Abstract
Streptococcus pneumoniae is the leading infectious cause of death in children in the world. However, the mechanisms that drive the progression from asymptomatic colonization to disease are poorly understood. Two virulence-associated genomic accessory regions (ARs) were deleted in a highly virulent serotype 1 clinical isolate (strain 4496) and examined for their contribution to pathogenesis. Deletion of a prophage encoding a platelet-binding protein (PblB) resulted in reduced adherence, biofilm formation, reduced initial infection within the lungs, and a reduction in the number of circulating platelets in infected mice. However, the region's overall contribution to the survival of mice was not significant. In contrast, deletion of the variable region of pneumococcal pathogenicity island 1 (vPPI1) was also responsible for a reduction in adherence and biofilm formation but also reduced survival and invasion of the pleural cavity, blood, and lungs. While the 4496ΔPPI1 strain induced higher expression of the genes encoding interleukin-10 (IL-10) and CD11b in the lungs of challenged mice than the wild-type strain, very few other genes exhibited altered expression. Moreover, while the level of IL-10 protein was increased in the lungs of 4496ΔPPI1 mutant-infected mice compared to strain 4496-infected mice, the levels of gamma interferon (IFN-γ), CXCL10, CCL2, and CCL4 were not different in the two groups. However, the 4496ΔPPI1 mutant was found to be more susceptible than the wild type to phagocytic killing by a macrophage-like cell line. Therefore, our data suggest that vPPI1 may be a major contributing factor to the heightened virulence of certain serotype 1 strains, possibly by influencing resistance to phagocytic killing.
INTRODUCTION
Streptococcus pneumoniae (the pneumococcus) is a significant cause of human morbidity and mortality and is a leading cause of pneumonia, bacteremia, meningitis, and otitis media (1). However, the mechanism by which the pathogen progresses from asymptomatic colonization to disease is poorly understood. Moreover, significant variations in virulence exist even between strains of the same serotype (2–5). For example, outbreaks of unusually severe invasive pneumococcal disease (IPD) caused by serotype 1 strains belonging to clonal lineage group B have been reported in parts of Africa (6, 7), while clustered cases of carriage without disease caused by serotype 1 strains belonging to clonal lineage A, which include the common clonal complex (CC) CC227, have been reported within Australia (8). Furthermore, disease caused by invasive lineage A strains has been reported to be of relatively low severity (9). These naturally occurring differences in virulence provide opportunities to investigate the molecular processes that drive progression to disease by comparing closely related invasive and noninvasive strains. In previous work, the pathogenesis of noninvasive and invasive serotype 1 clinical isolates was characterized in mice (10, 11). Highly virulent human strains 1861 (sequence type 3079 [ST3079] [CC217]) and 4496 (ST3018 [CC615]), belonging to serotype 1 lineages B and C, respectively, were found to rapidly invade and survive in the lungs, pleural cavity, and blood of mice. In contrast, human carriage strains, including strain 1 (ST304), were able to colonize the nasopharynx to an extent similar to that seen with the highly virulent strains but were rapidly cleared from the lungs and were not detected in either the pleural cavity or the blood. In addition, highly virulent strain 1861 induces a stronger type I interferon (IFN-1) response in the lungs shortly after challenge that facilitates the early stages of invasion of the pleural cavity and blood (10). Genomic comparisons identified 8 accessory regions (AR) >1 kb in length that were present in both highly virulent strains but were absent from the less virulent lineage A strains (11). These ARs include a region encoding a prophage (AR1), a specific variant of the variable region of pneumococcal pathogenicity island 1 (vPPI1) (AR3 and AR4), a region encoding a putative iron permease and DyP-type peroxidase (AR6), a region encoding an ABC-type transporter and ArsR family transcriptional regulator (AR7), and other regions consisting of sequences encoding putative metabolic enzymes and transcriptional regulators (AR2, AR5, and AR8) (11). In particular, the content of vPPI1 was shown to influence the relative levels of competitive fitness of D39 mutants in niches of the mouse associated with disease (11). In this study, a serotype 1-derived variant of vPPI1 encoding the PezAT toxin-antitoxin system, and a number of genes annotated as encoding metabolic enzymes such as neopullulanase, 3-hydroxyisobutyrate dehydrogenase, UDP-glucose 4-epimerase, prephenate dehydratase, and biotin carboxylase, was associated with increased invasive potential. In an earlier study, mice challenged with a TIGR4 mutant lacking pezT survived longer following intraperitoneal challenge than the wild type (12). Bioinformatic analyses show that, while the accessory component of vPPI1 is harbored by other strains and serotypes in publicly available databases, such as G54, MLV-016, and 11-BS70, these strains are all PezAT negative (11). Therefore, the configuration of vPPI1 under study appears to be more specific to lineage B and C serotype 1 strains, such as P1031 (ST303), PNI0373 (ST618), and NCTC7465 (ST615).
BLAST analysis of available S. pneumoniae genome sequences indicates that, as described above, the complete prophage of AR1 is present in the same lineage B and C serotype 1 strains as vPPI1, although portions of the phage are more widely distributed. The prophage includes a gene with sequence similarity to the platelet-binding protein gene (pblB) and a gene encoding an endolysin, both of which were shown to be required for the virulence of Streptococcus mitis in an animal model of infective endocarditis (13). In addition, the product of pblB has been shown to promote persistence within the murine nasopharynx and lungs of the serotype 14 ST46 clone as well as increased adherence to the lung and nasopharyngeal epithelium (14). However, the role that the phage plays in more highly virulent strains, such as serotype 1 clinical isolates, is not clear. Moreover, efforts to study the contributions that these ARs make to pathogenesis have to date been hampered by the genetically intractable nature of invasive serotype 1 isolates. In previous work, our laboratory was the first to construct isogenic mutants in the serotype 1 ST306 background (15). In the present study, we developed a technique to construct mutants in non-lineage A serotype 1 isolates and used it to remove the prophage and vPPI1 to determine the relative levels of phenotypic impact of these regions on the highly virulent serotype 1 4496 strain. This work documents some of the first evidence for a single region of the accessory genome contributing almost the entire difference in virulence between noninvasive and highly invasive isolates of the same serotype.
MATERIALS AND METHODS
Ethics statement.
This study was conducted in compliance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (7th edition, 2004) and the South Australian Animal Welfare Act of 1985. All animal experiments were approved by the Animal Ethics Committee of the University of Adelaide.
Bacterial strains and media.
Serotype 2 strain D39 (NCTC 7466) has been described previously (16). Strain 4496 is a highly virulent serotype 1 clinical isolate that has also been described previously (11, 17). Opaque-phase variants of all strains selected on Todd-Hewitt broth supplemented with 1% yeast extract (THY broth)-catalase plates (18) were used in all animal experiments. Strains were routinely grown in THY broth or C+Y medium (19) or on blood agar (BA). For animal inoculation, the bacteria were grown in serum broth (SB) (nutrient broth [10 g/liter peptone {Oxoid}, 10 g/liter Lab Lemco powder {Oxoid}, 5 g/liter NaCl] plus 10% [vol/vol] donor horse serum) to an absorbance at 600 nm (A600) of 0.16, which approximates 1 × 108 CFU/ml.
Construction of mutants in highly virulent serotype 1 strain 4496.
Transformation of 4496 requires chromosomal DNA as the template. Therefore, in order to construct the 4496ΔΦ and 4496ΔPPI1 mutants, the relevant mutations were first constructed in the more tractable S. pneumoniae D39 strain (i.e., D39ΔΦ and D39ΔPPI1 mutants). The erm-containing construct used to make the D39ΔPPI1 mutant was generated by overlap extension PCR, as previously described (20), using the relevant primers listed in Table 1 to amplify the flanking products from strain 4496 template DNA (primers 5 to 8) and primers J214 to J215 to amplify erm from pVA831. D39 was transformed with the product of overlap extension PCR, as previously described (21, 22), and selected on erythromycin-containing blood agar plates. In the case of the D39ΔΦ mutant, the wild-type strain does not carry the prophage, so the erm cassette was instead inserted into the position in the D39 chromosome that corresponded to the location of the prophage in strain 4496. The construct used to make the 4496ΔΦ mutant was generated by PCR amplification of flanking products from strain 4496 template DNA using primers 1 to 4 and the erm cassette as described above but using primers J293a and J215b (2). The amplified flanking products and erm cassette subsequently underwent restriction digestion using EagI-HF and XhoI-HF (NEB), followed by ligation. D39 was transformed with the resultant ligation product as described above. 4496 mutants were constructed in a 2-step process, commencing with transformation with genomic DNA (gDNA) from the appropriate D39 construct. After selection on BA plus 2 μg/ml erythromycin, genomic DNA was extracted from one of the transformants and used as the donor in a second round of transformation of strain 4496. The transformation procedure itself was a modification of that developed previously for type 1 ST306 (15). Strain 4496 was grown overnight on BA and then inoculated into a 1:1 mixture of C+Y medium and Dulbecco's modified Eagle's medium (DMEM; Gibco, Grand Island, NY) supplemented with 10% heat-inactivated fetal calf serum (FCS). After 1 h at 37°C, the culture was diluted into fresh medium and incubated for a further 2 h at 37°C. Competence-stimulating peptide 1 (CSP-1) was added to achieve a concentration of 50 ng/ml, followed 15 min later by donor DNA (approximately 1 μg). After a further 2 h of incubation at 37°C, transformation mixes were plated on BA plus erythromycin.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| 1 | GCAATACCTTTTACGAGGCTC |
| 2 | GGTGTAGATGTGTCAGACCAG |
| 3 | CTTCTTCGGCCGGTCTATAGTATACCCGACCTATC |
| 4 | GTATATCTCGAGCTTTTTCATAATAATCTCCCTATAG |
| 5 | GCGTATTAATCTCCAGTATGTC |
| 6 | TTGTTCATGTAATCACTCCTTCGAAGATTTTCTAGAGAATTTTC |
| 7 | CGGGAGGAAATAATTCTATGAGGTTCAAAATGTCCAGTT |
| 8 | CCTGATAATCTTCCTGCGTTG |
| J214 | GAAGGAGTGATTACATGAACAA |
| J215 | CTCATAGAATTATTTCCTCCCG |
| J293a | GATCATCGGCCGGGTTCGCGGGAAGTCTACTAAG |
| J215b(2) | TATATACTCGAGCTCATAGAATTATTTCCTCCCG |
A549 and Detroit 562 adherence assays.
A549 (human type II pneumocyte) and Detroit 562 (human nasopharyngeal carcinoma) cells were grown in DMEM and in a 1:1 mix of DMEM and Ham's F-12 nutrient mixture (Gibco), respectively, supplemented in both cases with 5% fetal bovine serum, 2 mM l-glutamine, 50 IU/ml penicillin, and 50 μg/ml streptomycin. Confluent monolayers in 24-well plates were washed with phosphate-buffered saline (PBS) and infected with pneumococci (approximately 5 × 105 CFU per well) in a 1:1 mixture of the respective culture medium (without antibiotics) and C+Y (pH 7.4). Plates were centrifuged at 500 × g for 5 min and then incubated at 37°C in 5% CO2 for 2.5 h. Monolayers were washed 3 times in PBS, and adherent bacteria were released by treatment with 100 μl trypsin–EDTA, followed by 400 μl 0.025% Triton X-100. Lysates were serially diluted and plated on BA to enumerate adherent bacteria.
Biofilm assays.
Biofilm formation was performed on immobilized A549 cells as previously described (23). Briefly, cells were grown until confluent in 12-well polystyrene plates and immobilized by fixation with 2% paraformaldehyde (Sigma). Immobilized cells were then washed thoroughly with sterile deionized water and supplemented with C+Y media containing 1 × 107 CFU/ml of each bacterial strain. Following incubation for 6 h, plates were sonicated for 3 s at 35 kHz in a Soniclean ultrasonic water bath (Soniclean, Thebarton, S.A., Australia). Biofilm cell counts (quantified as CFU per milliliter) were determined by dilution and plating of the resulting dispersed biofilm-derived bacterial suspensions.
Quantitation of CPS.
Capsular polysaccharide (CPS) samples were prepared by resuspending pneumococci grown in C+Y medium–PBS to an A600 of 0.5. Subsequent preparation and quantification of CPS were carried out using a uronic acid assay as described previously (24).
Quantitative Western blot analysis.
Strain 4496 and the 4496ΔPPI1 mutant were grown to an A600 of 0.2 in C+Y. Cultures were concentrated 10-fold and incubated at 37°C for 30 min in 1× PBS–0.1% sodium deoxycholate (Sigma-Aldrich). Protein concentrations were determined by the use of a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific), according to the manufacturer's instructions. Lysates containing 20 μg total protein were subjected to SDS-PAGE before transfer to nitrocellulose using an iBlot system (Life Technologies). Blots were probed with antigen-specific antisera, as described previously (25), prior to detection by the use of anti-mouse IRDye 800 and analysis using an Odyssey infrared imaging system (Li-COR). Band intensities were quantitated according to the manufacturer's instructions.
Animal studies.
Outbred 5-to-6-week-old female CD1 (Swiss) mice were used in all animal experiments. For intranasal (i.n.) challenge, mice were anesthetized by intraperitoneal (i.p.) injection of pentobarbital sodium (Nembutal; Rhone-Merieux) at a dose of 66 μg per g of body weight, followed by i.n. challenge with 50 μl of bacterial suspension containing approximately 1 × 107 CFU bacteria and SB. The challenge dose was confirmed retrospectively by serial dilution and plating on BA. For survival experiments, mice were monitored regularly for signs of illness and euthanized once moribund. All surviving mice were euthanized at 336 h postchallenge.
For the pathogenesis experiments, mice were euthanized by CO2 asphyxiation at the indicated time points. Blood was collected by syringe from the posterior vena cava. The pleural cavity was lavaged with 1 ml sterile PBS containing 2 mM EDTA introduced through the diaphragm. Pulmonary vasculature was perfused by infusion of sterile PBS through the heart. Lungs were subsequently excised into 2-ml vials containing 1 ml sterile PBS and 2.8-mm-diameter ceramic beads for CFU counts. To obtain unattached pneumococci, the nasopharynx was subjected to lavage by insertion of a 26-gauge needle sheathed in tubing into the tracheal end of the upper respiratory tract and injection of 1 ml 0.5% trypsin–1× PBS through the nasopharynx and collection from the nares. Additionally, the upper palate and nasopharynx were excised and placed into 2-ml vials containing 1 ml sterile PBS and 2.8-mm-diameter ceramic beads to obtain attached pneumococci. CFU counts for both the nasal wash and nasal tissue samples were combined to determine the total number of bacteria in the nasopharynx. Lung and nasopharyngeal tissues were homogenized using a Precellys 24 tissue homogenizer (Bertin Technologies) at 3 cycles of 30 s and 5,000 rpm. At each time point, a 40-μl aliquot of homogenate was serially diluted in SB and plated on BA to determine the number of CFU present. Aliquots (20 μl) of blood and pleural lavage samples were serially diluted and plated on BA to determine the number of CFU in these niches. Data were analyzed in GraphPad Prism using unpaired t tests, as described in figure legends.
Isolation of RNA.
Lungs were excised from resting (mock-infected) and S. pneumoniae-challenged mice (n = 4 per group), following perfusion as described above, and transferred to 2-ml vials containing 1 ml TRIzol reagent (Life Technologies) and 2.8-mm-diameter ceramic beads for immediate homogenization as described above. RNA was isolated from TRIzol-treated samples per the manufacturer's instructions. All RNA samples were purified using an RNeasy RNA minikit (Qiagen) per the manufacturer's instructions, including on-column DNase treatment (Qiagen) performed to remove any contaminating genomic DNA (gDNA) before use in quantitative arrays. Removal of gDNA was confirmed by PCR amplification of the gene encoding GAPDH (glyceraldehyde-3-phosphate dehydrogenase) with and without reverse transcriptase.
PCR arrays.
cDNA synthesis was carried out on the RNA extracted as described above using a RT2 First Strand kit (Qiagen). RNA was analyzed using a LightCycler 480 II system (Roche) by quantitative reverse transcription-PCR (qRT-PCR) and a RT2 Profiler PCR Array Mouse Innate and Adaptive Immune Responses kit (Qiagen), according to the instructions of the manufacturers. Data were analyzed using PCR Array data analysis software provided by the manufacturer.
Cytokine quantification by ELISA.
The concentrations of IFN-γ, IL-10, CXCL10, CCL2, and CCL4 were determined in lung homogenates (n = 5) using an enzyme-linked immunosorbent assay (ELISA) and a Quantikine ELISA system (R&D Systems), according to the manufacturer's instructions. Homogenized lung tissue at 6 h post-intranasal challenge was subjected to centrifugation at 21,000 × g for 5 min at 4°C. Supernatants were then assayed by ELISA. Absorbance readings were performed on a PHERAstar FS microplate reader (BMG Lab Tech). Calculations to determine the concentration of each cytokine were performed by regression analysis according to the kit manufacturer's instructions.
Cell culture and differentiation of THP-1 cells into a macrophage-like cell type.
All tissue culture media and reagents were obtained from Gibco. THP-1 cells (ATCC TIB-202) were grown in 95% air and 5% CO2 at 37°C in complete RPMI medium (RPMI medium with phenol red, supplemented with 10% FCS, 10 mM HEPES, 50 IU/ml penicillin, and 50 μg/ml streptomycin). Flasks (25 cm2) were seeded with 1 × 105 THP-1 cells and differentiated by adding phorbol 12-myristate 13-acetate (PMA) to reach a final concentration of 100 ng/ml and incubating for 3 days at 37°C in 95% air and 5% CO2. The differentiated cells attached to the plastic surface. Following the 3-day incubation, cells were washed twice with complete RPMI medium and then rested at 37°C in 95% air and 5% CO2 and complete RPMI medium for a further 3 days.
Macrophage killing assay.
Resting differentiated THP-1 cells were detached by using StemPro Accutase cell dissociation reagent (Gibco) and resuspended in fresh Hanks' balanced salt solution (HBSS). S. pneumoniae strains were freshly grown to mid-log phase and added to the macrophage culture (bacterium/macrophage ratio, 10:1) in a 1.5-ml tube. Survival of the internalized bacteria was determined after incubation of bacteria and macrophages for 2 h followed by incubation with antibiotics for different time periods. To evaluate the internalized bacteria, antibiotics (10 μg/ml penicillin and 200 μg/ml gentamicin) in HBSS were applied for 30 min at 37°C to kill extracellular bacteria. Samples at each time point were lysed with 0.025% trypsin, and bacterial CFU counts were determined on the blood agar plates. The survival rate was calculated from comparisons of the CFU count at each time point to the CFU count obtained at 30 min post-antibiotic treatment (assumed to be 100%). All the assays were performed in biological triplicates, and statistical significance was analyzed using a two-tailed unpaired t test.
RESULTS
Adherence and biofilm formation by 4496ΔΦ and 4496ΔPPI1 mutants.
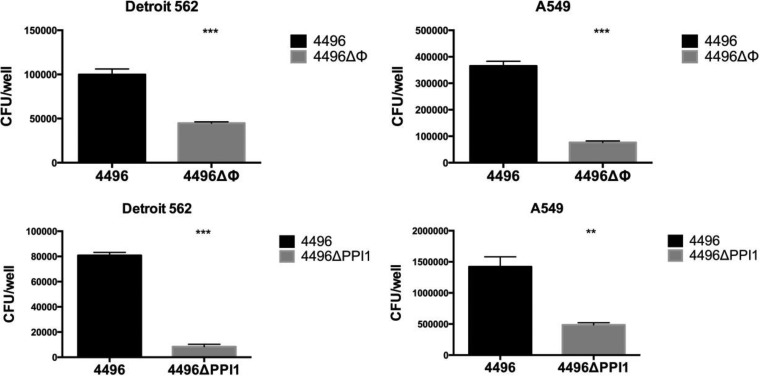
In order to assess the contribution that the PblB-encoding prophage and vPPI1 make to the virulence of strain 4496, isogenic mutants were constructed as described in Materials and Methods, generating 4496ΔΦ and 4496ΔPPI1 mutants, respectively. Mutant constructs were confirmed by PCR and sequence analysis (results not shown). In vitro growth rates of strain 4496 and the 4496ΔΦ and 4496ΔPPI1 mutant strains were compared; the results showed no differences in growth rates in either C+Y or SB media (data not shown). The relative capacities of the two mutants and the wild-type 4496 strain to adhere to A549 and Detroit 562 cells were then assayed (Fig. 1). Both mutants exhibited significantly lower adherence to both cell types than the wild type. For the 4496ΔΦ mutant strain, adherence to A549 cells was 20.9% of that of 4496, while adherence to Detroit 562 cells was 44.9% of that of the wild-type strain (P < 0.001 in both cases). For the 4496ΔPPI1 mutant strain, adherence to A549 cells was 34.2% of that of 4496, while adherence to Detroit 562 cells was only 10.4% of that of the wild-type strain (P < 0.01 and P < 0.001, respectively).
FIG 1.
The capacity of the 4496 strain and the 4496ΔΦ and 4496ΔPPI1 mutants to adhere to A549 and Detroit 562 cell monolayers was examined as described in Materials and Methods. Data are means ± standard errors of the means (SEM) of CFU/well for quadruplicate assays. Significant differences in adherence relative to that for the respective 4496 control are indicated as follows: **, P < 0.01; ***, P < 0.001 (Student's t test, two-tailed).
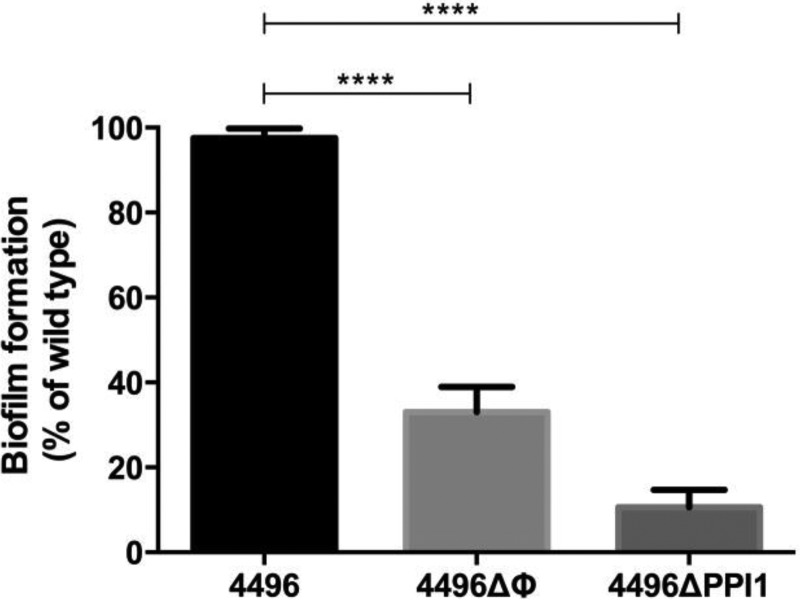
The ability of the 4496ΔΦ and 4496ΔPPI1 mutants to form a biofilm on fixed A549 cells was also compared with the biofilm-forming ability of the wild type (Fig. 2). Again, both mutants exhibited significantly lower biofilm formation capacity than 4496. Biofilm formation by the 4496ΔΦ and 4496ΔPPI1 mutants was only 30% and 10% of that of the wild type, respectively (P < 0.01 and P < 0.001, respectively).
FIG 2.
Biofilm formation by the 4496ΔΦ and 4496ΔPPI1 mutants relative to the 4496 strain. Biofilm formation on fixed A549 cells was determined after 6 h of incubation, as described in Materials and Methods. Data are expressed as means (± SEM) of percentages of that for wild type 4496. Statistical differences were analyzed by two-tailed unpaired t tests (****, P < 0.0001).
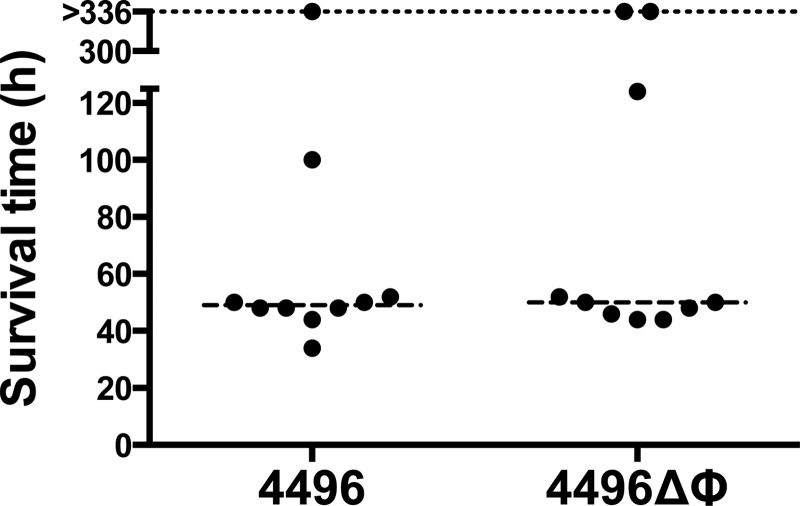
Virulence of the 4496ΔΦ mutant.
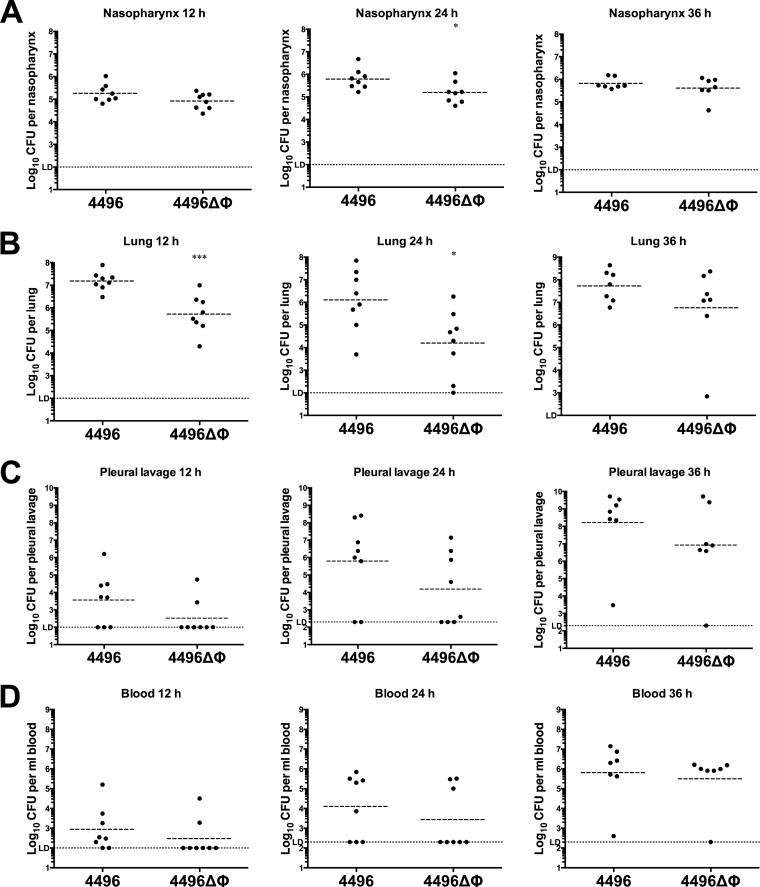
The survival times of mice following i.n. challenge with either the 4496 or 4496ΔΦ strain were initially compared (Fig. 3). However, there was no significant difference in survival rates or median survival times between the two groups. A more detailed comparison of the pathogenic profiles of the two strains was performed by quantifying the number of pneumococci present in samples from the nasopharynx, lungs, pleural lavage fluid, and blood at 12 h, 24 h, or 36 h post-i.n. challenge. The 4496ΔΦ strain colonized the nasopharynx to an extent similar to that shown by the wild type at both 12 h and 36 h. However, there was a small but significant reduction in nasal colonization by the 4496ΔΦ mutant at 24 h compared to the wild type (P < 0.05) (Fig. 4A). The number of mutant bacteria in the lungs was significantly lower than the number of wild-type bacteria at both 12 h and 24 h (P < 0.001 and P < 0.05, respectively) (Fig. 4B). However, numbers of the mutant bacteria in the lungs were not significantly different from those of the wild type by 36 h, suggesting that the pathogenic impact of the prophage is greatest during the initial stages of lung infection. There was no statistically significant difference between the numbers of mutant and wild-type bacteria in the pleural cavity or the blood at the three time points that were tested (Fig. 4C and D). Nevertheless, only 2 of the 8 mice infected with the 4496ΔΦ strain had detectable bacteria in either the pleural cavity or blood at 12 h compared with 5/8 and 6/8 for the respective compartments in mice infected with 4496. These findings may suggest that the 4496ΔΦ strain invades the blood more slowly than the wild-type strain.
FIG 3.
Survival times of mice challenged with the 4496 or 4496ΔΦ strain. Two groups of 10 mice were infected intranasally with 107 CFU of the 4496 or 4496ΔΦ strain. Survival times were recorded over a period of 336 h. Data represent survival times (in hours) for each mouse. The broken line denotes the median survival time for each group.
FIG 4.
Pathogenesis of the 4496 strain versus the 4496ΔΦ strain. Two groups of 8 mice were infected intranasally with 107 CFU of the 4496 or 4496ΔΦ strain. At 12 h, 24 h, or 36 h postchallenge, numbers of pneumococci in the nasopharynx (A), lungs (B), pleural cavity (C), and blood (D) were determined. Horizontal broken lines indicate the geometric mean bacterial load for each group. The horizontal dotted line indicates the limit of detection (LD). At the 12-h time point, this equates to 102 CFU in all niches; at 24 h and 36 h, this equates to 102 CFU in nasopharynx and lung and to 2 × 102 CFU in the pleural lavage fluid and blood. Statistical differences were analyzed by two-tailed unpaired t tests performed on log-transformed data (*, P < 0.05; ***, P < 0.001).
Impact of the PblB-encoding phage on circulating platelets.
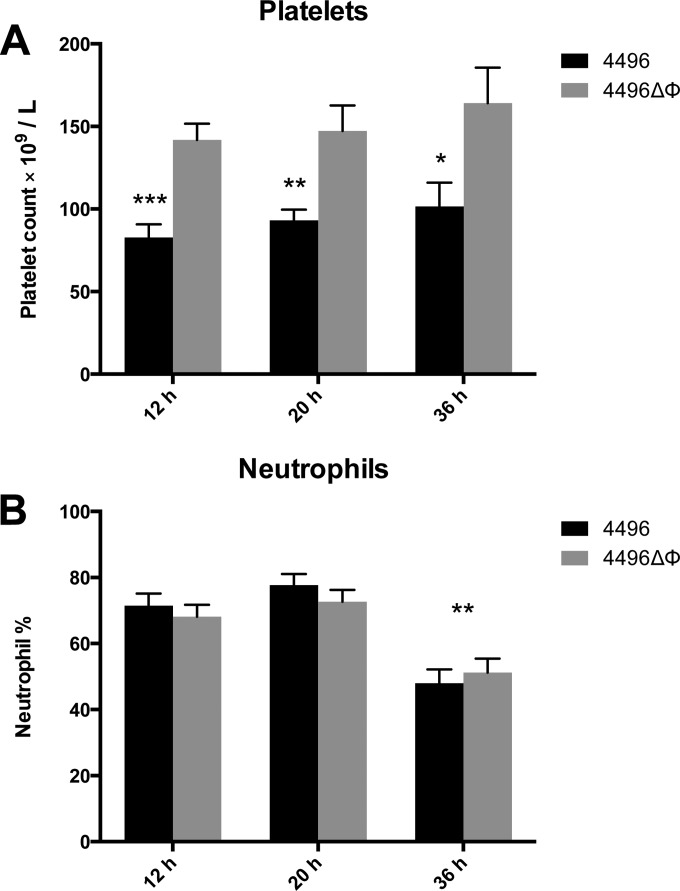
Previous work has shown that the protein encoded by other pblB genes mediates attachment to platelets (14, 26). Therefore, we investigated whether the ΔΦ mutation would lead to any change in the number of platelets in the peripheral circulation. The data presented are representative of the results of two separately conducted experiments. Blood was taken at 12 h, 20 h, and 36 h post-i.n. challenge. Platelet counts revealed a significant reduction in the number of circulating platelets in mice challenged with the wild type compared to the mutant at 12 h (P < 0.001), 20 h (P < 0.01), and 36 h (P < 0.05) (Fig. 5A). There was no change in the number of circulating platelets between time points for either challenge group, which suggests that the progression of bacteremia has no impact on the number of circulating platelets. No difference in the amount of platelet aggregation was observed in blood smears between strain 4496-infected and 4496ΔΦ mutant-infected mice (data not shown). Neutrophil counts were also performed at each time point. However, while overall neutrophil numbers as a proportion of white blood cells were elevated to an extent consistent with infection at all of the time points, there was no difference between the mutant and wild type (Fig. 5B). The proportion of neutrophils was significantly reduced in both challenge groups once mice had reached the fulminant stage of disease at 36 h compared to the two earlier time points (P < 0.001). However, the ΔΦ mutation had no detectable impact on the number of neutrophils as a proportion of total white blood cells during bacteremia.
FIG 5.
Platelet and neutrophil counts in the peripheral blood of strain 4496 and 4496ΔΦ mutant-infected mice. (A) Data are platelet counts ± SEM (n = 8 at 36 h; n = 10 at 12 h and 20 h) post-i.n. challenge. (B) Data are neutrophil counts (means ± SEM; n = 10) for each challenge group, expressed as percentages of total white blood cells. Statistical differences were analyzed by two-tailed unpaired t tests (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
PPI1 is important for the invasiveness of strain 4496.
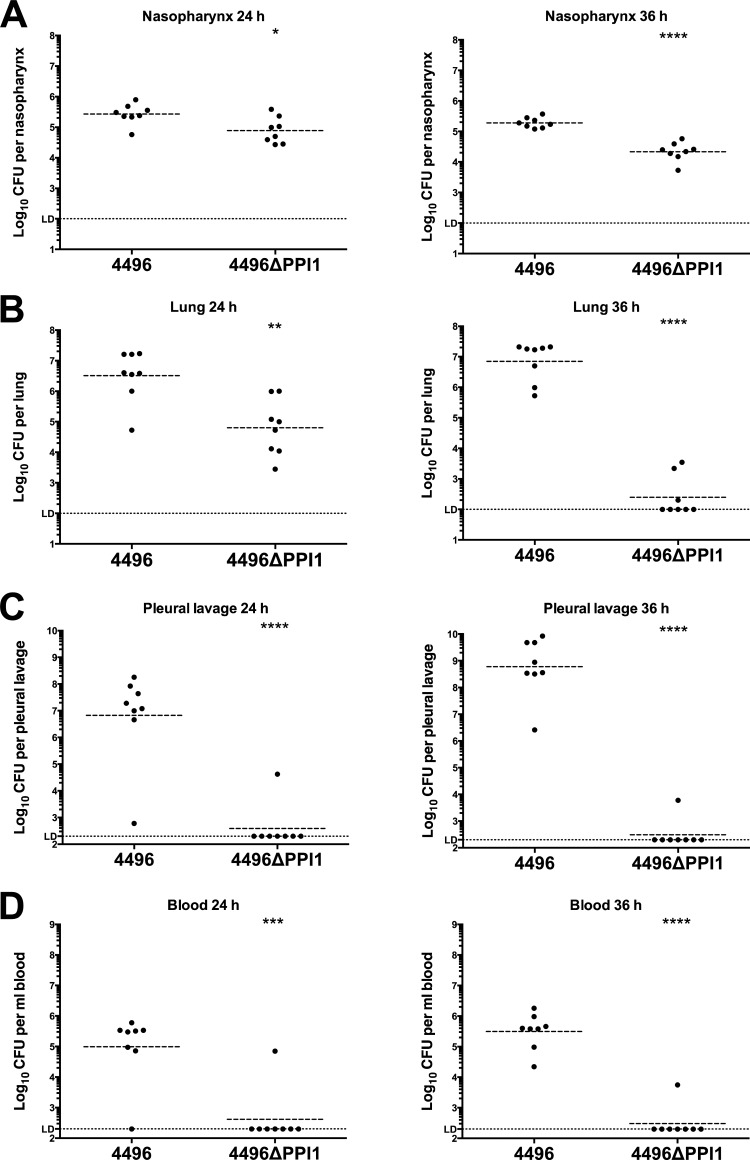
The pathogenic profiles of strain 4496 and the 4496ΔPPI1 mutant were compared by quantifying the number of pneumococci in the nasopharynx, lungs, pleural cavity, and blood at 24 h and 36 h post-i.n. challenge. A small but significant difference in nasopharyngeal colonization results was observed at both 24 h and 36 h (P < 0.05 and P < 0.00001, respectively), with 0.51- and 1-log10 lower geometric mean (GM) CFU counts for the mutant strain at the respective time points (Fig. 6A). However, differences in bacterial loads in the lungs were much more pronounced, with the GM CFU count for the 4496ΔPPI1 mutant at 24 h 1.7-log10 lower than for mice infected with wild-type 4496 (P < 0.01) (Fig. 6B). By 36 h, numbers of 4496ΔPPI1 mutant bacteria in the lungs had diminished further and were below the limit of detection in 5 of 8 mice, whereas lung bacterial loads in 4496-infected mice remained at approximately 107 CFU (P < 0.0001) (Fig. 6B). However, the differences in bacterial loads between strain 4496- and 4496ΔPPI1 mutant-infected mice were greatest in the pleural cavity and blood. In the former compartment, GM CFU counts for strain 4496 were approximately 107 and 109 at 24 h and 36 h, respectively. In stark contrast, only 1 of 8 4496ΔPPI1 mutant-infected mice had detectable CFU in the lungs at each of the two time points, and even in these 2 animals, bacterial loads were 2- and 5-log10-fold lower at 24 h and 36 h, respectively (P < 0.0001 at both time points) (Fig. 6C). Similarly, in the blood, 7 of 8 and 8 of 8 4496-infected mice were bacteremic at both 24 h and 36 h, respectively (GM CFU/ml, approximately 105 and 5 × 105, respectively), compared with 1 of 8 4496ΔPPI1 mutant-infected mice at both time points (P < 0.001 and 0.0001, respectively) (Fig. 6D). Therefore, we conclude that vPPI1 plays a critical role in the virulence of 4496 by enabling the strain to survive and proliferate in the lungs and then to invade the pleural cavity and blood.
FIG 6.
Pathogenesis of the 4496 strain versus the 4496ΔPPI1 mutant in mice. Two groups of 8 mice were infected intranasally with 107 CFU of the 4496 or 4496ΔPPI1 strain. At 24 h and 36 h postchallenge, numbers of pneumococci in the nasopharynx (A), lung (B), pleural cavity (C), and blood (D) were determined. Horizontal broken lines indicate the geometric mean bacterial load for each group. The horizontal dotted line indicates the limit of detection (LD). At the 12-h time point, this equates to 102 CFU in all niches; at 24 h and 36 h, this equates to 102 CFU in nasopharynx and lung and to 2 × 102 CFU in the pleural lavage fluid and blood. Statistical differences were analyzed by two-tailed unpaired t tests performed on log-transformed data (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Expression of known pneumococcal virulence factors by the 4496ΔPPI1 mutant strain.
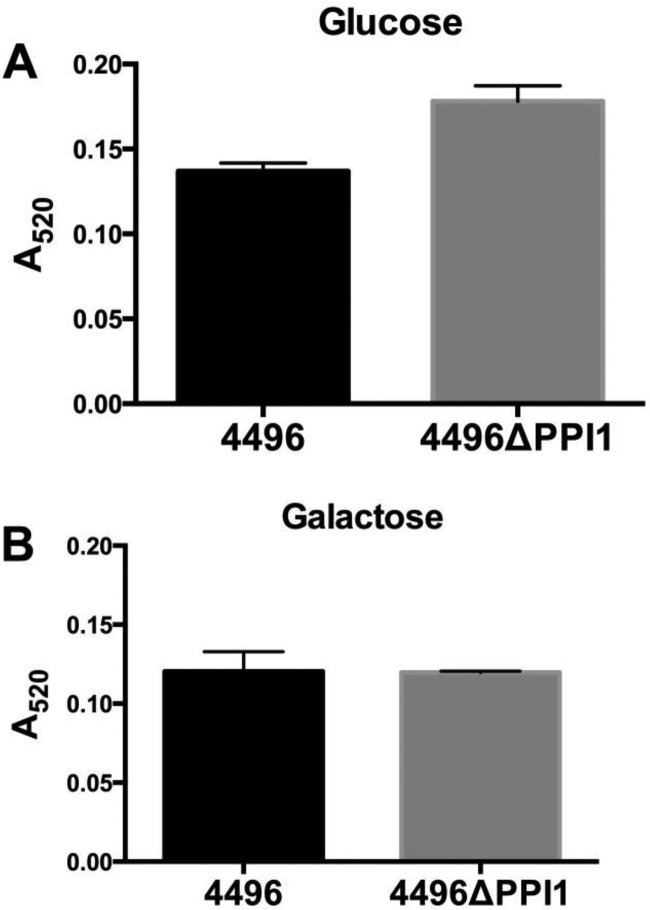
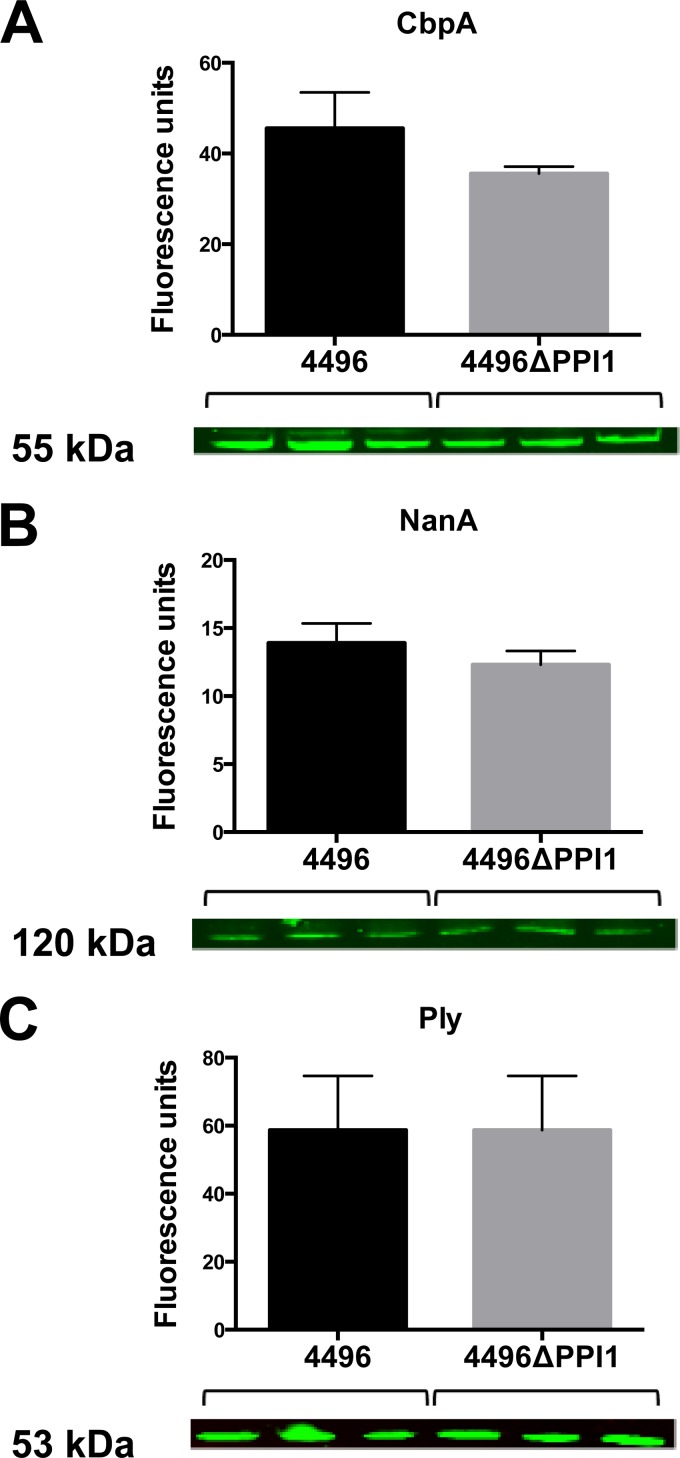
A number of the genes present within vPPI1 of strain 4496 exhibit sequence similarity to metabolic pathway genes (11). One such example is the UDP-glucose 4-epimerase gene galE1, which is responsible for the interconversion of UDP-glucose and UDP-galactose. Since UDP-glucose is a precursor for CPS synthesis, we investigated whether the loss of vPPI1 impacted in vitro CPS production by strain 4496 with either glucose or galactose as the carbon source. Using the uronic acid assay, we found no significant difference in the amount of capsule between the 4496 and 4496ΔPPI1 strains with either carbon source (Fig. 7). In order to determine whether the loss of these putative metabolic enzymes could have a secondary effect on the expression of other virulence factors, total levels of choline-binding protein A (CbpA), neuraminidase A (NanA), and pneumolysin (Ply) were compared between the 4496 strain and the 4496ΔPPI1 mutant strain by quantitative Western blot analysis (Fig. 8). These proteins were chosen because of their well-known roles in adherence to respiratory epithelial cells, nasopharyngeal colonization, and generation of inflammatory responses, particularly in the lung. No significant differences between the 4496 and 4496ΔPPI1 strains in the relative expression levels of CbpA, NanA, and Ply were detected.
FIG 7.
Capsular polysaccharide production by strain 4496 and the 4496ΔPPI1 mutant with either glucose or galactose as the carbon source. Total CPS production was quantitated colorimetrically using a uronic acid assay, as described in Materials and Methods. Data are means ± SEM of the results of A520 determinations for biological duplicates. Cells were grown in C+Y medium with either glucose (A) or galactose (B) as the carbon source. Differences between mean A520 values were analyzed using the two-tailed unpaired t test.
FIG 8.
Expression of key pneumococcal virulence factors determined by quantitative Western blot analysis. Expression levels of CbpA (A), NanA (B), and Ply (C) were compared for strain 4496 and the 4496ΔPPI1 mutant grown to mid-log phase in C+Y media using the relevant antisera. The size of the relevant protein is indicated. Data are means ± SEM (in fluorescence units) of the results of biological triplicates. Mean levels of expression were compared between strains for each target by unpaired two-tailed t tests (P < 0.05).
Immune response to strain 4496 and the 4496ΔPPI1 mutant in the lungs.
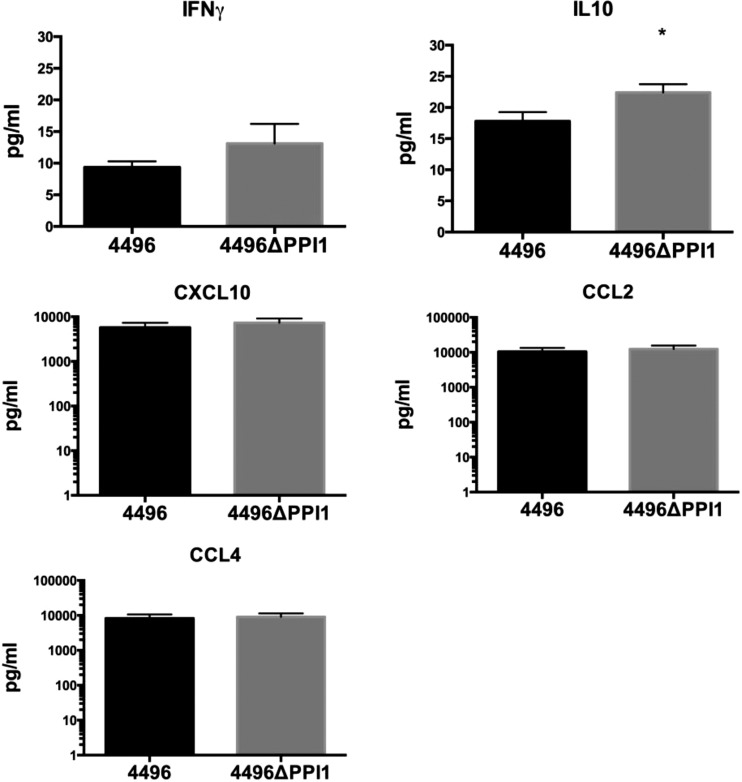
In previous work, the IFN-1 response was found to facilitate the early stages of invasion of the pleural cavity and represented the primary difference in the response to a noninvasive serotype 1 strain versus the response to a highly virulent serotype 1 strain (10). Therefore, we investigated the possibility that expression of vPPI1-carried genes might increase the strength of the IFN-1 response. The immune responses (at the transcriptional level) to the two strains were compared in the lungs at 6 h postchallenge, a time at which bacterial loads in the lungs of strain 4496-infected and 4496ΔPPI1 mutant-infected mice are similar (Fig. 9). RNA extracted from infected lungs was examined by real-time qRT-PCR analysis to compare transcription levels of 84 genes that represent the major pathways of the murine innate and adaptive immune response to microbial pathogens (data not shown). Six genes with significantly altered expression in the presence of the 4496ΔPPI1 mutant relative to strain 4496 were detected (Table 2). The genes encoding IL-10 (Il10), CD11b (Itgam), lymphocyte antigen 96 (Ly96), and Toll-like receptor 4 (Tlr4) exhibited increased expression in the presence of the 4496ΔPPI1 mutant compared to the wild type. The genes encoding lysozyme 2 (Lyz2) and retinoic acid receptor (RAR)-related orphan receptor gamma (RorC) exhibited decreased expression in the presence of the 4496ΔPPI1 mutant relative to the wild type. Therefore, the presence or absence of vPPI1 does not appear to contribute to any change in the IFN-1 response at the level of transcription. In addition to the transcriptional analyses, IFN-γ, IL-10, CXCL10, CCL2, and CCL4 were compared at the protein level in the lungs of infected mice at 6 h postchallenge (Fig. 10). These targets were chosen because they were differentially expressed in noninvasive versus invasive serotype 1 isolates in previous work (10). IL-10 expression was significantly increased in the lungs of mice challenged with the 4496ΔPPI1 mutant compared to those challenged with strain 4496 (P < 0.05). The higher level of IL-10 in the lungs of 4496ΔPPI1-infected mice is consistent with the increased Il10 transcription observed above. However, there were no significant differences in the levels of IFN-γ, CXCL10, CCL2, or CCL4 in the lungs between the two challenge groups.
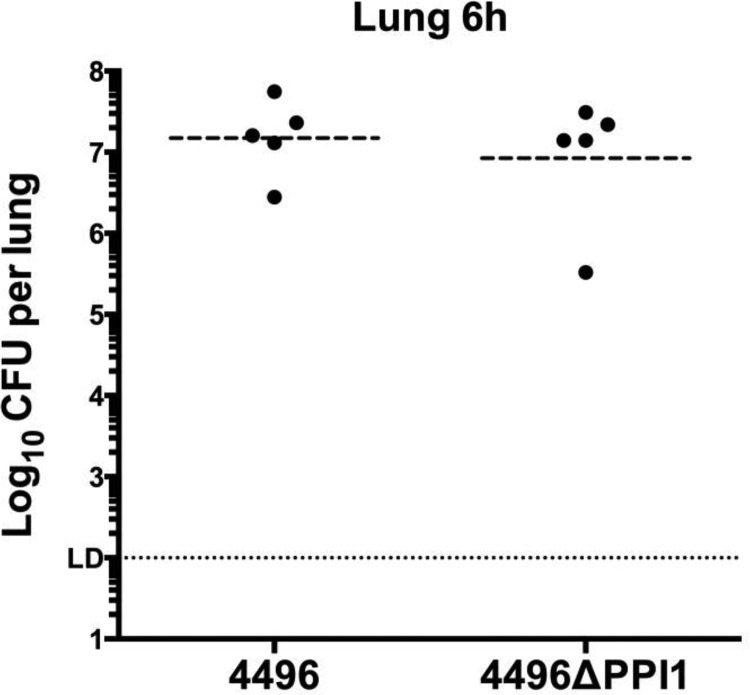
FIG 9.
Numbers of strain 4496 and 4496ΔPPI1 mutant pneumococci in the lungs at 6 h postchallenge. Two groups of 5 mice were infected intranasally with 107 CFU of the 4496 or 4496ΔPPI1 strain. At 6 h, numbers of pneumococci in the lungs were determined. Horizontal lines indicate the geometric mean bacterial load for each group. The horizontal dotted line indicates the limit of detection (LD), which equates to 102 CFU. Statistical differences were analyzed by two-tailed unpaired t tests performed on log-transformed data (P < 0.05).
TABLE 2.
Differentially expressed immune response genesa
| Gene | 4496ΔPPI1 mutant vs strain 4496 |
|
|---|---|---|
| Fold change | Significance | |
| Il10 | 3.5 | *** |
| Itgam | 3.4 | * |
| Ly96 | 2.5 | * |
| Lyz2 | −2.45 | * |
| RorC | −2.08 | ** |
| Tlr4 | 2.33 | * |
Expression levels of innate and adaptive immune response genes in the lungs of mice at 6 h postchallenge with the 4496ΔPPI1 or 4496 strain were assayed by PCR array as described in Materials and Methods. Data represent fold change in expression between 4496ΔPPI1 mutant-infected and strain 4496-infected mice. A positive value indicates higher expression in the mutant than in the wild type; a negative value indicates higher expression in the wild type than in the mutant. Only genes with statistically significant differences in expression are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (as determined by software provided by Qiagen).
FIG 10.
Concentrations of specific cytokines in lung homogenates. The concentrations of IFN-γ, IL-10, CXCL10, CCL2, and CCL4 were determined in the homogenized lung supernatants by ELISA at 6 h postchallenge. Data are the means ± SEM of the results determined for 5 mice per group in technical duplicates. The concentrations were calculated according to the instructions of the manufacturers by using regression analysis to determine a line of best fit from the relevant standards. Statistical differences were analyzed by two-tailed unpaired t tests (*, P < 0.05).
The 4496ΔPPI1 mutant is more susceptible than 4496 to killing by macrophage-like cells.
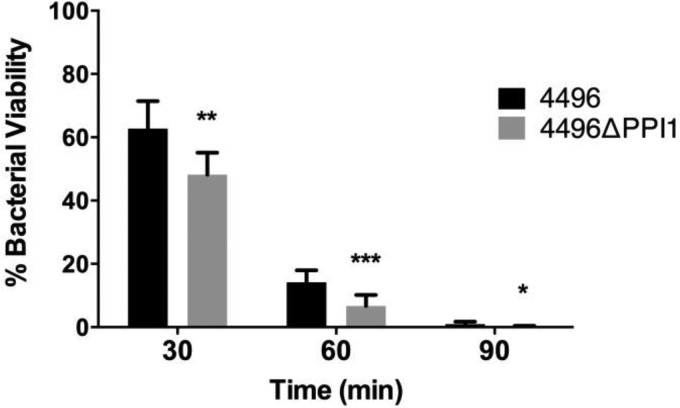
Levels of susceptibility to phagocytic killing by a macrophage-like cell line were compared between the 4496 and 4496ΔPPI1 strains. Following a 2-h coincubation, extracellular pneumococci were removed by antibiotic treatment for 30 min, and the number of viable intracellular pneumococci was quantified after a further 0, 30, 60, or 90 min of incubation. Data at 30, 60, and 90 min are expressed as a percentage of the data determined for intracellular bacteria at 0 min (Fig. 11). At 30 min, 62.7% of the strain 4496 bacteria remained viable compared to 48.1% of the 4496ΔPPI1 mutant bacteria (P < 0.01). At 60 min, 14.1% of the strain 4496 bacteria remained viable compared to 6.7% of the 4496ΔPPI1 mutant bacteria (P < 0.001), and at 90 min, 0.9% remained viable compared to 0.2% of the 4496ΔPPI1 mutant bacteria (P < 0.05). Therefore, the 4496ΔPPI1 mutant appeared to be more susceptible to phagocytic killing.
FIG 11.
Phagocytic killing of the 4496 strain versus the 4496ΔPPI1 mutant by a macrophage-like cell line. The viabilities of the 4496 strain and the 4496ΔPPI1 mutant inside a macrophage-like cell were compared following 2 h of coincubation between the macrophage-like cell line and either the 4496 strain or the 4496ΔPPI1 mutant initially at a 1:10 ratio. Viability was quantified at 30 min, 60 min, and 90 min following gentamicin treatment performed to remove extracellular pneumococci at 0 min. Data at 30 min, 60 min, and 90 min represent the means of the results of 3 separate experiments expressed as the percentages of intracellular pneumococci at 0 min. Statistical differences between the two strains were analyzed at each time point by two-tailed unpaired t tests (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
To date, very little is known about the association between the accessory genome of the pneumococcus and the various propensities of different strains to cause disease. In previous work, invasive and noninvasive serotype 1 clinical isolates were compared at the genomic level, and in terms of virulence profiles and the early immune response in mice, to help understand why some strains cause disease more readily than others. In the present study, a prophage encoding a putative platelet-binding protein and a variant of the vPPI1 were analyzed to determine their contribution to the heightened virulence of a highly invasive serotype 1 strain. The ΔΦ mutation was found to have a modest impact on the ability of strain 4496 to establish early infection within the lungs. However, this deficiency appeared to diminish with time. Moreover, no differences were observed in bacterial loads in the pleural cavity or blood at any of the time points tested. The reduced ability of the 4496ΔΦ mutant to adhere to live A549 cells and its reduced capacity to form a biofilm on fixed A549 monolayers are consistent with the reduced capacity of the mutant to establish infection within the lungs which was observed following intranasal challenge in mice. The impact of the ΔΦ mutation on adherence to Detroit 562 nasopharyngeal cells was less pronounced, which is consistent with the lesser, but still statistically significant, effect on colonization of the murine nasopharynx at 24 h. These findings are also consistent with the recently published report that phage-encoded PblB plays a role in adherence to the lung epithelium in serotype 14 pneumococci (14). The fact that a difference was observed in the present study only at early time points, whereas the mutant serotype 14 strain was reduced in numbers in the lung over longer time periods, may be a function of the distinct virulence profiles. Typically, serotype 14 strains are not highly virulent in mouse models, whereas type 1 strains such as 4496 may cause fulminant sepsis within 2 days of challenge. Interestingly, in our study significantly fewer platelets were found in the circulation of mice challenged with the wild type than in those challenged with the phage-deficient strain. While differences in platelet aggregation were not observed in blood smears, the PblB-encoding phage clearly has some impact on the number of circulating platelets, perhaps through its recently reported ability to mediate attachment to platelets (14). However, while the AR1 phage appears to have some impact on the early stages of lung infection by the serotype 1 strain, the mutant and the wild type progress to fulminant infection at similar rates. Therefore, AR1 on its own is unlikely to be responsible for the vast differences in virulence between the highly invasive strains and the less virulent lineage A serotype 1 strains.
In contrast to the 4496ΔΦ mutant, the 4496ΔPPI1 mutant was severely attenuated in its ability to persist within the lungs and invade and survive in either the pleural cavity or blood. However, the 4496ΔPPI1 mutant was attenuated only modestly in its ability to colonize the nasopharynx. It is interesting that the pathogenic profile of the 4496ΔPPI1 strain closely mirrored that of the naturally noninvasive type 1 lineage A strain studied in previously published work (10, 11). Thus, it appears that the particular variant of vPPI1 carried by the highly invasive serotype 1 strains may be a significant factor contributing to their heightened virulence in mice and perhaps to the unusually severe disease caused by closely related strains in humans. Notwithstanding the facts noted above, it is interesting that, while the IFN-1 response was found to play a role in the earliest stages of invasion of the pleural cavity by a closely related serotype 1 strain in previous work, vPPI1 plays no role in inducing this response. Instead, there were very few immune function genes that were differentially expressed between the two groups, which suggests that the contribution of vPPI1 to virulence has little to do with inducing a particular type of host response at the level of transcription. However, while the expression of IL-10 was also increased at the protein level, the levels of other important cytokines such as CXCL10, IFN-γ, CCL2, and CCL4 were unaltered by the loss of vPPI1. Increased expression of Il10 and Itgam in the presence of the 4496ΔPPI1 mutant compared to the wild type suggests possible alterations in neutrophil and macrophage recruitment into the lungs (27–29). In particular, increased Il10 expression suggests greater suppression of the inflammatory response to the 4496ΔPPI1 mutant, which may be a reflection of the fact the mutant is more readily cleared from the lungs than the wild type. This is consistent with the finding that the 4496ΔPPI1 mutant appeared to be significantly more susceptible than the wild type to in vitro phagocytic killing. However, what is apparent is that, while the 4496ΔPPI1 mutant may be more susceptible to phagocytosis, the differences, if any, in the immune responses to strain 4496 versus the 4496ΔPPI1 mutant appear to be more subtle than could be detected by the methods employed in this study. This hypothesis does not discount the importance of host factors in the development of disease caused by S. pneumoniae strains more characteristically associated with opportunistic infections (9). However, those strains that tend to behave as primary pathogens may drive much more of the invasive process by direct mechanisms unrelated to modulation of host innate or adaptive immune responses.
In this study, we have provided the first evidence linking unusually severe disease caused by non-lineage A serotype 1 S. pneumoniae isolates to a specific region of its accessory genome. While we showed that the PblB-encoding prophage within AR1 does appear to play some role in the earliest stages of infection, its contribution is modest compared to that of vPPI1. Thus, future study of the gene products encoded on vPPI1 in strain 4496 will provide important information on the mechanisms that drive the progression from colonization to severe invasive disease.
ACKNOWLEDGMENTS
J.C.P. is a National Health and Medical Research Council (NHMRC) Senior Principal Research Fellow; C.T. is an Australian Research Council DECRA Fellow.
REFERENCES
- 1.O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T; Hib and Pneumococcal Global Burden of Disease Study Team. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 2.Brueggemann AB, Griffiths DT, Meats E, Peto T, Crook DW, Spratt BG. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J Infect Dis 187:1424–1432. doi: 10.1086/374624. [DOI] [PubMed] [Google Scholar]
- 3.Brueggemann AB, Peto TE, Crook DW, Butler JC, Kristinsson KG, Spratt BG. 2004. Temporal and geographic stability of the serogroup-specific invasive disease potential of Streptococcus pneumoniae in children. J Infect Dis 190:1203–1211. doi: 10.1086/423820. [DOI] [PubMed] [Google Scholar]
- 4.Sandgren A, Sjostrom K, Olsson-Liljequist B, Christensson B, Samuelsson A, Kronvall G, Henriques Normark B. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J Infect Dis 189:785–796. doi: 10.1086/381686. [DOI] [PubMed] [Google Scholar]
- 5.Henriques-Normark B, Blomberg C, Dagerhamn J, Battig P, Normark S. 2008. The rise and fall of bacterial clones: Streptococcus pneumoniae. Nat Rev Microbiol 6:827–837. doi: 10.1038/nrmicro2011. [DOI] [PubMed] [Google Scholar]
- 6.Leimkugel J, Adams Forgor A, Gagneux S, Pfluger V, Flierl C, Awine E, Naegeli M, Dangy JP, Smith T, Hodgson A, Pluschke G. 2005. An outbreak of serotype 1 Streptococcus pneumoniae meningitis in northern Ghana with features that are characteristic of Neisseria meningitidis meningitis epidemics. J Infect Dis 192:192–199. doi: 10.1086/431151. [DOI] [PubMed] [Google Scholar]
- 7.Yaro S, Lourd M, Traore Y, Njanpop-Lafourcade BM, Sawadogo A, Sangare L, Hien A, Ouedraogo MS, Sanou O, Parent du Chatelet I, Koeck JL, Gessner BD. 2006. Epidemiological and molecular characteristics of a highly lethal pneumococcal meningitis epidemic in Burkina Faso. Clin Infect Dis 43:693–700. doi: 10.1086/506940. [DOI] [PubMed] [Google Scholar]
- 8.Smith-Vaughan H, Marsh R, Mackenzie G, Fisher J, Morris PS, Hare K, McCallum G, Binks M, Murphy D, Lum G, Cook H, Krause V, Jacups S, Leach AJ. 2009. Age-specific cluster of cases of serotype 1 Streptococcus pneumoniae carriage in remote indigenous communities in Australia. Clin Vaccine Immunol 16:218–221. doi: 10.1128/CVI.00283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sjöström K, Spindler C, Ortqvist A, Kalin M, Sandgren A, Kühlmann-Berenzon S, Henriques-Normark B. 2006. Clonal and capsular types decide whether pneumococci will act as a primary or opportunistic pathogen. Clin Infect Dis 42:451–459. doi: 10.1086/499242. [DOI] [PubMed] [Google Scholar]
- 10.Hughes CE, Harvey RM, Plumptre CD, Paton JC. 2014. Development of primary invasive pneumococcal disease caused by serotype 1 pneumococci is driven by early increased type I interferon response in the lung. Infect Immun 82:3919–3926. doi: 10.1128/IAI.02067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvey RM, Stroeher UH, Ogunniyi AD, Smith-Vaughan HC, Leach AJ, Paton JC. 2011. A variable region within the genome of Streptococcus pneumoniae contributes to strain-strain variation in virulence. PLoS One 6:e19650. doi: 10.1371/journal.pone.0019650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown JS, Gilliland SM, Spratt BG, Holden DW. 2004. A locus contained within a variable region of pneumococcal pathogenicity island 1 contributes to virulence in mice. Infect Immun 72:1587–1593. doi: 10.1128/IAI.72.3.1587-1593.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell J, Siboo IR, Takamatsu D, Chambers HF, Sullam PM. 2007. Mechanism of cell surface expression of the Streptococcus mitis platelet binding proteins PblA and PblB. Mol Microbiol 64:844–857. doi: 10.1111/j.1365-2958.2007.05703.x. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh YC, Lin TL, Lin CM, Wang JT. 2015. Identification of PblB mediating galactose-specific adhesion in a successful Streptococcus pneumoniae clone. Sci Rep 5:12265. doi: 10.1038/srep12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harvey RM, Hughes CE, Paton AW, Trappetti C, Tweten RK, Paton JC. 2014. The impact of pneumolysin on the macrophage response to Streptococcus pneumoniae is strain-dependent. PLoS One 9:e103625. doi: 10.1371/journal.pone.0103625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avery OT, Macleod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvey RM, Ogunniyi AD, Chen AY, Paton JC. 2011. Pneumolysin with low hemolytic activity confers an early growth advantage to Streptococcus pneumoniae in the blood. Infect Immun 79:4122–4130. doi: 10.1128/IAI.05418-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiser JN, Austrian R, Sreenivasan PK, Masure HR. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect Immun 62:2582–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacks S, Hotchkiss RD. 1960. A study of the genetic material determining an enzyme in Pneumococcus. Biochim Biophys Acta 39:508–518. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 20.Morona JK, Paton JC, Miller DC, Morona R. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol Microbiol 35:1431–1442. [DOI] [PubMed] [Google Scholar]
- 21.Giammarinaro P, Paton JC. 2002. Role of RegM, a homologue of the catabolite repressor protein CcpA, in the virulence of Streptococcus pneumoniae. Infect Immun 70:5454–5461. doi: 10.1128/IAI.70.10.5454-5461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin B, Garcia P, Castanie MP, Glise B, Claverys JP. 1995. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls an SOS regulon. Dev Biol Stand 85:293–300. [PubMed] [Google Scholar]
- 23.Vidal JE, Howery KE, Ludewick HP, Nava P, Klugman KP. 2013. Quorum-sensing systems LuxS/autoinducer 2 and Com regulate Streptococcus pneumoniae biofilms in a bioreactor with living cultures of human respiratory cells. Infect Immun 81:1341–1353. doi: 10.1128/IAI.01096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morona JK, Morona R, Paton JC. 2006. Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc Natl Acad Sci U S A 103:8505–8510. doi: 10.1073/pnas.0602148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAllister LJ, Ogunniyi AD, Stroeher UH, Leach AJ, Paton JC. 2011. Contribution of serotype and genetic background to virulence of serotype 3 and serogroup 11 pneumococcal isolates. Infect Immun 79:4839–4849. doi: 10.1128/IAI.05663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bensing BA, Siboo IR, Sullam PM. 2001. Proteins PblA and PblB of Streptococcus mitis, which promote binding to human platelets, are encoded within a lysogenic bacteriophage. Infect Immun 69:6186–6192. doi: 10.1128/IAI.69.10.6186-6192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadioglu A, De Filippo K, Bangert M, Fernandes VE, Richards L, Jones K, Andrew PW, Hogg N. 2011. The integrins Mac-1 and alpha4beta1 perform crucial roles in neutrophil and T cell recruitment to lungs during Streptococcus pneumoniae infection. J Immunol 186:5907–5915. doi: 10.4049/jimmunol.1001533. [DOI] [PubMed] [Google Scholar]
- 28.Kirby AC, Raynes JG, Kaye PM. 2006. CD11b regulates recruitment of alveolar macrophages but not pulmonary dendritic cells after pneumococcal challenge. J Infect Dis 193:205–213. doi: 10.1086/498874. [DOI] [PubMed] [Google Scholar]
- 29.Peñaloza HF, Nieto PA, Muñoz-Durango N, Salazar-Echegarai FJ, Torres J, Parga MJ, Alvarez-Lobos M, Riedel CA, Kalergis AM, Bueno SM. 2015. Interleukin-10 plays a key role in the modulation of neutrophils recruitment and lung inflammation during infection by Streptococcus pneumoniae. Immunology 146:100–112. doi: 10.1111/imm.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]