Abstract
Staphylococcus aureus is an important human pathogen that can cause two categories of severe infections. Acute infections are characterized by pronounced toxin production, while chronic infections often involve biofilm formation. However, it is poorly understood how S. aureus controls the expression of genes associated with acute versus biofilm-associated virulence. We here identified an AraC-type transcriptional regulator, Rsp, that promotes the production of key toxins while repressing major biofilm-associated genes and biofilm formation. Genome-wide transcriptional analysis and modeling of regulatory networks indicated that upregulation of the accessory gene regulator (Agr) and downregulation of the ica operon coding for the biofilm exopolysaccharide polysaccharide intercellular adhesin (PIA) were central to the regulatory impact of Rsp on virulence. Notably, the Rsp protein directly bound to the agrP2 and icaADBC promoters, resulting in strongly increased levels of the Agr-controlled toxins phenol-soluble modulins (PSMs) and alpha-toxin and reduced production of PIA. Accordingly, Rsp was essential for the development of bacteremia and skin infection, representing major types of acute S. aureus infection. Our findings give important insight into how S. aureus adapts the expression of its broad arsenal of virulence genes to promote different types of disease manifestations and identify the Rsp regulator as a potential target for strategies to control acute S. aureus infection.
INTRODUCTION
Staphylococcus aureus is an important human pathogen and one of the leading causes of morbidity and mortality from infectious diseases worldwide. It is especially notorious for health care-associated infections and the development of antibiotic-resistant strains, among which methicillin-resistant S. aureus (MRSA) is most infamous (1, 2).
Two of the most prominent features of S. aureus as a pathogen are the vast variety of infection types that it may cause and the large number of virulence determinants that it can produce. The latter are often characteristic for a specific type of infection. Importantly, acute S. aureus infections are commonly dependent on toxin production. For example, skin and blood infections, which are among the most frequent types of acute S. aureus infections, are dependent on the production of alpha-toxin and phenol-soluble modulins α (PSMα)—cytolytic toxins that are produced by virtually all S. aureus strains (3, 4). Of note, most S. aureus toxins are under positive control by the accessory gene regulator (Agr) quorum-sensing system (5).
Contrastingly, chronic types of S. aureus disease, such as infections on indwelling medical devices, endocarditis, or chronic wound infections, usually proceed with the involvement of biofilms (6). Biofilms are sticky, often surface-attached, agglomerations of bacteria with a distinct pattern of gene expression. Staphylococcal biofilm gene expression has been shown to include a general downregulation of cell processes associated with rapid cell growth but upregulation of other systems, such as the urease system, which is believed to be involved in pH homeostasis (7, 8). Notably, the biofilm mode of growth considerably increases resistance to antibiotics and the human immune system (9). The sticky matrix that holds cells together in a biofilm consists of a variety of polymeric molecules, such as polymer-forming proteins, polysaccharides, DNA, and teichoic acids (6). Although recent research has shown that it is not of universal importance and other biofilm factors may substitute for its absence (6), the biofilm exopolysaccharide polysaccharide intercellular adhesin (PIA, also called PNAG for poly-N-acetylglucosamine) is strongly associated with biofilm formation, biofilm-associated infection, and immune evasion (10–14). The toxin regulator Agr impacts biofilm formation by its negative impact on several surface proteins that facilitate initial attachment to host tissues, which, however, may be strongly strain specific (15). Furthermore, Agr contributes to biofilm development via its marked impact on the production of PSMs, which structure biofilms via their detergent effect, leading to cell-cell detachment (16). However, Agr has no effect on the production of most biofilm matrix molecules, notably including PIA (15).
In the present study, we searched for yet-unknown master regulators of toxin production using hemolytic capacity as a screen. We identified a regulatory protein, Rsp, a member of the family of AraC-type transcriptional regulators (17), which revealed a strong impact on hemolytic capacity, production of toxins, and pathogenicity in acute types of S. aureus infection. Notably, Rsp had strongly opposite effects on toxin versus biofilm production, which included the upregulation of Agr and downregulation of PIA expression by direct binding of Rsp to the respective promoters. Our study thus identified a master regulator that can manage the switch between gene expression patterns associated with acute versus chronic pathogenicity in S. aureus.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the Chinese Association for Laboratory Animal Sciences (CALAS) and approved by the ethics committee of Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China (protocol RJ-M-2013-0107).
Human red blood cells were isolated from healthy individuals in accordance with a protocol approved by the ethics committee of Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China (protocol RJ-H-2015-0031). All individuals gave informed consent prior to donating blood.
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are summarized in Table S1 in the supplemental material. Escherichia coli strain DH5α was used in the cloning experiments. S. aureus strain RN4220 was used as a gateway strain prior to the propagation of plasmids into S. aureus BD02-25 (MRSA, ST8, USA500) or MW2 (MRSA, ST1, USA400). Three hundred clinical S. aureus isolates used for rsp gene distribution analysis were randomly collected from patients with S. aureus infection in a comprehensive teaching hospital in Shanghai, China (Renji Hospital, affiliated with Shanghai Jiao Tong University) in 2014. Bacteria were routinely grown in tryptic soy broth (TSB, containing 0.25% glucose) or TSB agar plates at 37°C, unless otherwise indicated. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 10 μg/ml; erythromycin, 5 μg/ml; kanamycin 50 μg/ml.
Transposon mutagenesis of strain BD02-25.
A stationary-phase culture (24 h) of strain BD02-25 harboring the mariner-based transposon system pBTn (18) grown at 30°C in TSB (without glucose) containing 0.5% xylose, chloramphenicol, and erythromycin was diluted 1:100 into fresh TSB (without glucose) with 0.5% xylose and erythromycin, and the mixture was incubated at 42°C for 24 h. This was repeated twice with the antibiotics and once without the antibiotics. Afterwards, transposon insertion mutants (Ermr Cms) were selected by plating the Ermr colonies on TSB agar containing chloramphenicol. Clones with an Ermr Cms phenotype were then preserved in glycerol stocks at −80°C for future screening.
Screening of the transposon library by detection of hemolytic capacity on sheep blood agar plates.
A single clone was grown to mid-logarithmic growth phase (4 h), and equal amounts (2 μl) of cells were spotted onto sheep blood agar plates, incubated at 37°C for 24 h, and visually evaluated for zones of lysis.
Hemolysis assays.
Culture filtrates were collected from bacterial cultures grown for 12 h. Hemolytic activities were determined by incubating samples with human red blood cells (2% [vol/vol] in Dulbecco's phosphate-buffered saline [DPBS]) for 1 h at 37°C. Hemolysis was determined by measuring the optical density at 540 nm (OD540) using an enzyme-linked immunosorbent assay (ELISA) reader. The assay was performed in triplicate.
Arbitrary primed (inverse) PCR and nucleotide sequencing.
Arbitrary primed (inverse) PCR and nucleotide sequencing were performed as described before (18).
Screening for presence of the rsp gene by analytical PCR.
The S. aureus isolates were tested for the presence of the rsp gene by analytical PCR using the primers listed in Table S2 in the supplemental material, amplifying a 584-bp fragment. To confirm the identity of the rsp gene, several PCR products were randomly selected for sequencing using an ABI 3730XL DNA sequencer.
Allelic gene replacement by homologous recombination and genetic complementation.
To delete the rsp gene from the genomes of S. aureus BD02-25 and MW2 by homologous recombination, plasmid pKOR1 was used (19). DNA fragments for upstream and downstream sequences of rsp were PCR amplified from chromosomal DNA of strains BD02-25 or MW2, respectively, using the primers listed in Table S2 in the supplemental material. Then, overlap PCR with the resulting fragments as the templates and primers rsp-att1 and rsp-att2 was performed. The resulting PCR product with attB sites at the two ends was used for cloning into plasmid pKOR1, yielding plasmid pKOR1Δrsp. This plasmid was transferred via electroporation first to S. aureus RN4220 and then BD02-25 or MW2. Allelic replacement mutations were selected as described by Bae and Schneewind (19). Proper integration was verified by analytical PCR and sequencing of the genomic DNA at the borders of the PCR-derived regions. For genetic complementation, the rsp gene was amplified by PCR with primers Crsp-BamH and Crsp-EcoR and cloned in plasmid pRB473. Strains harboring only plasmid pRB473 were used as controls. The corresponding wild-type and isogenic gene deletion mutant pairs and the pRB473- and pRBrsp-harboring pairs did not show changes in growth (see Fig. S1 in the supplemental material). The agr operon was deleted using the same procedure as for rsp, with the primers shown in Table S2 in the supplemental material.
Semiquantitative biofilm assay.
Semiquantitative biofilm assays were performed as described elsewhere (20). Subsequently, cells were fixed by Bouin's fixative. The fixative was removed after 1 h, and wells were washed with phosphate-buffered saline (PBS). Cells in the wells were stained with crystal violet, and the floating stain was washed off with slowly running water. After drying, the stained biofilm was read with a MicroELISA autoreader (Bio-Rad) at 570 nm.
Biofilm formation using a Bioflux microfluidic flow cell system.
The BioFlux 1000z microfluidic system (Fluxion Biosciences, CA) was used to assess biofilm formation under flow conditions as described previously (21). To grow biofilms, the microfluidic channels were primed with TSB supplemented with 0.5% glucose at 2 dynes (dyn)/cm2 (1 dyn is 1 × 10−5 N) for 10 min. Each channel of a 48-well plate was coated with 20% platelet-poor human plasma in 50 mM carbonate buffer, pH 9.6, and incubated for 24 h at 4°C before biofilm assays were set up. The exponential-growth cultures were diluted 1:100 in TSB supplemented with 0.5% glucose. Bacterial suspensions were seeded at 0.2 dyn/cm2 for 5 s in all channels. The plate was then incubated at room temperature for 1 h to allow cells to adhere. Excess inoculums were removed, and 0.8 ml of TSB supplemented with 0.5% glucose was added to the input wells. Biofilms were grown at 37°C with a flow of fresh medium at a constant shear of 0.1 dyn/cm2. Images were taken every 10 min for 24 h at a magnification of ×10 under bright-field microscopy.
Mouse bacteremia and skin abscess models.
BALB/c female mice were used for the bacteremia model. Outbred, immunocompetent hairless mice were used for the abscess model. All mice were between 4 and 6 weeks of age at the time of use. S. aureus strains were grown to mid-exponential phase, washed once with sterile PBS, and then resuspended in PBS at 1 × 108 CFU/100 μl (bacteremia model) or 1 × 107 CFU/50 μl (abscess model). For the bacteremia model, we injected each mouse with 108 CFU of live S. aureus in 0.1 ml sterile saline into the retro-orbital vein. Control animals received sterile saline only. After inoculation, mouse health and disease advancement were monitored every 3 h for the first 24 h and then every 8 h for up to 168 h. Mice were euthanized immediately if they showed signs of respiratory distress, mobility loss, or inability to eat and drink. All surviving animals were euthanized at 168 h. For the abscess model, mice were anesthetized with isoflurane and inoculated with 50 μl PBS containing 107 CFU of live S. aureus or PBS alone in the right flank by subcutaneous injection. We examined test animals at 24-h intervals for a total of 14 days with a caliper. Length (L) and width (W) values were used to calculate the area of lesions (formula: area = L × W).
RNA-Seq experiments.
For high-throughput RNA sequencing (RNA-Seq) experiments, the comparisons were based on two biological replicates. Overnight cultures were diluted 1:100 into 50 ml of TSB and incubated at 37°C with shaking at 200 rpm until grown to mid-logarithmic growth phase (for the wild-type strain, to an OD600 of 1.768 and 1.781; for the rsp mutant strain, to an OD600 of 1.727 and 1.778). Cells were harvested and washed twice in 10 mM sodium phosphate buffer (pH 6.5). Total RNA was isolated using an RNeasy minikit (Qiagen) as recommended in a standard protocol. A total of 10 μg of each RNA sample was subjected to further purification to enrich the mRNA using a MicrobExpress kit (Ambion) according to the manufacturer's instructions. Each mRNA sample was suspended in 25 μl of RNA storage solution, and the quality of mRNA obtained was determined using an Agilent 2100 Bioanalyzer. Bacterial mRNA was fragmented using an RNA fragmentation kit (Ambion) to obtain fragments in the size range of 200 to 250 bp. Double-stranded cDNA was generated using the SuperScript Double-Stranded cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions. An Illumina Paired End Sample Prep kit was used to prepare the RNA-Seq library according to the manufacturer's instructions. All samples were sequenced using the Hiseq2000 (Illumina, CA) sequencer at Oebiotech Corp. (Shanghai). The RNA-Seq data were submitted to the NCBI Gene Expression Omnibus (GEO) archive (see below). Reads were aligned to S. aureus USA300_FPR3757 (RefSeq accession number NC_007793.1) using the Burrows-Wheeler Alignment tool (BWA). The RNA-Seq data analysis included the four stages as described by Qin et al. (22). Based on reads per kilobase of transcript per million mapped reads (RPKM) normalization, we performed analyses of differentially expressed genes. Genes with an adjusted P value of <0.05, a false discovery rate (FDR) of <0.001, and a fold change of >2 were identified as being differentially expressed.
Protein-protein interaction prediction.
In the first step, all differentially expressed genes identified by RNA-Seq were used to perform the gene ontology enrichment analysis with the functional annotation tool at the DAVID bioinformatics server (23, 24). The most relevant biological process (BP; gene ontology [GO] analysis), cell component (CC), molecular function (MF), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were associated with the query genes. Default settings with an Expression Analysis Systematic Explorer (EASE) score of 0.1 were used. In the second step, information on protein-protein interaction of the 501 up- and downregulated genes was obtained via the STRING database (version 9.1) (25); a default setting with a combined score of >0.4 but no text mining was applied. For our data set, few direct interactions existed among the query genes. The built-in one-step expand algorithm was used to discover potential indirect interactions. A 3-step extension of the network was performed. Eventually, an interactome of 114 downregulated and 35 upregulated genes was obtained for further model construction. In the third step, the network model was constructed with information gained in both the first and second steps with the cytoscape (26). For the virulence- and biofilm-focused network, the same procedure was used based on selected, differentially regulated biofilm- and virulence-related genes (ureA, ureB, ureC, ureD, ureE, ureF, fadD, fadE, icaA, atl, agrD, hla, and hld) and 8 of the 10 top Rsp-regulated KEGG pathways (see Table S3 in the supplemental material), using an EASE score of 0.03.
Prediction of the DNA-binding motif in Rsp and Rsp-binding sites in promoters.
As DNA binding motifs are commonly conserved, the conserved domains of different Rsp proteins were determined using the NCBI cdd database (27). DP-Bind was used to predict DNA-binding residues based on a combined algorithm of SVM, KLR, PLR. Predicted residue positions that are located in the conserved domain were assumed to be involved in the interaction with DNA (28). The Regulatory Sequence Analysis Tools (RSAT) were used to detect sites for regulatory protein binding in the promoter regions of the agrP2, icaADBC, and sarR promoters.
Quantitative RT-PCR.
Total RNA was isolated using the methods described above in “RNA-Seq Experiments.” cDNA was synthesized from total RNA using the QuantiTect reverse transcription (RT) system (Qiagen) according to the manufacturer's instructions. Oligonucleotide primers were designed using Primer Express and are listed in Table S2 in the supplemental material. The resulting cDNA and negative-control samples were amplified using the QuantiTect SYBR green PCR kit (Qiagen). Reactions were performed in a MicroAmp Optical 96-well reaction plate using a 7500 Sequence Detector (Applied Biosystems). Standard curves were determined for each gene, using purified chromosomal DNA at concentrations of 0.005 to 50 ng/ml. All qRT-PCR experiments were performed in duplicate or triplicate, with gyrB as control.
Overexpression and purification of Rsp.
The rsp gene was cloned and overexpressed as histidine residue-tagged (6×His tag) fusion. Primers for the amplification of the rsp gene from S. aureus strain BD02-25 genomic DNA are listed in Table S2 in the supplemental material. PCR products were purified, digested using SaII and PstI, and cloned into the His tag fusion vector pQE-9 (Qiagen). The recombinant vector pQE-rsp was transferred to E. coli strain XL1 blue (Qiagen) for propagating plasmids and then transferred to E. coli strain SG13009 [pREP4] (Qiagen) for overexpression. SG13009 (pQE-rsp) was grown at 37°C to an OD600 of 1.0. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM, the cultures were incubated for an additional 5 h, and cells were harvested. The 6His-Rsp fusion protein was purified on Ni-NTA agarose matrix columns by washing with 10 volumes of 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8.0, followed by elution with 5 volumes of 50 mM NaH2PO4, 300 mM NaCl, and 100 mM imidazole, pH 8.0. The purified fusion protein was concentrated in Centriprep-10 concentrators (Amicon) and dialyzed against 10 mM sodium phosphate buffer (pH 7.5) using PD-10 desalting columns (Amersham Biosciences). The size of the 6His-Rsp fusion protein was confirmed by SDS-PAGE. To generate mutant Rsp proteins, site-directed mutagenesis was performed as previously described (29). Each obtained plasmid was sequenced to confirm the accuracy of the mutations. Expression and purification protocols were as for wild-type 6His-Rsp. Of the three mutant proteins that we attempted to express and purify (see Tables S1 and S2 in the supplemental material), only the 191-192 mutant protein (i.e., containing mutations in positions 191 and 192) could be expressed and purified.
EMSA.
For electrophoretic mobility shift assays (EMSA), the agrP2, icaADBC, and sarR promoters were amplified with the primer pairs shown in Table S2 in the supplemental material. Approximately 0.5 μmol of the purified PCR products was biotinylated with a biotin 3′-end DNA labeling kit (Pierce Biotechnology). The biotinylated probes were purified and denatured at 94°C for 3 min and then slowly cooled to room temperature to allow proper annealing. The purified 6His-Rsp protein was incubated with biotin-labeled DNA probes (20 fmol each) for 20 min at room temperature in 20 μl of binding buffer (LightShift chemiluminescence EMSA kit; Pierce). As a control, the sample was incubated with an excess of unlabeled probe. A 6% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA (TBE) was prerun for 40 min, after which samples were loaded and electrophoresed at 100 V for 1.5 h on ice. The biotinylated probes were transferred to a nylon membrane (Amersham Biosciences) at 380 mA for 30 min. The transferred DNA was cross-linked to the membrane with UV light. The biotin-labeled DNA was detected with a LightShift chemiluminescence EMSA kit (Pierce).
PSM measurements.
PSMs in culture filtrates were measured as previously described using high-pressure liquid chromatography/electrospray ionization mass spectrometry (HPLC/ESI-MS) (30). PSM abundance is expressed as the combined intensity of the two main peaks detected in the ion chromatogram of the respective peak in the total ion chromatogram.
Detection of PIA by immune dot blot.
PIA was determined by immune dot blot assays with anti-PIA antiserum as previously described, after releasing PIA from the cell surface by boiling with 0.5 M EDTA (31). Signal intensities of the dots were measured by densitometry.
Detection of alpha-toxin and protein A by Western blotting.
Protein gels were blotted onto nitrocellulose membranes using an iBlot Dry blotting system (Life Technologies). Membranes were blocked in SuperBlock blocking buffer (Thermo Scientific) followed by incubation with anti-staphylococcal alpha-toxin rabbit serum (Sigma-Aldrich) in blocking buffer (1:1,000). After washing with TBST (Tris-buffered saline with 0.1% Tween 20), Cy5-conjugated goat anti-rabbit IgG (Life Technologies) was applied to the membrane in blocking buffer (1:4,000). Membranes were washed with TBST and scanned using a Typhoon Trio Plus (GE Healthcare Life Sciences) followed by quantification with ImageQuant TL software (GE Healthcare Life Sciences, Piscataway, NJ).
Statistics.
Statistical analysis was performed using Graph Pad Prism version 6.02. For the comparison of two groups, unpaired t tests were used; for three or more groups, 1-way or 2-way analysis of variance (ANOVA) was used, as appropriate. All error bars depict the standard errors of the means.
Nucleotide sequence accession number.
The RNA-Seq data obtained in this study were submitted to the NCBI Gene Expression Omnibus (GEO) archive (accession number GSE67344).
RESULTS
Identification of Rsp as a regulator of acute virulence.
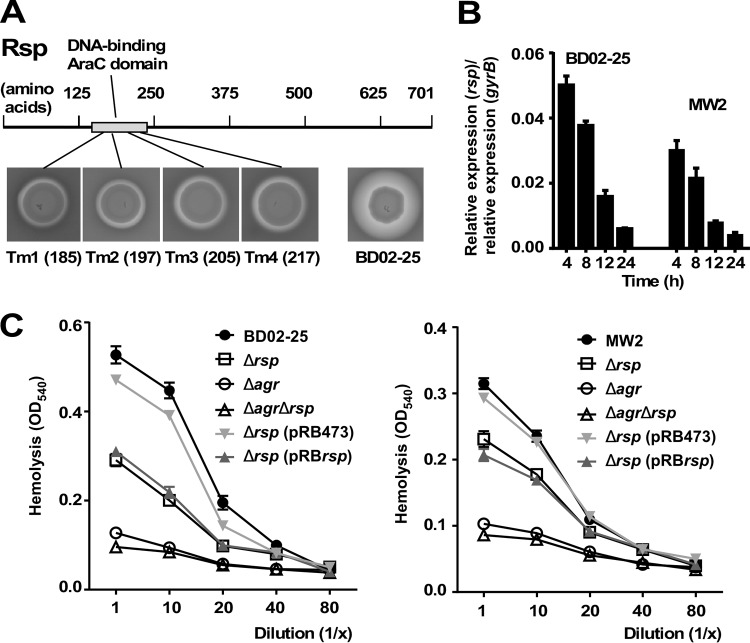
To detect yet-unidentified genes that influence the acute virulence of S. aureus, we performed mariner-based transposon mutagenesis and chose hemolytic capacity as readout of the screen. We used the highly virulent clinical MRSA strain BD02-25, which belongs to pulsed-field type USA500 and sequence type (ST) 8 (32). A large number of clones (∼10,000) were used for screening. We found four clones with considerably reduced hemolytic activity that had a single transposon insertion at four different locations within the rsp gene coding for a 701-amino-acid protein of the AraC family of transcriptional activators (Fig. 1A). AraC family proteins regulate diverse functions, including sugar catabolism, stress response, and virulence, and contain a characteristic 100-residue DNA binding domain (33), which is where all of the transposon insertions were located.
FIG 1.
Identification of Rsp in a transposon screen as regulator of hemolytic capacity and growth-dependent expression of Rsp. (A) In a transposon screen of ∼10,000 mutants in strain BD02-25, 4 mutants with decreased hemolytic capacity that mapped to the region encoding the DNA-binding domain of the Rsp protein were identified. Numbers refer to the sites of insertion (amino acid positions). (B) Growth-dependent expression of rsp determined by qRT-PCR in strains BD02-25 and MW2. (C) Hemolytic capacities of isogenic rsp and agr mutants and rsp agr double mutants of strains BD02-25 (left panel) and MW2 (right panel) and rsp-complemented and control strains. Error bars depict the standard errors of the means (±SEM).
Epidemiology and impact on basic virulence phenotypes.
To investigate the distribution of the Rsp-encoding gene in clinical S. aureus isolates, we used analytical PCR and tested 300 randomly selected S. aureus isolates from Renji Hospital, Shanghai, China: 98.7% (296/300) of the clinical S. aureus isolates carried the rsp gene.
To analyze basic characteristics of the rsp locus, we deleted the gene by allelic replacement from strains BD02-25 and MW2 (pulsed-field type USA400, ST1; another MRSA strain of clinical importance) (34) and complemented the resulting deletion mutants with a plasmid carrying the rsp gene. The rsp gene was most highly expressed during logarithmic growth phase and increasingly repressed during later stages of growth (Fig. 1B). Using the isogenic gene deletion mutants and genetic complementation strains, we confirmed that rsp strongly increased hemolytic capacity (Fig. 1C).
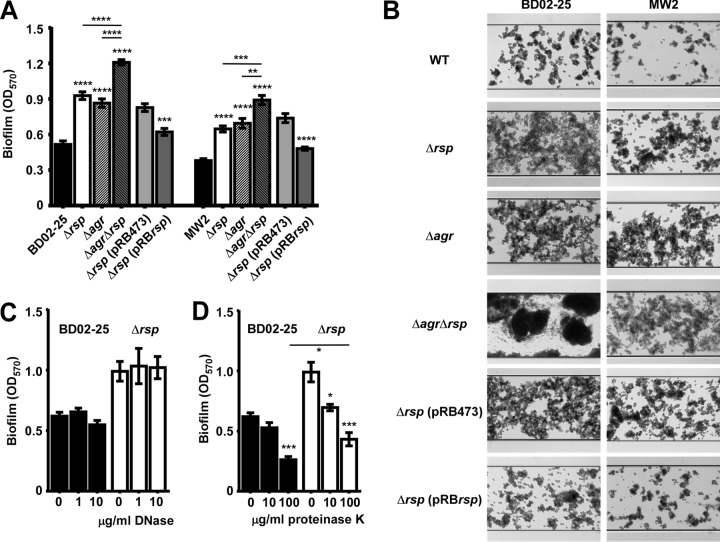
Contrastingly, we found that rsp significantly decreased biofilm formation as measured by semiquantitative microtiter plate assays (Fig. 2A) and a microfluidic flow cell system (Fig. 2B) in both strain backgrounds. The observed effects could be reversed by genetic complementation (Fig. 2A and B). Notably, a significant part of the increase in biofilm in the rsp mutant was independent of Agr. Biofilm formation in the BD02-25 wild-type and rsp mutant strains, in which biofilms were more pronounced than in the MW2 background, were barely affected by DNase (Fig. 2C), indicating that extracellular DNA (eDNA) was not a major component of the biofilms formed by those strains. In contrast, proteinase K digestion led to removal of a significant amount of biofilm (Fig. 2D), but even at the highest concentration of proteinase K used, we still observed significantly increased biofilm formation in the rsp mutant compared to the BD02-25 wild-type strain, indicating both protein- and non-protein-dependent (likely, exopolysaccharide-dependent) biofilms in the rsp mutant. This finding is in accordance with the observed Agr-independent contribution of Rsp to biofilm formation and the fact that the ica PIA biosynthesis genes are not regulated by Agr in S. aureus (15). Altogether, Rsp appeared to have a contrasting effect on virulence determinants associated with acute versus biofilm-associated infection.
FIG 2.
Impact of Rsp on biofilm formation. (A) Biofilm formation in microtiter plates. Data are from 6 independent measurements. The wild-type and isogenic mutant strain data were compared by 1-way ANOVA; data were obtained from plasmid-harboring control and rsp-complemented strains by unpaired t tests. Asterisks depict the statistical significance for comparison to the wild-type strain unless indicated by horizontal bars. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (B) Biofilm formation in microfluidic flow cells. (C, D) Degradation of BD02-25 and isogenic rsp mutant biofilms by DNase (C) and proteinase K (D). Comparisons were evaluated by 1-way ANOVA. Asterisks depict the statistical significance for the comparison to the bar labeled “0” (no enzyme added), unless indicated by horizontal bars. *, P < 0.05; ***, P < 0.001. Error bars depict ±SEM.
Rsp-regulated genes and biological processes.
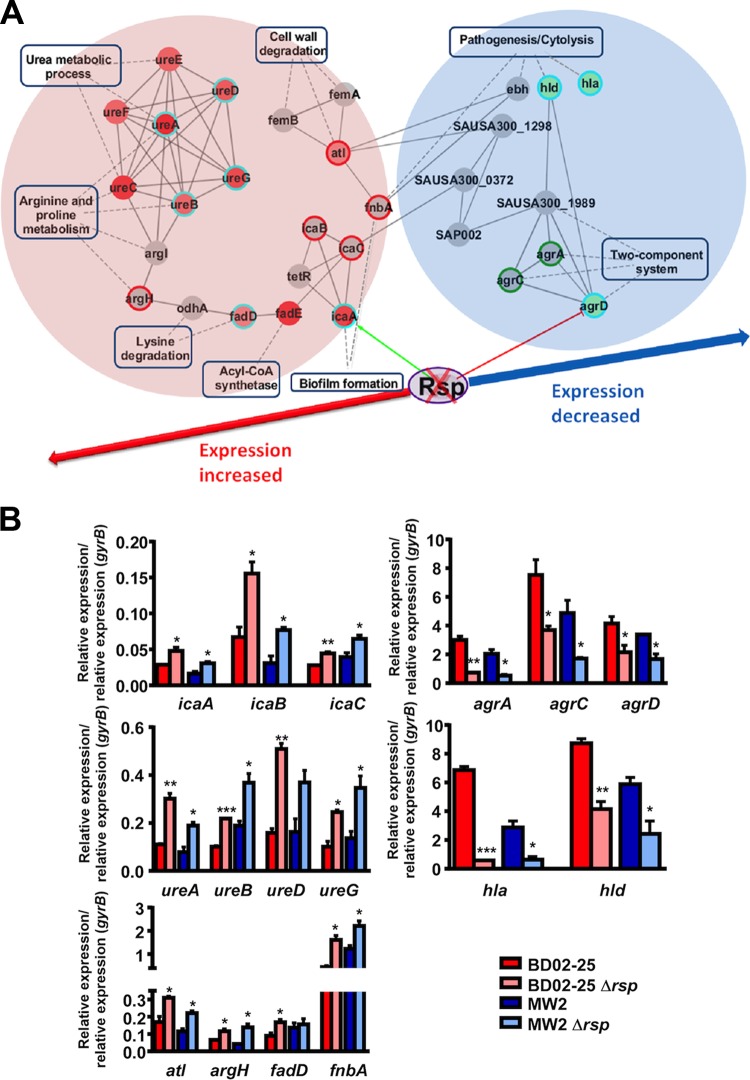
To characterize the genome-wide regulatory impact of rsp, we used RNA-Seq, which showed that at the peak of rsp expression at 4 h, rsp positively affected expression of 176 genes and negatively affected expression of 325 genes (see above for GEO accession number of deposited data). Among those, gene ontology analysis using the DAVID tool (23, 24) enriched 20 KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways as primarily regulated by rsp (using default settings, EASE score = 0.1; see Table S3 in the supplemental material). Information on protein-protein interaction was obtained via the STRING database (25). The center interactome was concentrated on 114 rsp-down- and 35 rsp-upregulated genes (see Table S4 in the supplemental material). The regulation network model (see Fig. S2 in the supplemental material) was constructed using cytoscape (26).
The enriched rsp-regulated genes contained several that were connected to the phenotypes that we had observed to be rsp dependent, namely, biofilm formation and hemolysis. To further confirm the impact of Rsp on those phenotypes and the associated genes, we first modeled an interaction network focusing on the respective related pathways (Fig. 3A). Then, we confirmed differential expression of the corresponding genes—those identified by RNA-Seq as well as related genes, such as those encoded in the same operon—by reverse transcription-quantitative PCR (qRT-PCR) using wild-type/Δrsp mutant pairs of strains BD02-25 and MW2 (Fig. 3B). These genes included, among others, those of the urease operon, the atl autolysin gene, and the ica operon encoding PIA biosynthesis factors, which were all downregulated by rsp. Upregulated genes included most notably those of the agr operon and the cytolysin genes hla (coding for alpha-toxin) and hld (coding for δ-toxin, a member of the PSM family). Of note, qRT-PCR results were consistent between the two S. aureus strains, underscoring that rsp-dependent gene regulation is not strain specific.
FIG 3.
Impact of Rsp on biofilm and virulence-related genes. (A) Rsp-regulated biofilm- and virulence-associated pathways and genes. Circle nodes represent genes, while rectangle nodes with a blue border indicate biological processes. For genes, borders of the nodes represent the type of gene regulation determined by RNA-Seq analysis (upregulated in red, downregulated in green). The centers of the nodes indicate the gene expression changes; color intensity is proportional to the level of regulation. The outer circles of the nodes indicate the gene expression changes as determined by qRT-PCR results (see panel B). The blue circle depicts corresponding results from RNA-Seq and qRT-PCR analyses. Directly related genes that did not show differential expression in the RNA-Seq analysis are shown in gray. Some of those were confirmed by qRT-PCR analysis to be regulated by rsp, and the outer circles of the nodes of those genes indicate the gene expression changes as determined by qRT-PCR results (upregulated in red, downregulated in green). Protein-protein interactions are depicted as gray solid lines. (B) qRT-PCR verification of differentially expressed genes in RNA-Seq analysis or relevant biological processes. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (wild-type strains versus isogenic mutants, t tests). Error bars depict ±SEM.
Altogether, these analyses indicated that Rsp represses active cell processes such as cell wall degradation and several metabolic pathways, including that of urease, which is similar to gene expression changes seen under the biofilm mode of growth. Contrastingly, Rsp increased the expression of genes that are connected to acute virulence, such as those involved in cytolysis, proteolysis, and two-component systems in charge of virulence gene expression.
Binding of Rsp to ica and agr promoters.
Next, we hypothesized that the pronounced regulatory impact of rsp on ica and agr transcript abundance occurs by direct binding of Rsp to the icaADBC and agrP2 promoters, respectively. The binding site prediction tool RSAT predicted a direct-repeat regulatory motif in the agrP2 promoter (TATAATGA_A) and a single palindromic binding motif in the icaADBC promoter (CCATATGG) (Fig. 4A and C; see also Fig. S3 in the supplemental material). To verify the predicted interaction of Rsp with those promoters, we expressed and purified His-tagged Rsp protein (see Fig. S4 in the supplemental material). DNA fragments containing the potential binding motifs were amplified and used for electrophoretic mobility shift assays (EMSAs) (Fig. 4B and D). The recombinant Rsp was able to bind to those DNA fragments already at low concentrations (≥30 μg/ml). Specificity of the interaction was demonstrated by competing with an excess of unlabeled DNA fragments of the promoters. Further substantiating the specificity of binding, a mutated His-tagged Rsp protein with an exchange of two amino acids in the DNA-binding site (positions 191 and 192, Asn-Phe to Ala-Ala) was not able to bind to the agrP2 or icaADBC promoters, even at the highest concentration used for native Rsp (235 μg/ml) (Fig. 4B and D). These findings showed that Agr expression and PIA biosynthesis are regulated by direct binding of Rsp to the respective promoter regions, which is in accordance with the role that our in silico analyses attributed to them. Furthermore, direct, Agr-independent regulation of the biofilm factor PIA by Rsp is in accordance with the fact that an rsp agr double mutant showed significantly increased biofilm formation compared to an agr mutant, both in the BD02-25 and MW2 backgrounds (Fig. 2A and B).
FIG 4.
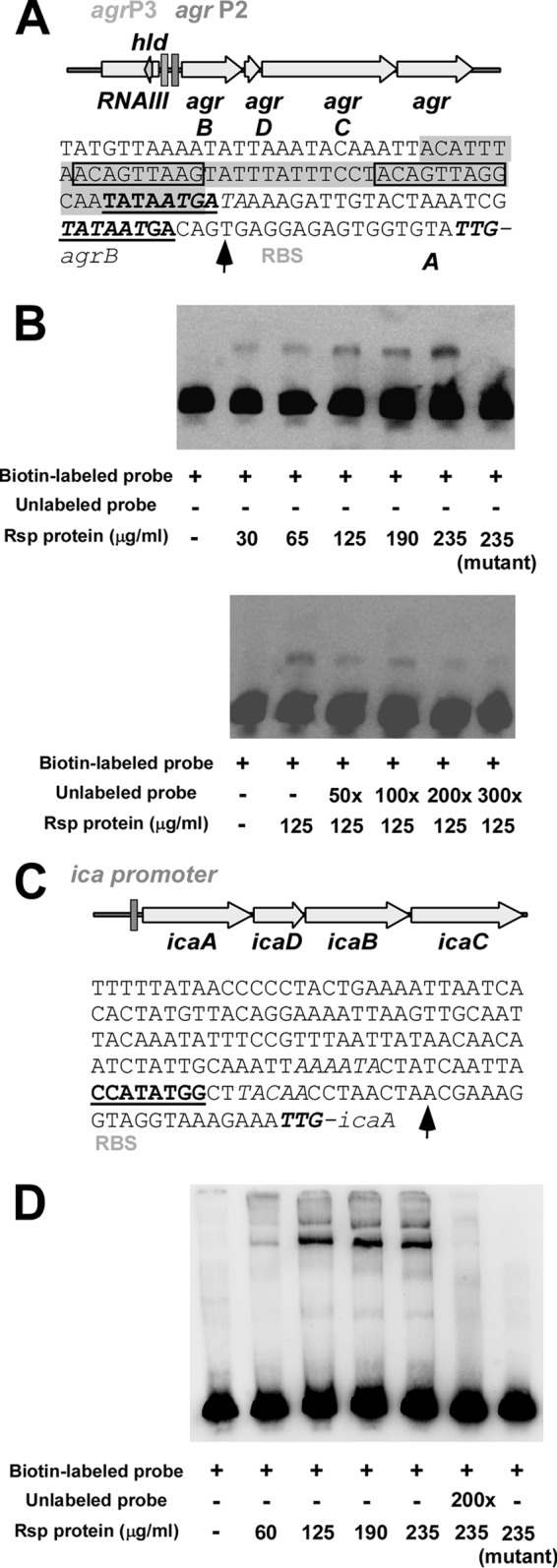
Binding of Rsp to agr and ica promoters. (A, C) Predicted binding sites (bold, underlined) in the agrP2 (A) and icaADBC (C) promoters. Prediction was performed using RSAT. The codon shown in bold or italic font is the start codon of the agrB or the icaA gene, respectively. RBS, ribosome binding site. Transcription start sites (43, 44) are indicated by an arrow. (Presumable) −10 and −35 regions are shown in italic font. In the agrP2 promoter, the shaded region is the AgrA binding region as predicted by Koenig et al. (44), and the direct repeats conforming to the consensus LytTR recognition sites are boxed. (B, D) Confirmation of binding of Rsp protein to DNA fragments comprising the predicted binding sites. Rsp protein was expressed as 6His-tagged fusion protein. The mutant Rsp protein was engineered to contain two alanine residues within the DNA binding site at positions 191 and 192 (instead of asparagine-phenylalanine).
However, many of the transcriptional changes that we found are likely secondary, such as those of genes that are under the control of Agr, and not due to direct binding of Rsp to the respective promoters. We analyzed binding of Rsp to the promoter region of sarR, another regulatory gene that showed rsp-dependent differential regulation in the RNA-Seq analysis, as an example. In contrast to the agrP2 and icaADBC promoter regions, the sarR promoter did not directly bind Rsp (see Fig. S5 in the supplemental material).
Impact of Rsp on virulence factor production.
To substantiate that the virulence- and biofilm-associated Rsp-dependent gene-regulatory changes that we observed translate to different production of the associated virulence factors, we determined the levels of production of PSMs (PSMα and PSMβ peptides, δ-toxin) by HPLC/MS, production of alpha-toxin and protein A by Western blotting, and production of PIA by immune dot blot assays using PIA-specific antiserum (Fig. 5). PSMα and PSMβ peptides showed significantly increased production in the BD02-25 wild-type strain compared to the corresponding Δrsp deletion strains; and genetic complementation of the Δrsp strains showed a significant increase of these peptides, demonstrating a strong impact of Rsp on PSM production. Results obtained for PSMα3 and PSMβ1 are shown in Fig. 5 as examples for the PSMα and PSMβ peptides, which are encoded by the PSMα and PSMβ operons, respectively, and whose respective products are linked in expression (35). Interestingly, differences in values for the δ-toxin were only slight in a comparison of the wild-type and Δrsp strains. This is potentially due to the fact that δ-toxin is present in very high concentrations and nontranscriptional factors may limit the further increase in secretion of this generally most abundant protein in the S. aureus culture filtrate in the wild-type versus Δrsp strains (36). However, there was a pronounced difference in δ-toxin levels in the plasmid-harboring complementation versus control strains, where overall PSM levels were lower (owing to less optimal growth conditions, with antibiotic present). Furthermore, as expected, alpha-toxin levels were significantly increased in the BD02-25 and rsp-expressing complemented strains compared to the respective strains lacking rsp. As predicted by the transcriptional effects of rsp on the ica genes, PIA production was influenced in the opposite fashion. Moreover, the Agr-downregulated surface protein A also showed the expected patters of Agr-mediated control by Rsp. These results demonstrated that the gene-regulatory changes of rsp-controlled virulence genes translate to the level of production of the associated virulence determinants. Furthermore, the reduced production levels of alpha-toxin and PSM explain well the difference in hemolytic capacity observed for the rsp transposon mutant and isogenic deletion strains, as these are the most important hemolytic toxins of S. aureus (37, 38). As alpha-toxin and PSMs are regulated by Agr, these results are also in accordance with the fact that hemolysis of an agr rsp double mutant was not further increased compared to that mediated by an agr mutant (Fig. 1C).
FIG 5.
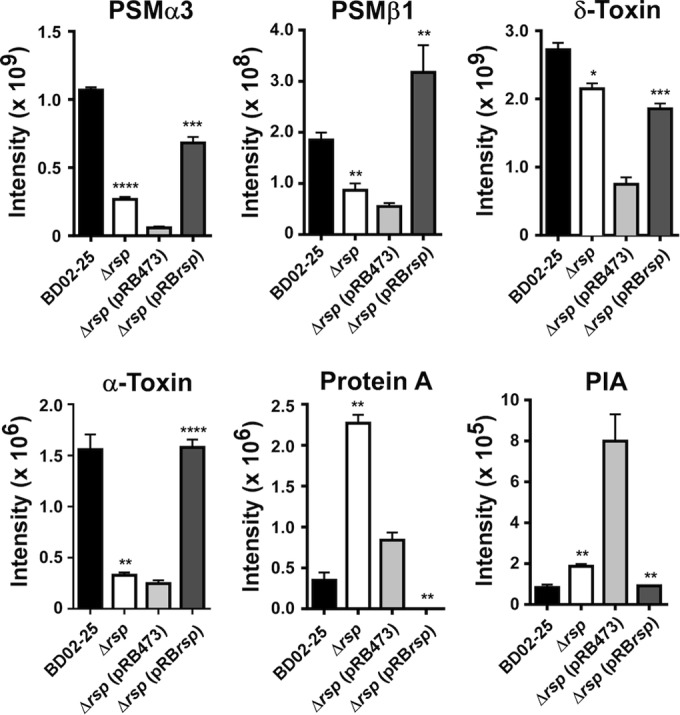
Rsp-dependent production of virulence determinants. Production of PSM peptides and δ-toxin was measured by HPLC/MS in culture filtrates (6 h of growth). Intensity values are from the integration of the two most abundant m/z peaks for every peptide. PSMα3 is shown as an example for PSMα peptides and PSMβ1 for PSMβ peptides. Protein A and alpha-toxin levels were measured by densitometry of Western blots of culture filtrate samples. Antibodies specific for alpha-toxin were used, and the nonspecific reactivity of the Fc part of the antibodies with protein A was used to detect and measure protein A. Protein A was measured in 6-h and alpha-toxin in 8-h culture filtrates. PIA production was determined by extracting PIA from the cell surface in cells from 6-h cultures, immunodot blot, and densitometry. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (wild-type strains versus isogenic mutants; complemented strain versus plasmid control, t tests). Error bars depict ±SEM.
Control of virulence in animal infection models.
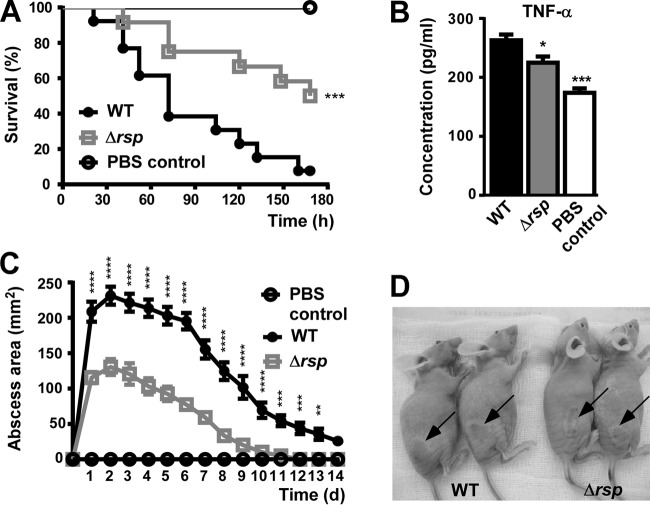
Finally, we evaluated the impact of rsp on virulence using murine models of bacteremia and skin infection, which are frequent manifestations of acute disease caused by S. aureus (1). We used the BD02-25 strain because of its pronounced virulence in those infection models (32). MW2, in contrast, is known to have limited virulence in murine infection models, which is associated with the low production of alpha-toxin in that strain (39). The BD02-25 wild-type strain was significantly more virulent in the bacteremia and skin infection models than was the Δrsp strain, as measured by skin abscess sizes and survival rates, respectively (Fig. 6A, C, and D). We also measured at the time of death the levels of tumor necrosis factor alpha (TNF-α) in blood as a marker of inflammation (Fig. 6B). TNF-α levels correlated well with the survival curves, inasmuch as its concentrations were significantly lower in the Δrsp strain group. These results demonstrated that Rsp significantly promotes acute forms of S. aureus disease, which is in accordance with our findings on the gene-regulatory impact of Rsp.
FIG 6.
Impact of Rsp on S. aureus infection. (A) Bacteremia model. Female BALB/c mice were injected with 108 CFU via retro-orbital injection. Survival curves were compared using log-rank (Mantel-Cox) tests. ***, P < 0.0001. (B) Concentration of TNF-α in mouse blood at the time of death (euthanasia). *, P < 0.05; ***, P < 0.001 (1-way ANOVA with Dunnett's posttest, comparison versus wild-type strain [WT]). (C) Abscess model. Outbred, immunocompetent hairless mice were injected subcutaneously with 107 live S. aureus isolates, and abscess or dermonecrosis areas were measured each day. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (2-way ANOVA with Tukey's posttest; only wild-type versus Δrsp mutant strain comparisons are depicted). (A to C) WT, wild-type strain BD02-25; Δrsp, isogenic rsp mutant. (D) Representative skin lesions on day 2. Arrows point to abscesses. Error bars depict ±SEM.
DISCUSSION
The broad spectrum of S. aureus infections can be divided in two main classes: (i) acute infections that are characterized by marked production of toxins and other proinflammatory and tissue-degrading molecules and (ii) chronic infections that often proceed with the development of biofilms as a long-term immune evasion strategy and without pronounced production of factors that aggressively damage host cells and tissues. These infection types require dedicated methods of therapeutic intervention. In particular, virulence-targeted approaches that are currently being evaluated are entirely different for those two types of infection, as they need to target toxin production versus biofilm formation.
The regulatory mechanisms that S. aureus uses to adapt gene expression to those categorically different modes of pathogenesis are poorly understood. In this study, we identified Rsp as a virtually omnipresent S. aureus regulator with a distinctly opposite impact on toxin versus biofilm gene expression and whose influence on toxin production, biofilm formation, and virulence in acute infection models followed that opposite mode of regulation (Fig. 7). In both animal models of acute S. aureus infection, namely, skin and blood infections, the Δrsp mutant strain caused significantly fewer signs of disease than did the isogenic wild-type strain, in accordance with the positive impact of Rsp on established mediators of acute S. aureus infection, such as alpha-toxin and PSMs (3–5), that we show in the present study.
FIG 7.
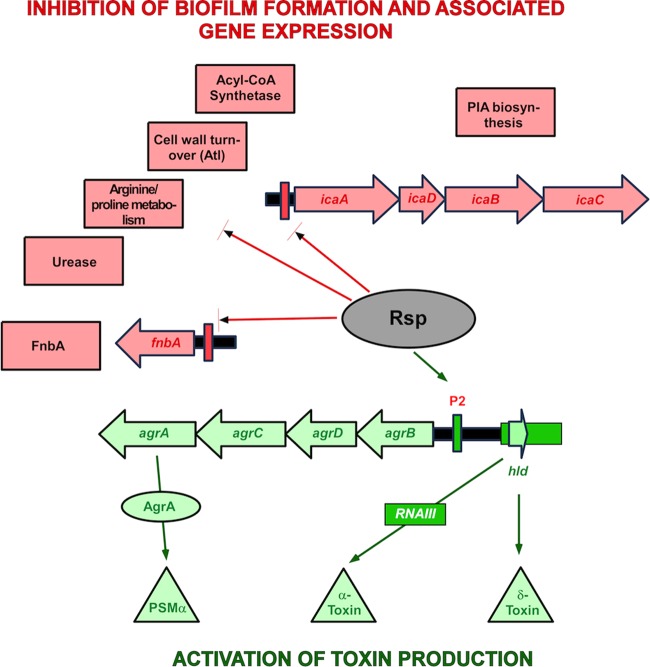
Model of Rsp's function in S. aureus virulence gene regulation. Bottom (green): Rsp positively controls the production of major S. aureus toxins by binding to the agrP2 promoter and activating Agr. PSMα peptides are directly controlled by binding of AgrA (45), while the production of other toxins is increased due to an increase of overall Agr activity and RNAIII production (46). Top (red): Rsp negatively regulates factors involved in biofilm formation. Production of PIA is decreased by binding of Rsp to the icaADBC promoter. According to our RNA-Seq results, a series of further biofilm-associated metabolic processes are also downregulated by Rsp. Finally, as shown by Lei et al. (41), Rsp negatively impacts production of FnbA by binding to the respective promoter. Acyl-CoA, acyl coenzyme A.
The direct binding yet opposite impact of Rsp on controlled target genes, such as shown here for Rsp's control of the agr versus ica promoters, is not surprising, as AraC-type transcriptional regulators such as Rsp are known to be able to elicit positive or negative control of expression by binding to DNA promoter sequences (40). What remains puzzling is that the RSAT-predicted binding motifs of the agrP2 and the ica promoters are different. Furthermore, we found no similar sequence in the fnbA promoter, which reportedly also binds Rsp (41), and RSAT predicted yet another binding motif within that promoter (TAAGATTGT). Moreover, the RSAT-predicted binding sequences in the agrP2 and ica promoter regions were not found anywhere else in the S. aureus genome except within open reading frames. Thus, whether other identified Rsp target genes are regulated by direct binding to possibly poorly conserved binding sites or are all secondary targets remains to be investigated. Clearly, the details of the Rsp-DNA interaction will require more in-depth investigation in the future. Finally, while we identified Agr, Agr-controlled toxins, and the PIA biosynthesis system as key targets of Rsp regulation, genes downregulated by Rsp also included atl and fnbA, in addition to metabolic processes associated with biofilm growth. Thus, Rsp impacts genes associated with biofilm formation of both PIA- and Atl/FnbA-dependent type of S. aureus biofilms (21), underscoring its general importance.
Of note, as reflected by the differential biofilm formation of agr mutants and agr rsp mutants, there are important differences that set Rsp-mediated control apart from that exerted by Agr, the main target of positive control by Rsp. First, the impact of Agr on biofilm formation is somewhat ambiguous. This is because it impacts biofilm development to a large extent through the control of PSM peptides, which are important for the development of a vital biofilm but whose absence leads to excessive growth of unstructured biofilms (16). Notably, Agr does not control expression of the main biofilm exopolysaccharide PIA (15). Furthermore, Agr does not negatively control biofilm-associated metabolic genes in a coherent fashion like Rsp (15). Moreover, Agr becomes active only at a certain level of cell density, while Rsp expression is maximal at earlier times during growth and thus at lower cell densities. In an in vivo situation, this difference would translate to an earlier onset of Rsp-mediated than an Agr-mediated control, with the impact of Rsp on Agr expression likely accelerating Agr activation.
Patterns of gene expression of staphylococci that are characteristic for biofilm formation are believed to be similar to those during asymptomatic colonization on mammalian epithelial surfaces, reflecting a generally nonaggressive mode of growth (42). The switch in gene expression that Rsp controls may thus also be interpreted as a response to the environmental changes that the bacteria encounter when breaching through the epithelial barrier, coming in contact with a wider array of host defense mechanisms and a generally more hostile environment. These considerations are of special importance considering that AraC-type regulators such as Rsp often sense environmental conditions by binding of small molecules that reflect those conditions (40). The identification of such a signaling molecule is difficult. However, future endeavors to identify a potential signal that binds Rsp may be facilitated by our findings that indicate that the gene regulatory changes controlled by Rsp, such as an upregulation of toxins and other aggressive virulence factors, are typical for the situation that the bacteria encounter during acute infection, that is, a low availability of nutrients and an abundance of host defense mechanisms.
Finally, we are not the first to report gene regulatory changes that are dependent on Rsp. Rsp was named in 2011 by Lei et al., who described it as an inhibitor of biofilm formation and attributed that phenotype to Rsp's impact on the expression of FnbA (41). We were able to confirm the impact of Rsp on fnbA expression in our qRT-PCR experiments, although it appeared limited in relative terms. The failure by Lei et al. to recognize the broad impact of Rsp on gene expression that became evident in our study may have been due to the fact that those authors did not analyze gene expression at the maximum of rsp expression. Furthermore, the microarray-based analysis that they performed may not have been as sensitive as our RNA-Seq-based study.
In conclusion, we here identified a master regulator of S. aureus' adaptation of gene expression to acute types of diseases. Our findings give important insight into how S. aureus controls important sets of virulence-associated genes and indicate that Rsp may be a target for drug development efforts directed against acute forms of S. aureus infection.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grants 81322025, 81171623, and 81371875), Shanghai Shuguang Talent Project (12SG03), the Shanghai Committee of Science and Technology, China (14140901000), and the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (81421001) (to M.L.) and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, the National Institutes of Health (ZIA AI000904; to M.O.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01088-15.
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Lowy FD. 2003. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest 111:1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobayashi SD, Malachowa N, Whitney AR, Braughton KR, Gardner DJ, Long D, Bubeck Wardenburg J, Schneewind O, Otto M, DeLeo FR. 2011. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J Infect Dis 204:937–941. doi: 10.1093/infdis/jir441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med 13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 5.Otto M. 2014. Staphylococcus aureus toxins. Curr Opin Microbiol 17:32–37. doi: 10.1016/j.mib.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otto M. 2008. Staphylococcal biofilms. Curr Top Microbiol Immunol 322:207–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Resch A, Rosenstein R, Nerz C, Gotz F. 2005. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol 71:2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Y, Sturdevant DE, Otto M. 2005. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J Infect Dis 191:289–298. doi: 10.1086/426945. [DOI] [PubMed] [Google Scholar]
- 9.Scherr TD, Heim CE, Morrison JM, Kielian T. 2014. Hiding in plain sight: interplay between staphylococcal biofilms and host immunity. Front Immunol 5:37. doi: 10.3389/fimmu.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol 178:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. 1999. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun 67:2627–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rupp ME, Ulphani JS, Fey PD, Mack D. 1999. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect Immun 67:2656–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun 67:5427–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, Otto M. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol 6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 15.Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M. 2011. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun 79:1927–1935. doi: 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Periasamy S, Joo HS, Duong AC, Bach TH, Tan VY, Chatterjee SS, Cheung GY, Otto M. 2012. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci U S A 109:1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. 1997. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev 61:393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Rigby K, Lai Y, Nair V, Peschel A, Schittek B, Otto M. 2009. Staphylococcus aureus mutant screen reveals interaction of the human antimicrobial peptide dermcidin with membrane phospholipids. Antimicrob Agents Chemother 53:4200–4210. doi: 10.1128/AAC.00428-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis 188:706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- 21.Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, Loftus BJ, Pier GB, Fey PD, Massey RC, O'Gara JP. 2012. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog 8:e1002626. doi: 10.1371/journal.ppat.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin N, Tan X, Jiao Y, Liu L, Zhao W, Yang S, Jia A. 2014. RNA-Seq-based transcriptome analysis of methicillin-resistant Staphylococcus aureus biofilm inhibition by ursolic acid and resveratrol. Sci Rep 4:5467. doi: 10.1038/srep05467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. 2007. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. 2007. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. 2013. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. 2007. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang S, Gou Z, Kuznetsov IB. 2007. DP-Bind: a web server for sequence-based prediction of DNA-binding residues in DNA-binding proteins. Bioinformatics 23:634–636. doi: 10.1093/bioinformatics/btl672. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Naismith JH. 2008. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joo HS, Otto M. 2014. The isolation and analysis of phenol-soluble modulins of Staphylococcus epidermidis. Methods Mol Biol 1106:93–100. doi: 10.1007/978-1-62703-736-5_7. [DOI] [PubMed] [Google Scholar]
- 31.Cramton SE, Gerke C, Gotz F. 2001. In vitro methods to study staphylococcal biofilm formation. Methods Enzymol 336:239–255. doi: 10.1016/S0076-6879(01)36593-X. [DOI] [PubMed] [Google Scholar]
- 32.Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. 2009. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A 106:5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tobes R, Ramos JL. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res 30:318–321. doi: 10.1093/nar/30.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.CDC. 1999. From the Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. JAMA 282:1123–1125. [PubMed] [Google Scholar]
- 35.Cheung GY, Joo HS, Chatterjee SS, Otto M. 2014. Phenol-soluble modulins—critical determinants of staphylococcal virulence. FEMS Microbiol Rev 38:698–719. doi: 10.1111/1574-6976.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chatterjee SS, Joo HS, Duong AC, Dieringer TD, Tan VY, Song Y, Fischer ER, Cheung GY, Li M, Otto M. 2013. Essential Staphylococcus aureus toxin export system. Nat Med 19:364–367. doi: 10.1038/nm.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung GY, Duong AC, Otto M. 2012. Direct and synergistic hemolysis caused by Staphylococcus phenol-soluble modulins: implications for diagnosis and pathogenesis. Microbes Infect 14:380–386. doi: 10.1016/j.micinf.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berube BJ, Bubeck Wardenburg J. 2013. Staphylococcus aureus alpha-toxin: nearly a century of intrigue. Toxins (Basel) 5:1140–1166. doi: 10.3390/toxins5061140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salgado-Pabon W, Schlievert PM. 2014. Models matter: the search for an effective Staphylococcus aureus vaccine. Nat Rev Microbiol 12:585–591. doi: 10.1038/nrmicro3308. [DOI] [PubMed] [Google Scholar]
- 40.Yang J, Tauschek M, Robins-Browne RM. 2011. Control of bacterial virulence by AraC-like regulators that respond to chemical signals. Trends Microbiol 19:128–135. doi: 10.1016/j.tim.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Lei MG, Cue D, Roux CM, Dunman PM, Lee CY. 2011. Rsp inhibits attachment and biofilm formation by repressing fnbA in Staphylococcus aureus MW2. J Bacteriol 193:5231–5241. doi: 10.1128/JB.05454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otto M. 2010. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol 5:183–195. doi: 10.1586/edm.10.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cue D, Lei MG, Lee CY. 2012. Genetic regulation of the intercellular adhesion locus in staphylococci. Front Cell Infect Microbiol 2:38. doi: 10.3389/fcimb.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK. 2004. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J Bacteriol 186:7549–7555. doi: 10.1128/JB.186.22.7549-7555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Queck SY, Jameson-Lee M, Villaruz AE, Bach TH, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell 32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12:3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.