Abstract
Urinary tract infections (UTIs) are a major burden to human health. The overwhelming majority of UTIs are caused by uropathogenic Escherichia coli (UPEC) strains. Unlike some pathogens, UPEC strains do not have a fixed core set of virulence and fitness factors but do have a variety of adhesins and regulatory pathways. One such UPEC adhesin is the nonfimbrial adhesin TosA, which mediates adherence to the epithelium of the upper urinary tract. The tos operon is AT rich, resides on pathogenicity island aspV, and is not expressed under laboratory conditions. Because of this, we hypothesized that tosA expression is silenced by H-NS. Lrp, based on its prominent function in the regulation of other adhesins, is also hypothesized to contribute to tos operon regulation. Using a variety of in vitro techniques, we mapped both the tos operon promoter and TosR binding sites. We have now identified TosR as a dual regulator of the tos operon, which could control the tos operon in association with H-NS and Lrp. H-NS is a negative regulator of the tos operon, and Lrp positively regulates the tos operon. Exogenous leucine also inhibits Lrp-mediated tos operon positive regulation. In addition, TosR binds to the pap operon, which encodes another important UPEC adhesin, P fimbria. Induction of TosR synthesis reduces production of P fimbria. These studies advance our knowledge of regulation of adhesin expression associated with uropathogen colonization of a host.
INTRODUCTION
Urinary tract infections (UTIs), which are among the most common bacterial infections of humans (1), can occur in otherwise healthy individuals when bacteria colonizing the gastrointestinal tract gain access to the periurethral area. Most individuals with UTIs develop an infection of the bladder, referred to as cystitis (1). However, the infecting bacterium may ascend the ureters to infect the kidneys (pyelonephritis) and, in some cases, enter the bloodstream leading to bacteremia and sometimes fatal urosepsis (1–4).
A diverse group of extraintestinal pathogenic Escherichia coli strains, referred to as uropathogenic E. coli (UPEC), cause the overwhelming majority of uncomplicated UTIs (2, 5). While numerous UPEC virulence factors have been identified, including adhesins, motility systems, toxins, and iron acquisition systems, a core set of virulence factors has not been strictly defined (6–8). However, it is critical to understand specific virulence factors and how they are regulated.
Previous work identified and characterized the E. coli repeats-in-toxin (RTX) nonfimbrial adhesin TosA (i.e., type one secretion as the predicted secretion mechanism) (7, 9–13). In particular, it was noted that tosA and the other tos operon genes have poor in vitro expression (9–11). TosA, a >250-kDa surface-exposed protein, mediates UPEC adherence to epithelial cells derived from the upper urinary tract (9). This is in contrast to a number of other RTX proteins, which are fully secreted into the extracellular milieu and act as toxins (14–18). We estimated that ∼32% of UPEC strains carry genes encoding TosA and its cognate type 1 secretion system, TosCBD (10). In strain CFT073, the tos operon resides on pathogenicity island aspV (PAI-aspV) (12). The tos operon, in addition to tosA and predicted cognate secretion system genes tosCBD, also contains the regulatory genes tosR, tosE, and tosF (9, 10). TosE and TosF together suppress motility (10), a feature also found in other adhesin operon regulators (19–21). TosR, a member of the PapB family, is a negative regulator of the tos operon (10).
PapB, the prototypical member of its family, is a well-characterized positive and negative transcriptional regulator of the pap operon (22–24) that encodes the structural and secretion machinery necessary for P-fimbria assembly (25, 26). P fimbriae are epidemiologically associated with UPEC strains (27) and have been shown to be important during experimental UTI (27–30). PapB mediates transcriptional regulation by binding within the DNA minor groove (31), which suggests that PapB might recognize structured DNA in a manner proposed for nucleoid-associated proteins (32–36). In addition, the well-known nucleoid-structuring protein Lrp also contributes to both positive and negative regulation of the pap operon (22, 37–39).
H-NS regulates the expression of many genes through binding structured AT-rich DNA sequences, compacting the bacterial chromosome into defined nucleoid macrodomains (40–44). PAIs are often identified by their AT richness (41, 43–45), and AT-rich genes and PAIs are often silenced by H-NS (41, 43, 44). In addition, H-NS also contributes to negative regulation of adhesin operons and dual regulation of motility operons (38, 46–58). Indeed, PapB was previously suggested to mediate positive regulation of the pap operon by anti-silencing H-NS repression (58).
Lrp and H-NS are key regulators associated with a variety of other genes, including those coding for adhesins in addition to P fimbriae (19, 22, 37-39, 43, 46-57, 59-67). In agreement with this, others have noted the possibility that Lrp and H-NS antagonize the activity of each other or could interact together to potentiate gene regulation (35, 38, 59, 68). This type of regulation resembles a regulatory switch, in which one nucleoid-structuring protein switches in predominance at key regulatory elements to perturb gene regulation. This switch may be mediated by varying the protein composition during different growth phases (i.e., Lrp levels increase during the mid-exponential phase and decrease thereafter) (40, 69). However, in the case of the pap operon, switch regulation may not be mediated by direct antagonism between H-NS and Lrp (38), but indirect regulation could be possible. It is unknown whether H-NS and Lrp switch regulation is responsible for tos operon regulation and whether H-NS and Lrp regulation of the tos operon is direct or indirect.
Whether TosR, like PapB, might function in the capacity of an activator in addition to repressing the tos operon was previously unknown. Thus, in this study, we examined the capacity of TosR to serve as both a tos operon activator under certain conditions and as a repressor under others. Additionally, we propose that an H-NS and Lrp regulatory switch, similar to the one described above, is responsible for tos operon regulation. We also examined the capacity of TosR to negatively regulate production of P fimbria. To our knowledge, TosA has become the first nonfimbrial adhesin and RTX protein to be fully integrated into the network underlying the reciprocal regulation between different adhesins and between adhesins and motility systems. This cross-regulation also suggests that hierarchical regulation of adhesins and motility is much broader than previously thought.
MATERIALS AND METHODS
Bacterial strains.
A phage transductant of the original tosR deletion mutation (10) was unmarked using the FLP recombinase as previously described (10). The Δhns Δlrp CFT073 strain was engineered through phage-mediated transduction of a previous CFT073 Δlrp mutation (19) into a previous lambda Red-engineered CFT073 Δhns mutant unmarked as described above. Transductants were selected for on lysogeny broth (LB) agar (10 g/liter tryptone, 5 g/liter yeast extract, 0.5 g/liter NaCl, 15 g/liter agar) containing kanamycin (25 μg/ml). Deletion mutations were verified by PCR.
Engineered plasmids.
Untagged tosR and lrp genes were cloned into pBAD-myc-HisA (Invitrogen) as previously described (10). The pBAD-tosR-His6 and pRS551-Ptos-lacZ were previously engineered (10). The pBAD empty vector, pBAD-tosR, pBAD-tosR-His6, and pBAD-lrp constructs were maintained in LB (10 g/liter tryptone, 5 g/liter yeast extract, 0.5 g/liter NaCl) containing 100 μg/ml ampicillin, while the pBAD-lrp construct was also maintained in M9 medium (12.8 g/liter Na2HPO4·7H2O, 3 g/liter KH2PO4, 0.5 g/liter NaCl, 1.0 g/liter NH4Cl, 2 mM MgSO4, 0.4% glycerol, 0.1 mM CaCl2) containing ampicillin (100 μg/ml) (CFT073 ΔtosR strain). In addition, the pRS551-Ptos-lacZ and pRS551 empty vector constructs were maintained in LB containing ampicillin (50 μg/ml), except as noted below.
5′ RACE.
Plasmid pRS551-Ptos-lacZ was transformed into CFT073 ΔlacZ and CFT073 ΔtosR ΔlacZ and maintained in LB containing ampicillin (100 μg/ml). The 5′ random amplification of cDNA ends (RACE) procedure was performed similar to previous methods (70). cDNA was produced using the lacZ cDNA primer listed in Table 1 and SuperScript II reverse transcriptase as previously described (10). Input RNAs were hydrolyzed by adding NaOH (final concentration 0.16 mM) and boiled for 10 min. This reaction was neutralized by the addition of HCl (0.16 mM). A 3′ linker, listed in Table 1, was ligated to the cDNA described above using T4 RNA ligase (New England BioLabs). After ligation, the enzyme was inactivated by incubation at 65°C for 20 min. The first-round nested PCR was performed with the forward linker primer and lacZ nested primer 1 listed in Table 1. The second-round nested PCR was performed with the forward linker primer and lacZ nested primer 2. The resulting PCR fragment from the second round of nested PCR was sequenced.
TABLE 1.
Primers used in this study
| Primera | Sequence (5′ → 3′) |
|---|---|
| lacZ cDNA (R) | GCGGATTGACCGTAATGGGATAGGT |
| 3′ linker | TTTAGTGAGGGTTAATAAGCGGCCGCGTCGTGACTGGGAGCGC |
| Linker forward (F) | GCCGCTTATTAACCCTCACTAAA |
| lacZ nested primer 1 (R) | GACGACGACAGTATCGGCCTCAGGAAG |
| lacZ nested primer 2 (R) | CATTCAGGCTGCGCAACTGTTGGGAAGG |
| Ptos13 (F) | AAGTTTTGGGGTGCAGTCCAC |
| Ptos13 (R) | AAAAAGTGAAATCTCAAAACAAAAAAT |
| Ptos34 (F) | TAATATAGATATTATCTGCATATAA |
| Ptos34 (R) | TACTAGAGATTACATCTAAAAAATT |
| Ptos57 (F) | TTAGATAAAAACCCTACAGAGAAGT |
| Ptos57 (R) | CTGTATATGATCTGCCATACCATTACACAT |
| PpapBA (F) | CTCACTGTAACAAAGTTTCTTCGAATA |
| PpapBA (R) | GTTTCCCCCTTCTGTCGGGCCCCTG |
| lacZ (F) | GCGAATACCTGTTCCGTCATAGCG |
| lacZ (R) | CATCGCCAATCCACATCTGTGAAAG |
Orientations are indicated in parentheses (F, forward; R, reverse).
EMSAs.
tosR-His6 was induced in wild-type CFT073 and extracted using a QIAexpressionist protocol (Qiagen) and Ni-nitrilotriacetic acid (NTA) agarose (Invitrogen) as previously described (10). Input DNAs for electrophoretic mobility shift assays (EMSAs) were generated using the Ptos13, Ptos34, Ptos57, PpapBA, and lacZ primers listed in Table 1. Input DNAs were terminally labeled with a digoxigenin-11-ddUTP (DIG-ddUTP) using a 2nd-generation DIG gel shift kit (Roche Applied Science) as described previously (10). DNA binding reactions and detection of shifted DNA fragments were performed using a modified Roche DIG shift kit protocol and anti-DIG–alkaline phosphatase detection antibody (Roche Applied Science) as previously described (10), with the exception that between 400 nM and 4 μM TosR-His6 was used in each DNA binding reaction; between 2 pg/μl (PpapBA and lacZ) and 10 pg/μl (Ptos13, Ptos34, and Ptos57) DIG-ddUTP labeled fragments were used in the DNA binding reactions.
Western blots.
To detect TosA from induced overexpression constructs, pBAD-tosR, pBAD-tosR-His6, and pBAD-lrp were transformed into various CFT073 backgrounds and induced in LB containing 0, 0.06, 0.6, or 10 mM l-arabinose (pBAD-tosR, pBAD-tosR-His6, and the pBAD empty vector) or 0, 0.6, 1.2, 2.4, or 10 mM l-arabinose (pBAD-lrp) for 4 h. Four hours was chosen to allow E. coli to transit through the exponential phase, to ensure a high titer to maximize the likelihood of observing TosA, TosR, and PapA among bacterial cells in the culture, and to avoid prolonged incubation of the cultures within the stationary phase. Prior to induction, overnight bacterial cultures were diluted 1:100 (the CFT073 wild-type strain) and 1:40 (the CFT073 Δlrp and Δhns strains). The pBAD-lrp construct transformed into CFT073 ΔtosR was induced for 4.5 h with 0, 0.6, 1.2, or 10 mM l-arabinose in M9 minimal medium either containing 10 mM l-leucine or no exogenous l-leucine. Prior to induction, CFT073 ΔtosR harboring pBAD-lrp was cultured overnight in LB, pelleted at 6,000 × g, washed in M9 medium, and diluted 1:20. Total proteins from the inductions were collected in 10 mM HEPES (pH 8.3 to 8.9), quantified with a Pierce (BCA) protein assay kit (Thermo Scientific), and assayed by Western blotting with polyclonal anti-TosA antibodies or an anti-His6 antibody (Invitrogen) as previously described (10). To detect PapA, total proteins were assayed as described above in a CFT073 wild-type background harboring pBAD-tosR-His6, induced in LB containing 0, 0.6, or 10 mM l-arabinose; the only exception was that polyclonal anti-PapA antibodies (Rockland) were used in place of anti-TosA antibodies.
To detect TosA from the CFT073 wild-type strain and CFT073 Δhns, Δlrp, and Δhns Δlrp mutants, each background construct was cultured in LB for approximately 2.5 h to the exponential phase (A600 ≈ 0.3 to 0.5). Prior to being cultured to the exponential phase, CFT073 wild-type and Δlrp overnight cultures were diluted 1:100 in LB, and Δhns and Δhns Δlrp mutant overnight cultures were diluted 1:40 prior to being cultured in LB. Total proteins were collected in 10 mM HEPES (pH 8.3 to 8.9), quantified with a Pierce BCA protein assay kit (Thermo Scientific), and assayed by Western blotting with polyclonal anti-TosA antibodies as described above.
Promoter activity assay.
Promoter activities from the pRS551-Ptos-lacZ or pRS551 empty construct, transformed into wild-type CFT073 or the Δhns, Δlrp, and Δhns Δlrp mutant strains, were determined using a modified Miller assay as previously described (10). The modification to the Miller assay was the use of β-methylumbelliferyl β-d-galacopyranoside (0.5 mg/ml) as the substrate instead of o-nitrophenyl-β-galactoside.
Growth curves.
Overnight cultures of E. coli CFT073 harboring pRS551-Ptos-lacZ were diluted 1:100 (wild-type and Δlrp mutant strains) and 1:40 (Δhns and Δhns Δlrp mutant strains) into LB (10 g/liter tryptone, 5 g/liter NaCl) containing ampicillin (50 μg/ml). Constructs were cultured at 37°C for 24 h in a Bioscreen C automated growth curve system, with A600 readings recorded every 15 min.
RESULTS
The tos operon promoter is located upstream of the tos operon regulator gene, tosR.
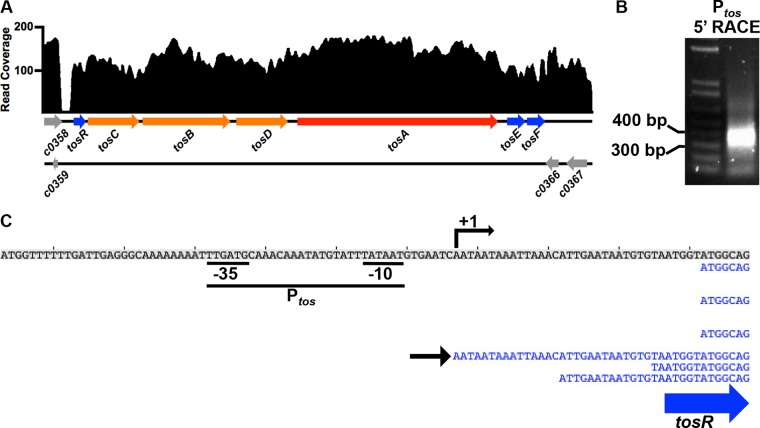
Our previous work localized the tos operon promoter (Ptos) to a 630-bp sequence upstream of tosR (10). To determine the precise location of Ptos and map associated promoter elements, we conducted both analysis of transcriptome sequencing (RNA-Seq [not strand specific]) and 5′ rapid amplification of cDNA ends (RACE). Mapping normalized tos operon cDNA reads obtained from E. coli CFT073, cultured in human urine (unpublished data), we predicted the tos operon transcriptional start site to be 23 bp upstream of tosR, based on the presence of a gap between tosR and the upstream open reading frames (ORFs) c0358 and c0359 (Fig. 1A). However, transcripts from genes encoded on the opposite DNA strand (c0366 and c0367) at the 3′ end of the tos operon make it difficult to predict transcriptional termination sites, as the RNA-Seq technique employed here is not strand specific. For verification of the predicted transcriptional start site, 5′ RACE was performed on transcripts expressed from the pRS551-Ptos-lacZ transcriptional fusion, used to ensure a high concentration of transcripts containing the tos operon start site. Following two rounds of nested PCR on cDNAs with a 3′ linker of a known sequence ligated to this segment, we amplified a PCR product of approximately 344 bp (Fig. 1B), which was consistent with the transcriptional start site obtained from the RNA-Seq analysis.
FIG 1.
Ptos is predicted to be located upstream of tosR. (A) The tos operon is presented along with a log-transformed cDNA read plot corresponding to cDNAs obtained from the tos operon of UPEC strain CFT073 cultured in filter-sterilized human urine. The scale indicates mapped reads. Two parallel lines below the read plot represent the two strands of DNA, and the directions of the arrows represent the strand on which the indicated genes are carried. Only a partial sequence of c0358 is depicted in the read plot. (B) Resolution of the 5′ RACE products obtained from transcripts expressed from the vector pRS551-Ptos-lacZ yields a product between the indicated 300- and 400-bp size markers. (C) Mapped cDNAs (in blue) are depicted below the top shaded DNA sequence; a blue arrow indicates the location of tosR. A black arrow to the left depicts the most upstream read obtained from the RNA-Seq experiment, which was also precisely the same sequence identified from sequencing of the PCR product obtained in panel B. An angled black arrow indicates the transcriptional start site of the tos operon. The predicted −35 and −10 sequences of the tos operon promoter, Ptos, are depicted.
Sequencing of the 5′ RACE PCR product identified the distal 5′ sequence (transcriptional start site) identical to that of the RNA-Seq analysis, which in turn allowed us to map a modified σ70 promoter upstream of the transcriptional start site (Fig. 1C). This promoter shows 67% identity (TTGAtg) to the canonical σ70 −35 sequence and 100% identity to the canonical σ70 −10 sequence (TATAAT). Consistent with other σ70 promoters (71, 72), these −35 and −10 sequences are separated by 16 nucleotides. The transcriptional start site is 7 bp downstream from the end of last nucleotide −10 sequence, a spacing also consistent with σ70 promoters (72). In addition, the first base in the predicted transcript, adenosine, is also typical of many transcriptional start sites (73, 74). However, a putative ribosome-binding site upstream of the predicted TosR translational start site could not be clearly identified, which could suggest that TosR translation is inefficient.
TosR is both a positive and negative regulator of the tos operon.
We have previously identified a repressor function for TosR (10). As the location of the previously identified TosR-binding site (10) is not near Ptos (160 bp upstream of the newly identified promoter), we predicted that there could be additional, weaker binding sites near the tos operon promoter. To test this prediction, we performed an electrophoretic mobility shift assay (EMSA) on digoxigenin (DIG)-labeled DNA fragments of Ptos containing the strong TosR-binding site, an intergenic region between the strong TosR binding site and Ptos, and a region containing Ptos (Fig. 2A and B). As expected, we found that the region of Ptos containing the strong TosR-binding site had a reduced electrophoretic mobility (i.e., was shifted) when incubated with TosR. Additionally, we found that TosR shifted the Ptos fragment containing the tos operon promoter. As the latter fragment was almost fully shifted only at the highest levels of TosR (4 μM), we also reasoned that TosR weakly binds this region, compared with the strong binding site previously identified. At least 50 bp separates the strong and weak TosR binding sites in Ptos (Fig. 2B). In addition, it is possible that TosR has some affinity for AT-rich sequences, as is also the case for other PapB family members (31). This is supported by the observation that TosR slightly shifts the intergenic region between the strong and weak binding sites (i.e., the intensity of the Ptos34 unshifted fragment is lower at the highest TosR concentration). However, affinity for AT-rich sequences alone cannot explain all of the TosR binding activity, as AT-rich regions of Ptos failed to be effective competitors for TosR binding to the strong binding site in the vicinity of Ptos (10). This does not rule out the possibility that TosR recognizes a structural element, especially as another promoter, PpapBA (see Fig. S1A and S1B in the supplemental material), is regulated by the prototype member of the PapB family (22–24). In agreement with this, BLASTN revealed no significant sequence similarity between the weak and strong TosR binding sites.
FIG 2.
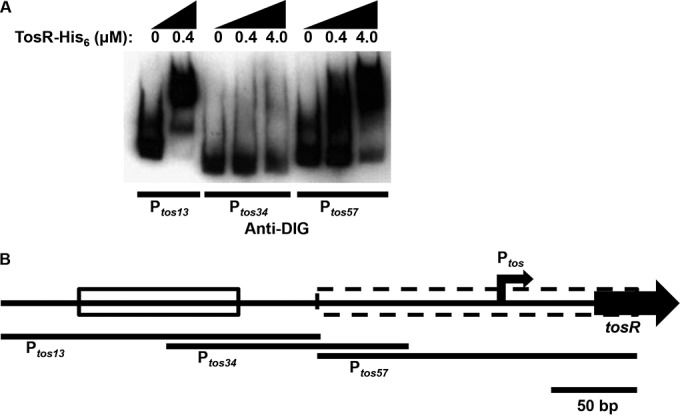
An EMSA indicates that TosR binds Ptos at promoter distal and proximal positions and with various strengths. (A) The indicated amounts of TosR-His6 were incubated with terminally DIG-labeled Ptos fragments. Shifted and unshifted DNA fragments were detected with an anti-DIG antibody. The EMSA is representative of two independent experiments. (B) A schematic of Ptos region indicates the positions of the Ptos fragments used for the above EMSA, the location of the operon promoter (angled black arrow), the stronger TosR-His6 binding site distal to the promoter (solid box), and the weaker TosR-His6 binding site proximal to the promoter (dashed box).
Other PapB family members have been described as dual regulators of their cognate operons (22–24). Thus, based on the various degrees of TosR binding strengths for sites in the vicinity of Ptos, we speculated that TosR could also have an additional positive regulatory function on the tos operon. To test whether TosR could induce expression of the tos operon, we used a pBAD-tosR-His6 construct and a pBAD-tosR untagged construct to assay TosA synthesis at various tosR induction levels. Using a Western blot of proteins from whole-cell preparations obtained from these pBAD overexpression constructs, we found that TosA levels are inversely related to induced tosR levels (Fig. 3). For induced levels of TosR below the detectable limit of our anti-His6 antibody, we observed high levels of TosA synthesis; with high levels of TosR, detectable with anti-His6 antibody, TosA levels were low. Likewise, these functions appear independent of the presence of the His6 tag, as both tagged and untagged TosR proteins yielded similar results. Expression was also independent of the presence of arabinose alone, as an empty pBAD vector failed to regulate TosA synthesis. A similar result was also observed when tosR was induced in another urinary tract isolate background, ABU 83972, harboring the tos operon (not shown). It is important to note, however, that each of these findings is based on overexpression of TosR. Therefore, it may be the case that additional regulators supplement TosR-mediated activation and repression under native conditions. However, it is also important to note that E. coli CFT073 has no arabinose utilization gene mutations, and arabinose will be metabolized during these assays, which may contribute to the absence of TosR at some induction levels. Induction from the pBAD vector may also be subject to the “all-or-nothing” phenomenon (75–77). Titration from this vector may, therefore, be limited (78), especially when considering both the all-or-nothing phenomenon and arabinose utilization.
FIG 3.
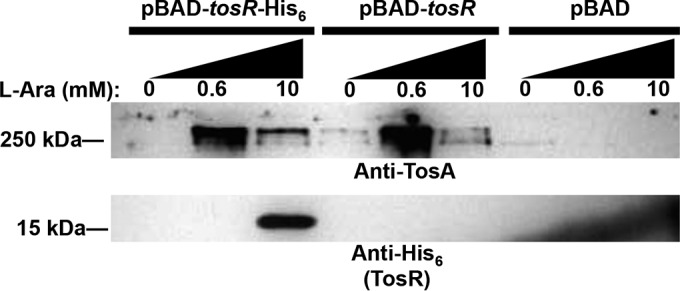
TosR is a dual positive and negative regulator of TosA. Western blots using polyclonal anti-TosA antibodies or an anti-His6 antibody were performed to detect TosA (>250 kDa) or TosR (∼15 kDa). Total proteins for the Western blot were obtained from UPEC strain CFT073 harboring the indicated pBAD constructs induced with the indicated concentrations of l-arabinose. Equal amounts of proteins were loaded, as determined using a Pierce BCA protein assay kit. The Western blot is representative of two independent experiments.
Nucleoid-structuring proteins contribute to TosR regulation of the tos operon.
The tos operon is localized to the PAI-aspV pathogenicity island in UPEC strain CFT073 (12). It is well accepted that genes on PAIs and other AT-rich sequences are often bound and regulated by nucleoid-structuring proteins, including H-NS and Lrp (41, 43, 44, 67, 79). A 400-bp region containing Ptos is AT rich (74%). In addition, both of these nucleoid-structuring proteins regulate the expression of many genes, including adhesin and flagellar genes (19, 22, 37–39, 43, 46–57, 59–67). To determine whether or not a 240-bp AT-rich region near Ptos is similarly curved to an analogous region in PpapBA, suggesting Lrp and H-NS-mediated nucleoid structuring, we utilized a web-based tool (http://www.lfd.uci.edu/∼gohlke/dnacurve/) to predict DNA curvature. We found that both regions predicted a similar curved architecture (see Fig. S1A and S1B in the supplemental material), which suggests Lrp and H-NS could regulate the tos operon. To further predict whether H-NS and Lrp bind to Ptos, we examined this sequence for putative H-NS and Lrp binding sites (see Fig. S2 in the supplemental material). There are four clusters of putative Lrp binding sites (GN2–3TTT), based on PpapBA (80), downstream and partially overlapping the predicted strong TosR binding site and upstream and partially overlapping the predicted weak TosR binding site. One of the predicted Lrp binding sites also overlaps Ptos. In addition, there are also two putative high-affinity H-NS binding sites with 80% (aCaATAAATT) and 70% (ataATAAATT) identity to a sequence with known high affinity for H-NS (81, 82) located upstream of the weak TosR binding site and downstream of Ptos, near the predicted transcriptional start site.
To determine whether H-NS and Lrp do indeed regulate the tos operon, we performed Western blots on proteins from whole-cell preparations obtained from CFT073 Δhns and Δlrp backgrounds (Fig. 4A). TosA levels were dramatically increased in the Δhns background compared to wild-type CFT073, suggesting that H-NS could function as a negative regulator of the tos operon. However, it is important to note that H-NS perturbs the expression of a number of different genes (22, 43, 46–57); therefore, it remains unclear if additional regulators supplement H-NS-mediated negative regulation of the tos operon. The loss of Lrp failed to increase tos operon expression, as was observed for loss of H-NS.
FIG 4.
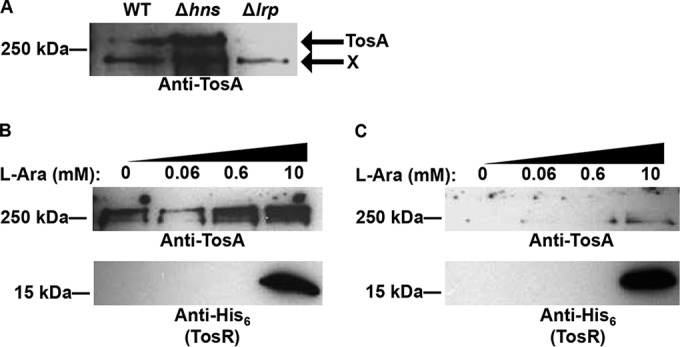
TosR-mediated negative and positive regulation is perturbed in the Δhns and Δlrp backgrounds. (A) A Western blot using polyclonal anti-TosA antibodies was performed on total proteins obtained from the indicated CFT073 backgrounds. Bands corresponding to TosA are indicated in the figure, and a nonspecific band is indicated with an X. The Western blot is representative of two independent experiments. Equal amounts of proteins were loaded, as determined using a Pierce BCA protein assay kit. Western blots were also performed as described above using polyclonal anti-TosA antibodies or an anti-His6 antibody in the CFT073 Δhns (B) or Δlrp (C) background harboring pBAD-tosR-His6 induced with the indicated concentrations of l-arabinose. The Western blot is representative of two independent experiments.
With respect to the multitude of PapB family members, however, it is not always obvious how nucleoid structure and the cognate PapB family members integrate to govern expression of the adhesin. To determine whether H-NS and Lrp contribute to TosR regulation of the tos operon, we performed the same pBAD-tosR-His6 overexpression experiment described above in the CFT073 Δhns (Fig. 4B) and Δlrp (Fig. 4C) backgrounds. As described above, loss of hns resulted in increased TosA synthesis, but high levels of TosR did not decrease TosA levels in the CFT073 Δhns background. Conversely, we found that the TosR-mediated activation was dependent on Lrp; no change in TosA levels could be detected regardless of TosR level in the CFT073 Δlrp background. Likewise, a shift to a lower antibiotic concentration for the Δhns and Δlrp backgrounds harboring pBAD-tosR-His6 does not perturb TosR regulation itself; the lower antibiotic concentration did not perturb TosR-mediated regulation in the wild-type E. coli CFT073 background (data not shown). As for H-NS, Lrp is also a global regulator (19, 22, 37–39, 59–67). Therefore, it remains unclear if additional gene products also supplement Lrp- and TosR-mediated positive regulation of the tos operon.
Induction of lrp expression is sufficient to drive TosA synthesis.
Observing that Lrp is required for tos operon expression, we tested whether exogenous expression of Lrp alone would be sufficient to induce tos operon expression. To determine whether Lrp acts as an activator of the tos operon, as is the case with the pap operon (22, 37, 38), we performed a pBAD-lrp overexpression experiment in wild-type CFT073. Western blotting of whole-cell proteins from this overexpression construct revealed that low levels of lrp induction increased TosA levels (Fig. 5). In turn, high levels of lrp induction diminished TosA levels. However, this effect was dependent on the presence of TosR (see Fig. S3 in the supplemental material). Taken together, Lrp appears to be an activator of the tos operon, but it can also contribute to tos operon repression in the presence of TosR. It is important to note, however, that the same caveats of lrp overexpression should also be considered, as with tosR overexpression described above.
FIG 5.

Lrp is a positive regulator of TosA. A Western blot using polyclonal anti-TosA antibodies was performed on total proteins obtained from CFT073 harboring pBAD-lrp and induced with the indicated concentrations of l-arabinose. Bands corresponding to TosA are indicated in the figure. Equal amounts of proteins were loaded, as determined using a Pierce BCA protein assay kit. The Western blot is representative of two independent experiments.
Some genes regulated by Lrp are positively or negatively regulated by exogenous leucine (63–65, 67). To test whether exogenous leucine positively or negatively regulates the tos operon, we performed our pBAD-lrp overexpression assay in the CFT073 ΔtosR background in M9 minimal medium with and without exogenous leucine (10 mM). The CFT073 ΔtosR background was chosen over wild-type CFT073 due to the fact that pBAD-lrp was unable to induce expression of the tos operon in M9 minimal medium (data not shown). In the CFT073 ΔtosR background, induction of lrp expression resulted in higher TosA levels only in the absence of exogenous leucine (see Fig. S4 in the supplemental material). This demonstrates that Lrp-mediated positive regulation of the tos operon is subject to regulation by leucine.
An H-NS and Lrp regulatory switch drives tos operon transcriptional regulation.
It has been previously proposed and noted that Lrp might act to anti-silence H-NS repression (35, 38, 59, 68). We hypothesize, therefore, that an H-NS and Lrp regulation switch (i.e., predominance of either H-NS or Lrp) explains the observed regulation of the tos operon. In particular, if H-NS-mediated negative regulation is abolished, Lrp-mediated positive regulation of the tos operon would no longer be required in the Δhns background. To test this hypothesis, we performed a Western blot on a CFT073 Δhns Δlrp mutant (Fig. 6A). Loss of Lrp in the Δhns background only slightly decreased TosA levels, compared to the Δhns background alone. This further strengthens the premise that Lrp is a positive regulator of the tos operon, as even these slightly reduced levels were much higher than the nearly undetectable levels of TosA in the CFT073 wild-type background. Coupled with the finding that TosR regulation is abolished in the Δlrp background, these results suggest that an H-NS and Lrp regulation switch likely contributes to tos operon regulation, and TosR has a function within this regulatory switch. It is important to note, however, that both H-NS and Lrp are global regulators (19, 22, 37–39, 59–67). Therefore, it cannot be ruled out that regulation between H-NS and Lrp, at Ptos is indirect.
FIG 6.
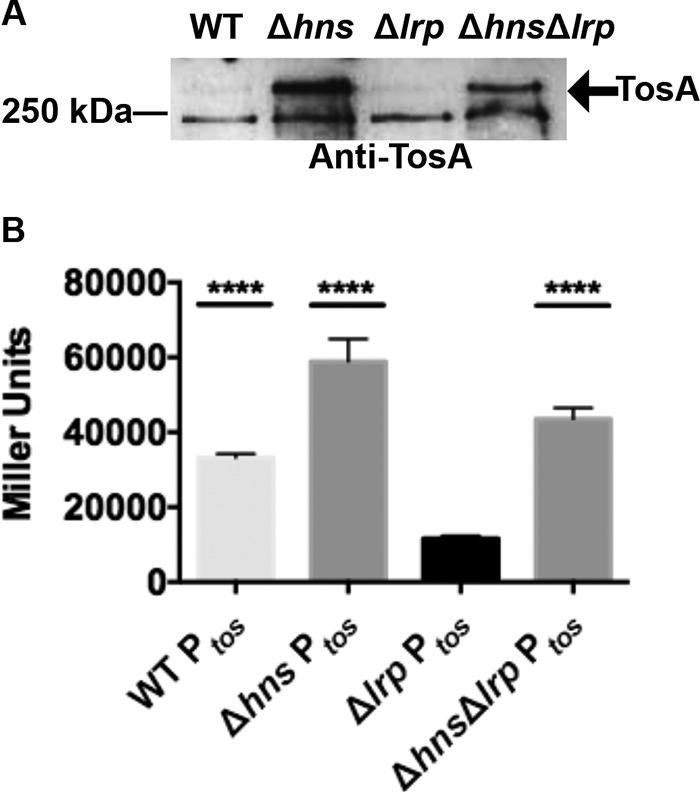
Lrp is not required for Ptos transcriptional activation in the Δhns background. (A) A Western blot using polyclonal anti-TosA antibodies was performed on total proteins obtained from the indicated CFT073 backgrounds. Equal amounts of proteins were loaded, as determined using a Pierce BCA protein assay kit. (B) A Miller assay was performed using β-galactosidase translated from the lacZ gene of the pRS551-Ptos-lacZ vector harbored in the indicated backgrounds. Bars represent mean values of Miller units obtained from two biological replicates with two technical replicates each. Error bars represent 1 standard deviation around the mean, and **** represents P values of <0.0001 obtained by comparing lacZ expression from pRS551-Ptos-lacZ construct harbored in the respective mutant or wild-type CFT073 background with the Δlrp background (determined using analysis of variance [ANOVA] followed by Tukey's multiple-comparison test).
Furthermore, we tested whether the proposed H-NS and Lrp regulatory switch would function at the transcriptional level at Ptos. To determine whether Ptos is transcriptionally responsive to H-NS and Lrp, we measured the activity of our pRS551-Ptos-lacZ transcriptional fusion in both wild-type CFT073 and CFT073 nucleoid-structuring mutants using a Miller assay (Fig. 6B). As previously observed (10), the Ptos promoter showed high activity in wild-type CFT073. Additionally, Ptos showed high activity in the CFT073 Δhns mutant. Ptos promoter activity was greatly reduced in the CFT073 Δlrp background. In the CFT073 Δhns Δlrp background, however, Ptos activity was restored to slightly higher than wild-type levels. No growth differences were observed among bacterial strains harboring pRS551 (see Fig. S5 in the supplemental material). These findings suggest that native Lrp levels induce Ptos on the pRS551 construct by overcoming H-NS-mediated negative regulation. Thus, regulation of the tos operon by the H-NS and Lrp regulatory switch is at the transcriptional level. From its Ptos DNA binding activities and association with H-NS and Lrp regulation of the tos operon, we further suggest that TosR also transcriptionally regulates the tos operon.
TosR is a negative regulator of the pap2 operon, another component of the H-NS and Lrp regulatory switch.
Both H-NS and Lrp are global regulators that affect the expression of a variety of genes, including adhesin and flagellar genes (19, 22, 37–39, 43, 46–57, 59–67). It is, therefore, not surprising that fimbrial regulators associated with H-NS and Lrp could also participate in cross-regulation between adherence and motility genes (62, 83–86). Both PapB (P-fimbrial operon) and FocB (F1C fimbrial operon) share approximately 80% amino acid sequence identity and regulate their respective pap and foc operons. Cross-regulation between PapB and FocB is a well-characterized phenomenon (84). In contrast, TosR has only 28% amino acid sequence identity to PapB. To determine whether TosR can also regulate the pap operon, we performed our pBAD-tosR-His6 overexpression assay and Western blotting with an anti-PapA antibody (Fig. 7A). With increased TosR levels, PapA2 levels were decreased.
FIG 7.
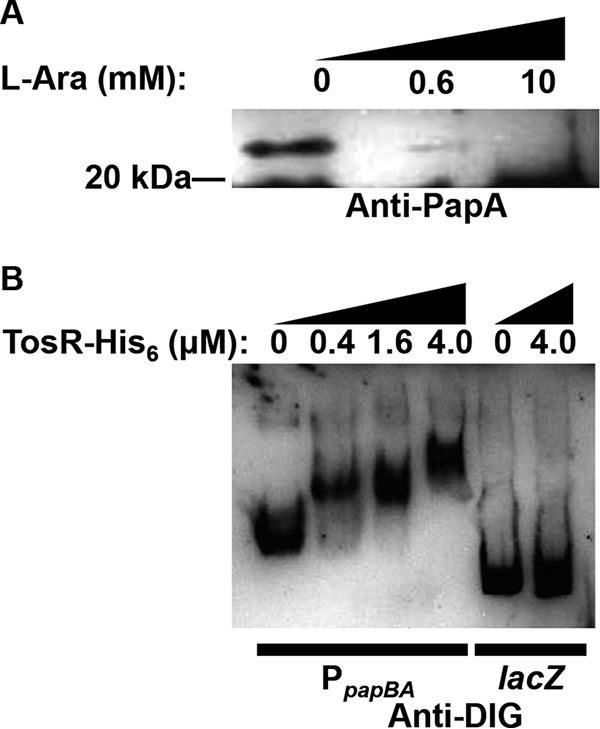
TosR negatively regulates P-fimbria synthesis. (A) A Western blot using polyclonal anti-PapA antibodies to detect PapA2 (∼23 kDa) was performed on total proteins obtained from CFT073 harboring pBAD-tosR-His6 and induced with the indicated concentrations of l-arabinose. This blot is representative of two biological replicates. Equal amounts of proteins were loaded, as determined using a Pierce BCA protein assay kit. (B) The indicated amounts of TosR-His6 were treated along with terminally DIG-labeled PpapBA or lacZ fragments. Shifted and unshifted DNA fragments were detected using an anti-DIG antibody. The EMSA is representative of two independent experiments.
Previous work characterizing PapB and FocB cross-regulation explored the abilities of both proteins to mediate this regulation through binding to PpapBA and PfocBA (84). To determine whether TosR might also mediate pap operon cross-regulation through binding PpapBA, we performed an EMSA on a DIG-labeled PpapBA fragment and, as a control, a fragment of lacZ (Fig. 7B). TosR shifted the PpapBA fragment but failed to shift the control lacZ fragment. Thus, we conclude that despite markedly low amino acid identity between PapB and TosR, TosR mediates negative regulation of the pap operon through specific binding of PpapBA. Intriguingly, like the weak Ptos binding site, BLASTN reveals no substantial sequence homology with the strong Ptos binding site and PpapBA.
DISCUSSION
Here, we present a model of tos operon regulation involving PapB family member TosR and two global gene regulators, H-NS and Lrp (Fig. 8). TosR is a positive and negative transcriptional regulator of the tos operon. This regulation is predicted to be mediated through differential binding of the chromosomal region containing Ptos, the tos operon promoter, by TosR. The global regulator H-NS transcriptionally silences expression of the tos operon, while another global regulator, Lrp, overcomes H-NS silencing to mediate positive regulation of the tos operon. When TosR levels are low, TosR promotes Lrp-mediated positive regulation of the tos operon. However, when TosR levels are high, TosR promotes H-NS-mediated negative regulation of the tos operon. We also predict that H-NS and Lrp interact either directly or indirectly to modify tos operon positive regulation. Additionally, TosR also negatively regulates expression of the P-fimbrial (pap) operon.
FIG 8.
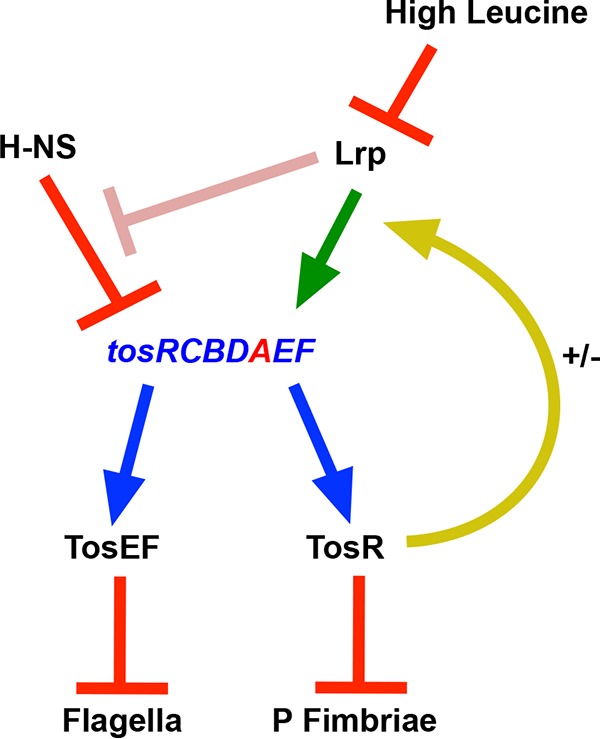
Model of tos operon regulation and its involvement in reciprocal regulation of adhesins and flagella. The tos operon is indicated by blue text, and tosA is represented by red text. A blue arrow indicates that TosR, TosE, and TosF are translated from genes transcriptionally linked to the tos operon. Under typical laboratory conditions, H-NS silences expression of the tos operon (red bar). At low and high concentrations, respectively, TosR both positively and negatively (+/−) regulates (yellow arrow) Lrp-mediated positive regulation of the tos operon (green arrow). Lrp may also directly or indirectly relieve H-NS negative regulation of the tos operon (pink bar). In turn, TosR also negatively regulates P-fimbria synthesis (red bar), and together the cognate regulators TosE and TosF negatively regulate flagellum synthesis (red bar). High levels of leucine, in addition, negatively regulate Lrp-mediated positive regulation of the tos operon (red bar).
Using RNA-Seq and 5′ RACE, we identified the transcriptional start site of the tos operon 23 bp upstream of tosR and Ptos 30 bp upstream of tosR. The promoter sequence has only a few modifications from the canonical σ70, which include two base substitutions from the canonical −35 sequence and spacing between the −35 and −10 sequences 1 bp shorter than that for the average σ70 promoter (71, 72). Additionally, the first base of the transcript, adenosine, is also typical of other σ70 promoters (73, 74), and the spacing between the promoter and the start of the transcript (7 bp) is also observed with other σ70 promoters (72). Thus, together these results suggest that Ptos could be a strong promoter, which is consistent with our previous observation of strong activity from the pRS551-Ptos-lacZ transcriptional fusion (10). This finding, however, is confounded by the weak expression of the tos operon previously observed (9–11), which points to negative transcriptional regulation at Ptos by other proteins as a possible mechanism of tos operon regulation.
Our group previously reported that the tos operon is repressed when cultured under laboratory conditions (LB broth, both aerated and static, at 37°C) (9–11). Some of this negative regulation was attributed to TosR (10). However, other PapB family members act as dual regulators (both activator and repressor) of their cognate operons (22–24). We found this was also the case for TosR, which shows a reciprocal relationship with TosA levels: if TosR levels are low, TosA levels are high, and when TosR levels are high, TosA levels are significantly reduced. Thus, TosR is a dual regulator of the tos operon. We predict that at least some of this differential behavior is mediated through TosR binding to two sites within Ptos—one site when TosR levels are low (strong binding site) and the other when TosR levels are higher (weak binding site).
Regulation of the tos operon not only involves TosR but also includes both H-NS and Lrp. We predict that TosR positive regulation of the tos operon, when TosR levels are low, may be mediated through an alleviation of negative regulation by H-NS, thereby promoting Lrp-mediated positive regulation. In terms of the predicted H-NS binding site upstream of Ptos (see Fig. S2 in the supplemental material), we speculate that TosR binding either displaces an H-NS filament or prevents further H-NS polymerization at this site, both known mechanisms of overcoming H-NS silencing (87). Subsequently, this activity may allow Lrp to bind to predicted binding sites in the vicinity of Ptos to promote positive regulation. This model is further supported by TosR-mediated positive regulation of the tos operon no longer being required in the Δhns background. Furthermore, Lrp-mediated positive regulation is no longer necessary in the Δhns background, and TosR regulation is abolished in the Δlrp background. Lrp alone is also sufficient to promote expression of the tos operon, especially in the absence of leucine, which further supports our prediction of Lrp-mediated positive regulation of the tos operon. Thus, in terms of the predicted Lrp binding sites (see Fig. S2) in the vicinity of Ptos and previous work on Lrp by others (88–90), we speculate that Lrp binding may facilitate RNA polymerase contact with either Lrp itself or additional unknown elements near Ptos. Similarly, we propose that TosR-negative regulation of the tos operon, when TosR levels are high, may be mediated through interference of Lrp-promoted positive regulation. This effect is also predicted to be dependent on H-NS. It is speculated, in terms of the second predicted TosR and H-NS binding sites (see Fig. S2) in the vicinity of Ptos, that TosR binding to this site occludes Lrp binding to the predicted binding sites above and subsequent occlusion of RNA polymerase from Ptos at the second predicted H-NS binding site near Ptos. Further support for this conclusion comes from the finding that TosR is no longer a negative regulator of the tos operon in a Δhns background. Also by integrating the two predicted H-NS binding sites into this regulation model, it is possible that bridging at these sites could promote negative regulation (87, 91–94).
From the strong activity of Ptos in a Δhns Δlrp background, we also predict that an H-NS and Lrp regulation switch is responsible for much of the tos operon regulation. The switch may act through alteration of the predominance of H-NS and Lrp regulation at Ptos, which is consistent with modulation of nucleoid levels of associated proteins during different growth phases (40, 69). However, as H-NS and Lrp are pleiotropic regulators (19, 22, 37–39, 43, 46–57, 59–67), this switch may also be through indirect interactions. To emphasize the possibility that H-NS and Lrp indirectly interact, a pink bar is depicted in our model (Fig. 8). In agreement with this idea of a switch between the two nucleoid proteins, the strong decrease in Ptos activity observed with the loss of Lrp is increased when H-NS is absent. This leads us to predict that Lrp functions to overcome H-NS negative regulation of the tos operon, consistent with our belief that an H-NS and Lrp switch governs tos operon expression. It is also an intriguing possibility that the same switch is similar to a previous description of nucleoid contributions to reciprocal regulation of adherence and motility (54). However, whether this H-NS and Lrp regulation switch is mediated by direct antagonism of each component or indirect effects will need to be examined further.
We note that the estimated leucine content of pooled human urine (∼0.01 mM) (95) is much lower than that of LB (∼8 mM) (96). This suggests that UPEC would adjust gene expression to accommodate the low leucine levels found in human urine. Thus, growth in an environment with relatively low levels of leucine is an environmental stress encountered by UPEC during an infection. As exogenous Lrp does not positively regulate the tos operon in the presence of high leucine levels, we also conclude that a low leucine level is an environmental cue that upregulates the tos operon. We also propose that the presence of higher levels of leucine in LB at least partially accounts for poor tos operon expression when cultured in this medium.
As evident from the variety of genes, including those localized to adhesin operons in addition to flagellum-mediated motility genes regulated by H-NS and Lrp (19, 22, 37–39, 43, 46–57, 59–67), cross-regulation appears to be a feature of this regulatory switch (62, 83–86). We found that TosR is a negative regulator of P-fimbria production. We predict that this negative regulation is potentiated through TosR binding to PpapBA, the pap operon promoter. This is a surprising finding in that the previously well-described cross-regulation between PapB family members occurred between PapB and FocB, which shared 80% amino acid sequence identity (84). TosR and PapB share only 28% amino acid sequence identity. We believe that these results have important implications for studying adhesin expression. Further work should explore whether TosR, like PapB and FocB, also regulates FimA and FocA levels (84). Nevertheless, it is now our conclusion that such cross-regulation between PapB family members and different types of adhesins (i.e., fimbrial and nonfimbrial adhesins) is a broader phenomenon than previously thought. Thus, a more detailed exploration of adhesin cross-regulation, especially between unrelated or poorly related adhesins and adhesin regulators, should be undertaken to gain a more accurate picture of microbial adhesin regulation. These future explorations should include determination of whether reciprocal regulation between adhesins is an important fitness trait during infection. For example, P fimbria and TosA both make contributions during experimental UTI (27–30), but it is unknown whether TosR inactivation could suppress a tosA mutation through allowing UPEC to continue to synthesize P fimbria instead of simultaneously inhibiting P-fimbria production and attempting to produce a defective TosA adhesin.
Finally, previous work (10) has already established that TosEF, expressed when the tos operon is expressed, negatively regulate FliC levels. Together with the TosR findings presented above, we have also found that tos operon regulation participates in reciprocal regulation of adherence and motility. It is intriguing to note that a protein encoded by the terminal gene of the pap operon, papX, suppresses motility in UPEC strain CFT073 (19–21). Future work may thus also explore whether overexpression of lrp in tosEF and papX mutant constructs decreases motility. Taken together, these results could delineate the function of the H-NS and Lrp regulatory switch in reciprocal regulation and during infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christopher Alteri, Sargurunathan Subashchandrabose, David Friedman, Sébastien Crépin, and Chelsie Armbruster for insightful discussions about our work.
This work was funded by National Institutes of Health Public Health Service grant AI059722 (to H.L.T.M.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01302-15.
REFERENCES
- 1.Foxman B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am J Med 113(Suppl 1A):5S–13S. [DOI] [PubMed] [Google Scholar]
- 2.Foxman B. 2010. The epidemiology of urinary tract infection. Nat Rev Urol 7:653–660. doi: 10.1038/nrurol.2010.190. [DOI] [PubMed] [Google Scholar]
- 3.Kalra OP, Raizada A. 2009. Approach to a patient with urosepsis. J Glob Infect Dis 1:57–63. doi: 10.4103/0974-777X.52984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikaheimo R, Siitonen A, Karkkainen U, Mustonen J, Heiskanen T, Makela PH. 1994. Community-acquired pyelonephritis in adults: characteristics of E. coli isolates in bacteremic and non-bacteremic patients. Scand J Infect Dis 26:289–296. doi: 10.3109/00365549409011797. [DOI] [PubMed] [Google Scholar]
- 5.Stamm WE, Hooton TM. 1993. Management of urinary tract infections in adults. N Engl J Med 329:1328–1334. doi: 10.1056/NEJM199310283291808. [DOI] [PubMed] [Google Scholar]
- 6.Nielubowicz GR, Mobley HL. 2010. Host-pathogen interactions in urinary tract infection. Nat Rev Urol 7:430–441. doi: 10.1038/nrurol.2010.101. [DOI] [PubMed] [Google Scholar]
- 7.Vigil PD, Stapleton AE, Johnson JR, Hooton TM, Hodges AP, He Y, Mobley HL. 2011. Presence of putative repeat-in-toxin gene tosA in Escherichia coli predicts successful colonization of the urinary tract. mBio 2:e00066–11. doi: 10.1128/mBio.00066-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spurbeck RR, Dinh PC Jr, Walk ST, Stapleton AE, Hooton TM, Nolan LK, Kim KS, Johnson JR, Mobley HL. 2012. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect Immun 80:4115–4122. doi: 10.1128/IAI.00752-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vigil PD, Wiles TJ, Engstrom MD, Prasov L, Mulvey MA, Mobley HL. 2012. The repeat-in-toxin family member TosA mediates adherence of uropathogenic Escherichia coli and survival during bacteremia. Infect Immun 80:493–505. doi: 10.1128/IAI.05713-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engstrom MD, Alteri CJ, Mobley HL. 2014. A conserved PapB family member, TosR, regulates expression of the uropathogenic Escherichia coli RTX nonfimbrial adhesin TosA while conserved LuxR family members TosE and TosF suppress motility. Infect Immun 82:3644–3656. doi: 10.1128/IAI.01608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vigil PD, Alteri CJ, Mobley HL. 2011. Identification of in vivo-induced antigens including an RTX family exoprotein required for uropathogenic Escherichia coli virulence. Infect Immun 79:2335–2344. doi: 10.1128/IAI.00110-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd AL, Henderson TA, Vigil PD, Mobley HL. 2009. Genomic islands of uropathogenic Escherichia coli contribute to virulence. J Bacteriol 191:3469–3481. doi: 10.1128/JB.01717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welch RA, Burland V, Plunkett G III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A 99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wandersman C, Delepelaire P. 1990. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc Natl Acad Sci U S A 87:4776–4780. doi: 10.1073/pnas.87.12.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser P, Sakamoto H, Bellalou J, Ullmann A, Danchin A. 1988. Secretion of cyclolysin, the calmodulin-sensitive adenylate cyclase-haemolysin bifunctional protein of Bordetella pertussis. EMBO J 7:3997–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boardman BK, Satchell KJ. 2004. Vibrio cholerae strains with mutations in an atypical type I secretion system accumulate RTX toxin intracellularly. J Bacteriol 186:8137–8143. doi: 10.1128/JB.186.23.8137-8143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linhartova I, Bumba L, Masin J, Basler M, Osicka R, Kamanova J, Prochazkova K, Adkins I, Hejnova-Holubova J, Sadilkova L, Morova J, Sebo P. 2010. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev 34:1076–1112. doi: 10.1111/j.1574-6976.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fullner KJ, Mekalanos JJ. 2000. In vivo covalent cross-linking of cellular actin by the Vibrio cholerae RTX toxin. EMBO J 19:5315–5323. doi: 10.1093/emboj/19.20.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simms AN, Mobley HL. 2008. Multiple genes repress motility in uropathogenic Escherichia coli constitutively expressing type 1 fimbriae. J Bacteriol 190:3747–3756. doi: 10.1128/JB.01870-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simms AN, Mobley HL. 2008. PapX, a P fimbrial operon-encoded inhibitor of motility in uropathogenic Escherichia coli. Infect Immun 76:4833–4841. doi: 10.1128/IAI.00630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reiss DJ, Mobley HL. 2011. Determination of target sequence bound by PapX, repressor of bacterial motility, in flhD promoter using systematic evolution of ligands by exponential enrichment (SELEX) and high throughput sequencing. J Biol Chem 286:44726–44738. doi: 10.1074/jbc.M111.290684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernday A, Krabbe M, Braaten B, Low D. 2002. Self-perpetuating epigenetic pili switches in bacteria. Proc Natl Acad Sci U S A 99(Suppl 4):S16470–S16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baga M, Goransson M, Normark S, Uhlin BE. 1985. Transcriptional activation of a pap pilus virulence operon from uropathogenic Escherichia coli. EMBO J 4:3887–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forsman K, Goransson M, Uhlin BE. 1989. Autoregulation and multiple DNA interactions by a transcriptional regulatory protein in E. coli pili biogenesis. EMBO J 8:1271–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauer FG, Remaut H, Hultgren SJ, Waksman G. 2004. Fiber assembly by the chaperone-usher pathway. Biochim Biophys Acta 1694:259–267. doi: 10.1016/j.bbamcr.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Hull RA, Gill RE, Hsu P, Minshew BH, Falkow S. 1981. Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect Immun 33:933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kallenius G, Mollby R, Svenson SB, Helin I, Hultberg H, Cedergren B, Winberg J. 1981. Occurrence of P-fimbriated Escherichia coli in urinary tract infections. Lancet 2:1369–1372. [DOI] [PubMed] [Google Scholar]
- 28.Buckles EL, Luterbach CL, Wang X, Lockatell CV, Johnson DE, Mobley HL, Donnenberg MS. 2015. Signature-tagged mutagenesis and co-infection studies demonstrate the importance of P fimbriae in a murine model of urinary tract infection. Pathog Dis 73:ftv014. doi: 10.1093/femspd/ftv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagberg L, Hull R, Hull S, Falkow S, Freter R, Svanborg Eden C. 1983. Contribution of adhesion to bacterial persistence in the mouse urinary tract. Infect Immun 40:265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane MC, Mobley HL. 2007. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int 72:19–25. doi: 10.1038/sj.ki.5002230. [DOI] [PubMed] [Google Scholar]
- 31.Xia Y, Forsman K, Jass J, Uhlin BE. 1998. Oligomeric interaction of the PapB transcriptional regulator with the upstream activating region of pili adhesin gene promoters in Escherichia coli. Mol Microbiol 30:513–523. doi: 10.1046/j.1365-2958.1998.01080.x. [DOI] [PubMed] [Google Scholar]
- 32.Sette M, Spurio R, Trotta E, Brandizi C, Brandi A, Pon CL, Barbato G, Boelens R, Gualerzi CO. 2009. Sequence-specific recognition of DNA by the C-terminal domain of nucleoid-associated protein H-NS. J Biol Chem 284:30453–30462. doi: 10.1074/jbc.M109.044313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang CC, Nash HA. 1989. The interaction of E. coli IHF protein with its specific binding sites. Cell 57:869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]
- 34.Stella S, Cascio D, Johnson RC. 2010. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev 24:814–826. doi: 10.1101/gad.1900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pul U, Wurm R, Lux B, Meltzer M, Menzel A, Wagner R. 2005. LRP and H-NS—cooperative partners for transcription regulation at Escherichia coli rRNA promoters. Mol Microbiol 58:864–876. doi: 10.1111/j.1365-2958.2005.04873.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q, Calvo JM. 1993. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J 12:2495–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braaten BA, Nou X, Kaltenbach LS, Low DA. 1994. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell 76:577–588. doi: 10.1016/0092-8674(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 38.van der Woude MW, Kaltenbach LS, Low DA. 1995. Leucine-responsive regulatory protein plays dual roles as both an activator and a repressor of the Escherichia coli pap fimbrial operon. Mol Microbiol 17:303–312. doi: 10.1111/j.1365-2958.1995.mmi_17020303.x. [DOI] [PubMed] [Google Scholar]
- 39.Khandige S, Kronborg T, Uhlin BE, Moller-Jensen J. 2015. sRNA-mediated regulation of P-fimbriae phase variation in uropathogenic Escherichia coli. PLoS Pathog 11:e1005109. doi: 10.1371/journal.ppat.1005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dorman CJ. 2013. Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat Rev Microbiol 11:349–355. doi: 10.1038/nrmicro3007. [DOI] [PubMed] [Google Scholar]
- 41.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog 2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rimsky S, Zuber F, Buckle M, Buc H. 2001. A molecular mechanism for the repression of transcription by the H-NS protein. Mol Microbiol 42:1311–1323. [DOI] [PubMed] [Google Scholar]
- 43.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 44.Dorman CJ. 2007. H-NS, the genome sentinel. Nat Rev Microbiol 5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 45.Lloyd AL, Rasko DA, Mobley HL. 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol 189:3532–3546. doi: 10.1128/JB.01744-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White-Ziegler CA, Villapakkam A, Ronaszeki K, Young S. 2000. H-NS controls pap and daa fimbrial transcription in Escherichia coli in response to multiple environmental cues. J Bacteriol 182:6391–6400. doi: 10.1128/JB.182.22.6391-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donato GM, Lelivelt MJ, Kawula TH. 1997. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J Bacteriol 179:6618–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jordi BJ, Dagberg B, de Haan LA, Hamers AM, van der Zeijst BA, Gaastra W, Uhlin BE. 1992. The positive regulator CfaD overcomes the repression mediated by histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J 11:2627–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsen A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. 1993. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol 7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 50.Torres AG, Slater TM, Patel SD, Popov VL, Arenas-Hernandez MM. 2008. Contribution of the Ler- and H-NS-regulated long polar fimbriae of Escherichia coli O157:H7 during binding to tissue-cultured cells. Infect Immun 76:5062–5071. doi: 10.1128/IAI.00654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller CM, Dobrindt U, Nagy G, Emody L, Uhlin BE, Hacker J. 2006. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J Bacteriol 188:5428–5438. doi: 10.1128/JB.01956-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertin P, Terao E, Lee EH, Lejeune P, Colson C, Danchin A, Collatz E. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J Bacteriol 176:5537–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol 181:7500–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez-Antonio A, Janga SC, Thieffry D. 2008. Functional organisation of Escherichia coli transcriptional regulatory network. J Mol Biol 381:238–247. doi: 10.1016/j.jmb.2008.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Girgis HS, Liu Y, Ryu WS, Tavazoie S. 2007. A comprehensive genetic characterization of bacterial motility. PLoS Genet 3:1644–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ko M, Park C. 2000. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J Bacteriol 182:4670–4672. doi: 10.1128/JB.182.16.4670-4672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krin E, Danchin A, Soutourina O. 2010. RcsB plays a central role in H-NS-dependent regulation of motility and acid stress resistance in Escherichia coli. Res Microbiol 161:363–371. doi: 10.1016/j.resmic.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 58.Forsman K, Sonden B, Goransson M, Uhlin BE. 1992. Antirepression function in Escherichia coli for the cAMP-cAMP receptor protein transcriptional activator. Proc Natl Acad Sci U S A 89:9880–9884. doi: 10.1073/pnas.89.20.9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McFarland KA, Lucchini S, Hinton JC, Dorman CJ. 2008. The leucine-responsive regulatory protein, Lrp, activates transcription of the fim operon in Salmonella enterica serovar Typhimurium via the fimZ regulatory gene. J Bacteriol 190:602–612. doi: 10.1128/JB.01388-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braaten BA, Platko JV, van der Woude MW, Simons BH, de Graaf FK, Calvo JM, Low DA. 1992. Leucine-responsive regulatory protein controls the expression of both the pap and fan pili operons in Escherichia coli. Proc Natl Acad Sci U S A 89:4250–4254. doi: 10.1073/pnas.89.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huisman TT, Bakker D, Klaasen P, de Graaf FK. 1994. Leucine-responsive regulatory protein, IS1 insertions, and the negative regulator FaeA control the expression of the fae (K88) operon in Escherichia coli. Mol Microbiol 11:525–536. doi: 10.1111/j.1365-2958.1994.tb00333.x. [DOI] [PubMed] [Google Scholar]
- 62.Casadesus J, Low D. 2006. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev 70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gally DL, Rucker TJ, Blomfield IC. 1994. The leucine-responsive regulatory protein binds to the fim switch to control phase variation of type 1 fimbrial expression in Escherichia coli K-12. J Bacteriol 176:5665–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Calvo JM, Matthews RG. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev 58:466–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roesch PL, Blomfield IC. 1998. Leucine alters the interaction of the leucine-responsive regulatory protein (Lrp) with the fim switch to stimulate site-specific recombination in Escherichia coli. Mol Microbiol 27:751–761. doi: 10.1046/j.1365-2958.1998.00720.x. [DOI] [PubMed] [Google Scholar]
- 66.Shimada T, Saito N, Maeda M, Tanaka K, Ishihama A. 15 July 2015. Expanded roles of leucine-responsive regulatory protein in transcription regulation of the Escherichia coli genome: genomic SELEX screening of the regulation targets. mGen 1. doi: 10.1099/mgen.0.000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cho BK, Barrett CL, Knight EM, Park YS, Palsson BO. 2008. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc Natl Acad Sci U S A 105:19462–19467. doi: 10.1073/pnas.0807227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Will WR, Lu J, Frost LS. 2004. The role of H-NS in silencing F transfer gene expression during entry into stationary phase. Mol Microbiol 54:769–782. doi: 10.1111/j.1365-2958.2004.04303.x. [DOI] [PubMed] [Google Scholar]
- 69.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol 181:6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Troutt AB, McHeyzer-Williams MG, Pulendran B, Nossal GJ. 1992. Ligation-anchored PCR: a simple amplification technique with single-sided specificity. Proc Natl Acad Sci U S A 89:9823–9825. doi: 10.1073/pnas.89.20.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hawley DK, McClure WR. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res 11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harley CB, Reynolds RP. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res 15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu CW, Goldthwait DA. 1969. Studies of nucleotide binding to the ribonucleic acid polymerase by a fluorescence technique. Biochemistry 8:4450–4458. doi: 10.1021/bi00839a034. [DOI] [PubMed] [Google Scholar]
- 74.Mendoza-Vargas A, Olvera L, Olvera M, Grande R, Vega-Alvarado L, Taboada B, Jimenez-Jacinto V, Salgado H, Juarez K, Contreras-Moreira B, Huerta AM, Collado-Vides J, Morett E. 2009. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One 4:e7526. doi: 10.1371/journal.pone.0007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smolke CD, Khlebnikov A, Keasling JD. 2001. Effects of transcription induction homogeneity and transcript stability on expression of two genes in a constructed operon. Appl Microbiol Biotechnol 57:689–696. doi: 10.1007/s002530100812. [DOI] [PubMed] [Google Scholar]
- 76.Khlebnikov A, Risa O, Skaug T, Carrier TA, Keasling JD. 2000. Regulatable arabinose-inducible gene expression system with consistent control in all cells of a culture. J Bacteriol 182:7029–7034. doi: 10.1128/JB.182.24.7029-7034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Siegele DA, Hu JC. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc Natl Acad Sci U S A 94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morra R, Shankar J, Robinson CJ, Halliwell S, Butler L, Upton M, Hay S, Micklefield J, Dixon N. Dual transcriptional-translational cascade permits cellular level tuneable expression control. Nucleic Acids Res, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cui Y, Wang Q, Stormo GD, Calvo JM. 1995. A consensus sequence for binding of Lrp to DNA. J Bacteriol 177:4872–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nou X, Braaten B, Kaltenbach L, Low DA. 1995. Differential binding of Lrp to two sets of pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli. EMBO J 14:5785–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. 2007. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat Struct Mol Biol 14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 82.Lang B, Blot N, Bouffartigues E, Buckle M, Geertz M, Gualerzi CO, Mavathur R, Muskhelishvili G, Pon CL, Rimsky S, Stella S, Babu MM, Travers A. 2007. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res 35:6330–6337. doi: 10.1093/nar/gkm712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morschhauser J, Vetter V, Emody L, Hacker J. 1994. Adhesin regulatory genes within large, unstable DNA regions of pathogenic Escherichia coli: cross-talk between different adhesin gene clusters. Mol Microbiol 11:555–566. doi: 10.1111/j.1365-2958.1994.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 84.Lindberg S, Xia Y, Sonden B, Goransson M, Hacker J, Uhlin BE. 2008. Regulatory Interactions among adhesin gene systems of uropathogenic Escherichia coli. Infect Immun 76:771–780. doi: 10.1128/IAI.01010-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Holden NJ, Uhlin BE, Gally DL. 2001. PapB paralogues and their effect on the phase variation of type 1 fimbriae in Escherichia coli. Mol Microbiol 42:319–330. doi: 10.1046/j.1365-2958.2001.02656.x. [DOI] [PubMed] [Google Scholar]
- 86.van der Woude MW, Low DA. 1994. Leucine-responsive regulatory protein and deoxyadenosine methylase control the phase variation and expression of the sfa and daa pili operons in Escherichia coli. Mol Microbiol 11:605–618. doi: 10.1111/j.1365-2958.1994.tb00340.x. [DOI] [PubMed] [Google Scholar]
- 87.Stoebel DM, Free A, Dorman CJ. 2008. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 88.Paul L, Blumenthal RM, Matthews RG. 2001. Activation from a distance: roles of Lrp and integration host factor in transcriptional activation of gltBDF. J Bacteriol 183:3910–3918. doi: 10.1128/JB.183.13.3910-3918.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paul L, Mishra PK, Blumenthal RM, Matthews RG. 2007. Integration of regulatory signals through involvement of multiple global regulators: control of the Escherichia coli gltBDF operon by Lrp, IHF, Crp, and ArgR. BMC Microbiol 7:2. doi: 10.1186/1471-2180-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weyand NJ, Braaten BA, van der Woude M, Tucker J, Low DA. 2001. The essential role of the promoter-proximal subunit of CAP in pap phase variation: Lrp- and helical phase-dependent activation of papBA transcription by CAP from −215. Mol Microbiol 39:1504–1522. doi: 10.1046/j.1365-2958.2001.02338.x. [DOI] [PubMed] [Google Scholar]
- 91.Navarre WW, McClelland M, Libby SJ, Fang FC. 2007. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev 21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 92.Dame RT, Wyman C, Goosen N. 2000. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res 28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dame RT. 2005. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol Microbiol 56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 94.Arold ST, Leonard PG, Parkinson GN, Ladbury JE. 2010. H-NS forms a superhelical protein scaffold for DNA condensation. Proc Natl Acad Sci U S A 107:15728–15732. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Armbruster CE, Hodges SA, Mobley HL. 2013. Initiation of swarming motility by Proteus mirabilis occurs in response to specific cues present in urine and requires excess l-glutamine. J Bacteriol 195:1305–1319. doi: 10.1128/JB.02136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol 189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.