced by the reactivity when rP8 was used to coat the plate well. In contrast, nearly all recombinant proteins representing the surface-displayed glycolytic enzymes of M. pneumoniae (except PdhC) demonstrated the ability to bind A549 cells (Fig. 6C). As expected, when the recombinant proteins were used to coat the wells, reactivity with each specific antiserum was at least at OD ≥ 0.5. Additionally, the binding of recombinant proteins to A549 cells can be reduced significantly by preincubation of proteins with corresponding antisera, confirming the specificity of protein-cell interaction (data not shown). Furthermore, preincubation of A549 cells with anti-Plg resulted in significantly decreased binding of seven of eight recombinant proteins with reduction rates between 12% (Pgm) and 73% (Tkt). Only the interaction of protein PdhC was not influenced (Fig. 6D), which had low adherence to A549 cells.
FIG 6.
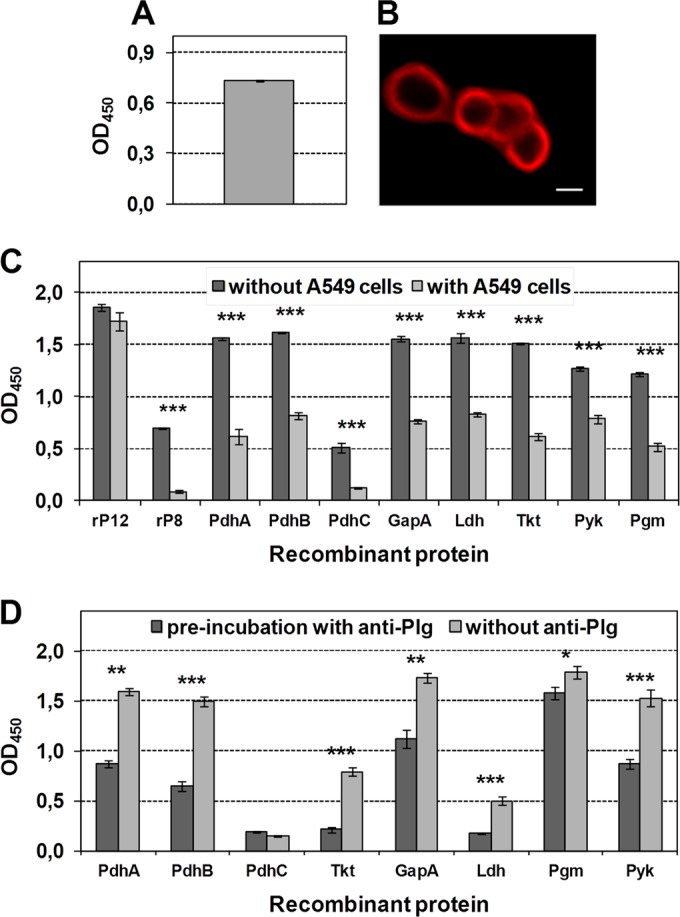
Presence of Plg on the surface of human lung epithelial cells and binding of recombinant proteins to cells. (A) Reactivity of immobilized A549 cells with rabbit anti-human Plg by ELISA. The means and standard deviations of eight replicates are shown. (B) Immunofluorescence of fixed A549 cells after incubation with anti-Plg and TRITC-labeled anti-rabbit IgG. Scale bar, 10 μm. (C) ELISA reactivity of immobilized recombinant proteins (without cells) compared to immobilized A549 cells, followed by incubation with recombinant proteins (with cells). All wells were incubated with guinea pig antisera to surface-localized glycolytic enzymes and HRP-conjugated anti-guinea pig IgG. The C-terminal (rP12) and central (rP8) parts of P1 adhesin were used as controls for a cell-binding and a non-cell-binding protein (26, 38). ***, P < 0.001. (D) Binding of recombinant proteins to immobilized A549 cells preincubated with anti-Plg or PBS. **, P < 0.01; *, P < 0.05. Experiments were carried out in duplicate. The data represent means and standard deviations of eight replicates of a single experiment (A, C, and D).
Activation of human Plg in the presence of recombinant proteins and degradation of human fibrinogen.
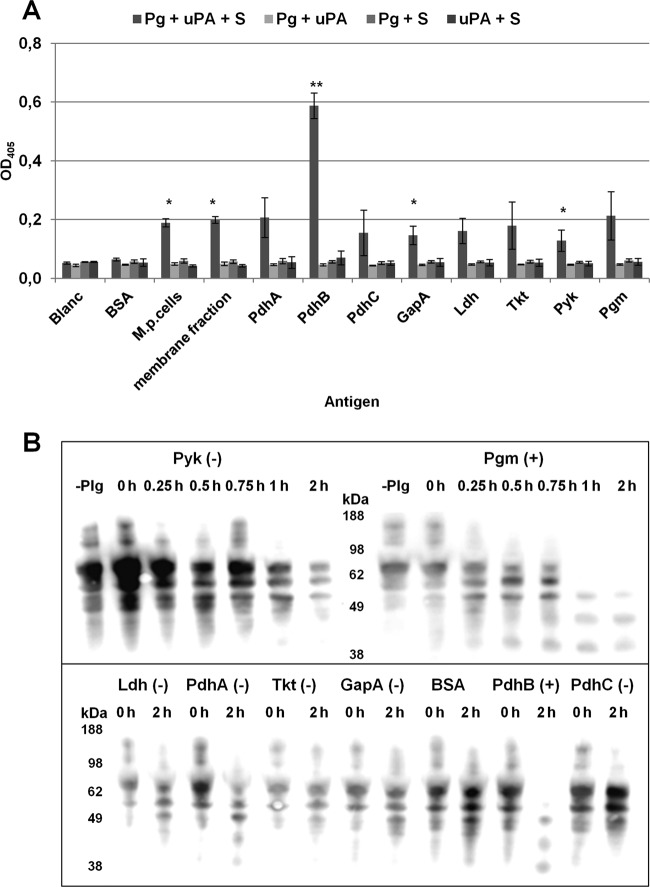
Plasmin activator (uPA) is a serine protease that converts Plg to plasmin by proteolytic cleavage at its Arg561-Val562 site. Plasmin activity can be detected by measuring the cleavage of substrate d-valyl-leucyl-lysine-p-nitroanilide dihydrochloride. The accessibility of Plg to uPA is a prerequisite for the conversion of Plg to proteolytically active plasmin. We demonstrated that Plg bound to M. pneumoniae proteins is accessible to uPA and is processed to active plasmin, whereas no cleavage of the chromogenic substrate was observed when BSA was used as a negative protein control (Fig. 7A). In comparison to controls lacking one of the components Plg, uPA, or the plasmin-specific substrate, significant elevated activity was measured in wells containing all these components and M. pneumoniae cells, proteins of the membrane-associated fraction, as well as recombinant proteins PdhB, GapA, and Pyk, respectively. It should be noted that the relatively high binding of Plg to recombinant protein PdhB (see Fig. S2 in the supplemental material) is correlated with the remarkable enzymatic activity of the Plg/PdhB complex, indicating an influence of the amount of bound Plg on plasmin generation.
FIG 7.
Activation of human Plg in the presence of recombinant proteins and degradation of human fibrinogen. (A) Conversion of Plg bound to M. pneumoniae cells, to the membrane fraction of total M. pneumoniae proteins, and to different recombinant proteins in the presence of urokinase (uPA) and the plasmin-specific substrate d-Val-Leu-Lys-p-nitroanilide dihydrochloride (S) to plasmin was detected by measuring the absorbance at 405 nm. Controls lacking one of the three components—Plg, uPA, and S—were included. BSA was used as negative control. Bars represent means and standard deviations of three independent experiments with eight replicates each. Statistically significant differences to the negative control (BSA) are shown. **, P < 0.01; *, P < 0.05. (B) Time-dependent degradation of fibrinogen was investigated by incubation of immobilized recombinant proteins and BSA (negative control) with Plg. After washing, uPA and fibrinogen were added, and degradation products were determined with goat anti-fibrinogen and HRP-conjugated anti-goat IgG. Pyk and Pgm are shown as examples to demonstrate the absence of degradation and time-dependent degradation of fibrinogen (upper panel). The influence of other surface-displayed glycolytic enzymes and BSA on fibrinogen is presented for time points at 0 and 2 h (lower panel). +, Degradation of fibrinogen; −, no degradation.
Plasmin activity can also be demonstrated by detecting the degradation of fibrinogen which is digested by plasmin. Using Western blotting, time-dependent digestion of fibrinogen was shown for recombinant proteins PdhB and Pgm, whereas fibrinogen remained unaltered in the presence of PdhA, PdhC, Pyk, Ldh, GapA, Tkt, and the negative control BSA (Fig. 7B).
DISCUSSION
The presence of different glycolytic enzymes on the surfaces of a broad range of pathogenic, opportunistic, and commensal bacteria and their involvement not only in metabolic pathways but also in further interactions with the host suggest a common aspect of communication between microbes and the environment (11, 14, 41). Especially in mycoplasmas, bacteria with reduced genomes, protein multifunction may be a way to use limited genomic resources more effectively to sustain the parasitic lifestyle in defined hosts. Nevertheless, despite the fact that surface display of microbial components is an important precondition for interaction with infected tissue, evidence for the moonlighting function of a glycolytic enzyme requires identification of factor(s) on the host site. Among these, Plg is regarded as an important component that influences various factors of the human fibrinolysis system; it is believed to play a part in the dissemination of microorganisms by activation of Plg and the subsequent proteolytic activity of emerging plasmin (15, 16). Binding of glycolytic enzymes to Plg seems to be a common aspect since it has been described for many bacterial proteins, including glycolytic enzymes (41). Plg harbors lysine-binding kringle domains (15), which interact with defined sequences of bacterial proteins, as reported for Eno of S. pneumoniae (42, 43). However, interaction with Plg was demonstrated not only for enolases but also for other glycolytic enzymes (41). Additionally, Plg binding without the influence of lysine residues was confirmed (25, 44). Regarding M. pneumoniae, binding of human Plg to different SDS-PAGE-separated and blotted total proteins was shown in previous reports, demonstrating bands with a molecular mass between 35 and 60 kDa (25, 26). The Plg interaction of all eight surface-associated glycolytic enzymes in the present study might imply that the association with Plg is a multifactorial process with special importance for colonization and/or that binding of metabolic proteins to Plg is not highly specific. The latter aspect is supported by the result that only four of the Plg-binding proteins (PdhB, Tkt, Pyk, and Pgm) are able to activate Plg and two (PdhB and Pgm) to degrade human fibrinogen.
Our study shows that there are differences in Plg-binding characteristics between the surface-localized glycolytic enzymes of M. pneumoniae (Fig. 6B and C). Interaction of all proteins can be decreased by the lysine-like ligand ACA, which binds to the kringle domains K1, K2, K4, and K5 of Plg (15). Interestingly, all proteins showed a persistent interaction with Plg in the presence of ACA, indicating that some of this interaction is lysine independent. After the addition of different salts, an influence of ionic forces on Plg binding between 9% (PdhA) and 57% (Tkt) was found. As regards the interaction of protein PdhB with Plg, various concentrations of salts resulted in an absence of inhibition. In preliminary experiments we also investigated the interaction of a defined recombinant glycolytic enzyme with Plg preincubated with an alternative surface-localized glycolytic enzyme able to bind Plg. Masking of binding sites resulted in all cases in a persistent Plg interaction which is low for PdhC and high for Pgm and PdhB (see Fig. S3 in the supplemental material). Hence, Plg binding of surface-displayed M. pneumoniae proteins might be attributable to different biochemical principles.
In many cases, evidence for the moonlighting role of glycolytic enzymes of a bacterial species resulted from proteomic studies. Here, we used a targeted search for these proteins by including the complete set of glycolytic enzymes of M. pneumoniae. Available genomic data and progress in specific tools to characterize mycoplasma proteins, such as multiple mutation reaction (36), allow this approach. However, determination of surface localization of cytosolic proteins by light microscopic and fluorescence-activated cell sorting-based methods is difficult in the case of relatively small mycoplasmas. In addition, differences in the concentrations of glycolytic enzymes, which can be expected in preparations of total proteins, further complicate their detection. Interestingly, the results of colony blot, digestion with chymotrypsin, and immunofluorescence analyses led to comparable data, allowing reliable conclusions with respect to localization. It should be noted that processes such as glycosylation of proteins of interest or lysis of bacteria could influence these results. Furthermore, after investigation of the cytosolic and membrane fractions of M. pneumoniae proteins, further glycolytic enzymes were detected among membrane-associated proteins. In agreement with the results of protein characterization after Triton X-100 fractionation (28), Tkt, GapA, Ldh, PdhA, and PdhB were confirmed as surface located, whereas other proteins detected in the Triton X-100-insoluble fraction, such as Fba and Pta, are not. The presence of membrane-associated but not surface-displayed glycolytic enzymes, as reported for PdhD (26), may explain this discrepancy. The biological role of such proteins remains unclear.
The result of the complete survey of glycolytic enzymes in M. pneumoniae has extended the spectrum of known surface-displayed members of this class of proteins. Some of the proteins have been found as surface localized in other species. As an example, GapA is among the proteins which have been known for many years and in numerous species to be a multitasking surface protein (41). Other proteins are rarely described as surface displayed. Antibodies specific to PdhA, PdhB, and PdhC were detected in sera of animals infected with M. bovis (45). PdhB was shown to be surface localized in Lactobacillus plantarum and interacts with human fibronectin (46). Tkt was found to be antigenic among membrane-associated proteins of Mycoplasma mycoides subsp. capri (47). Pgm and Pyk were confirmed in different bacterial species and in Candida albicans as surface localized and partly involved in interactions with host factors (48–52). To our knowledge, the surface localization of Ldh has been reported for the first time. In contrast, unlike many other mycoplasma and nonmycoplasma species, Eno of M. pneumoniae was found to be a cytosolic protein. These results may indicate species-specific processes affecting the transport of glycolytic enzymes to the cell surface and may confirm the suitability of the approach selected to use recombinant proteins and polyclonal antibodies for detection of surface-displayed proteins.
As described for different bacterial species, the excretion of cytosolic proteins is a further aspect of interaction with the host (53). The presence of all glycolytic enzymes of M. pneumoniae investigated in the present study in the concentrated supernatant of bouillon cultures of different ages was tested and was not detected in the secretome (see Fig. S4 in the supplemental material). In consequence, surface-located proteins are not released by the lysis of cells, and interaction with host factors is linked to the functional and specific anchoring of these enzymes in the mycoplasma membrane. This hypothesis is supported by the fact that only two glycolytic enzymes with confirmed surface localization (PdhA and PdhC) induce the production of specific antibodies in an animal infected with M. pneumoniae via the intranasal route and that chymotrypsin treatment of mycoplasma cells does not result in complete digestion of proteins analyzed (see Fig. S5 in the supplemental material). These results indicated that the extent of the accessible parts of surface-displayed glycolytic enzymes seems to be protein dependent.
Future studies will have to clarify whether surface-located glycolytic enzymes of M. pneumoniae are able to interact with other host factors. Until now, GapA has also been confirmed as a binding partner of human fibrinogen (24) and PdhB as interacting with fibronectin (27). The ability of microbial proteins to bind with more than one host factor has been demonstrated for different glycolytic enzymes (41). Since the prediction of moonlighting proteins by methods of bioinformatics is still difficult (54), experiments are necessary to provide proof of binding to further factors such as vitronectin, laminin, or components of the host complement system (41, 55). Results of experiments using A549 cells preincubated with anti-Plg (Fig. 6D) showed that binding of recombinant surface-located proteins is reduced but not blocked completely. Interaction of glycolytic enzymes with alternative host factors on the surface of human cells may cause the remaining association.
In conclusion, the results of the present study confirm that eight glycolytic enzymes of M. pneumoniae are surface-displayed proteins that are able to interact with human Plg. Moreover, since four proteins activate Plg, we postulate that these interactions contribute to the successful colonization of the human respiratory tract by a common pathogen with limited genome.
Supplementary Material
ACKNOWLEDGMENTS
The study was supported by a grant from the Deutsche Forschungsgemeinschaft (German Research Society; DU 1280/1-1).
We thank S. Gäbler for technical assistance and R. Herrmann (ZMBH, University of Heidelberg, Heidelberg, Germany) for providing the rabbit antiserum against the TX-100-insoluble protein fraction of M. pneumoniae.
Funding Statement
The funder had no influence on the data or the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01071-15.
REFERENCES
- 1.Atkinson TP, Balish MF, Waites KB. 2008. Epidemiology, clinical manifestations, pathogenesis, and laboratory detection of Mycoplasma pneumoniae infections. FEMS Microbiol Rev 32:956–973. doi: 10.1111/j.1574-6976.2008.00129.x. [DOI] [PubMed] [Google Scholar]
- 2.Jain S, Williams DJ, Arnold SR, Ampofo K, Bramley AM, Reed C, Stockmann C, Anderson EJ, Grijalva CG, Self WH, Zhu Y, Patel A, Hymas W, Chappell JD, Kaufman RA, Kan JH, Dansie D, Lenny N, Hillyard DR, Haynes LM, Levine M, Lindstrom S, Winchell JM, Katz JM, Erdman D, Schneider E, Hicks LA, Wunderink RG, Edwards KM, Pavia AT, McCullers JA, Finelli L, CDC EPIC Study Team. 2015. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med 372:835–845. doi: 10.1056/NEJMoa1405870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Baum H, Welte T, Marre R, Suttorp N, Lück C, Ewig S. 2009. Mycoplasma pneumoniae pneumonia revisited within the German competence network for community-acquired pneumonia (CAPNETZ). BMC Infect Dis 9:62. doi: 10.1186/1471-2334-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumke R, Schnee C, Pletz MW, Rupp J, Jacobs E, Sachse K, Rohde G, Capnetz Study Group. 2015. Mycoplasma pneumoniae and Chlamydia spp. infection in community-acquired pneumonia, Germany, 2011-2012. Emerg Infect Dis 21:426–434. doi: 10.3201/eid2103.140927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narita M. 2010. Pathogenesis of extrapulmonary manifestations of Mycoplasma pneumoniae infection with special reference to pneumonia. J Infect Chemother 16:162–169. doi: 10.1007/s10156-010-0044-X. [DOI] [PubMed] [Google Scholar]
- 6.Kühner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, Yamada T, Maier T, Bader S, Beltran-Alvarez P, Castaño-Diez D, Chen WH, Devos D, Güell M, Norambuena T, Racke I, Rybin V, Schmidt A, Yus E, Aebersold R, Herrmann R, Böttcher B, Frangakis AS, Russell RB, Serrano L, Bork P, Gavin AC. 2009. Proteome organization in a genome-reduced bacterium. Science 326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 7.Hasselbring BM, Jordan JL, Krause RW, Krause DC. 2006. Terminal organelle development in the cell wall-less bacterium Mycoplasma pneumoniae. Proc Natl Acad Sci U S A 103:16478–16483. doi: 10.1073/pnas.0608051103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kannan TR, Baseman JB. 2006. ADP-ribosylating and vacuolating cytotoxin of Mycoplasma pneumoniae represents unique virulence determinant among bacterial pathogens. Proc Natl Acad Sci U S A 103:6724–6729. doi: 10.1073/pnas.0510644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hames C, Halbedel S, Hoppert M, Frey J, Stülke J. 2009. Glycerol metabolism is important for cytotoxicity of Mycoplasma pneumoniae. J Bacteriol 191:747–753. doi: 10.1128/JB.01103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pancholi V, Chhatwal GS. 2003. Housekeeping enzymes as virulence factors for pathogens. Int J Med Microbiol 293:391–401. doi: 10.1078/1438-4221-00283. [DOI] [PubMed] [Google Scholar]
- 11.Henderson B, Martin A. 2011. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun 79:3476–3491. doi: 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pancholi V, Fischetti VA. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate dehydrogenase with multiple binding activity. J Exp Med 176:415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Chen H, Xia Y, Cui J, Gu Z, Song Y, Chen YQ, Zhang H, Chen W. 2013. How are the non-classically secreted bacterial proteins released into the extracellular milieu? Curr Microbiol 67:688–695. doi: 10.1007/s00284-013-0422-6. [DOI] [PubMed] [Google Scholar]
- 14.Wang G, Xia Y, Cui J, Gu Z, Song Y, Chen YQ, Chen H, Zhang H, Chen W. 2013. The roles of moonlighting proteins in bacteria. Curr Issues Mol Biol 16:15–22. [PubMed] [Google Scholar]
- 15.Sanderson-Smith ML, De Oliveira DMP, Ranson M, McArthur JD. 2012. Bacterial plasminogen receptors: mediators of a multifaceted relationship. J Biomed Biotechnol 2012:272148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raymond BA, Djordjevic S. 2015. Exploitation of plasmin(ogen) by bacterial pathogens of veterinary significance. Vet Microbiol 178:1–13. doi: 10.1016/j.vetmic.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Song Z, Li Y, Liu Y, Xin J, Zou X, Sun W. 2012. α-Enolase, an adhesion-related factor of Mycoplasma bovis. PLoS One 7:e38836. doi: 10.1371/journal.pone.0038836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, Yu S, Shen X, Chen D, Qiu X, Song C, Ding C. 2011. The Mycoplasma gallisepticum α-enolase is cell surface-exposed and mediates adherence by binding to chicken plasminogen. Microb Pathog 51:285–290. doi: 10.1016/j.micpath.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Hoelzle LE, Hoelzle K, Helbling M, Aupperle H, Schoon HA, Ritzmann M, Heinritzi K, Felder KM, Wittenbrink MW. 2007. MSG1, a surface-localised protein of Mycoplasma suis is involved in the adhesion to erythrocytes. Microbes Infect 9:466–474. doi: 10.1016/j.micinf.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Schreiner SA, Sokoli A, Felder KM, Wittenbrink MM, Schwarzenbach S, Guhl B, Hoelzle K, Hoelzle LE. 2012. The surface-localized α-enolase of Mycoplasma suis is an adhesion protein. Vet Microbiol 156:88–95. doi: 10.1016/j.vetmic.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Bao S, Guo X, Yu S, Ding J, Tan L, Zhang F, Sun Y, Qiu X, Chen G, Ding C. 2014. Mycoplasma synoviae enolase is a plasminogen/fibronectin binding protein. BMC Vet Res 25:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yavlovich A, Rechnitzer H, Rottem S. 2007. α-Enolase resides on the cell surface of Mycoplasma fermentans and binds plasminogen. Infect Immun 75:5716–5719. doi: 10.1128/IAI.01049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alvarez RA, Blaylock MW, Baseman JB. 2003. Surface localized glyceraldehyde-3-phosphate dehydrogenase of Mycoplasma genitalium binds mucin. Mol Microbiol 48:1417–1425. doi: 10.1046/j.1365-2958.2003.03518.x. [DOI] [PubMed] [Google Scholar]
- 24.Dumke R, Hausner M, Jacobs E. 2011. Role of Mycoplasma pneumoniae glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in mediating interactions with the human extracellular matrix. Microbiology 157:2328–2338. doi: 10.1099/mic.0.048298-0. [DOI] [PubMed] [Google Scholar]
- 25.Thomas C, Jacobs E, Dumke R. 2013. Characterization of pyruvate dehydrogenase subunit B and enolase as plasminogen-binding proteins in Mycoplasma pneumoniae. Microbiology 159:352–365. doi: 10.1099/mic.0.061184-0. [DOI] [PubMed] [Google Scholar]
- 26.Gründel A, Friedrich K, Pfeiffer M, Jacobs E, Dumke R. 2015. Subunits of the pyruvate dehydrogenase cluster of Mycoplasma pneumoniae are surface-displayed proteins that bind and activate human plasminogen. PLoS One 10:e126600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dallo SF, Kannan TR, Blaylock MW, Baseman JB. 2002. Elongation factor Tu and E1 β subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol Microbiol 46:1041–1051. doi: 10.1046/j.1365-2958.2002.03207.x. [DOI] [PubMed] [Google Scholar]
- 28.Regula JT, Boguth G, Görg A, Hegermann J, Mayer F, Frank R, Herrmann R. 2001. Defining the mycoplasma “cytoskeleton”: the protein composition of the Triton X-100 insoluble fraction of the bacterium Mycoplasma pneumoniae determined by 2-D gel electrophoresis and mass spectrometry. Microbiology 147:1045–1057. doi: 10.1099/00221287-147-4-1045. [DOI] [PubMed] [Google Scholar]
- 29.Ling E, Feldman G, Portnoi M, Dagan R, Overweg K, Mulholland F, Chalifa-Caspi V, Wells J, Mizrachi-Nebenzahl Y. 2004. Glycolytic enzymes associated with the cell surface of Streptococcus pneumoniae are antigenic in humans and elicit protective immune responses in the mouse. Clin Exp Immunol 138:290–298. doi: 10.1111/j.1365-2249.2004.02628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De la Paz Santangelo M, Gest PM, Guerin ME, Coincon M, Pham H, Ryan G, Puckett SE, Spencer JS, Gonzalez-Juarrero M, Daher R, Lenaerts AJ, Schnappinger D, Therisod M, Ehrt S, Sygusch J, Jackson M. 2011. Glycolytic and non-glycolytic functions of Mycobacterium tuberculosis fructose-1,6-bisphosphate aldolase, an essential enzyme produced by replicating and non-replicating bacilli. J Biol Chem 18:40219–40231. doi: 10.1074/jbc.M111.259440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kainulainen V, Loimaranta V, Pekkala A, Edelman S, Antikainen J, Kylväjä R, Laaksonen M, Laakkonen L, Finne J, Korhonen TK. 2012. Glutamine synthetase and glucose-6-phosphate isomerase are adhesive moonlighting proteins of Lactobacillus crispatus released by epithelial cathelicidin LL-37. J Bacteriol 194:2509–2519. doi: 10.1128/JB.06704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez CM, Wallich R, Riesbeck K, Skerka C, Zipfel PF. 2014. Candida albicans uses the surface protein Gpm1 to attach to human endothelial cells and to keratinocytes via the adhesive protein vitronectin. PLoS One 9:e90796. doi: 10.1371/journal.pone.0090796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidl SR, Gronau K, Pietack N, Hecker M, Becher D, Stülke J. 2010. The phosphoproteome of the minimal bacterium Mycoplasma pneumoniae: analysis of the complete known Ser/Thr kinome suggests the existence of novel kinases. Mol Cell Proteomics 9:1228–1242. doi: 10.1074/mcp.M900267-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dandekar T, Huynen M, Regula JT, Ueberle B, Zimmermann CU, Andrade MA, Doerks T, Sánchez-Pulido L, Snel B, Suyama M, Yuan YP, Herrmann R, Bork P. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res 28:3278–3288. doi: 10.1093/nar/28.17.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL. 2010. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hames C, Halbedel S, Schilling O, Stülke J. 2005. Multiple-mutation reaction: a method for simultaneous introduction of multiple mutations into the glpK gene of Mycoplasma pneumoniae. Appl Environ Microbiol 71:4097–4100. doi: 10.1128/AEM.71.7.4097-4100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proft T, Herrmann R. 1994. Identification and characterization of hitherto unknown Mycoplasma pneumoniae proteins. Mol Microbiol 13:337–348. doi: 10.1111/j.1365-2958.1994.tb00427.x. [DOI] [PubMed] [Google Scholar]
- 38.Schurwanz N, Jacobs E, Dumke R. 2009. A strategy to create chimeric proteins derived from functional adhesin regions of Mycoplasma pneumoniae for vaccine development. Infect Immun 77:5007–5015. doi: 10.1128/IAI.00268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandes LG, Vieira ML, Kirchgatter K, Alves IJ, de Morais ZM, Vasconcellos SA, Romero EC, Nascimento AL. 2012. Omp1 is an extracellular matrix- and plasminogen-interacting protein of Leptospira spp. Infect Immun 80:3679–3692. doi: 10.1128/IAI.00474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koenigs A, Hammerschmidt C, Jutras BL, Pogoryelov D, Barthel D, Skerka C, Kugelstadt D, Wallich R, Stevenson B, Zipfel PF, Kraiczy P. 2013. BBA70 of Borrelia burgdorferi is a novel plasminogen-binding protein. J Biol Chem 288:25229–12243. doi: 10.1074/jbc.M112.413872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kainulainen V, Korhonen TK. 2014. Dancing to another tune: adhesive moonlighting proteins in bacteria. Biology 3:178–204. doi: 10.3390/biology3010178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergmann S, Wild D, Diekmann O, Frank R, Bracht D, Chhatwal GS, Hammerschmidt S. 2003. Identification of a novel plasmin(ogen)-binding motif in surface displayed alpha-enolase of Streptococcus pneumoniae. Mol Microbiol 49:411–423. doi: 10.1046/j.1365-2958.2003.03557.x. [DOI] [PubMed] [Google Scholar]
- 43.Derbise A, Song YP, Parikh S, Fischetti VA, Pancholi V. 2004. Role of the C-terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen-binding activities of group A streptococci. Infect Immun 72:94–105. doi: 10.1128/IAI.72.1.94-105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crowe JD, Sievwright IK, Auld GC, Moore NR, Gow NA, Booth NA. 2003. Candida albicans binds human plasminogen: identification of eight plasminogen-binding proteins. Mol Microbiol 47:1637–1651. doi: 10.1046/j.1365-2958.2003.03390.x. [DOI] [PubMed] [Google Scholar]
- 45.Sun Z, Fu P, Wei K, Zhang H, Zhang Y, Xu J, Jiang F, Liu X, Xu W, Wu W. 2014. Identification of novel immunogenic proteins from Mycoplasma bovis and establishment of an indirect ELISA based on recombinant E1 beta subunit of the pyruvate dehydrogenase complex. PLoS One 9:e88328. doi: 10.1371/journal.pone.0088328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vastano V, Salzillo M, Siciliano RA, Muscariello L, Sacco M, Marasco R. 2014. The E1 beta-subunit of pyruvate dehydrogenase is surface-expressed in Lactobacillus plantarum and binds fibronectin. Microbiol Res 169:121–127. doi: 10.1016/j.micres.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 47.Corona L, Cillara G, Tola S. 2013. Proteomic approach for identification of immunogenic proteins of Mycoplasma mycoides subsp. capri. Vet Microbiol 167:434–439. doi: 10.1016/j.vetmic.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 48.Kinnby B, Booth NA, Svensäter G. 2008. Plasminogen binding by oral streptococci from dental plaque and inflammatory lesions. Microbiology 154:924–931. doi: 10.1099/mic.0.2007/013235-0. [DOI] [PubMed] [Google Scholar]
- 49.Wu Z, Zhang W, Lu C. 2008. Immunoproteomic assay of surface proteins of Streptococcus suis serotype 9. FEMS Immunol Med Microbiol 53:52–59. doi: 10.1111/j.1574-695X.2008.00401.x. [DOI] [PubMed] [Google Scholar]
- 50.Katakura Y, Sano R, Hashimoto T, Ninomiya K, Shioya S. 2010. Lactic acid bacteria display on the cell surface cytosolic proteins that recognize yeast mannan. Appl Microbiol Biotechnol 86:319–326. doi: 10.1007/s00253-009-2295-y. [DOI] [PubMed] [Google Scholar]
- 51.Blom AM, Bergmann S, Fulde M, Riesbeck K, Agarwal V. 2014. Streptococcus pneumoniae phosphoglycerate kinase is a novel complement inhibitor affecting the membrane attack complex formation. J Biol Chem 289:32499–32511. doi: 10.1074/jbc.M114.610212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez CM, Wallich R, Riesbeck K, Skerka C, Zipfel PF. 2014. Candida albicans uses the surface protein Gpm1 to attach to human endothelial cells and to keratinocytes via the adhesive protein vitronectin. PLoS One 9:e90796. doi: 10.1371/journal.pone.0090796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Götz F, Yu W, Dube L, Prax M, Ebner P. 2015. Excretion of cytosolic proteins (ECP) in bacteria. Int J Med Microbiol 305:230–237. doi: 10.1016/j.ijmm.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 54.Beynon RJ, Hammond D, Harman V, Woolerton Y. 2014. Can bioinformatics help in the identification of moonlighting proteins? Biochem Soc Trans 42:1698–1703. doi: 10.1042/BST20140277. [DOI] [PubMed] [Google Scholar]
- 55.Singh B, Su YC, Riesbeck K. 2010. Vitronectin in bacterial pathogenesis: a host protein used in complement escape and cellular invasion. Mol Microbiol 78:545–560. doi: 10.1111/j.1365-2958.2010.07373.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.