Abstract
Chlamydia spp. are ubiquitous, obligate, intracellular Gram-negative bacterial pathogens that undergo a unique biphasic developmental cycle transitioning between the infectious, extracellular elementary body and the replicative, intracellular reticulate body. The primary Chlamydia species associated with human disease are C. trachomatis, which is the leading cause of both reportable bacterial sexually transmitted infections and preventable blindness, and C. pneumoniae, which infects the respiratory tract and is associated with cardiovascular disease. Collectively, these pathogens are a significant source of morbidity and pose a substantial financial burden on the global economy. Past efforts to elucidate virulence mechanisms of these unique and important pathogens were largely hindered by an absence of genetic methods. Watershed studies in 2011 and 2012 demonstrated that forward and reverse genetic approaches were feasible with Chlamydia and that shuttle vectors could be selected and maintained within the bacterium. While these breakthroughs have led to a steady expansion of the chlamydial genetic tool kit, there are still roads left to be traveled. This minireview provides a synopsis of the currently available genetic methods for Chlamydia along with a comparison to the methods used in other obligate intracellular bacteria. Limitations and advantages of these techniques will be discussed with an eye toward the methods still needed, and how the current state of the art for genetics in obligate intracellular bacteria could direct future technological advances for Chlamydia.
INTRODUCTION
Chlamydia spp. are ubiquitous bacterial pathogens that infect a wide variety of animals, including a number of economically important species, as well as humans (1). The Chlamydia spp. of primary concern to human health are C. trachomatis and C. pneumoniae (C. psittaci and C. abortus are serious, but rare, zoonotic pathogens). C. trachomatis serovars D to K and L1 to L3 are the leading cause of reportable, bacterial sexually transmitted infections (STIs) both in the United States and worldwide (2, 3). The number of cases in the United States exceeds one million per year and is likely two- to threefold higher due to asymptomatic infections, particularly in women. Chlamydial STIs in women are highly significant, as infection can lead to pelvic inflammatory disease, life-threatening ectopic pregnancy, or infertility (4). C. trachomatis serovars A to C are responsible for the ocular infection trachoma, which is a leading cause of preventable blindness in underdeveloped regions of the world (5). While C. pneumoniae is primarily a respiratory pathogen that causes pneumonia, infections are associated with an increased risk for the development of cardiovascular disease which remains the leading cause of mortality in the United States (6). Collectively, these pathogens represent a substantial burden on the world economy and human health.
One of the major impediments confronting chlamydial researchers has been a paucity of genetic methods for the creation of mutant strains necessary for the thorough dissection of chlamydial biology in relation to pathogenesis. This limitation has hindered the development of vaccines and therapeutics. The failure to successfully adapt genetic tools widely used in other bacteria for Chlamydia can be largely attributed to its obligate, intracellular lifestyle and unique developmental cycle (7, 8). These bacteria transition between an extracellular, infectious form known as the elementary body (EB) and an intracellular, replicative form termed the reticulate body (RB). Infection is initiated upon binding of the EB to the surface of a cell followed by internalization where the EB resides in a host-derived membrane vesicle termed the inclusion. Within the inclusion, the EB differentiates into the RB, which replicates for approximately 20 h before the population begins an asynchronous transition back into the EB form. After 48 to 72 h, the EBs escape from the cell via cell lysis or a process known as inclusion extrusion (9). During differentiation and growth, gene expression and protein profiles proceed through virus-like early, middle, and late phases of expression. The overall timing of differentiation and cell exit varies between species and is further affected by the cell line used for growth (10–13).
As an obligate, intracellular bacterium, Chlamydia spp. possess reduced genomes ranging from ∼1 to 1.5 Mbp that show little evidence of recent acquisitions of foreign DNA including the lack of restriction modification systems, the presence of few transposons and phages or their remnants, and only a single known horizontally acquired resistance marker (14). The only identified drug resistance marker is tetC, which encodes a tetracycline efflux pump and is found in C. suis (15). Analysis of chlamydial genomes also reveals the presence of Chlamydia-conserved histone-like proteins known to complex with the chromosomal DNA in EBs, an absence of DNA methyltransferases including DNA adenosine methyltransferase (DAM) and DNA cytosine methyltransferase (DCM), and a cryptic plasmid present in most, but not all, species (14, 16, 17). Collectively, the intracellular lifestyle and physiology of Chlamydia present a myriad of road blocks to the development of genetic systems. These road blocks include the following: (i) resolving which bacterial form would be amenable to manipulation, as the EB is relatively metabolically inert and has a highly cross-linked outer membrane presenting a possible barrier to DNA entry, while the RB form is shielded by the host cell, inclusion, and bacterial membranes; (ii) discerning when to apply antibiotic selection in regard to the length of time required for phenotypic conversion after introduction of resistance markers; (iii) determining which antibiotics would provide robust selection, while being cell permeable, safe for the host cell, and not restricted due to clinical usage; (iv) assessing whether Chlamydia would be able to recognize foreign promoters and express foreign genes; and (v) assessing whether plasmid-free species could maintain extrachromosomal DNA. Consequently, the development of genetic methods for Chlamydia has lagged well behind other bacteria though certainly not for a lack of effort. Multiple methods for introduction of chlamydial DNA and cryptic-plasmid-based shuttle vectors were attempted including transfection of infected HeLa cells, conjugal mating with Escherichia coli, calcium phosphate and natural transformation, electroporation, microinjection of RB-stage inclusions, and delivery via a ballistic gun (unpublished).
THE EARLY YEARS
Introduction of foreign DNA into bacteria by horizontal gene transfer occurs via three mechanisms: (i) transduction, in which DNA is delivered by a bacteriophage, (ii) conjugation, in which DNA is directly transferred between bacteria via a conjugal mating bridge, and (iii) transformation, whereby “naked” DNA from the environment is taken up by bacteria. Transformation can occur naturally if the bacteria are naturally competent or artificially via chemical transformation or electroporation. For a review on horizontal gene transfer, readers are referred to Soucy et al. (18).
Genetic work with Chlamydia was initiated by various research groups that characterized the cryptic plasmids found in C. trachomatis and C. psittaci in the mid-1980s (19–22). Variation between the C. trachomatis and C. psittaci cryptic plasmids was identified using hybridization and DNA restriction digestion analysis. These studies also identified the need for an Escherichia coli-Chlamydia shuttle vector, as the cryptic plasmid could not be maintained in E. coli. Full nucleotide sequences of the cryptic plasmids from C. trachomatis serovar E and C. trachomatis serovar L1 determined via Sanger sequencing were reported in 1987 and 1988, respectively (Table 1 summarizes major developments in chlamydial genetics) (21, 22). Such studies typically lay the foundation necessary for the development of shuttle vectors through identification of plasmid replication and maintenance mechanisms. However, due to lack of homology with previously sequenced plasmids, little information on cryptic plasmid replication and maintenance were gained from sequence analysis. More-recent studies, highlighted below, have determined that the cryptic plasmid carries eight open reading frames (ORFs), of which Pgp1 (putative helicase), Pgp2, Pgp6, and Pgp8 (putative integrase/recombinase, only the coding sequence is required) are essential for plasmid maintenance, while Pgp4, which regulates expression of pgp3 and chromosomal genes including glgA, along with Pgp3, Pgp5, and Pgp7 are dispensable (23, 24).
TABLE 1.
Summary of genetic advances for Chlamydia
| Genetic advance and species and serovar or strain(s) | Methoda | Comment(s) | Reference |
|---|---|---|---|
| Lateral gene transfer | |||
| C. trachomatis L1 | Homologous recombination, lateral gene transfer | Lateral gene transfer and homologous recombination between the same strain | 37 |
| C. trachomatis, C. muridarum, or C. suis | Homologous recombination, lateral gene transfer | Transfer of the tet(C) allele between species | 39 |
| Transformation | |||
| C. trachomatis E | Shuttle vector, electroporation, EB | First transformation of Chlamydia; plasmid was unstable | 25 |
| C. psittaci 6BC | Homologous recombination, suicide vector, electroporation, EB | Demonstrated feasibility of artificial transformation and homologous recombination | 43 |
| C. trachomatis L2 and L2(25667R), a plasmid-free strain | Shuttle vector, chemical transformation, EB | First demonstration of stable plasmid maintenance; use of bla in C. trachomatis serovars D to K is restricted by NIH regulations | 26 |
| C. trachomatis L2 | Shuttle vectors, chemical transformation, EB | MCSb with and without incDEFG promoter and terminator; fluorescent protein expression | 48 |
| Shuttle vector, chemical transformation, EB | Tet-inducible promoter | 53 | |
| C. trachomatis L2(25667R), a plasmid-free strain | Shuttle vector, chemical transformation, EB | In vitro deletion analysis of all plasmid ORFs; pgp1, pgp2, pgp6, and pgp8 are essential for maintenance | 24 |
| C. trachomatis L2 | Group II intron (targeted insertions [TargeTron]), suicide vector, chemical transformation, EB | Development of a targeted gene insertion method for reverse genetic approaches | 75 |
| Chemical mutagenesis | |||
| C. trachomatis D | Targeting-induced local lesions in genome (TILLING), chemical-based (EMS) mutagenesis, infected cells (RB stage) | Reverse genetic approach; CELI digestion of PCR amplicons used to identify mutant for phenotype analysis | 28 |
| C. trachomatis L2 | Chemical-based (EMS) mutagenesis, followed by lateral gene transfer and homologous recombination, infected cells (RB stage) | Forward genetic approach; high-content chemical mutagenesis and plaque phenotype screening, followed by genome sequencing and genotype/phenotype linkage via lateral gene transfer and homologous recombination | 69 |
| EMS mutagenesis; lateral gene transfer and homologous recombination; shuttle vector (chemical transformation, EB) | Complementation of tepP mutant using a shuttle vector | 56 | |
| Chemical-based (EMS and ENU) mutagenesis approach for forward and reverse genetics | Mutant library construction coupled with full library sequencing; library can be searched for desired gene mutations (reverse genetics) or phenotype screening (forward genetics) | 70 | |
| Dendrimer-enabled transformation | |||
| C. trachomatis K | Gene knockdown via antisense oligonucleotides, dendrimer-enabled transformation, infected cells (RB stage) | Targeted gene knockdown in C. trachomatis using antisense oligomers | 79 |
| Other | |||
| C. trachomatis L2 | Axenic medium | Effort to develop axenic medium; seminal event in Coxiella genetics | 84 |
The method of genetic manipulation and the form of the bacterium (EB or RB) used with the method.
MCS, multiple-cloning site.
For Chlamydia, the first publication reporting the creation of a shuttle vector would come in 1994 (25). The chloramphenicol-selectable E. coli-C. trachomatis shuttle vector used by Tam et al. (25) had a C. trachomatis serovar E cryptic plasmid backbone and carried the cat resistance gene. C. trachomatis serovar E transformants created through electroporation of the EB form with the shuttle vector possessed chloramphenicol acetyltransferase activity and were resistant to low levels of chloramphenicol (5 μg/ml). However, the plasmid was transient and was rarely maintained beyond four passages in cell culture. Recent data provide some insight into this failed attempt, as the recombinant plasmid lacked a 500-bp region within pgp1, which we now know to be essential for plasmid maintenance. Had serendipity interceded in choosing a different region for insertion of the E. coli vector into the cryptic plasmid, stable transformation may have been achieved much earlier. It would take another 17 years for a stable shuttle vector system to be developed for Chlamydia (26). In the interim, the small genome size of Chlamydia coupled with rapid developments in sequencing technology would set the stage for the genomics era.
ADOLESCENCE: THE GENOMICS ERA
The genome sequence of a C. trachomatis serovar D strain was published in 1998 and would prove to be the first of many (17). As sequencing technology rapidly improved, a field starved for genetics took advantage of the lone available tool to gain insight into the DNA sequences underpinning chlamydial pathogenesis. Currently, more than 150 genome sequences are available for Chlamydia covering all known species and including numerous clinical C. trachomatis strains. These efforts revealed that while Chlamydia spp. are closely related and exhibit strong genome synteny, key differences were present, and these differences were speculatively linked to tissue and host tropisms displayed both across and within species (14). In particular, researchers sought to explain differences in strain virulence through sequence variations affecting metabolic pathways, putative virulence factors, and overall coding capacity. As a prime example, defects in tryptophan synthesis between C. trachomatis ocular and genital serovars should enable genital serovars to synthesize tryptophan from indole (27). This would allow for rescue of growth in the face of gamma interferon (IFN-γ)-stimulated depletion of tryptophan (if indole is provided) for genital serovars, but not ocular serovars. The necessity of the trpBA genes for survival of genital serovars in the presence of IFN-γ would eventually be demonstrated by Kari et al. using a chemical mutagenesis-based reverse genetic approach (28).
A second major outcome of the genomics era was a more thorough appreciation for the potential virulence gene repertoire of Chlamydia including the type III secretion system (previously reported, but not completely mapped [29]), the presence of a family of polymorphic membrane proteins (Pmps), various inclusion membrane proteins (Incs), proteases, etc. The massive sequencing effort of numerous research groups had presented the field with a slew of potential virulence factors and new hypotheses to test, but no methods to perform targeted mutagenesis experiments that could readily be performed in other pathogens. In comparison, by the late 1990s and early 2000s, gene inactivation by homologous recombination and transposon mutagenesis was being undertaken in other obligate intracellular bacteria (30–34). Table 2 summarizes the genetic tools available for other obligate intracellular bacteria.
TABLE 2.
Genetic tools available for other obligate intracellular bacteria
| Organism(s) | Methoda | Reference(s) |
|---|---|---|
| Anaplasma phagocytophilum and A. marginale | Transposon mutagenesis (Himar1); electroporation | 86–88 |
| Coxiella burnetii | Shuttle vector; homologous recombination (single crossover) and episomal maintenance; Himar1 transposon mutagenesis; electroporation (preaxenic medium) | 89, 90 |
| Axenic medium | 83, 85 | |
| Shuttle vector (IncQ broad-host-range plasmid); Himar1 and Tn7 transposon mutagenesis; homologous recombination (sacB counterselection and Cre-lox-mediated genome editing); electroporation | 91–96 | |
| Ehrlichia chaffeensis | Group II intron (TargeTron) and transposon mutagenesis (Himar1); electroporation | 77 |
| Rickettsia prowazekii, R. typhi, R. conorii, R. monacensis, R. rickettsii, R. bellii | Homologous recombination (single and double crossover); EZ::TN and Himar1 transposon mutagenesis; shuttle vectors; group II intron (TargeTron); peptide nucleic acids (antisense; translation inhibition); electroporation | 30–34, 78, 97–105 |
Methods have not been validated for all species listed in each category.
Analysis of individual genes, particularly ompA and the pmp family members, had hinted that DNA exchange and recombination occurred within the C. trachomatis serovars, but direct experimental evidence was lacking (35, 36). In 2007, Demars et al. (37) asked a straightforward question—was C. trachomatis capable of lateral gene transfer and homologous recombination? Using marked strains of C. trachomatis L1 carrying spontaneous mutations conferring resistance to ofloxacin, trimethoprim, rifampin, or lincomycin, Demars et al. coinfected cells with strains carrying single markers and then selected for both markers. Dual-resistant strains emerged at rates higher than expected for spontaneous mutants and carried the parental mutations, consistent with recombination. This report was followed shortly by a series of studies using similar coinfection strategies to demonstrate that lateral gene transfer and homologous recombination could occur not only between serovars but also between species (38–40). Of particular note, Suchland et al. found that the tet(C) allele from C. suis could be transferred among C. suis, C. trachomatis, and C. muridarum, indicating that horizontal gene transfer and recombination could lead to the spread of resistance (39). These later studies were particularly convincing, as recombinant strains were mapped via whole-genome sequencing, which has also been used to support recombination between clinical strains in vivo (41).
The recognition that lateral gene transfer and homologous recombination could be performed in cell culture and that recombinants could be selected using resistance markers derived from spontaneous mutations would prove critical for developing chemical mutagenesis-based forward and reverse genetic approaches. It still remains unclear how DNA is transferred between strains, although natural transformation is the best supported mechanism. Chlamydia spp. lack genes for conjugation, and the currently known chlamydiaphages all have genomes of less than 4.8 kbp, making them too small to account for the documented transfer of DNA segments exceeding 100 kbp (38, 42). Of relevance for future genetic methods, the small genome size of the chlamydiaphage also makes them unlikely to be useful for transduction approaches (for a review of chlamydiaphage biology, see reference 42).
THE AGE OF DIRECTED MUTAGENESIS
In 2009, Binet and Maurelli reported the first example of targeted mutagenesis in Chlamydia providing proof of principle that Chlamydia could be artificially transformed with foreign DNA and that integration of DNA into the chromosome could be selected (43). Using a suicide plasmid, a 16S rRNA allele carrying mutations introduced by site-directed mutagenesis to impart resistance to kasugamycin and spectinomycin was introduced into C. psittaci via electroporation, and double-resistant mutant strains were obtained. An important feature of the Binet and Maurelli study (43) was the ability to select for recombinant bacteria immediately after transformation via plaque assay, which permitted method optimization. Variables assessed included DNA methylation status, concentration of DNA, number of bacteria, and DNA form (circular or linear). For example, unmethylated plasmid DNA isolated from an hsdS-, dam-, and dcm-deficient E. coli strain was found to be the most efficient for obtaining recombinants. Most current methods for transformation of shuttle vectors continue to use unmethylated DNA, although the efficiency of methylated versus unmethylated DNA for plasmid maintenance has not been reported. This is partly due to the inability to immediately plaque plasmid transformants using the current selection markers, preventing calculation of transformation efficiency. As the Binet and Maurelli study selected for recombinants and not transformants, transformation frequency could not be directly assessed. Development of selection protocols allowing for first passage mutant isolation such as plaque assay or flow cytometry (43, 44) will facilitate expansion of genetic methods by allowing for optimization approaches as exemplified by Binet and Maurelli (43). For example, when transposon mutagenesis methods are established, saturation of the genome will require either efficient transformation or repeated transformations with poorly efficient methods.
SHUTTLE VECTORS
After the breakthrough report of Binet and Maurelli (43), another 3 years would pass before the landmark study by Wang et al. detailing the development of a stable E. coli-C. trachomatis shuttle vector (26). Two different shuttle vectors were created: pBR325::L2 based on the C. trachomatis L2 cryptic plasmid (45) and pGFP::SW2 based on the cryptic plasmid from a Swedish clinical C. trachomatis E isolate carrying a deletion in CDS1 (pgp7) and a 44-bp duplication in CDS3 (pgp1) (46). These vectors were used to transform cryptic-plasmid-positive and cryptic-plasmid-negative C. trachomatis L2 strains. Significant results from this study include the following. (i) A CaCl2 chemical transformation method for EBs was described, and this method serves as the basis for the majority of subsequently published transformation protocols. (ii) It was demonstrated that penicillin (via the bla resistance marker) can be used to select for transformants. (iii) Both plasmid-negative and plasmid-positive C. trachomatis isolates can be transformed. (iv) Selection for the shuttle vector will cure the strain of its native cryptic plasmid. (v) Foreign promoters and genes (cat, bla, and gfp) can be expressed in C. trachomatis. It should be noted that while chloramphenicol resistance was demonstrated with the transformants (the pGFP::SW2 vector carries both bla and Φcat-gfp), penicillin was used for initial selection. Xu et al. later demonstrated that chloramphenicol could be used for initial selection using a modified pGFP::SW2 vector lacking the bla gene (47). This finding is relevant, as the use of bla-mediated resistance is not permitted in C. trachomatis serovars D to K in the United States without special approval from the National Institutes of Health.
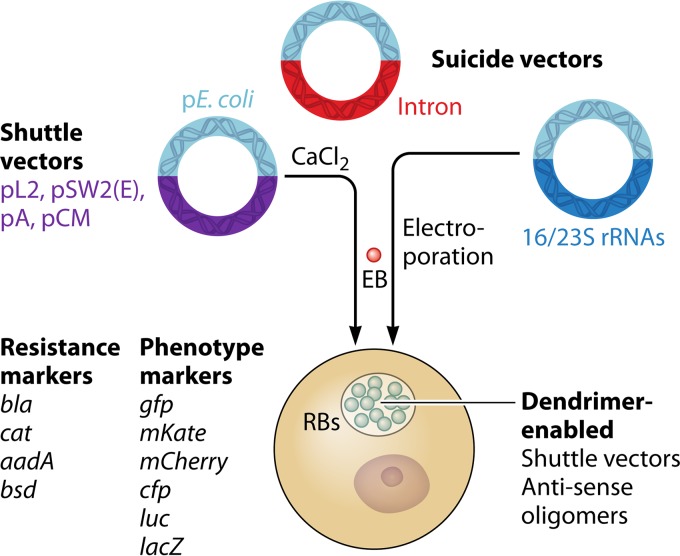
Following the seminal report from Wang et al. (26), numerous reports of modified shuttle-vector systems began to appear in the literature. These vectors used either the pSW2 or pL2 cryptic plasmid backbone reported by Wang et al. (26) and increased the utility of the shuttle vector through the following: (i) development of an inducible TetR-based promoter system; (ii) introduction of multiple cloning sites with and without promoters to allow for constitutive expression or regulation by native promoters; (iii) expression of a variety of fluorescent proteins including green fluorescent protein (GFP), mCherry, cyan fluorescent protein (CFP), and mKate to facilitate live cell imaging and easier identification of transformants; and (iv) adaptation of systems for measuring protein translocation (23, 48–54). In addition, bsd (conferring resistance to blasticidin) was validated providing a third selection marker (55). These tools have enabled previously impossible research such as complementation of chromosomal mutants and analysis of type III effectors using C. trachomatis rather than surrogate bacterial systems (49, 51, 56). The ability to freely manipulate the cryptic plasmid backbone in shuttle vectors also led to a more thorough understanding of cryptic plasmid genes required for plasmid maintenance as well as virulence (24, 57–59). Vector platforms and transformation approaches are summarized in Fig. 1.
FIG 1.
Summary of transformation methods available for Chlamydia. The vector backbones drawn in light blue represent regions based on E. coli plasmids (pE. coli). The suicide vectors are used for insertional mutagenesis (intron) or allelic exchange (16/23S rRNA vector). Available markers for selection of transformants and markers that provide a trackable phenotype are listed to the left of the cell diagram.
As the shuttle vector repertoire expanded, it became apparent that the pSW2- and pL2-based plasmids were not compatible with all Chlamydia spp. Work by Song et al. demonstrated that C. trachomatis serovar A, C. muridarum, and C. trachomatis serovar L2 could be transformed only with shuttle vectors possessing their own cryptic plasmid as a backbone (60). Subsequently, Wang et al. determined that the plasmid tropism between C. muridarum and C. trachomatis is due, in part, to CDS2 (pgp8) (61). These results indicate that species-specific shuttle vectors may be important for expanding genetic methods into other C. trachomatis serovars and Chlamydia spp. (58). However, some caveats to plasmid tropism exist. pGFP::SW2, a serovar E cryptic-plasmid-based shuttle vector, is stable in C. trachomatis serovars L2, D, and F and a C. trachomatis L2 cryptic-plasmid-based shuttle vector was reported to be stable in C. pneumoniae (26, 50, 52, 55). The latter study used a dendrimer-DNA complex to facilitate selection-free transformation (50). These varied reports make it clear that while our understanding of chlamydial plasmid biology has greatly improved in the age of shuttle vectors, much remains unknown. Consequently, it is crucial that experiments performed with recombinant plasmid strains be matched with strains carrying the empty vector rather than matched to strains lacking a plasmid or carrying the native cryptic plasmid.
While the robust chemical transformation protocol pioneered by Wang et al. (26) and the rapidly expanding shuttle vector repertoire allows the field to push the boundaries of chlamydial genetics, some limitations remain. The identification of multiple resistance markers will be essential for allowing gene complementation along with construction of strains carrying multiple mutations. As these new resistance markers are assessed, it is necessary to be mindful of spontaneous mutation rates that lead to resistance as well as the differential susceptibility of species to antibiotics. These problems are highlighted by failed attempts to utilize the rifampin resistance marker arr-2 for selection in C. trachomatis L2 and the natural resistance of C. muridarum to ofloxacin (62, 63). In addition, the mechanism of drug action may also impact the efficacy of selection. While the bla marker is prohibited for use in certain serovars, it has proven robust and is the marker of choice for most researchers. The robustness of bla-mediated resistance may be due, in part, to the fact that β-lactams appear to be bacteriostatic for Chlamydia inducing a persistent growth state rather than death (64). This may allow for increased time for phenotypic conversion, resulting in an increased rate of transformant recovery. It will also be important to further flesh out how protocols and vectors affect transformation efficiencies, particularly when attempting to maximize transformation efficiencies for random insertional mutagenesis methods such as transposon mutagenesis. The easiest method to measure transformation efficiencies is through plaque assay (65). In this assay, a confluent host cell monolayer is infected with EBs, and then an agarose overlay is added to the infected cells. Upon cell lysis and release of EBs, only cells adjacent to the infected cells can be infected due to the agarose overlay. Repeated rounds of development lead to a localized clearing of the monolayer referred to as a plaque. Plaque assay is also a useful method for obtaining clonal strains, although fluorescence-activated cell sorting methods appear promising for recombinant strains expressing fluorescent proteins (44). Drugs such as blasticidin and zeocin, which are toxic to both host cells and Chlamydia, impede the use of the plaque assay (55). In addition, chloramphenicol at high doses or prolonged treatment periods can also be toxic for the host cell (66).
TARGETING THE CHROMOSOME
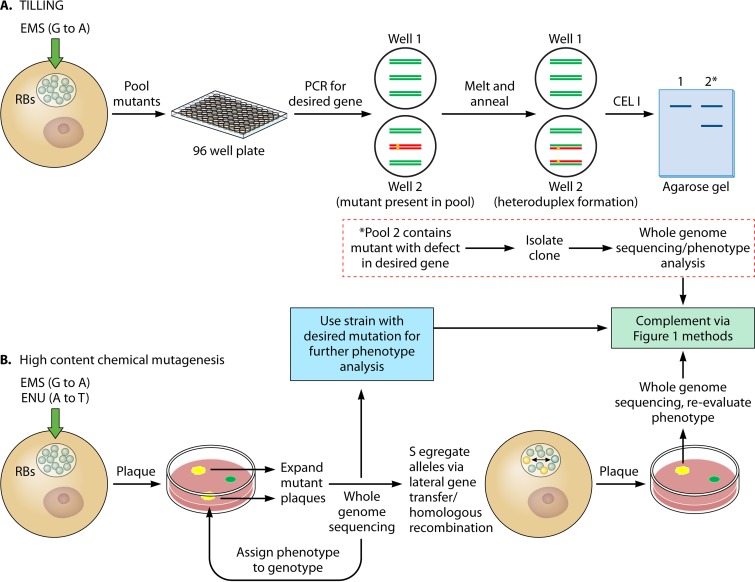
Accompanying the description of a shuttle vector for C. trachomatis by Wang et al. (26) were two studies reporting the use of ethyl methanesulfonate (EMS) (an alkylating agent that causes G-to-A transition mutations) for chemical mutagenesis of C. trachomatis. In the first study, Kari et al. used low levels of EMS targeting a mutation rate of one base per genome to generate a library of C. trachomatis mutants (28). The library strains were then pooled and assessed for the desired mutation using PCR, followed by CELI digestion. This method is known as targeting-induced local lesions in genome, or TILLING (Fig. 2A). To detect mutants, PCR amplicons from each pool are heat denatured and then allowed to anneal. If a pool contains wild-type and mutant strains, a heteroduplex will form due to a base mismatch, which allows for cleavage by CELI and detection of the cleaved product by DNA gel electrophoresis. The desired clones can then be obtained from the mutant library by PCR screening of clones obtained through limiting dilution or plaque assays followed by sequencing to map the exact mutation. Mutant phenotypes can then be assessed to determine gene function. Once the EMS mutant library pools are established, one can search for all desired mutants within the pool. Limitations of this method include extensive hands-on work to both create the library and identify mutants along with back-end whole-genome sequencing to confirm the clonal isolate contains a mutation only in the gene of interest. This reverse genetic approach has been used to study the function of trpB and pmpD in C. trachomatis and has been adapted for use with C. muridarum (28, 67, 68).
FIG 2.
Chemical mutagenesis methods used for TILLING (A) or forward genetic approaches (B) are shown. The green and red lines in the wells in the TILLING section represent wild-type and mutant amplicons, respectively (see the text). Both methods require whole-genome sequencing to map mutations and may require lateral gene transfer and homologous recombination between the mutant (yellow) and parental (green) strains to isolate mutant alleles. Chemical mutants (and intron-generated mutants) can be complemented using shuttle vectors (Fig. 1).
EMS has also proven to be a useful tool for forward genetics in C. trachomatis. Using increased amounts of EMS compared to those used by Kari et al. (28) to generate 3 to 20 mutations per genome, Nguyen and Valdivia (69) prepared a library of mutants that were then assessed for growth defects via automated analysis of plaque morphology (Fig. 2B). Strains giving rise to altered plaques were isolated, expanded in cell culture, and genotyped using whole-genome sequencing. As strains contained multiple mutations, lateral gene transfer and homologous recombination as initially described by Demars et al. (37) was then used to link gene lesions with specific phenotypes. This high-content mutagenesis approach has been further expanded by Kokes et al. (70) using either EMS or N-ethyl-N-nitrosourea (ENU) (alkylating agent leading to A-to-T transversion mutation) to mutate C. trachomatis, creating a collection of 934 mutant strains carrying 6 to 25 mutations. Strains were screened via plaque morphology, and whole-genome sequencing was used to map all mutated alleles within the pools of mutant strains. The availability of this sequence data allows mutants of interest to be selected and assessed for their phenotype following segregation of the allele using lateral gene transfer and homologous recombination.
Chemical mutagenesis is clearly a powerful tool for discerning the roles of genes in the biology of Chlamydia. A wide range of mutants have been obtained using these approaches including numerous strains with nonsense mutations that will be particularly useful for studying the impact of gene loss on strain growth in cell culture and animal models. It is important to note that nonsense mutations are less susceptible to polar effects observed with random insertional approaches such as transposon mutagenesis. Chemical mutants can be complemented using the various existing shuttle vectors, and these approaches should be readily adaptable to other Chlamydia spp. The primary weaknesses lie in the cost and effort associated with mutant generation and genotype analysis and the need to perform lateral gene transfer to isolate specific mutant alleles when using large amounts of EMS or ENU. In addition, isolation of chemically generated mutants via plaque assay selects against mutants that have lost the ability to form plaques, and screening for plaque morphology variants may miss strains that are competent for growth in cell culture but would be attenuated in an animal model. The latter limitation would be difficult to overcome with a chemical mutagenesis approach, but it could be dealt with using transposon mutagenesis approaches such as signature-tagged mutagenesis to screen mutant pools for genes required for in vitro growth versus in vivo growth (71, 72).
Currently, the only method available for generating targeted chromosomal mutations in C. trachomatis utilizes a mobile group II intron, Ll.LtrB, originally isolated from Lactococcus lactis and marketed as TargeTron by Sigma-Aldrich and by TargeTronics, LLC (73). Targeting the intron to a gene of interest is performed using PCR to mutate bases within the 5′ region of the intron to allow for hybridization with the target gene. The required mutations are identified using a proprietary algorithm. With the assistance of an intron-encoded protein, LtrA (expressed independently from the intron), the intron is spliced into the target gene creating an insertional gene mutant (74). The intron is introduced into C. trachomatis on a suicide vector, and selection of insertion is performed using ampicillin (the intron carries a bla marker). Johnson and Fisher initially validated TargeTron in C. trachomatis L2 through insertional inactivation of incA (75). Intron insertion is stable in the absence of selection, and at least one insertion site has been identified in each C. trachomatis ORF. Recently, a second selection marker, aadA (spectinomycin resistance), was validated for intron-based mutagenesis in C. trachomatis and has been shown to allow creation of dual insertion mutants (62, 76). As the intron can carry cargo, this approach could also be used for complementation approaches or to insert foreign genes into the chromosome. The major limitation of this approach holds true for all gene insertion methods, namely, the possibility of polar effects. This can make it difficult to distinguish if the resulting phenotype is due to inactivation of a downstream gene (addressable by complementation). Additionally, failure to generate a mutant could result from improper transformation/intron function or polar effects acting on an essential gene, rather than the essentiality of the targeted gene. The Ll.LtrB group II intron has recently been modified to allow for Cre-mediated recombination, which could be adapted for genome editing or marker recycling, and a second intron-based method using an EcI5 intron has been developed that is more efficient than the LtrB intron (not evaluated in Chlamydia) (74). The TargeTron system has also been used in the obligate intracellular pathogens Rickettsia rickettsii and Ehrlichia chaffeensis (77, 78).
FUTURE PROSPECTS
The field of chlamydial genetics has seen major advances over the past 5 years. We can now generate targeted mutants, carry out large-scale chemical mutagenesis for forward and reverse genetic approaches, express foreign and recombinant genes using various shuttle vectors, and we are positioned to begin generating conditional lethal mutants by combining inducible promoter systems with chemical or targeted mutagenesis. The latter approach may prove to be vital for studying many chlamydial genes that may be essential due to the reduced genomes and obligate intracellular lifestyle of these pathogens (17). Alternative approaches available for studying essential genes in Chlamydia include regulated overexpression of wild-type genes or dominant-negative mutant genes along with further development of antisense RNA approaches described by Mishra et al. (79). In addition, validation of fluorescent protein expression in C. trachomatis coupled with detection of fluorescent EBs via flow cytometry (or enumeration in plaque assay) should allow for optimization of transformation procedures as new methods of mutagenesis are explored (44). Along this line, transformation via electroporation as utilized by Binet and Maurelli (and the method of choice for transformation in other obligate, intracellular bacteria) is worth revisiting to assess efficiency versus chemical transformation (43). Collectively, the currently available tools have provided important insights into chlamydial pathogenesis that could not have been ascertained as recently as 4 years ago, such as elucidating the function of the secreted protease, CPAF (chlamydial protease-like activity factor) (80).
However, the chlamydial field still lacks numerous genetic methods available for other bacterial pathogens. Major needs include methods for allelic exchange and identification of broad-host-range plasmids to allow for complementation and/or expression of recombinant genes without loss or alteration of native cryptic plasmids as well as use of a single shuttle plasmid across species/strains. Another major limitation is the absence of transposon mutagenesis systems for both random mutagenesis and targeted delivery of genes to the chromosome as exemplified by the Tn7 platform (81, 82). For many of these approaches, we can look to other obligate intracellular bacteria for inspiration (Table 2). A plethora of tools are available for manipulating Coxiella burnetii, the majority of which were established after the development of axenic growth conditions for the bacterium (83). Axenic growth conditions would likely yield similar fruit for Chlamydia (84). In addition, transposon mutagenesis has been successfully employed in Rickettsia spp., E. chaffeensis, Anaplasma spp., and C. burnetii prior to establishment of axenic growth conditions supporting the feasibility of transposon mutagenesis for Chlamydia (Table 2). While it remains impossible to predict the future, it seems clear that in regard to chlamydial genetics, it will be bright.
ACKNOWLEDGMENTS
We thank Bethany Rader (Southern Illinois University) for critical review of the manuscript and the reviewers for their numerous helpful comments and critiques.
REFERENCES
- 1.Horn M. 2008. Chlamydiae as symbionts in eukaryotes. Annu Rev Microbiol 62:113–131. doi: 10.1146/annurev.micro.62.081307.162818. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2014. Sexually transmitted diseases surveillance 2013. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/std/stats13/default.htm. [Google Scholar]
- 3.World Health Organization. 2015. Trachoma fact sheet 382. World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs382/en/. [Google Scholar]
- 4.Darville T. 2013. Recognition and treatment of chlamydial infections from birth to adolescence. Adv Exp Med Biol 764:109–122. [DOI] [PubMed] [Google Scholar]
- 5.Hu VH, Holland MJ, Burton MJ. 2013. Trachoma: protective and pathogenic ocular immune responses to Chlamydia trachomatis. PLoS Negl Trop Dis 7:e2020. doi: 10.1371/journal.pntd.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell LA, Rosenfeld ME. 2015. Infection and atherosclerosis development. Arch Med Res 46:339–350. doi: 10.1016/j.arcmed.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdelrahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol Rev 29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Bastidas RJ, Elwell CA, Engel JN, Valdivia RH. 2013. Chlamydial intracellular survival strategies. Cold Spring Harb Perspect Med 3:a010256. doi: 10.1101/cshperspect.a010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hybiske K, Stephens RS. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci U S A 104:11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albrecht M, Sharma CM, Dittrich MT, Muller T, Reinhardt R, Vogel J, Rudel T. 2011. The transcriptional landscape of Chlamydia pneumoniae. Genome Biol 12:R98. doi: 10.1186/gb-2011-12-10-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci U S A 100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyairi I, Mahdi OS, Ouellette SP, Belland RJ, Byrne GI. 2006. Different growth rates of Chlamydia trachomatis biovars reflect pathotype. J Infect Dis 194:350–357. doi: 10.1086/505432. [DOI] [PubMed] [Google Scholar]
- 13.Saka HA, Thompson JW, Chen YS, Kumar Y, Dubois LG, Moseley MA, Valdivia RH. 2011. Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol Microbiol 82:1185–1203. doi: 10.1111/j.1365-2958.2011.07877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voigt A, Schofl G, Saluz HP. 2012. The Chlamydia psittaci genome: a comparative analysis of intracellular pathogens. PLoS One 7:e35097. doi: 10.1371/journal.pone.0035097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dugan J, Rockey DD, Jones L, Andersen AA. 2004. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob Agents Chemother 48:3989–3995. doi: 10.1128/AAC.48.10.3989-3995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hackstadt T, Baehr W, Ying Y. 1991. Chlamydia trachomatis developmentally regulated protein is homologous to eukaryotic histone H1. Proc Natl Acad Sci U S A 88:3937–3941. doi: 10.1073/pnas.88.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 18.Soucy SM, Huang J, Gogarten JP. 2015. Horizontal gene transfer: building the web of life. Nat Rev Genet 16:472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 19.Joseph T, Nano FE, Garon CF, Caldwell HD. 1986. Molecular characterization of Chlamydia trachomatis and Chlamydia psittaci plasmids. Infect Immun 51:699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer L, Falkow S. 1986. A common plasmid of Chlamydia trachomatis. Plasmid 16:52–62. doi: 10.1016/0147-619X(86)90079-X. [DOI] [PubMed] [Google Scholar]
- 21.Hatt C, Ward ME, Clarke IN. 1988. Analysis of the entire nucleotide sequence of the cryptic plasmid of Chlamydia trachomatis serovar L1. Evidence for involvement in DNA replication. Nucleic Acids Res 16:4053–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sriprakash KS, Macavoy ES. 1987. Characterization and sequence of a plasmid from the trachoma biovar of Chlamydia trachomatis. Plasmid 18:205–214. doi: 10.1016/0147-619X(87)90063-1. [DOI] [PubMed] [Google Scholar]
- 23.Gong S, Yang Z, Lei L, Shen L, Zhong G. 2013. Characterization of Chlamydia trachomatis plasmid-encoded open reading frames. J Bacteriol 195:3819–3826. doi: 10.1128/JB.00511-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, Watkins H, Zhou B, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD. 2013. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect Immun 81:636–644. doi: 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam JE, Davis CH, Wyrick PB. 1994. Expression of recombinant DNA introduced into Chlamydia trachomatis by electroporation. Can J Microbiol 40:583–591. doi: 10.1139/m94-093. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caldwell HD, Wood H, Crane D, Bailey R, Jones RB, Mabey D, Maclean I, Mohammed Z, Peeling R, Roshick C, Schachter J, Solomon AW, Stamm WE, Suchland RJ, Taylor L, West SK, Quinn TC, Belland RJ, McClarty G. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J Clin Invest 111:1757–1769. doi: 10.1172/JCI17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Virok D, Rajaram K, Endresz V, McClarty G, Nelson DE, Caldwell HD. 2011. Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci U S A 108:7189–7193. doi: 10.1073/pnas.1102229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsia RC, Pannekoek Y, Ingerowski E, Bavoil PM. 1997. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol Microbiol 25:351–359. doi: 10.1046/j.1365-2958.1997.4701834.x. [DOI] [PubMed] [Google Scholar]
- 30.Rachek LI, Tucker AM, Winkler HH, Wood DO. 1998. Transformation of Rickettsia prowazekii to rifampin resistance. J Bacteriol 180:2118–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Troyer JM, Radulovic S, Azad AF. 1999. Green fluorescent protein as a marker in Rickettsia typhi transformation. Infect Immun 67:3308–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rachek LI, Hines A, Tucker AM, Winkler HH, Wood DO. 2000. Transformation of Rickettsia prowazekii to erythromycin resistance encoded by the Escherichia coli ereB gene. J Bacteriol 182:3289–3291. doi: 10.1128/JB.182.11.3289-3291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Renesto P, Gouin E, Raoult D. 2002. Expression of green fluorescent protein in Rickettsia conorii. Microb Pathog 33:17–21. doi: 10.1006/mpat.2002.0508. [DOI] [PubMed] [Google Scholar]
- 34.Qin A, Tucker AM, Hines A, Wood DO. 2004. Transposon mutagenesis of the obligate intracellular pathogen Rickettsia prowazekii. Appl Environ Microbiol 70:2816–2822. doi: 10.1128/AEM.70.5.2816-2822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomes JP, Bruno WJ, Borrego MJ, Dean D. 2004. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J Bacteriol 186:4295–4306. doi: 10.1128/JB.186.13.4295-4306.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomes JP, Nunes A, Bruno WJ, Borrego MJ, Florindo C, Dean D. 2006. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J Bacteriol 188:275–286. doi: 10.1128/JB.188.1.275-286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demars R, Weinfurter J, Guex E, Lin J, Potucek Y. 2007. Lateral gene transfer in vitro in the intracellular pathogen Chlamydia trachomatis. J Bacteriol 189:991–1003. doi: 10.1128/JB.00845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeMars R, Weinfurter J. 2008. Interstrain gene transfer in Chlamydia trachomatis in vitro: mechanism and significance. J Bacteriol 190:1605–1614. doi: 10.1128/JB.01592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suchland RJ, Sandoz KM, Jeffrey BM, Stamm WE, Rockey DD. 2009. Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob Agents Chemother 53:4604–4611. doi: 10.1128/AAC.00477-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeffrey BM, Suchland RJ, Eriksen SG, Sandoz KM, Rockey DD. 2013. Genomic and phenotypic characterization of in vitro-generated Chlamydia trachomatis recombinants. BMC Microbiol 13:142. doi: 10.1186/1471-2180-13-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeffrey BM, Suchland RJ, Quinn KL, Davidson JR, Stamm WE, Rockey DD. 2010. Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect Immun 78:2544–2553. doi: 10.1128/IAI.01324-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pawlikowska-Warych M, Sliwa-Dominiak J, Deptula W. 2015. Chlamydial plasmids and bacteriophages. Acta Biochim Pol 62:1–6. doi: 10.18388/abp.2014_764. [DOI] [PubMed] [Google Scholar]
- 43.Binet R, Maurelli AT. 2009. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc Natl Acad Sci U S A 106:292–297. doi: 10.1073/pnas.0806768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vromman F, Laverriere M, Perrinet S, Dufour A, Subtil A. 2014. Quantitative monitoring of the Chlamydia trachomatis developmental cycle using GFP-expressing bacteria, microscopy and flow cytometry. PLoS One 9:e99197. doi: 10.1371/journal.pone.0099197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomson NR, Holden MT, Carder C, Lennard N, Lockey SJ, Marsh P, Skipp P, O'Connor CD, Goodhead I, Norbertzcak H, Harris B, Ormond D, Rance R, Quail MA, Parkhill J, Stephens RS, Clarke IN. 2008. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res 18:161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unemo M, Seth-Smith HM, Cutcliffe LT, Skilton RJ, Barlow D, Goulding D, Persson K, Harris SR, Kelly A, Bjartling C, Fredlund H, Olcen P, Thomson NR, Clarke IN. 2010. The Swedish new variant of Chlamydia trachomatis: genome sequence, morphology, cell tropism and phenotypic characterization. Microbiology 156:1394–1404. doi: 10.1099/mic.0.036830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu S, Battaglia L, Bao X, Fan H. 2013. Chloramphenicol acetyltransferase as a selection marker for chlamydial transformation. BMC Res Notes 6:377. doi: 10.1186/1756-0500-6-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agaisse H, Derre I. 2013. A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS One 8:e57090. doi: 10.1371/journal.pone.0057090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerard HC, Mishra MK, Mao G, Wang S, Hali M, Whittum-Hudson JA, Kannan RM, Hudson AP. 2013. Dendrimer-enabled DNA delivery and transformation of Chlamydia pneumoniae. Nanomedicine 9:996–1008. doi: 10.1016/j.nano.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Mueller KE, Fields KA. 2015. Application of beta-lactamase reporter fusions as an indicator of effector protein secretion during infections with the obligate intracellular pathogen Chlamydia trachomatis. PLoS One 10:e0135295. doi: 10.1371/journal.pone.0135295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Persson K, Bjartling C, Clarke IN. 2013. Genetic transformation of a clinical (genital tract), plasmid-free isolate of Chlamydia trachomatis: engineering the plasmid as a cloning vector. PLoS One 8:e59195. doi: 10.1371/journal.pone.0059195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickstrum J, Sammons LR, Restivo KN, Hefty PS. 2013. Conditional gene expression in Chlamydia trachomatis using the Tet system. PLoS One 8:e76743. doi: 10.1371/journal.pone.0076743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campbell J, Huang Y, Liu Y, Schenken R, Arulanandam B, Zhong G. 2014. Bioluminescence imaging of Chlamydia muridarum ascending infection in mice. PLoS One 9:e101634. doi: 10.1371/journal.pone.0101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding H, Gong S, Tian Y, Yang Z, Brunham R, Zhong G. 2013. Transformation of sexually transmitted infection-causing serovars of Chlamydia trachomatis using blasticidin for selection. PLoS One 8:e80534. doi: 10.1371/journal.pone.0080534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen YS, Bastidas RJ, Saka HA, Carpenter VK, Richards KL, Plano GV, Valdivia RH. 2014. The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Pathog 10:e1003954. doi: 10.1371/journal.ppat.1003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Y, Zhang Q, Yang Z, Conrad T, Liu Y, Zhong G. 2015. Plasmid-encoded Pgp5 is a significant contributor to Chlamydia muridarum induction of hydrosalpinx. PLoS One 10:e0124840. doi: 10.1371/journal.pone.0124840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Chen C, Gong S, Hou S, Qi M, Liu Q, Baseman J, Zhong G. 2014. Transformation of Chlamydia muridarum reveals a role for Pgp5 in suppression of plasmid-dependent gene expression. J Bacteriol 196:989–998. doi: 10.1128/JB.01161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, Huang Y, Yang Z, Sun Y, Gong S, Hou S, Chen C, Li Z, Liu Q, Wu Y, Baseman J, Zhong G. 2014. Plasmid-encoded Pgp3 is a major virulence factor for Chlamydia muridarum to induce hydrosalpinx in mice. Infect Immun 82:5327–5335. doi: 10.1128/IAI.02576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song L, Carlson JH, Zhou B, Virtaneva K, Whitmire WM, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD. 2014. Plasmid-mediated transformation tropism of chlamydial biovars. Pathog Dis 70:189–193. doi: 10.1111/2049-632X.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Cutcliffe LT, Skilton RJ, Ramsey KH, Thomson NR, Clarke IN. 2014. The genetic basis of plasmid tropism between Chlamydia trachomatis and Chlamydia muridarum. Pathog Dis 72:19–23. doi: 10.1111/2049-632X.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lowden NM, Yeruva L, Johnson CM, Bowlin AK, Fisher DJ. 2015. Use of aminoglycoside 3′ adenyltransferase as a selection marker for Chlamydia trachomatis intron-mutagenesis and in vivo intron stability. BMC Res Notes 8:570. doi: 10.1186/s13104-015-1542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandoz KM, Rockey DD. 2010. Antibiotic resistance in Chlamydiae. Future Microbiol 5:1427–1442. doi: 10.2217/fmb.10.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skilton RJ, Cutcliffen LT, Barlow D, Wang Y, Salim O, Lambden PR, Clarke IN. 2009. Penicillin induced persistence in Chlamydia trachomatis: high quality time lapse video analysis of the developmental cycle. PLoS One 4:e7723. doi: 10.1371/journal.pone.0007723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Banks J, Eddie B, Schachter J, Meyer KF. 1970. Plaque formation by Chlamydia in L cells. Infect Immun 1:259–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKee EE, Ferguson M, Bentley AT, Marks TA. 2006. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob Agents Chemother 50:2042–2049. doi: 10.1128/AAC.01411-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kari L, Southern TR, Downey CJ, Watkins HS, Randall LB, Taylor LD, Sturdevant GL, Whitmire WM, Caldwell HD. 2014. Chlamydia trachomatis polymorphic membrane protein D is a virulence factor involved in early host-cell interactions. Infect Immun 82:2756–2762. doi: 10.1128/IAI.01686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rajaram K, Giebel AM, Toh E, Hu S, Newman JH, Morrison SG, Kari L, Morrison RP, Nelson DE. 2015. Mutational analysis of the Chlamydia muridarum plasticity zone. Infect Immun 83:2870–2881. doi: 10.1128/IAI.00106-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen BD, Valdivia RH. 2012. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc Natl Acad Sci U S A 109:1263–1268. doi: 10.1073/pnas.1117884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kokes M, Dunn JD, Granek JA, Nguyen BD, Barker JR, Valdivia RH, Bastidas RJ. 2015. Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia. Cell Host Microbe 17:716–725. doi: 10.1016/j.chom.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 72.Merrell DS, Hava DL, Camilli A. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol 43:1471–1491. doi: 10.1046/j.1365-2958.2002.02857.x. [DOI] [PubMed] [Google Scholar]
- 73.Zhong J, Karberg M, Lambowitz AM. 2003. Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res 31:1656–1664. doi: 10.1093/nar/gkg248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Enyeart PJ, Mohr G, Ellington AD, Lambowitz AM. 2014. Biotechnological applications of mobile group II introns and their reverse transcriptases: gene targeting, RNA-seq, and non-coding RNA analysis. Mob DNA 5:2. doi: 10.1186/1759-8753-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson CM, Fisher DJ. 2013. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS One 8:e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thompson CC, Griffiths C, Nicod SS, Lowden NM, Wigneshweraraj S, Fisher DJ, McClure MO. 2015. The Rsb phosphoregulatory network controls availability of the primary sigma factor in Chlamydia trachomatis and influences the kinetics of growth and development. PLoS Pathog 11:e1005125. doi: 10.1371/journal.ppat.1005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheng C, Nair AD, Indukuri VV, Gong S, Felsheim RF, Jaworski D, Munderloh UG, Ganta RR. 2013. Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog 9:e1003171. doi: 10.1371/journal.ppat.1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Noriea NF, Clark TR, Hackstadt T. 2015. Targeted knockout of the Rickettsia rickettsii OmpA surface antigen does not diminish virulence in a mammalian model system. mBio 6:e00323-15. doi: 10.1128/mBio.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mishra MK, Gerard HC, Whittum-Hudson JA, Hudson AP, Kannan RM. 2012. Dendrimer-enabled modulation of gene expression in Chlamydia trachomatis. Mol Pharm 9:413–421. doi: 10.1021/mp200512f. [DOI] [PubMed] [Google Scholar]
- 80.Snavely EA, Kokes M, Dunn JD, Saka HA, Nguyen BD, Bastidas RJ, McCafferty DG, Valdivia RH. 2014. Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog Dis 71:336–351. doi: 10.1111/2049-632X.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi KH, Kim KJ. 2009. Applications of transposon-based gene delivery system in bacteria. J Microbiol Biotechnol 19:217–228. [DOI] [PubMed] [Google Scholar]
- 82.Waddell CS, Craig NL. 1988. Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes Dev 2:137–149. doi: 10.1101/gad.2.2.137. [DOI] [PubMed] [Google Scholar]
- 83.Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, Porcella SF, Heinzen RA. 2009. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci U S A 106:4430–4434. doi: 10.1073/pnas.0812074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. 2012. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc Natl Acad Sci U S A 109:19781–19785. doi: 10.1073/pnas.1212831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Omsland A, Beare PA, Hill J, Cockrell DC, Howe D, Hansen B, Samuel JE, Heinzen RA. 2011. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol 77:3720–3725. doi: 10.1128/AEM.02826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crosby FL, Wamsley HL, Pate MG, Lundgren AM, Noh SM, Munderloh UG, Barbet AF. 2014. Knockout of an outer membrane protein operon of Anaplasma marginale by transposon mutagenesis. BMC Genomics 15:278. doi: 10.1186/1471-2164-15-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Felsheim RF, Chavez AS, Palmer GH, Crosby L, Barbet AF, Kurtti TJ, Munderloh UG. 2010. Transformation of Anaplasma marginale. Vet Parasitol 167:167–174. doi: 10.1016/j.vetpar.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Felsheim RF, Herron MJ, Nelson CM, Burkhardt NY, Barbet AF, Kurtti TJ, Munderloh UG. 2006. Transformation of Anaplasma phagocytophilum. BMC Biotechnol 6:42. doi: 10.1186/1472-6750-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beare PA, Howe D, Cockrell DC, Omsland A, Hansen B, Heinzen RA. 2009. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J Bacteriol 191:1369–1381. doi: 10.1128/JB.01580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suhan ML, Chen SY, Thompson HA. 1996. Transformation of Coxiella burnetii to ampicillin resistance. J Bacteriol 178:2701–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. 2011. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2:e00175-11. doi: 10.1128/mBio.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beare PA, Larson CL, Gilk SD, Heinzen RA. 2012. Two systems for targeted gene deletion in Coxiella burnetii. Appl Environ Microbiol 78:4580–4589. doi: 10.1128/AEM.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beare PA, Sandoz KM, Omsland A, Rockey DD, Heinzen RA. 2011. Advances in genetic manipulation of obligate intracellular bacterial pathogens. Front Microbiol 2:97. doi: 10.3389/fmicb.2011.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen C, Banga S, Mertens K, Weber MM, Gorbaslieva I, Tan Y, Luo ZQ, Samuel JE. 2010. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci U S A 107:21755–21760. doi: 10.1073/pnas.1010485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martinez E, Cantet F, Fava L, Norville I, Bonazzi M. 2014. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PLoS Pathog 10:e1004013. doi: 10.1371/journal.ppat.1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Voth DE, Beare PA, Howe D, Sharma UM, Samoilis G, Cockrell DC, Omsland A, Heinzen RA. 2011. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol 193:1493–1503. doi: 10.1128/JB.01359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baldridge GD, Burkhardt N, Herron MJ, Kurtti TJ, Munderloh UG. 2005. Analysis of fluorescent protein expression in transformants of Rickettsia monacensis, an obligate intracellular tick symbiont. Appl Environ Microbiol 71:2095–2105. doi: 10.1128/AEM.71.4.2095-2105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baldridge GD, Burkhardt NY, Oliva AS, Kurtti TJ, Munderloh UG. 2010. Rickettsial ompB promoter regulated expression of GFPuv in transformed Rickettsia montanensis. PLoS One 5:e8965. doi: 10.1371/journal.pone.0008965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Burkhardt NY, Baldridge GD, Williamson PC, Billingsley PM, Heu CC, Felsheim RF, Kurtti TJ, Munderloh UG. 2011. Development of shuttle vectors for transformation of diverse Rickettsia species. PLoS One 6:e29511. doi: 10.1371/journal.pone.0029511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Clark TR, Ellison DW, Kleba B, Hackstadt T. 2011. Complementation of Rickettsia rickettsii RelA/SpoT restores a nonlytic plaque phenotype. Infect Immun 79:1631–1637. doi: 10.1128/IAI.00048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Clark TR, Lackey AM, Kleba B, Driskell LO, Lutter EI, Martens C, Wood DO, Hackstadt T. 2011. Transformation frequency of a mariner-based transposon in Rickettsia rickettsii. J Bacteriol 193:4993–4995. doi: 10.1128/JB.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Driskell LO, Yu XJ, Zhang L, Liu Y, Popov VL, Walker DH, Tucker AM, Wood DO. 2009. Directed mutagenesis of the Rickettsia prowazekii pld gene encoding phospholipase D. Infect Immun 77:3244–3248. doi: 10.1128/IAI.00395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu ZM, Tucker AM, Driskell LO, Wood DO. 2007. Mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl Environ Microbiol 73:6644–6649. doi: 10.1128/AEM.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oliver JD, Burkhardt NY, Felsheim RF, Kurtti TJ, Munderloh UG. 2014. Motility characteristics are altered for Rickettsia bellii transformed to overexpress a heterologous rickA gene. Appl Environ Microbiol 80:1170–1176. doi: 10.1128/AEM.03352-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pelc RS, McClure JC, Kaur SJ, Sears KT, Rahman MS, Ceraul SM. 2015. Disrupting protein expression with peptide nucleic acids reduces infection by obligate intracellular Rickettsia. PLoS One 10:e0119283. doi: 10.1371/journal.pone.0119283. [DOI] [PMC free article] [PubMed] [Google Scholar]