Abstract
Ehrlichia chaffeensis invades and survives in phagocytes by modulating host cell processes and evading innate defenses, but the mechanisms are not fully defined. Recently we have determined that E. chaffeensis tandem repeat proteins (TRPs) are type 1 secreted effectors involved in functionally diverse interactions with host targets, including components of the evolutionarily conserved Wnt signaling pathways. In this study, we demonstrated that induction of host canonical and noncanonical Wnt pathways by E. chaffeensis TRP effectors stimulates phagocytosis and promotes intracellular survival. After E. chaffeensis infection, canonical and noncanonical Wnt signalings were significantly stimulated during early stages of infection (1 to 3 h) which coincided with dephosphorylation and nuclear translocation of β-catenin, a major canonical Wnt signal transducer, and NFATC1, a noncanonical Wnt transcription factor. In total, the expression of ∼44% of Wnt signaling target genes was altered during infection. Knockdown of TRP120-interacting Wnt pathway components/regulators and other critical components, such as Wnt5a ligand, Frizzled 5 receptor, β-catenin, nuclear factor of activated T cells (NFAT), and major signaling molecules, resulted in significant reductions in the ehrlichial load. Moreover, small-molecule inhibitors specific for components of canonical and noncanonical (Ca2+ and planar cell polarity [PCP]) Wnt pathways, including IWP-2, which blocks Wnt secretion, significantly decreased ehrlichial infection. TRPs directly activated Wnt signaling, as TRP-coated microspheres triggered phagocytosis which was blocked by Wnt pathway inhibitors, demonstrating a key role of TRP activation of Wnt pathways to induce ehrlichial phagocytosis. These novel findings reveal that E. chaffeensis exploits canonical and noncanonical Wnt pathways through TRP effectors to facilitate host cell entry and promote intracellular survival.
INTRODUCTION
Ehrlichia chaffeensis is an obligately intracellular bacterium responsible for the emerging life-threatening human zoonosis human monocytotropic ehrlichiosis (HME) (1). E. chaffeensis selectively infects mononuclear phagocytes and resides in early-endosome-like membrane-bound vacuoles (1). The mechanisms by which E. chaffeensis enters host cells, establishes persistent infection, and avoids host defenses are not completely understood but occur through functionally relevant host-pathogen interactions involving secreted ehrlichial tandem repeat protein (TRP) effectors that are posttranslationally modified by ubiquitin (Ub) and the small ubiquitin-like modifier (SUMO) (2–5). E. chaffeensis TRPs interact with a diverse group of human proteins associated with major cellular processes, including transcription, translation, protein trafficking, cell signaling, cytoskeleton organization, and apoptosis, indicating that they play a role in manipulating these important cellular processes to facilitate infection (6–8).
E. chaffeensis TRPs were first recognized as antigens that elicit strong protective antibody responses during infection that are directed at continuous species-specific epitopes in tandem repeat regions (9–12). Subsequently, our understanding of the functional role of TRPs as effectors in pathobiology has been advanced through studies that have defined specific TRP-host protein and DNA interactions (4, 13). Notably, E. chaffeensis TRP120 and TRP32 interact with numerous host proteins and genes associated with the canonical and noncanonical Wnt signaling pathways. One of TRP32-interacting targets, deleted-in-azoospermia-associated protein 2 (DAZAP2), is a highly conserved protein that modulates gene transcription driven by Wnt/β-catenin signaling effector T-cell factors (TCFs), and knockdown of host DAZAP2 by small interfering RNA (siRNA) reduced the E. chaffeensis load in infected cells (7, 14). Several other TRP120-interacting host proteins, such as AT-rich interactive domain 1B (ARID1B), lysine (K)-specific demethylase 6B (KDM6B), interferon regulatory factor 2 binding protein 2 (IRF2BP2), protein phosphatase 3 regulatory subunit B alpha (PPP3R1), and vacuolar protein sorting 29 homolog (Saccharomyces cerevisiae) (VPS29), are also involved in Wnt pathway signaling (6). Moreover, TRP120 binds host genes associated with Wnt signaling pathways, such as Wnt, Dishevelled (Dvl), and nuclear factor of activated T cells (NFAT) (15). Thus, E. chaffeensis appears to exploit Wnt pathways through TRP-Wnt signaling proteins and by modulating the expression of Wnt pathway genes via TRP transcriptional modulation.
Wnt signaling was initially studied for its role in carcinogenesis but has more recently been recognized for its central role in embryonic development, differentiation, cell proliferation, cell motility, cell polarity, and adult tissue homeostasis (16, 17). The importance of Wnt signaling has been demonstrated by mutations that lead to a variety of diseases, including breast and prostate cancer, glioblastoma, diabetes, and others (18, 19). Wnt signaling pathways are highly evolutionarily conserved (20, 21). Thus far, three Wnt pathways have been characterized: the canonical Wnt/β-catenin pathway and two noncanonical β-catenin-independent pathways (Wnt/Ca2+ and Wnt/planar cell polarity [PCP]). Wnt signaling is activated by the binding of a Wnt ligand to a Frizzled (Fzd) receptor (22, 23). In the canonical Wnt/β-catenin pathway, activated Fzd heterodimerizes with lipoprotein receptor-related protein (LRP) to recruit and activate Dishevelled (Dvl), which subsequently recruits the protein complex containing axis inhibitor (Axin), adenomatous polyposis coli (APC), casein kinase 1 (CK1), and glycogen synthase kinase 3 (GSK3), leading to the inhibition of phosphorylation of β-catenin by these kinases. Unphosphorylated β-catenin accumulates and subsequently translocates to the nucleus, where it associates with TCF/lymphoid-enhancing factor (LEF) family transcription factors to induce the expression of Wnt target genes (23–25). The noncanonical Wnt/Ca2+ pathway signals through heterotrimeric G proteins, which further activates phospholipase C (PLC), leading to intracellular Ca2+ release and activation of calcium/calmodulin-dependent protein kinase II (CaMKII), calcineurin, and protein kinase C (PKC) (17). These processes can stimulate NFAT and other transcription factors such as cyclic AMP (cAMP) response element binding protein (CREB) (17, 26). The noncanonical Wnt/PCP pathway involves the Rho and Rac GTPases, Rho kinase (ROCK), and c-Jun N-terminal kinase (JNK) and regulates cytoskeletal reorganization, cell motility, and tissue polarity (17, 27). More recent studies have suggested that the activity of Wnt ligands and their binding to Fzd receptors depend on the cellular context; thus, Wnt and Fzd proteins cannot be rigorously subdivided according to the pathways they induce (28, 29). However, Wnt3a and Wnt5a are more commonly associated with canonical and noncanonical Wnt signaling, respectively (30).
Recently, the role of the Wnt pathway in phagocytosis of microorganisms has been demonstrated (31, 32). The Wnt ligand receptor (Wnt5a-Fzd5) signaling in macrophages was reported to promote phagocytosis of bacteria and enhanced survival through lipid rafts with Rac1–phosphatidylinositol 3-kinase (PI3K)–IκB kinase (IKK) activation (31). Others have demonstrated that Salmonella activates host β-catenin signaling; moreover, Salmonella type 3 secreted effectors AvrA and SopB can individually activate Wnt/β-catenin signaling in epithelial cells, leading to increases of stem cells and proliferative cells and transdifferentiation of primed epithelial cells into M cells, respectively, to promote intestinal invasion (33–37). Wnt5a is known to activate canonical and noncanonical Wnt pathways and is linked to cytoskeletal modulation and proinflammatory cytokine activation, e.g., in human macrophages stimulated by Mycobacterium (38–42).
E. chaffeensis binding and entry are known to involve one or more glycosylphosphatidylinositol (GPI)-anchored proteins associated with caveolae at the cell surface, inducing receptor-mediated phagocytosis that triggers Wnt signaling-like events, including transglutamination, tyrosine phosphorylation, phospholipase Cγ2 (PLC-γ2) activation, inositol-(1,4,5)-trisphosphate (IP3) production, and intracellular calcium release (43, 44). In addition, multiple studies have shown the importance of E. chaffeensis TRP120 in ehrlichial binding and internalization (45, 46); however, the specific cellular pathways exploited to mediate invasion and intracellular survival have not been defined. In this study, we demonstrated that host Wnt signaling pathways are exploited for ehrlichial internalization and infection and that ehrlichial TRPs are directly involved.
MATERIALS AND METHODS
Cell culture and cultivation of E. chaffeensis.
Human cervical epithelial adenocarcinoma cells (HeLa, from ATCC) were propagated in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT). Human monocytic leukemia cells (THP-1) were propagated in RPMI medium 1640 with l-glutamine and 25 mM HEPES buffer (Invitrogen), supplemented with 1 mM sodium pyruvate, 2.5 g/liter d-(+)-glucose (Sigma, St. Louis, MO), and 10% fetal bovine serum. E. chaffeensis (Arkansas strain) was cultivated in THP-1 cells as previously described (47).
Inhibitors, siRNAs, and antibodies.
Wnt signaling pathway inhibitors included pyrvinium pamoate, IWP-2, BAY11-7082, and NSC23766 (Sigma) and KN93, SB202190, TBCA, FH535, and LY294002 (Calbiochem/EMD, Billerica, MA). The lipid raft-disrupting agent nystatin was from Sigma. Human DKK3, Dvl2, Fzd9, Jun, NFATC1 (NFAT2), NFATC3 (NFAT4), PP2B-Aα (calcineurin PPP3CA), PP2B-Aβ (calcineurin PPP3CB), TCF4, Wnt6, Wnt10a, and control-A siRNAs were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Validated siRNAs of human β-catenin, Wnt3a, Wnt5a, and LRP6 and endoribonuclease-prepared siRNAs (esiRNAs) of human ARID1B, KDM6B, IRF2BP2, PPP3R1, and VPS29 were from Sigma. Human Fzd5, Akt, CKIε, CKII, CaMKII, IKK, PI3K, and RhoA siRNAs were obtained from GE Dharmacon (Lafayette, CO). Alexa Fluor 488-labeled negative siRNA was from Qiagen (Germantown, MD). Rabbit and mouse anti-TRP32 antibodies have been described previously (12). Other antibodies used in this study were mouse anti-human α-tubulin, NFATC1 (Santa Cruz), and β-catenin (Pierce, Rockford, IL) and rabbit anti-human Fzd9 (Pierce), phospho-β-catenin, Dvl2 (Cell Signaling, Beverly, MA), and Jun (Santa Cruz).
PCR array.
The RT2 Profiler PCR arrays (version 4.0; SABiosciences, Valencia, CA) were used, including human Wnt signaling pathway plus PCR array and human Wnt signaling target PCR array (see Fig. S1 in the supplemental material and the SABiosciences website for gene list and functional gene grouping). The human Wnt signaling pathway plus PCR array profiles the expression of 84 genes related to Wnt-mediated signal transduction, including Wnt signaling ligands, receptors, and regulators, as well as downstream signaling molecules and target proteins for all three Wnt pathways. The array uses experimentally derived signature biomarker genes along with classification algorithms to generate the pathway activity score and determines whether Wnt pathway activity is activated or repressed in experimental samples. The human Wnt signaling target PCR array profiles the expression of 84 key genes responsive to Wnt signal transduction, including Wnt signaling pathway transcription factors and highly relevant target genes, to analyze Wnt pathway status. PCR array analyses were performed according to the PCR array handbook from the manufacturer. In brief, uninfected and E. chaffeensis-infected THP-1 cells at different intervals postinfection (p.i.) were collected, and total RNA was purified using the RNeasy minikit (Qiagen). During RNA purification, on-column DNA digestion was performed using the RNase-free DNase set (Qiagen). The concentration and purity were determined by measuring the absorbance using a NanoDrop 100 spectrophotometer (Thermo Scientific, West Palm Beach, FL), and rRNA band integrity was verified by running an aliquot of each RNA sample on an RNA FlashGel (Lonza, Rockland, ME). Genomic DNA was eliminated, and cDNA was synthesized from 0.5 μg of total RNA using the RT2 first-strand kit (Qiagen). Real-time PCR was performed using the RT2 Profiler PCR array in combination with RT2 SYBR green master mix (Qiagen) on a Mastercycler EP Realplex2 S (Eppendorf, Germany). Cycling conditions were as follows: 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. The real-time cycler software RealPlex 1.5 (Eppendorf) was used for PCR and data collection. The baseline was set automatically, the threshold was defined manually, and then the threshold cycle (CT) for each well was calculated by RealPlex. The threshold was set in the proper location and at the same level for all PCR arrays in the same analysis so that the values from the positive PCR control (PPC) assays on all arrays were between 18 CT and 22 CT. The CT values for all wells were exported for analysis using Web-based PCR array data analysis software (version 3.5; SABiosciences). PCR array quality checks were performed by the software before data analysis, including PCR array reproducibility, reverse transcription efficiency control (RTC), human genomic DNA contamination control (HGDC), and PPC.
Western immunoblotting.
The THP-1 cell lysates were prepared using CytoBuster protein extraction reagent (Novagen/EMD, Gibbstown, NJ), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to a nitrocellulose membrane. Western immunoblotting was performed with horseradish peroxidase-labeled goat anti-rabbit or -mouse IgG (heavy and light chains) conjugate (Kirkegaard & Perry Laboratories, Gaithersburg, MD) and SuperSignal West Dura chemiluminescent substrate (Thermo Scientific).
Immunofluorescence microscopy.
Uninfected or E. chaffeensis-infected THP-1 cells at different intervals p.i. were collected, and the indirect immunofluorescent-antibody assay was performed as previously described (6), except that anti-β-catenin or NFATC1 antibody (1:100) and anti-TRP32 antibody (1:10,000) were used.
Reporter assay.
The activity of the noncanonical Wnt/Ca2+ signal pathway (NFAT-mediated transcription) was monitored using the Cignal NFAT reporter (Qiagen). Briefly, HeLa cells (2 × 104/well) were seeded in a 96-well black plate, and the following day cells were transfected with NFAT reporter plasmid using Lipofectamine 3000 (Invitrogen) and incubated overnight again. The cell-free E. chaffeensis (multiplicity of infection [MOI] = ∼50) or uninfected THP-1 control was added to transfected HeLa cells. Firefly and Renilla luciferase activities of each well were measured at different time points using the Dual-Glo luciferase assay system (Promega, Madison, WI) and a Veritas microplate luminometer (Turner Biosystems, Sunnyvale, CA). Firefly luciferase activity was normalized with Renilla luciferase.
RNA interference.
THP-1 cells (1 × 105/well on a 96-well plate) were transfected with 5 pmol human siRNA using Lipofectamine 3000 (Invitrogen). A control-A siRNA consisting of a scrambled sequence was used as a negative control, and an Alexa Fluor 488-labeled negative siRNA was used as a control to monitor transfection efficiency. At 1 day posttransfection, the cells were synchronously infected by cell-free E. chaffeensis at an MOI of ∼50, and they were collected at 1 day and 2 days p.i. for quantitative PCR (qPCR) and Western blotting to determine knockdown levels.
Quantification of E. chaffeensis by qPCR.
THP-1 cells were pelleted, washed with phosphate-buffered saline (PBS), lysed in SideStep lysis and stabilization buffer (Agilent, Santa Clara, CA) for 30 min at room temperature, and analyzed for bacterial load using real-time qPCR. Amplification of the integral ehrlichial gene dsb was performed using Brilliant II SYBR green master mix (Agilent), 200 nM forward primer (5′-GCTGCTCCACCAATAAATGTATCCCT-3′), and 200 nM reverse primer (5′-GTTTCATTAGCCAAGAATTCCGACACT-3′). The qPCR thermal cycling protocol (denaturation at 95°C for 10 min and then 40 cycles of 95°C for 30 s, 58°C for 1 min, and 72°C for 1 min) was performed on the Mastercycler EP Realplex2 S (Eppendorf). A standard plasmid, pBAD-dsb, was constructed by cloning the ehrlichial dsb gene using the TOPO TA cloning kit (Invitrogen). The plasmid copy number for the standards was calculated using the following formula: plasmid copy/μl = [(plasmid concentration g/μl)/(plasmid length in base pairs × 660)] × 6.022 × 1023. The absolute E. chaffeensis dsb copy number in the cells was determined against the standard curve, or the fold change of dsb copy number relative to the control was normalized to qPCR-detected levels of the host genomic glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene.
Small-molecule inhibitor treatment.
THP-1 cells were plated in FBS-free medium and treated with inhibitor or dimethyl sulfoxide (DMSO) control at 3 h before infection with cell-free E. chaffeensis at an MOI of ∼50, or cells were infected and then treated with inhibitor or DMSO at 3 h p.i. Percentages of infected cells were monitored daily over 3 days by Diff-Quik staining and counting of 100 cells. Bright-field images of these slides were collected on an Olympus BX61 epifluorescence microscope using a color camera. Initially, inhibitors were tested at the concentrations 5- to 10-fold higher than the 50% inhibitory concentration (IC50) or Ki as provided by the manufacturer. If the bacteria were inhibited at this concentration, MICs were then determined by using 2-fold serial dilutions. The MIC was defined as the minimum inhibitor concentration required for inhibition of the growth of the bacteria compared to that of the control (without the inhibitor) at day 3. An infected culture without the inhibitor served as a positive growth control, and an uninfected cell culture served as a negative control. All experiments were repeated three times to confirm results.
Phagocytosis of microspheres.
Expression and purification of E. chaffeensis tandem repeat proteins (TRPs) has been described previously (9, 10, 12). FluoSpheres sulfate microspheres (1.0 μm, yellow-green fluorescent; Invitrogen) (10 μl, ∼3.6 × 108 beads) were washed with 40 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 6.1) and then incubated with 15 μg of TRP protein in 500 μl MES buffer at room temperature for 1 h with mixing at 20 rpm. The coated beads were collected by centrifugation, washed twice in MES buffer, resuspended in RPMI medium, and then gently sonicated to disperse the beads. The efficiency of bead coating was confirmed by dot blot assay. TRP-coated or control thioredoxin protein-coated beads were added to THP-1 cells at a multiplicity of approximately 50 beads per cell and incubated for 2 h at 37°C with 5% CO2. Unbound beads were removed by washing and low-speed centrifugation three times, and then cells were observed using an Olympus IX71 inverted fluorescence microscope. Fluorescence intensity was measured using a FluoroSkan fluorometer (Thermo Scientific). For estimating inhibition of phagocytosis, designated inhibitors were added 2 h before addition of coated beads.
Measurement of Ca2+.
Intracellular Ca2+ release was measured in THP-1 cells using an acetoxymethyl (AM) ester derivative of fluo-3 (fluo-3/AM) (PromoKine, Heidelberg, Germany) as previously described (43). Fluo-3-preloaded cells were treated with cell-free E. chaffeensis (MOI = ∼50), TRP-coated beads (50 beads/cell), or a control (cell-free uninfected THP-1 or thioredoxin-coated beads) and mixed briefly. Emission at 525 nm after excitation at 488 nm was recorded every 15 s for 40 min using a FluoroSkan fluorometer (Thermo Scientific). Nonfluorescent polystyrene latex beads (LB-11; Sigma) were coated with TRPs as described above.
Statistics.
The statistical differences between experimental groups were assessed with the two-tailed Student t test, and significance was indicated by a P value of <0.05.
RESULTS
Gene activity analysis of the Wnt signaling pathway in E. chaffeensis-infected host cells by PCR array.
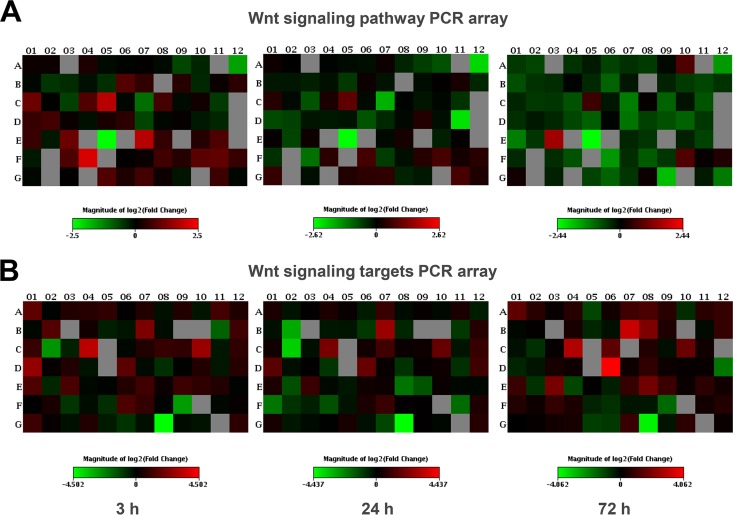
To define the impact of E. chaffeensis on host canonical and noncanonical Wnt signaling, the gene activity of the Wnt signaling pathway in E. chaffeensis-infected THP-1 cells at different time points (1 h, 3 h, 8 h, 24 h, and 72 h) p.i. was assessed by qPCR array analysis (Table 1). Compared to that in uninfected cells (under basal conditions), the Wnt signaling pathway activity in THP-1 cells was stimulated significantly by E. chaffeensis early (at 3 h p.i.), whereas the activity was repressed significantly at late infection (72 h p.i.). However, there was no significant change in pathway activity observed in E. chaffeensis-infected THP-1 cells at 1 h, 8 h, and 24 h p.i. relative to the uninfected/basal level. Heat maps of gene expression of Wnt signaling pathways at 3 h, 24 h, and 72 h p.i. also show overall more gene upregulation (red) at 3 h and downregulation (green) at 72 h than at 24 h (Fig. 1A).
TABLE 1.
Activity scores for the Wnt signaling pathway in E. chaffeensis-infected host cells as determined by PCR array analysis
| Time point (h p.i.) | Pathway activity scorea | P value |
|---|---|---|
| 1 | −0.06 | 0.42 |
| 3 | 0.49 | 0.01 |
| 8 | −0.06 | 0.42 |
| 24 | −0.06 | 0.42 |
| 72 | −0.37 | 0.04 |
A positive activity score of >0.3 or <−0.3 with a P value of <0.05 indicates significant stimulation or repression of pathway activity.
FIG 1.
Heat maps of gene expression of the Wnt signaling pathway in THP-1 cells at 3, 24, and 72 h postinfection with E. chaffeensis determined with PCR arrays. Each individual well represents an individual gene (see Fig. S1 in the supplemental material for gene tables and line graphs of important gene expression changes). The scale bar shows color-coded differential expression levels compared to those in uninfected cells. Red and green indicate upregulation and downregulation, respectively. (A) Wnt signaling pathway PCR array. (B) Wnt signaling target PCR array.
Among 84 host genes associated with Wnt signaling pathways assessed by PCR array analysis (see Fig. S1A [top panel] in the supplemental material for a gene table), 18 (21% of the total) genes exhibited significant differences in expression for at least one time point after E. chaffeensis infection, either upregulation (fold regulation/fold change of >2) or downregulation (fold regulation of <−2 [fold change of <0.5]), compared to uninfected cells, including Wnt ligands, receptors, inhibitors, a transcription factor, and targets of Wnt signaling pathways (Table 2). Figure S1A in the supplemental material (bottom panel) shows the expression levels of seven Wnt component/target genes at different time points p.i., including Wnt ligands Wnt6 and Wnt10a, Frizzled receptors Fzd5 and Fzd9, transcription factor 7 (TCF7), and the Wnt targets FOS-like antigen 1 (FOSL1) and MYC. Expression profiles of these important genes of Wnt pathways were captured in the global pathway analysis results, which identified significant global upregulation of Wnt pathway at 3 h p.i. and downregulation at 72 h p.i.
TABLE 2.
Functionally categorized Wnt signaling genes with significant changes of expression in E. chaffeensis-infected host cells determined by Wnt pathway PCR array analysis
| Role in Wnt pathway | Gene | Fold regulationa at: |
||||
|---|---|---|---|---|---|---|
| 1 h | 3 h | 8 h | 24 h | 72 h | ||
| Inhibitor | DKK3 | −4.6 | −3.0 | −8.5 | −4.6 | −2.8 |
| PRICKLE1 | −2.0 | −1.6 | −1.3 | −1.3 | −2.2 | |
| SFRP4 | −1.3 | −1.3 | −1.7 | −5.1 | −1.4 | |
| Ligand | WNT10A | 3.0 | 3.5 | 1.3 | −1.1 | −1.3 |
| WNT5B | −1.1 | 1.7 | −1.8 | −2.2 | −1.3 | |
| WNT6 | 3.5 | 4.4 | 1.1 | 1.4 | −1.7 | |
| WNT7B | 1.1 | 1.0 | −1.2 | 1.6 | −2.7 | |
| Receptor | FZD5 | 1.7 | 2.0 | 2.0 | 1.3 | −1.3 |
| FZD7 | −1.0 | −1.6 | −2.1 | −1.8 | −1.4 | |
| FZD9 | 3.0 | 3.5 | 2.8 | 2.0 | 1.5 | |
| VANGL2 | 1.8 | 2.1 | −1.5 | 1.1 | 2.9 | |
| Transcription factor | TCF7 | −1.1 | 1.5 | 1.0 | 1.0 | −2.3 |
| Target | FOSL1 | 2.1 | 1.7 | 1.6 | 1.1 | −1.9 |
| JUN | −1.8 | −2.1 | −3.3 | −3.6 | −2.3 | |
| WISP1 | −6.4 | −5.7 | −6.2 | −6.2 | −5.4 | |
| CHSY1 | 2.6 | −1.0 | 1.1 | 1.5 | 1.1 | |
| MYC | −1.5 | −1.1 | −1.1 | −1.2 | −3.5 | |
| SKP2 | −1.6 | −1.0 | 1.3 | 1.2 | −2.2 | |
Fold regulation of >2 indicates a significant upregulation, and fold regulation of <−2 indicates a significant downregulation.
Expression analysis of Wnt signaling target genes in E. chaffeensis-infected host cells by PCR array.
We further examined the expression of a large number of genes that are targets of canonical/noncanonical Wnt signaling in order to demonstrate the Wnt pathway regulation and identify specific genes that are most affected with Ehrlichia-induced Wnt signaling. Figure 1B shows heat maps of gene expression of Wnt signaling targets at 3 h, 24 h, and 72 h p.i., and Table 3 shows functionally categorized target genes with significant changes of expression for at least one time point (3 h, 8 h, 24 h, or 72 h p.i.) in infected cells compared to that in uninfected cells. Among 84 host genes responsive to Wnt signaling (see Fig. S1B [top panel] in the supplemental material for a gene table), 37 genes (44%) showed significant differences in expression level for at least one time point after infection, either upregulation (fold regulation of >2) or downregulation (fold regulation of <−2]). Among these 37 genes with significant differences of expression, 19 genes (51% of 37) were upregulated, 13 genes (35%) were downregulated, and 5 genes (14%) were upregulated and downregulated at different time points. The functional categories represented by these 37 Wnt signaling target genes include development and differentiation, calcium binding and signaling, adhesion, migration, cell cycle, proteolysis, signal transduction, and transcription factors. Notably, in the categories of development and differentiation, calcium binding and signaling, and migration, about half (51%, 46%, and 52%, respectively) of the genes examined were modulated by E. chaffeensis. Expression levels of six host target genes, those for cyclin D1 (CCND1), fibroblast growth factor 9 (FGF9), fibronectin 1 (FN1), MET proto-oncogene (MET), matrix metallopeptidase 2 (MMP2), and secreted frizzled-related protein 2 (SFRP2), are shown at different time points p.i. as determined with the Wnt signaling target PCR array (see Fig. S1B [bottom panel] in the supplemental material). Consistent with the observed activities of the Wnt pathway, these Wnt signaling target genes had a similar expression pattern, with relatively high expression at 3 h and 8 h p.i. and relatively low expression at 72 h p.i. Modulation of many Wnt target genes in the host cell during E. chaffeensis infection supports the regulation of Wnt signaling by E. chaffeensis.
TABLE 3.
Functionally categorized Wnt signaling target genes with significant changes of expression in E. chaffeensis-infected host cells determined by Wnt target PCR array analysis
| Role (no. of genes tested) | Genea | Fold regulationb at: |
|||
|---|---|---|---|---|---|
| 3 h | 8 h | 24 h | 72 h | ||
| Development and differentiation (45) | ANTXR1 | 1.6 | 2.1 | 1.4 | 1.5 |
| BMP4 | −1.2 | 1.2 | −1.1 | 2.3 | |
| CCND1 | 2.3 | 2.0 | 1.4 | 1.6 | |
| CDH1 | 2.7 | 2.6 | −8.0 | 1.0 | |
| DAB2 | −1.2 | −1.1 | −1.6 | 4.0 | |
| EFNB1 | 2.1 | 1.4 | 1.5 | 1.8 | |
| EGR1 | −6.3 | −9.1 | −12.3 | −1.7 | |
| FGF9 | 1.5 | 2.3 | 2.1 | 1.3 | |
| FGF20 | 9.9 | 5.3 | 4.3 | 7.1 | |
| FN1 | 1.9 | 2.1 | 1.1 | 1.4 | |
| FST | 6.8 | 4.5 | 3.4 | 3.1 | |
| GDNF | 6.1 | −1.2 | 2.9 | −1.5 | |
| IL-6 | 3.0 | 2.8 | 3.4 | 16.7 | |
| MET | 1.6 | 2.2 | 2.1 | −3.7 | |
| MMP9 | 2.4 | 2.7 | 2.0 | 3.7 | |
| NANOG | −1.1 | −2.3 | −1.3 | 1.0 | |
| NRP1 | 2.2 | 1.8 | 1.1 | 1.8 | |
| NTRK2 | 1.2 | −4.2 | −4.1 | 3.5 | |
| PDGFRA | 2.1 | 1.2 | −2.8 | 1.9 | |
| SIX1 | 1.8 | 2.1 | 2.0 | −1.2 | |
| SOX2 | −6.9 | −6.8 | 1.0 | −3.2 | |
| T (Brachyury) | −1.1 | 1.3 | −3.7 | 1.2 | |
| VEGFA | −1.2 | −1.7 | −2.3 | 1.9 | |
| Calcium binding and signaling (13) | CCND1 | 2.3 | 2.0 | 1.4 | 1.6 |
| CUBN | 4.6 | 7.5 | 6.6 | 9.3 | |
| MMP2 | 2.6 | 2.6 | 1.4 | 1.9 | |
| MMP7 | −1.6 | −2.6 | −2.5 | −1.7 | |
| MMP9 | 2.4 | 2.7 | 2.0 | 3.7 | |
| PTGS2 | −1.2 | −1.9 | −2.6 | 1.4 | |
| Adhesion (9) | ABCB1 | 3.1 | 2.4 | 1.4 | 2.8 |
| CDH1 | 2.7 | 2.6 | −8.0 | 1.0 | |
| Migration (21) | EFNB1 | 2.1 | 1.4 | 1.5 | 1.8 |
| FGF7 | −1.1 | −3.3 | 1.2 | 2.8 | |
| FN1 | 1.9 | 2.1 | 1.1 | 1.4 | |
| GDNF | 6.1 | −1.2 | 2.9 | −1.5 | |
| IL-6 | 3.0 | 2.8 | 3.4 | 16.7 | |
| MMP9 | 2.4 | 2.7 | 2.0 | 3.7 | |
| NRP1 | 2.2 | 1.8 | 1.1 | 1.8 | |
| PDGFRA | 2.1 | 1.2 | −2.8 | 1.9 | |
| PPAP2B | 1.0 | −2.3 | −4.2 | 1.4 | |
| SIX1 | 1.8 | 2.1 | 2.0 | −1.2 | |
| VEGFA | −1.2 | −1.7 | −2.3 | 1.9 | |
| Cell cycle (13) | CCND1 | 2.3 | 2.0 | 1.4 | 1.6 |
| MYC | −1.1 | −1.1 | −1.2 | −3.5 | |
| PTGS2 | −1.2 | −1.9 | −2.6 | 1.4 | |
| SOX2 | −6.9 | −6.8 | 1.0 | −3.2 | |
| Proteolysis (5) | DPP10 | −3.3 | −1.7 | −2.0 | −1.1 |
| MMP2 | 2.6 | 2.6 | 1.4 | 1.9 | |
| MMP7 | −1.6 | −2.6 | −2.5 | −1.7 | |
| MMP9 | 2.4 | 2.7 | 2.0 | 3.7 | |
| Signal transduction (23) | AXIN2 | 1.8 | −1.1 | 1.1 | −2.2 |
| BMP4 | −1.2 | 1.2 | −1.1 | 2.3 | |
| FGF9 | 1.5 | 2.3 | 2.1 | 1.3 | |
| FST | 6.8 | 4.5 | 3.4 | 3.1 | |
| SFRP2 | 2.6 | 3.1 | 1.4 | −1.7 | |
| PPAP2B | 1.0 | −2.3 | −4.2 | 1.4 | |
| WISP1 | −5.7 | −6.2 | −6.2 | −5.4 | |
| Transcription factors (22) | EGR1 | −6.3 | −9.1 | −12.3 | −1.7 |
| MYC | −1.1 | −1.1 | −1.2 | −3.5 | |
| NANOG | −1.1 | −2.3 | −1.3 | 1.0 | |
| SIX1 | 1.8 | 2.1 | 2.0 | −1.2 | |
| SOX2 | −6.9 | −6.8 | 1.0 | −3.2 | |
| T (Brachyury) | −1.1 | 1.3 | −3.7 | 1.2 | |
| TCF7 | 1.5 | 1.0 | 1.0 | −2.3 | |
A few genes are classified into more than one category.
Fold regulation of >2 indicates a significant upregulation, and fold regulation of <−2 indicates a significant downregulation.
E. chaffeensis infection suppresses phosphorylation of host β-catenin and mediates nuclear translocation of β-catenin and NFATC1.
β-Catenin and NFATC1 are important signal transducers/transcription factors involved in Wnt/β-catenin and Wnt/Ca2+-regulated gene transcription, respectively. Activation of Wnt signaling pathways results in the dephosphorylation and translocation of β-catenin and NFATC1 into the nucleus. Therefore, we examined the phosphorylation of β-catenin and the localization of β-catenin and NFATC1 after ehrlichial infection by Western blotting and immunofluorescence microscopy. Phosphorylation of β-catenin was reduced at 1 h after infection and inhibited almost completely at 3 h (Fig. 2A). Moreover, a dramatic redistribution of β-catenin and NFATC1 proteins to the nucleus was observed shortly after infection with E. chaffeensis. In uninfected THP-1 cells, β-catenin was diffusely distributed mainly in the cytoplasm and associated with the cell membrane, but in E. chaffeensis-infected cells as early as 1 h p.i., β-catenin translocated to the nucleus, exhibiting a punctate distribution. By 20 h p.i., the majority of the β-catenin was redistributed to the cytoplasm (Fig. 2B). Similarly, NFATC1 was diffusely distributed mainly in the cytoplasm of uninfected THP-1 cells but was translocated to the nucleus as early as 1 h p.i. Notably, there was nearly complete translocation of NFATC1 to the nucleus at 3 h, but by 20 h p.i., NFATC1 was observed mainly in the cytoplasm (Fig. 2B). The inhibition of phosphorylation of β-catenin and translocation of β-catenin and NFATC1 proteins to the nucleus between 1 and 3 h p.i. was consistent with PCR array analysis identifying activation of Wnt component and target genes at 1 to 3 h after E. chaffeensis infection.
FIG 2.
E. chaffeensis upregulates Wnt signaling. (A) Western blots show that phosphorylation of β-catenin is inhibited at 1 h and 3 h postinfection with E. chaffeensis. (B) Wnt signal transducer β-catenin and transcription factor NFATC1 translocate to the nucleus in THP-1 cells at 1 h and 3 h postinfection with E. chaffeensis. DAPI (4′,6′-diamidino-2-phenylindole) staining shows the nucleus. Bars, 10 μm. (C) E. chaffeensis infection activates an NFAT reporter. The cell-free E. chaffeensis or uninfected THP-1 control was added to NFAT reporter-transfected HeLa cells. Firefly and Renilla luciferase activities of each well were measured at different time points posttreatment. RLU, relative luciferase units of firefly that were normalized against Renilla luciferase activity. The experiment was repeated three times, and the values are means ± standard deviations (*, P < 0.05).
E. chaffeensis infection activates an NFAT reporter.
Wnt signaling PCR arrays analyze the expression of a panel of genes related to all three Wnt pathways, but the pathway activities determined by PCR array are based on the well-studied canonical Wnt signaling pathway; therefore, the Cignal NFAT reporter was used to examine the activity of the noncanonical Wnt/Ca2+ signal pathway (NFAT-mediated transcription). Compared to the uninfected control, stimulation of reporter signal was not observed at 3 h p.i., but the reporter activity increased at 8 h p.i. and was significantly upregulated by 24 h p.i. (Fig. 2C), suggesting that E. chaffeensis activates the noncanonical Wnt/Ca2+ signal pathway but that complete activation occurs later than that of the canonical Wnt/β-catenin pathway. In addition, the activation of the Wnt/Ca2+ signal pathway at the transcriptional and translational levels (8 to 24 h) occurs later than translocation of the NFAT protein into the nucleus (1 to 3 h).
Knockdown of Wnt signaling pathway components influences ehrlichial infection of macrophages.
We further confirmed the role of host Wnt signaling pathways in ehrlichial infection by RNA interference. In total, 23 siRNAs were used to target important components of Wnt signaling pathways, such as Wnt ligands, receptors and coreceptors, regulators, transcription factors, and Wnt targets (Fig. 3A). The siRNAs of some host genes modulated during E. chaffeensis infection, as determined by PCR array analysis, were also included. The decrease of most proteins (17 proteins at 1 day p.i. and 20 proteins at 2 days p.i.) decreased E. chaffeensis infection significantly, indicating that Wnt signaling plays a role in E. chaffeensis infection and survival. An exception was knockdown of DKK3 (at 1 day p.i.), which increased the ehrlichial load. Notably, DKK3 (Dickkopf homolog 3 from Xenopus laevis) is an antagonist of the canonical Wnt signaling pathway (48), thus further supporting the importance of Wnt signaling in ehrlichial survival. The inhibition of infection by siRNAs of Wnt signaling pathways occurred at 1 and/or 2 days p.i. Protein expression of three genes, those for Fzd9, Dvl2, and Jun, was reduced in specific siRNA-transfected cells compared with the unrelated control siRNA-transfected cells (Fig. 3B).
FIG 3.
Knockdown of a Wnt signaling pathway component or target influences ehrlichial infection of macrophages. THP-1 cells were transfected with a specific or control siRNA and then infected with E. chaffeensis. (A) Bacterial numbers were determined by qPCR at 1 day and 2 days p.i. All experiments were repeated three times, and the values are means ± standard deviations (*, P < 0.05). (B) Western blotting confirmed the reduction of Fzd9, Dvl2, and Jun proteins at 2 days p.i.
Small-molecule inhibitors of Wnt signaling pathways repress E. chaffeensis infection of host cells.
To confirm the important role of host Wnt signaling pathways in E. chaffeensis infection, the effects of various Wnt signaling pathway inhibitors on ehrlichial infection were examined (Table 4). Five inhibitors were found to have a significant impact on ehrlichial infection without apparent toxicity to host cells, i.e., CK1α/GSK3 activator (Akt inhibitor) pyrvinium, CaMKII inhibitor KN93, inhibitor of Wnt production II IWP-2, CK1δ/ε inhibitor SB202190, and CK2 inhibitor III TBCA (Fig. 4A to C). Pyrvinium, KN93, and IWP-2 were highly potent inhibitors of ehrlichial infection and could block the infection almost completely despite addition of inhibitor 3 h before or after ehrlichial infection. Two inhibitors, SB202190 and TBCA, reduced ehrlichial infection significantly. Addition of inhibitor SB202190 or TBCA 3 h before or after ehrlichial infection did not make significant difference in reduction of the bacterial load, suggesting that inhibitors SB202190 and TBCA function after ehrlichial infection and thus that CK1δ/ε and CK2 of the canonical Wnt pathway are not important for ehrlichial internalization. Notably, the three most potent inhibitors pyrvinium, KN93, and IWP-2 target the canonical Wnt pathway, the noncanonical Wnt/Ca2+ pathway, and Wnt ligand secretion, respectively, indicating the importance of both canonical and noncanonical Wnt signaling pathways and Wnt ligand secretion for ehrlichial survival. Moreover, MICs of pyrvinium, KN93, and IWP-2 for ehrlichial infection were determined to be 20 nM, 4 μM, and 0.3 μM, respectively, by using 2-fold serial dilutions (Table 4). In addition, we found that three other inhibitors, β-catenin/TCF inhibitor FH535, PI3K inhibitor LY294002, and IKK inhibitor BAY11-7082, could influence ehrlichial infection, they but exhibited toxicity to host cells after treatment for more than 1 day (data not shown).
TABLE 4.
Small-molecule inhibitors of Wnt pathways, their target proteins, and concentrations used in this study
| Pathway/process | Inhibitor | Targeta | Concn (μM) |
|---|---|---|---|
| Canonical Wnt pathway | FH535 | β-Catenin/TCF | 10 |
| Pyrvinium pamoate | CK1α or Akt (activate GSK3) | 0.02 | |
| SB202190 | CK1δ/ε | 1.4 | |
| TBCA | CK2 | 0.5 | |
| Noncanonical Wnt pathways | BAY11-7082 | IKK | 10 |
| KN93 | CaMKII | 4 | |
| LY294002 | PI3K | 10 | |
| NSC23766 | Rac1 | 50 | |
| Wnt ligand secretion | IWP-2 | PORCN | 0.3 |
All targets except CK1α are inhibited.
FIG 4.
Wnt signaling pathway inhibitors reduce ehrlichial infection of host cells. (A) Percentages of infected cells were determined by Diff-Quik staining and counting of 100 cells at 3 days p.i. Inhibitors were added 3 h before or after infection. An infected cell culture with the vehicle (DMSO) only served as a positive control, and an uninfected culture served as a negative control. Results are from three independent experiments, and the values are means ± standard deviations (*, P < 0.05; **, P < 0.01). (B) Trypan blue staining was performed to assess host cell viability and graphed as a percentage of the total cell count for THP-1 cells exposed to 72-h treatments with vehicle or inhibitor. (C) Bright-field images (magnification, ×40) of Diff-Quik-stained samples collected at 3 days p.i. demonstrate decreased numbers of infected cells following treatment with Wnt signaling pathway inhibitors pyrvinium and SB202190, as examples. Bars, 10 μm. Concentrations used for inhibitors were as follows: 20 nM pyrvinium, 4 μM KN93, 0.3 μM IWP-2, 0.5 μM TBCA, and 1.4 μM SB202190.
E. chaffeensis TRP120 interacts with host targets involved in Wnt signaling that influence infection.
Since E. chaffeensis TRPs have been identified as bacterial effector proteins and interact with multiple host proteins involved in Wnt signaling, we confirmed the role of these host proteins in ehrlichial infection by RNA interference. Similar to our previous result for TRP32-interacting protein DAZAP2, knockdown of five TRP120-interacting host proteins, i.e., ARID1B, KDM6B, IRF2BP2, PPP3R1, and VPS29, influenced E. chaffeensis infection of macrophages significantly (Table 5). The bacterial load in all specific siRNA-transfected cells decreased at both 1 day and 2 days p.i. (fold regulation of <−2), except that the bacterial load in ARID1B siRNA-transfected cells increased. ARID1B is an AT-rich DNA-interacting domain-containing protein and a component of the SWI/SNF chromatin-remodeling complex, and it has been reported to interact with β-catenin to suppress Wnt signaling (49). The results suggest that TRP120 plays a major role in influencing Wnt pathway activation by interacting with these Wnt pathway regulators during E. chaffeensis infection.
TABLE 5.
Fold regulation of E. chaffeensis bacterial load in THP-1 cells transfected with siRNA specific for TRP120-interacting host targets involved in Wnt signaling relative to control siRNA at 1 and 2 days p.i., as determined by qPCR
| siRNA target | Target property/function in Wnt signaling | Fold regulationa at: |
|
|---|---|---|---|
| 1 day | 2 days | ||
| ARID1B | Interacts with β-catenin to suppress Wnt signaling | 2.3 | 1.5 |
| KDM6B | β-Catenin binding; activates Wnt3 or DKK1 to stimulate or suppress Wnt signaling at different stages | −4.3 | −4.3 |
| IRF2BP2 | Interacts with NFATC2 to repress its transcriptional activity | −4.5 | −3.9 |
| PPP3R1 | Calcineurin regulatory subunit 1; calcium ion and calmodulin binding; calcium-dependent protein phosphatase activity; NFAT protein import into nucleus | −4.0 | −4.0 |
| VPS29 | Retrograde transport of proteins from endosomes to the trans-Golgi network; Wnt ligand biogenesis, secretion, and trafficking | −9.8 | −4.5 |
dsb gene copy number normalized to the GAPDH gene; the average is shown (n = 2).
E. chaffeensis TRPs stimulate phagocytosis of macrophages, and Wnt pathway inhibitors reduce the stimulation.
Wnt signaling is known to influence cytoskeletal changes, cell polarity, and phagocytosis (32, 50). To determine if E. chaffeensis tandem repeat proteins (TRPs) activated the Wnt pathway to induce phagocytosis, we coated microspheres with recombinant TRPs and examined their uptake. Phagocytosis of TRP120-coated microspheres by the THP-1 cells was significantly increased compared with that by the control protein (TRP120 N-terminal region [TRP120N])-coated microspheres (Fig. 5A). Similarly, TRP32- or TRP47-coated microspheres were also phagocytosed, but less efficiently than TRP120. Next we examined the roles of specific TRP120 domains in inducing Wnt-directed phagocytosis. Truncated TRP120TRC (containing all tandem repeats and the C terminus), TRP120-2R (containing two tandem repeats), and TRP120C (containing the C terminus) stimulated phagocytosis, but less efficiently than full-length TRP120. The N-terminal region of TRP120 was not phagocytosed suggesting that TR and C-terminal domains both contribute to TRP-induced phagocytosis (Fig. 5B). To examine the role of Wnt pathways associated with TRP-induced phagocytosis, we tested Wnt pathway inhibitors (Table 4). Noncanonical Wnt pathway inhibitors, including IKK inhibitor BAY11-7082 and PI3K inhibitor LY94002, significantly reduced the TRP-coated microsphere uptake, supporting an important role for one or more of these noncanonical Wnt pathways in TRP-induced phagocytosis (Fig. 5A and C). TRP120-induced microsphere internalization was also reduced after treatment with various Wnt pathway inhibitors, including CKIα/GSK3β activator (Akt inhibitor) pyrvinium, β-catenin/TCF inhibitor FH535, CaMKII inhibitor KN93, and Rac1 inhibitor NSC23766, suggesting that all three canonical and noncanonical Wnt pathways are involved in TRP-induced phagocytosis. However, the canonical Wnt pathway inhibitors CK1δ/ε inhibitor (SB202190) and CK2 inhibitor III (TBCA) did not reduce TRP microsphere internalization significantly, consistent with our data shown in Fig. 4A. Predictably, there was dramatic inhibition of TRP-mediated phagocytosis by the lipid raft-disrupting agent nystatin (10 μM), similar to that with the most potent noncanonical Wnt pathway inhibitors. An inhibitor of Wnt ligand biogenesis and secretion, IWP-2, did not inhibit the phagocytosis mediated by TRP, suggesting that TRP-induced microsphere internalization occurs independent of Wnt ligand secretion (Fig. 5C). These results demonstrate the importance of Wnt signaling pathways in ehrlichial internalization and highlight the roles of PI3K, IKK, Rac1, CK1α/GSK3β/Akt, β-catenin/TCF, and CaMKII as well as lipid rafts in TRP-induced phagocytosis.
FIG 5.
E. chaffeensis tandem repeat proteins stimulate phagocytosis and intracellular Ca2+ release in macrophages, and Wnt pathway inhibitors reduce the stimulation. (A) Compared with control TRP120N-coated beads, TRP120-coated beads were phagocytosed by the THP-1 cells. After treatment with Wnt pathway inhibitor Bay11-7082 (10 μM), the promotion of bead internalization by TRP120 was largely reduced. (B) Efficiency of promotion of bead internalization by different tandem repeat proteins and domains. (C) Effect of different Wnt pathway inhibitors on the promotion of bead internalization by TRP120. All experiments were repeated three times, and the values are means ± standard deviations (*, P < 0.05). (D) E. chaffeensis TRPs stimulate Ca2+ release. Ca2+ release was measured in THP-1 cells using fluo-3/AM. Fluo-3-preloaded cells were treated with cell-free E. chaffeensis (MOI = ∼50), TRP-coated beads (50 beads/cell), or a control (cell-free uninfected THP-1 or thioredoxin-coated beads) and mixed briefly, and emission at 525 nm after excitation at 488 nm was recorded every 20 s.
E. chaffeensis TRPs stimulate intracellular Ca2+ release.
Activation of the noncanonical Wnt/Ca2+ pathway leads to increased intracellular Ca2+ release and activation of CaMKII and calcineurin followed by NFAT and other transcription factors. A Ca2+ increase was detected in THP-1 cells infected with E. chaffeensis or treated with beads coated with a mixture of TRP32, TRP47, and TRP120. Consistent with a previous study (43), a rapid increase of Ca2+ was detected to about 80 s after cell-free E. chaffeensis was added. Similarly, TRP-coated beads also caused a rapid increase of Ca2+ to about 60 s, although the Ca2+ concentration was lower than that stimulated by Ehrlichia. As controls, uninfected THP-1 cell lysate or control beads did not induce Ca2+ release, indicating that Ehrlichia and TRPs stimulate intracellular Ca2+ release associated with E. chaffeensis entry (Fig. 5D). In addition, Ca2+ release by a single TRP-coated bead was not detected (data not shown), suggesting that all three TRPs are needed for Ca2+ release or that the TRP mixture caused more Ca2+ release.
DISCUSSION
Wnt signaling involves a complex signaling network of ligands, receptors, kinases, transcription factors, and other molecules that are associated with three distinct but interconnected Wnt pathways. In the eukaryotic cell, Wnt pathways control cellular proliferation, differentiation, and cell polarity, and recent reports have linked microbes and host Wnt signaling. Microbe activation of Wnt pathways has been linked to phagocytosis of bacteria and viruses, differentiation of cells to promote entry, expression of proinflammatory responses to microbial pathogen-associated molecular patterns, and intracellular bacterial survival (31, 32, 36, 38). Previously, we have defined interactions between ehrlichial TRP effectors and host DNA/proteins, including Wnt pathway components, regulators, and transcriptional regulators of Wnt pathway gene expression (6, 7, 15). This study extends these interactions and demonstrates that canonical and noncanonical Wnt pathway activation occurs during E. chaffeensis infection. In this study, we demonstrate that host Wnt signaling pathways play important roles in ehrlichial internalization and infection and that ehrlichial TRPs mediate the invasion and survival through activation and modulation of Wnt signaling pathways.
In order to identify temporal regulation of Wnt pathways and the specific Wnt signaling components and target genes induced by E. chaffeensis infection, we examined expression of Wnt pathway components and target genes using PCR arrays. Pathway activity scores demonstrated early activation and late repression of Wnt pathways, with significant changes in the expression level of Wnt ligands, receptors, inhibitors, signaling molecules, and transcription factors associated with both the canonical and noncanonical Wnt pathways. Gene transcription analysis indicated that a Wnt pathway(s) was activated at a very early stage of infection (3 h), suggesting involvement of the pathway in the internalization of bacteria, consistent with a previous report that the Wnt signaling in macrophages promoted phagocytosis of bacteria and enhanced survival (31). Ehrlichial entry occurs within 1 h to 3 h p.i. (45, 51), and the resulting activation of Wnt pathways at 3 h p.i. is consistent with the time frame of the entry process. Early activation of canonical and noncanonical Wnt pathways was confirmed by the dephosphorylation of β-catenin and nuclear translocation of β-catenin and NFATC1. Translocation of β-catenin and NFATC1 was also detected at 1 h p.i. These findings support that host Wnt signaling pathways are activated during Ehrlichia entry. Conversely, Wnt pathway repression was observed 72 h after infection in which the bacteria have completed a replication cycle and exit the host cell (51), suggesting that the Wnt pathway(s) is downregulated during the exit phase. Significant changes in pathway activity at other time points (1 h, 8 h, and 24 h) were not detected, and this finding may be associated with the limitation of signature biomarker genes that our PCR array selects, since different components may be regulated by Ehrlichia at different time points in a different way. In addition, we used the NFAT reporter assay to demonstrate that E. chaffeensis activates the noncanonical Wnt/Ca2+ pathway directly. The inconsistency in time points between NFAT nuclear translocation (1 to 3 h p.i.) and NFAT reporter expression (8 to 24 h p.i.) may be due to the later occurrence of gene expression than nuclear translocation of NFAT or may be caused by different cell lines and cell context.
Genes with significant changes of expression after ehrlichial infection (Table 2) are included in both canonical and noncanonical Wnt pathways. For example, Wnt5b ligand has been found to activate Wnt/PCP and Wnt/Ca2+ pathways and also to be associated with β-catenin (52–54); similarly, receptor Fzd5 is commonly linked to Wnt5a and Wnt/PCP pathways, but prototype noncanonical Wnt5a has been reported to signal to β-catenin in the presence of overexpressed Fzd5 (55). Therefore, it is predicted that these Wnt pathway components/targets with significant changes of expression play important roles during Ehrlichia entry/survival, and all three Wnt signaling pathways are utilized by E. chaffeensis. This is further supported by functionally categorized gene expression changes of Wnt signaling targets after ehrlichial infection, including Wnt targets involved in development and differentiation, calcium binding, and signaling and migration, as demonstrated by the Wnt target PCR array (Table 3). Wnt pathways are involved in these important cellular processes, suggesting that the regulation of canonical and noncanonical Wnt pathways is important for ehrlichial infection.
Experiments using small interfering RNAs and small-molecule inhibitors support the conclusion that canonical and noncanonical Wnt signaling pathways are activated during ehrlichial infection of host cells. Knockdown of some important components of Wnt signaling pathways, including Wnt ligands, receptors and coreceptors, signaling molecules, kinases, transcription factors, and targets as well as TRP120-interacting proteins, influenced E. chaffeensis infection significantly, indicating the importance of these proteins and canonical/noncanonical Wnt pathway activation during E. chaffeensis infection. Specifically, highly significant reductions of infection were observed at both 1 and 2 days p.i. after knockdown of ligand Wnt5a or its receptor Fzd5, suggesting that Wnt5a-Fzd5 signaling is very important for Ehrlichia survival after internalization. This is consistent with a previous report that Wnt5a-Fzd5 signaling reduced bacterial killing by macrophages (31). However, we found by gene expression analysis that Fzd5, but not Wnt5a, was upregulated by Ehrlichia, suggesting that Ehrlichia regulates the expression of Wnt5a receptor instead of Wnt5a itself, although Wnt5a does appear to be important for bacterial survival after entry. Wnt5a can bind to different classes of receptors and coreceptors, but Wnt5a signals primarily through the noncanonical pathway, where it mediates cell proliferation, adhesion, and movement (56). However, the role of Wnt5a in canonical signaling is still unclear, due to its complex nature. Depending on the receptor availability, Wnt5a can activate or inhibit the canonical Wnt signaling pathway (57). Moreover, Wnt5a has recently emerged as a macrophage effector molecule that triggers inflammation (58, 59). Another protein whose knockdown reduced the infection highly significantly at 1 day p.i. was low-density lipoprotein receptor-related protein 6 (LRP6), a coreceptor with Frizzled for Wnt to transmit the canonical Wnt signaling. Macrophages expressing receptors containing the cytoplasmic tail of LRP were able to bind and internalize sheep red blood cells, and LRP6 was reported to internalize with caveolin and interact with axin to promote the accumulation of β-catenin (60, 61), so LRP6 may be involved in both Ehrlichia internalization and survival. Knockdown of Akt reduced the infection very significantly at both 1 and 2 days p.i. Akt, also known as protein kinase B (PKB), is a serine/threonine-specific protein kinase that is involved in both canonical and noncanonical Wnt pathways. Akt plays a key role in multiple cellular processes such as cell survival, cell proliferation, cell migration, transcription, and apoptosis, and it is also associated with cytoskeleton reorganization and phagocytosis of bacteria by macrophages (62, 63); therefore, Akt is predicted to be involved in both Ehrlichia internalization and survival. In our study, the only siRNA which increased the ehrlichial load was DKK3, consistent with the fact that it is an antagonist of the canonical Wnt pathway.
TRP120-interacting host proteins ARID1B, KDM6B, IRF2BP2, PPP3R1, and VPS29 are involved in both canonical/noncanonical Wnt signaling pathways and Wnt ligand secretion and also appear to play important roles during ehrlichial infection. For example, KDM6B is a histone H3 lysine demethylase with an important gene regulatory role in development and physiology. It has been reported not only to interact with β-catenin and contribute to β-catenin-dependent promoter activation but also to upregulate Wnt3 expression and cause activation of Wnt signaling (64, 65). PPP3R1 is calcineurin subunit B type 1, an important component of the Wnt/Ca2+ pathway with Ca2+/calmodulin binding activity. Therefore, TRP120 interaction with KDM6B and PPP3R1 would be predicted to promote positive regulation of Wnt/β-catenin and the Wnt/Ca2+ pathway, respectively, to facilitate ehrlichial survival. VPS29 is a component of a large retromer complex which is involved in retrograde transport of Wnt carrier protein Wntless (Wls) from endosomes to the trans-Golgi network and is required for Wnt secretion (66). TRP120 may interact with VPS29 to assist the recycling of Wls protein and thus promote the secretion of Wnt ligands and activation of Wnt pathways during ehrlichial infection.
The siRNA experiments revealed that a reduction in E. chaffeensis load occurs at different stages of infection for different host targets, suggesting that the role of these targets varies at different points during infection. For example, Wnt3a, Wnt10a, and calcineurin PPP3CB siRNAs reduced the bacterial load significantly at 2 days p.i. but not significantly at 1 day, suggesting that these components may be not involved in bacterial internalization, or perhaps they play a larger role in Wnt signaling associated with bacterial survival rather than entry. In contrast, Fzd9 and DKK3 siRNAs reduced/increased the bacterial load significantly at 1 day p.i. but not significantly at 2 days, suggesting that they are more important for the bacterial entry or early stage. In addition, knockdown of some Wnt pathway components had a dramatic effect on the ehrlichial load, suggesting that they play key roles in the Ehrlichia-Wnt axis. For example, knockdown of Wnt5a, Fzd5, or Akt reduced the infection very significantly at both 1 and 2 days p.i., suggesting that they are critical components of Wnt signaling exploited by Ehrlichia survival; however, we cannot exclude the possibility that siRNAs may have different knockdown efficiency. Unlike the effect of some small-molecule inhibitors, the reduction of a single target protein by RNA interference could not abolish the ehrlichial growth completely. This may result from the incomplete knockdown of target proteins, as was demonstrated by Western blotting, or redundancy in Wnt signaling.
Several Wnt signaling pathway inhibitors reduced ehrlichial infection significantly, three of which, pyrvinium pamoate, KN93, and IWP-2, were highly effective. Pyrvinium was first found to activate CK1α, but it has been recently reported that it promotes Akt downregulation and GSK3 activation rather than activates CK1 (67, 68). Despite discrepancies in the understood mechanism of action, pyrvinium ultimately inhibits Wnt/β-catenin signaling. However, Akt also interacts with Wnt/Ca2+ and Wnt/PCP pathways, so pyrvinium may also influence noncanonical Wnt pathways. KN93 selectively binds to the calmodulin binding site of CaMKII and prevents the association of calmodulin with CaMKII, and it thus inhibits the Wnt/Ca2+ pathway (69); IWP-2 inhibits the cellular Wnt processing and secretion via selective blockage of PORCN-mediated Wnt palmitoylation (70). Two canonical Wnt pathway inhibitors, SB202190 and TBCA, which inhibit CK1δ/ε and CK2, respectively, were less effective. These inhibitors act on all three Wnt signaling pathways and Wnt secretion and thus reveal the importance of both canonical and noncanonical Wnt signaling pathways, especially CK1α/GSK3/Akt, CaMKII, and Wnt ligands, in ehrlichial infection. CK1δ/ε and CK2 of the canonical Wnt pathway were not important for ehrlichial internalization, since addition of inhibitor SB202190 or TBCA before or after ehrlichial infection did not make a significant difference in reduction of the bacterial load. Pyrvinium, KN93, and IWP-2 could block the infection almost completely, so the roles of CK1α/GSK3/Akt, CaMKII, and Wnt ligand secretion in ehrlichial internalization could not be verified by this experiment only. Wnt signaling pathway inhibitors have potential for ehrlichiosis treatment, but further study is needed to understand the importance of specific Wnt pathway components in ehrlichial pathobiology.
Consistent with recent reports (31, 44), our experiments indicate that inhibitors of PI3K, IKK, and lipid rafts reduced TRP-mediated phagocytosis significantly. Activation of Rac1-PI3K-IKK perhaps stimulates the assembly of scavenger receptors at lipid rafts and supports cytoskeletal rearrangements required for phagocytosis (71–73). In addition, inhibitors of Akt, Rac1, β-catenin/TCF, and CaMKII, but not CK1δ/ε and CK2, also reduced TRP-induced phagocytosis. Therefore, Wnt pathway components PI3K, IKK, Rac1, CK1α/GSK3β/Akt, β-catenin/TCF, and CaMKII appear to be important in signaling that leads to ehrlichial internalization. In contrast, CK1δ/ε and CK2 are not involved, consistent with our other experimental results. Based on the classification of these components, we may also define TRP-induced phagocytosis primarily as a noncanonical mode of Wnt signaling (most likely Wnt/PCP signaling), similar to Wnt5a-induced phagocytosis, although canonical Wnt signaling may be involved and influence phagocytosis. Interactions between E. chaffeensis TRPs and components/regulators of the Wnt signaling pathway appear to directly activate Wnt signaling. However, in contrast to the recent report demonstrating that Wnt5a triggered phagocytosis of a bacterium (31), the Wnt production inhibitor IWP-2 could not inhibit the phagocytosis mediated by TRP, suggesting that Wnt ligands, such as Wnt5a, may promote Wnt signaling activation to promote ehrlichial survival after internalization but are not involved in TRP-induced phagocytosis. Thus, it appears that Ehrlichia internalization is independent of Wnt ligand secretion, although further experimentation is needed to confirm this conclusion.
To date, the molecular mechanisms by which Ehrlichia enters and modulates host cells are not well understood (4, 6–8), but our findings regarding Wnt pathways are consistent with previous reports that described some characteristics of cellular signaling upon internalization of ehrlichiae (43, 44). E. chaffeensis enters the monocyte through lipid raft-caveola-mediated endocytosis, where it can reside within a cytoplasmic vacuole that resembles an early endosome, preventing lysosomal fusion and protecting it from killing (44). TRP120 is an ehrlichial surface protein preferentially expressed on dense-core cells, and it has been found to interact with host proteins involved in cytoskeleton organization (6). It has also been associated with ehrlichial binding and internalization (45, 46). We found that ehrlichial TRPs stimulate phagocytosis by macrophages and that Wnt pathway inhibitors inhibit phagocytosis of TRP-coated microspheres. Thus, ehrlichial TRPs appear to be agonists that directly activate Wnt signaling pathways of the host to induce internalization and facilitate intracellular survival. However, the underlying molecular mechanism remains unclear. Our data suggest that TRP120 induces phagocytosis more efficiently than other TRPs and that TR and C-terminal domains of TRP120 mediate TRP-induced phagocytosis. These domains associated with internalization were not examined in other TRPs but are predicted to be similar. Notably, another study reported that E. chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X to trigger entry via CD147 and hnRNP-K recruitment and actin mobilization (74, 75). The domains in TRPs responsible for internalization are structurally distinct from EtpE and are not predicted to interact with DNase X. Therefore, TRP-induced entry may involve a distinct TRP-interacting Wnt surface protein that remains to be identified, rather than DNase X. Figure 6 shows a proposed model of E. chaffeensis TRP-mediated activation of canonical and noncanonical Wnt signaling pathways. TRP effector proteins bind unidentified cell receptors and activate Dvl protein and other downstream signaling molecules of Wnt pathways identified in this study, resulting in regulation of gene transcription and cytoskeletal reorganization/phagocytosis. Further study is needed to determine the receptor involved, the roles of TRP-interacting proteins, and the relationship between TRP and EtpE in ehrlichial binding and internalization.
FIG 6.
Proposed schematic diagram showing E. chaffeensis TRP-mediated activation of canonical and noncanonical Wnt signaling pathways. TRP effector proteins bind unidentified cell receptors and activate Dvl protein, which is the hub molecule of all Wnt pathways. Dvl protein activates other signaling molecules of Wnt pathways in the cytosol, including many kinases and phosphatases, resulting in regulation of gene transcription by transcription factors, such as TCF and NFAT, and cytoskeletal reorganization and phagocytosis. TRPs also interact with direct components or regulators (shown in green) of Wnt pathways to regulate Wnt signaling.
Our study establishes for the first time an obligately intracellular pathogen-directed Wnt pathway-induced mechanism responsible for invasion and persistence and will provide insight into mechanisms utilized by intracellular pathogens to invade host cells. Further research will elucidate and define the mechanisms and specific interactions between TRPs and Wnt signaling pathways.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI105536 and AI106859 and by funding from the Clayton Foundation for Research (to Jere W. McBride).
Funding Statement
The funders had no role in study design, data collection and interpretation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01289-15.
REFERENCES
- 1.Paddock CD, Childs JE. 2003. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin Microbiol Rev 16:37–64. doi: 10.1128/CMR.16.1.37-64.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rikihisa Y. 2010. Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat Rev Microbiol 8:328–339. doi: 10.1038/nrmicro2318. [DOI] [PubMed] [Google Scholar]
- 3.McBride JW, Walker DH. 2011. Molecular and cellular pathobiology of Ehrlichia infection: targets for new therapeutics and immunomodulation strategies. Expert Rev Mol Med 13:e3. doi: 10.1017/S1462399410001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunphy PS, Luo T, McBride JW. 2013. Ehrlichia moonlighting effectors and interkingdom interactions with the mononuclear phagocyte. Microbes Infect 15:1005–1016. doi: 10.1016/j.micinf.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunphy PS, Luo T, McBride JW. 2014. Ehrlichia chaffeensis exploits host SUMOylation pathways to mediate effector-host interactions and promote intracellular survival. Infect Immun 82:4154–4168. doi: 10.1128/IAI.01984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo T, Kuriakose JA, Zhu B, Wakeel A, McBride JW. 2011. Ehrlichia chaffeensis TRP120 interacts with a diverse array of eukaryotic proteins involved in transcription, signaling, and cytoskeleton organization. Infect Immun 79:4382–4391. doi: 10.1128/IAI.05608-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo T, McBride JW. 2012. Ehrlichia chaffeensis TRP32 interacts with host cell targets that influence intracellular survival. Infect Immun 80:2297–2306. doi: 10.1128/IAI.00154-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakeel A, Kuriakose JA, McBride JW. 2009. An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signaling, transcriptional regulation, and vesicle trafficking. Infect Immun 77:1734–1745. doi: 10.1128/IAI.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doyle CK, Nethery KA, Popov VL, McBride JW. 2006. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect Immun 74:711–720. doi: 10.1128/IAI.74.1.711-720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo T, Zhang X, McBride JW. 2009. Major species-specific antibody epitopes of the Ehrlichia chaffeensis p120 and E. canis p140 orthologs in surface-exposed tandem repeat regions. Clin Vaccine Immunol 16:982–990. doi: 10.1128/CVI.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuriakose JA, Zhang X, Luo T, McBride JW. 2012. Molecular basis of antibody mediated immunity against Ehrlichia chaffeensis involves species-specific linear epitopes in tandem repeat proteins. Microbes Infect 14:1054–1063. doi: 10.1016/j.micinf.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo T, Zhang X, Wakeel A, Popov VL, McBride JW. 2008. A variable-length PCR target protein of Ehrlichia chaffeensis contains major species-specific antibody epitopes in acidic serine-rich tandem repeats. Infect Immun 76:1572–1580. doi: 10.1128/IAI.01466-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakeel A, Zhu B, Yu XJ, McBride JW. 2010. New insights into molecular Ehrlichia chaffeensis-host interactions. Microbes Infect 12:337–345. doi: 10.1016/j.micinf.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukas J, Mazna P, Valenta T, Doubravska L, Pospichalova V, Vojtechova M, Fafilek B, Ivanek R, Plachy J, Novak J, Korinek V. 2009. Dazap2 modulates transcription driven by the Wnt effector TCF-4. Nucleic Acids Res 37:3007–3020. doi: 10.1093/nar/gkp179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu B, Kuriakose JA, Luo T, Ballesteros E, Gupta S, Fofanov Y, McBride JW. 2011. Ehrlichia chaffeensis TRP120 binds a G+C-rich motif in host cell DNA and exhibits eukaryotic transcriptional activator function. Infect Immun 79:4370–4381. doi: 10.1128/IAI.05422-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nusse R, van Ooyen A, Cox D, Fung YK, Varmus H. 1984. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature 307:131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- 17.Komiya Y, Habas R. 2008. Wnt signal transduction pathways. Organogenesis 4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Logan CY, Nusse R. 2004. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 19.Klaus A, Birchmeier W. 2008. Wnt signalling and its impact on development and cancer. Nat Rev Cancer 8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 20.Nusse R, Varmus HE. 1992. Wnt genes Cell 69:1073–1087. [DOI] [PubMed] [Google Scholar]
- 21.Nusse R, Varmus H. 2012. Three decades of Wnts: a personal perspective on how a scientific field developed. EMBO J 31:2670–2684. doi: 10.1038/emboj.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habas R, Dawid IB. 2005. Dishevelled and Wnt signaling: is the nucleus the final frontier? J Biol 4:2. doi: 10.1186/jbiol22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao TP, Kuhl M. 2010. An updated overview on Wnt signaling pathways: a prelude for more. Circ Res 106:1798–1806. doi: 10.1161/CIRCRESAHA.110.219840. [DOI] [PubMed] [Google Scholar]
- 24.Staal FJ, Clevers H. 2000. Tcf/Lef transcription factors during T-cell development: unique and overlapping functions. Hematol J 1:3–6. doi: 10.1038/sj.thj.6200001. [DOI] [PubMed] [Google Scholar]
- 25.MacDonald BT, Tamai K, He X. 2009. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugimura R, Li L. 2010. Noncanonical Wnt signaling in vertebrate development, stem cells, and diseases. Birth Defects Res C Embryo Today 90:243–256. doi: 10.1002/bdrc.20195. [DOI] [PubMed] [Google Scholar]
- 27.Gordon MD, Nusse R. 2006. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem 281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 28.Niehrs C. 2012. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol 13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 29.Willert K, Nusse R. 2012. Wnt proteins. Cold Spring Harb Perspect Biol 4:a007864. doi: 10.1101/cshperspect.a007864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kikuchi A, Yamamoto H, Sato A, Matsumoto S. 2011. New insights into the mechanism of Wnt signaling pathway activation. Int Rev Cell Mol Biol 291:21–71. doi: 10.1016/B978-0-12-386035-4.00002-1. [DOI] [PubMed] [Google Scholar]
- 31.Maiti G, Naskar D, Sen M. 2012. The Wingless homolog Wnt5a stimulates phagocytosis but not bacterial killing. Proc Natl Acad Sci U S A 109:16600–16605. doi: 10.1073/pnas.1207789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu F, Zhang X. 2013. The Wnt signaling pathway is involved in the regulation of phagocytosis of virus in Drosophila. Sci Rep 3:2069. doi: 10.1038/srep02069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Hobert ME, Rao AS, Neish AS, Madara JL. 2004. Bacterial activation of beta-catenin signaling in human epithelia. Am J Physiol Gastrointest Liver Physiol 287:G220–227. doi: 10.1152/ajpgi.00498.2003. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Lu R, Wu S, Sun J. 2010. Salmonella regulation of intestinal stem cells through the Wnt/beta-catenin pathway. FEBS Lett 584:911–916. doi: 10.1016/j.febslet.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu R, Liu X, Wu S, Xia Y, Zhang YG, Petrof EO, Claud EC, Sun J. 2012. Consistent activation of the beta-catenin pathway by Salmonella type-three secretion effector protein AvrA in chronically infected intestine. Am J Physiol Gastrointest Liver Physiol 303:G1113–1125. doi: 10.1152/ajpgi.00453.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tahoun A, Mahajan S, Paxton E, Malterer G, Donaldson DS, Wang D, Tan A, Gillespie TL, O'Shea M, Roe AJ, Shaw DJ, Gally DL, Lengeling A, Mabbott NA, Haas J, Mahajan A. 2012. Salmonella transforms follicle-associated epithelial cells into M cells to promote intestinal invasion. Cell Host Microbe 12:645–656. doi: 10.1016/j.chom.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Lu R, Wu S, Zhang YG, Xia Y, Liu X, Zheng Y, Chen H, Schaefer KL, Zhou Z, Bissonnette M, Li L, Sun J. 2014. Enteric bacterial protein AvrA promotes colonic tumorigenesis and activates colonic beta-catenin signaling pathway. Oncogenesis 3:e105. doi: 10.1038/oncsis.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blumenthal A, Ehlers S, Lauber J, Buer J, Lange C, Goldmann T, Heine H, Brandt E, Reiling N. 2006. The Wingless homolog WNT5A and its receptor Frizzled-5 regulate inflammatory responses of human mononuclear cells induced by microbial stimulation. Blood 108:965–973. doi: 10.1182/blood-2005-12-5046. [DOI] [PubMed] [Google Scholar]
- 39.George SJ. 2008. Wnt pathway: a new role in regulation of inflammation. Arterioscler Thromb Vasc Biol 28:400–402. doi: 10.1161/ATVBAHA.107.160952. [DOI] [PubMed] [Google Scholar]
- 40.Pereira C, Schaer DJ, Bachli EB, Kurrer MO, Schoedon G. 2008. Wnt5A/CaMKII signaling contributes to the inflammatory response of macrophages and is a target for the antiinflammatory action of activated protein C and interleukin-10. Arterioscler Thromb Vasc Biol 28:504–510. doi: 10.1161/ATVBAHA.107.157438. [DOI] [PubMed] [Google Scholar]
- 41.Kim J, Kim J, Kim DW, Ha Y, Ihm MH, Kim H, Song K, Lee I. 2010. Wnt5a induces endothelial inflammation via beta-catenin-independent signaling. J Immunol 185:1274–1282. doi: 10.4049/jimmunol.1000181. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q, Symes AJ, Kane CA, Freeman A, Nariculam J, Munson P, Thrasivoulou C, Masters JR, Ahmed A. 2010. A novel role for Wnt/Ca2+ signaling in actin cytoskeleton remodeling and cell motility in prostate cancer. PLoS One 5:e10456. doi: 10.1371/journal.pone.0010456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin M, Zhu MX, Rikihisa Y. 2002. Rapid activation of protein tyrosine kinase and phospholipase C-gamma2 and increase in cytosolic free calcium are required by Ehrlichia chaffeensis for internalization and growth in THP-1 cells. Infect Immun 70:889–898. doi: 10.1128/IAI.70.2.889-898.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin M, Rikihisa Y. 2003. Obligatory intracellular parasitism by Ehrlichia chaffeensis and Anaplasma phagocytophilum involves caveolae and glycosylphosphatidylinositol-anchored proteins. Cell Microbiol 5:809–820. doi: 10.1046/j.1462-5822.2003.00322.x. [DOI] [PubMed] [Google Scholar]
- 45.Popov VL, Yu X, Walker DH. 2000. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb Pathog 28:71–80. doi: 10.1006/mpat.1999.0327. [DOI] [PubMed] [Google Scholar]
- 46.Kumagai Y, Matsuo J, Hayakawa Y, Rikihisa Y. 2010. Cyclic di-GMP signaling regulates invasion by Ehrlichia chaffeensis of human monocytes. J Bacteriol 192:4122–4133. doi: 10.1128/JB.00132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuriakose JA, Miyashiro S, Luo T, Zhu B, McBride JW. 2011. Ehrlichia chaffeensis transcriptome in mammalian and arthropod hosts reveals differential gene expression and post transcriptional regulation. PLoS One 6:e24136. doi: 10.1371/journal.pone.0024136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin DT, Wu W, Li M, Wang QE, Li H, Wang Y, Tang Y, Xing M. 2013. DKK3 is a potential tumor suppressor gene in papillary thyroid carcinoma. Endocr Relat Cancer 20:507–514. doi: 10.1530/ERC-13-0053. [DOI] [PubMed] [Google Scholar]
- 49.Vasileiou G, Ekici AB, Uebe S, Zweier C, Hoyer J, Engels H, Behrens J, Reis A, Hadjihannas MV. 2015. Chromatin-remodeling-factor ARID1B represses Wnt/beta-catenin signaling. Am J Hum Genet 97:445–456. doi: 10.1016/j.ajhg.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai SL, Chien AJ, Moon RT. 2009. Wnt/Fz signaling and the cytoskeleton: potential roles in tumorigenesis. Cell Res 19:532–545. doi: 10.1038/cr.2009.41. [DOI] [PubMed] [Google Scholar]
- 51.Zhang JZ, Popov VL, Gao S, Walker DH, Yu XJ. 2007. The developmental cycle of Ehrlichia chaffeensis in vertebrate cells. Cell Microbiol 9:610–618. doi: 10.1111/j.1462-5822.2006.00812.x. [DOI] [PubMed] [Google Scholar]
- 52.Fazzi R, Pacini S, Carnicelli V, Trombi L, Montali M, Lazzarini E, Petrini M. 2011. Mesodermal progenitor cells (MPCs) differentiate into mesenchymal stromal cells (MSCs) by activation of Wnt5/calmodulin signalling pathway. PLoS One 6:e25600. doi: 10.1371/journal.pone.0025600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeshita A, Iwai S, Morita Y, Niki-Yonekawa A, Hamada M, Yura Y. 2014. Wnt5b promotes the cell motility essential for metastasis of oral squamous cell carcinoma through active Cdc42 and RhoA. Int J Oncol 44:59–68. doi: 10.3892/ijo.2013.2172. [DOI] [PubMed] [Google Scholar]
- 54.Kim MK, Maeng YI, Sung WJ, Oh HK, Park JB, Yoon GS, Cho CH, Park KK. 2013. The differential expression of TGF-beta1, ILK and wnt signaling inducing epithelial to mesenchymal transition in human renal fibrogenesis: an immunohistochemical study. Int J Clin Exp Pathol 6:1747–1758. [PMC free article] [PubMed] [Google Scholar]
- 55.He X, Saint-Jeannet JP, Wang Y, Nathans J, Dawid I, Varmus H. 1997. A member of the Frizzled protein family mediating axis induction by Wnt-5A. Science 275:1652–1654. doi: 10.1126/science.275.5306.1652. [DOI] [PubMed] [Google Scholar]
- 56.Bhatt PM, Malgor R. 2014. Wnt5a: a player in the pathogenesis of atherosclerosis and other inflammatory disorders. Atherosclerosis 237:155–162. doi: 10.1016/j.atherosclerosis.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mikels AJ, Nusse R. 2006. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol 4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J, Chang W, Jung Y, Song K, Lee I. 2012. Wnt5a activates THP-1 monocytic cells via a beta-catenin-independent pathway involving JNK and NF-kappaB activation. Cytokine 60:242–248. doi: 10.1016/j.cyto.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 59.Schaale K, Neumann J, Schneider D, Ehlers S, Reiling N. 2011. Wnt signaling in macrophages: augmenting and inhibiting mycobacteria-induced inflammatory responses. Eur J Cell Biol 90:553–559. doi: 10.1016/j.ejcb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 60.Patel M, Morrow J, Maxfield FR, Strickland DK, Greenberg S, Tabas I. 2003. The cytoplasmic domain of the low density lipoprotein (LDL) receptor-related protein, but not that of the LDL receptor, triggers phagocytosis. J Biol Chem 278:44799–44807. doi: 10.1074/jbc.M308982200. [DOI] [PubMed] [Google Scholar]
- 61.Yamamoto H, Komekado H, Kikuchi A. 2006. Caveolin is necessary for Wnt-3a-dependent internalization of LRP6 and accumulation of beta-catenin. Dev Cell 11:213–223. doi: 10.1016/j.devcel.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 62.Follo MY, Manzoli L, Poli A, McCubrey JA, Cocco L. 2015. PLC and PI3K/Akt/mTOR signalling in disease and cancer. Adv Biol Regul 57:10–16. doi: 10.1016/j.jbior.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 63.Forsberg M, Blomgran R, Lerm M, Sarndahl E, Sebti SM, Hamilton A, Stendahl O, Zheng L. 2003. Differential effects of invasion by and phagocytosis of Salmonella typhimurium on apoptosis in human macrophages: potential role of Rho-GTPases and Akt. J Leukoc Biol 74:620–629. doi: 10.1189/jlb.1202586. [DOI] [PubMed] [Google Scholar]
- 64.Jiang W, Wang J, Zhang Y. 2013. Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res 23:122–130. doi: 10.1038/cr.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pereira F, Barbachano A, Silva J, Bonilla F, Campbell MJ, Munoz A, Larriba MJ. 2011. KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells. Hum Mol Genet 20:4655–4665. doi: 10.1093/hmg/ddr399. [DOI] [PubMed] [Google Scholar]
- 66.Lorenowicz MJ, Macurkova M, Harterink M, Middelkoop TC, de Groot R, Betist MC, Korswagen HC. 2014. Inhibition of late endosomal maturation restores Wnt secretion in Caenorhabditis elegans vps-29 retromer mutants. Cell Signal 26:19–31. doi: 10.1016/j.cellsig.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 67.Thorne CA, Hanson AJ, Schneider J, Tahinci E, Orton D, Cselenyi CS, Jernigan KK, Meyers KC, Hang BI, Waterson AG, Kim K, Melancon B, Ghidu VP, Sulikowski GA, LaFleur B, Salic A, Lee LA, Miller DM III, Lee E. 2010. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1alpha. Nat Chem Biol 6:829–836. doi: 10.1038/nchembio.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Venerando A, Girardi C, Ruzzene M, Pinna LA. 2013. Pyrvinium pamoate does not activate protein kinase CK1, but promotes Akt/PKB down-regulation and GSK3 activation. Biochem J 452:131–137. doi: 10.1042/BJ20121140. [DOI] [PubMed] [Google Scholar]
- 69.Sumi M, Kiuchi K, Ishikawa T, Ishii A, Hagiwara M, Nagatsu T, Hidaka H. 1991. The newly synthesized selective Ca2+/calmodulin dependent protein kinase II inhibitor KN-93 reduces dopamine contents in PC12h cells. Biochem Biophys Res Commun 181:968–975. doi: 10.1016/0006-291X(91)92031-E. [DOI] [PubMed] [Google Scholar]
- 70.Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, Roth MG, Amatruda JF, Chen C, Lum L. 2009. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol 5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin HL, Janmey PA. 2003. Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol 65:761–789. doi: 10.1146/annurev.physiol.65.092101.142517. [DOI] [PubMed] [Google Scholar]
- 72.Lafont F, van der Goot FG. 2005. Bacterial invasion via lipid rafts. Cell Microbiol 7:613–620. doi: 10.1111/j.1462-5822.2005.00515.x. [DOI] [PubMed] [Google Scholar]
- 73.Seveau S, Tham TN, Payrastre B, Hoppe AD, Swanson JA, Cossart P. 2007. A FRET analysis to unravel the role of cholesterol in Rac1 and PI 3-kinase activation in the InlB/Met signalling pathway. Cell Microbiol 9:790–803. doi: 10.1111/j.1462-5822.2006.00832.x. [DOI] [PubMed] [Google Scholar]
- 74.Mohan Kumar D, Yamaguchi M, Miura K, Lin M, Los M, Coy JF, Rikihisa Y. 2013. Ehrlichia chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X and trigger entry into mammalian cells. PLoS Pathog 9:e1003666. doi: 10.1371/journal.ppat.1003666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mohan Kumar D, Lin M, Xiong Q, Webber MJ, Kural C, Rikihisa Y. 2015. EtpE binding to DNase X induces ehrlichial entry via CD147 and hnRNP-K recruitment, followed by mobilization of N-WASP and actin. mBio 6:e01541. doi: 10.1128/mBio.01541-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.