Abstract
Bacterial adherence determines the virulence of many human-pathogenic bacteria. Experimental approaches elucidating this early infection event in greater detail have been performed using mainly methods of cellular microbiology. However, in vitro infections of cell monolayers reflect the in vivo situation only partially, and animal infection models are not available for many human-pathogenic bacteria. Therefore, ex vivo infection of human organs might represent an attractive method to overcome these limitations. We infected whole human umbilical cords ex vivo with Bartonella henselae or Acinetobacter baumannii under dynamic flow conditions mimicking the in vivo infection situation of human endothelium. For this purpose, methods for quantifying endothelium-adherent wild-type and trimeric autotransporter adhesin (TAA)-deficient bacteria were set up. Data revealed that (i) A. baumannii binds in a TAA-dependent manner to endothelial cells, (ii) this organ infection model led to highly reproducible adherence rates, and furthermore, (iii) this model allowed to dissect the biological function of TAAs in the natural course of human infections. These findings indicate that infection models using ex vivo human tissue samples (“organ microbiology”) might be a valuable tool in analyzing bacterial pathogenicity with the capacity to replace animal infection models at least partially.
INTRODUCTION
The analysis of adhesion to host cells is crucial for the elucidation of bacterial infection mechanisms. Animal infection models or cellular microbiology-based approaches are well established, and many important findings have thereby been gathered. By way of example, the protein injection machineries and the autotransporter adhesins (TAA) of Bartonella henselae and Acinetobacter baumannii have been elucidated in that manner (1–3). Although these methods are well established and widely accepted, they still are hampered by several limitations: animal models might reflect the course of human infections only partially (4) or might not even be available for many pathogens (e.g., B. henselae). Cellular infection models allow very detailed analyses of the interaction of bacteria with host cells under standardized conditions. However, they are usually done statically using cell monolayers, and this mimics the complexity of in vivo infection situation only to some extent. Methods to overcome the species barrier in infection models and to analyze host-pathogen interactions close to the dynamic human situation are urgently needed. Ex vivo infection models, using human organ grafts, might represent an attractive alternative or addition to animal or cellular microbiology experiments.
B. henselae causes cat scratch disease and endocarditis, whereas immunosuppressed individuals can suffer from vasculoproliferative disorders such as bacillary angiomatosis. B. henselae is believed to be an endotheliotropic pathogen. Bartonella adhesin A (BadA) has been identified as the key factor involved in the adherence to endothelial cells (ECs) and to extracellular matrix (ECM) components and in the induction of an angiogenic host cell response (2, 5–7). A. baumannii has become an emerging nosocomial pathogen worldwide, mainly in intensive care units. This bacterium is characterized by long-term survival on abiotic surfaces and multidrug resistance to antibiotics (8). After colonization of patients, A. baumannii is able to cause bloodstream (including endothelium) infections, urinary tract infections, and ventilator-associated pneumonia (9). Although bloodstream infections with A. baumannii are well described (10, 11), nearly nothing is known about the molecular mechanisms underlying the interplay between bacteria and ECs. Different outer membrane-associated proteins are known for Acinetobacter adhesion to abiotic surfaces, ECMs, and human host cells for clinical and environmental isolates (12–16). In this context, the Acinetobacter trimeric autotransporter adhesin (Ata) is supposed to play a key role in infection processes and adhesion to host cells (3, 17). Unfortunately, not much is known about Ata-dependent interaction of A. baumannii with host cells.
The first and often decisive step in bacterial infections is the adherence to the host. TAAs are widely represented in alpha-, beta- and gammaproteobacteria and play an important role in bacterial pathogenicity (18). They are designated autotransporters (or type V secretion systems), as they can self-export across the bacterial outer membrane in the absence of ATP (19). TAAs build a characteristic trimeric, “lollipop”-like surface structure and share a modular organization composed of different domains (“membrane anchor,” “stalk,” “neck,” and “head”), which are conserved modules present in nearly all TAAs (20). Both BadA of B. henselae and Ata of A. baumannii belong to the group of TAAs and share a similar domain structure architecture. BadA consists of 3,082 amino acids (aa) with a mass of 328 kDa per monomer. The repetitive 22 neck-stalk elements define the enormous length of BadA (∼240 nm) (2). Ata is encoded by a 6,786-bp gene. The protein is built up of 2,260 aa and has a mass of 230 kDa per monomer. Characteristic features of Ata are the double-head domain and a long N-terminal signal peptide (see Fig. S1 in the supplemental material).
The aim of this study was to elucidate whether an organ infection model using human umbilical cords might represent a valuable tool to analyze TAA-dependent host adherence of B. henselae or A. baumannii. The infection procedures were performed under dynamic blood flow mimicking conditions, and methods for the quantification of EC-adherent B. henselae and A. baumannii were designed (fluorometry and quantitative real-time PCR [qRT-PCR]).
MATERIALS AND METHODS
Bacterial strains.
B. henselae was grown for 4 days in Schneider's medium supplemented with 10% fetal calf serum (FCS) and 5% sucrose at 37°C in a humidified atmosphere with 5% CO2 (21). B. henselae lacking BadA (BadA−) was grown in the presence of 30 μg/ml kanamycin (Km). Expression analyses for B. henselae wild-type and B. henselae BadA− have been performed earlier (2, 22). Stock suspensions were prepared by freezing the bacteria in lysogeny broth (LB) containing 20% glycerol at −80°C. The number of viable bacteria per aliquot was determined by counting the CFU in serial dilutions from the frozen stocks, which were cultivated on Columbia blood agar (CBA) plates for 14 days. A. baumannii was grown in 2× yeast extract-tryptone (YT) medium and incubated at 37°C and 180 rpm overnight. The optical density at 600 nm (OD600) was adjusted to 0.05 in 2× YT medium, and bacteria were regrown to an OD600 of 0.2 (1.0 × 108 CFU/ml) for infection experiments. Strains used in this study are summarized in Table 1.
TABLE 1.
Bacterial strains used in this study
| Strain | Characteristics | Reference(s) or source |
|---|---|---|
| B. henselae wild type (BadA+) | Strain Marseille, patient isolate, early passage, expressing BadA | 2, 40 |
| B. henselae BadA− | Strain Marseille, transposon mutant, BadA-deficient, Kmr | 2 |
| A. baumannii ATCC 19606 wild type | Type strain, isolated from humans, expressing Ata | Gene Bank accession no. ACQB00000000.1 |
| A. baumannii ATCC 19606 Δata | ata markerless deleted mutant (positions 315,815–322,591) | This study |
| E. coli DH5α | Host strain used for cloning | Biochrom |
Construction of A. baumannii Δata.
The construction of an ata deletion mutant of A. baumannii ATCC 19606 was performed according to a method by Stahl and Averhoff (23), using the plasmids and primers listed in Tables 2 and 3, respectively. We constructed a vector containing a 1.5-kbp fragment both up- and downstream from the ata gene. For this, up- and downstream fragments (here termed up-fragment and down-fragment, respectively) were amplified from genomic DNA using primers ata_up_PstI_fwd/ata_up_BamHI_rev and ata_down_BamHI-fwd/ata_down_NotI_rev. Both fragments were digested (up-fragment, PstI/BamHI; down-fragment, BamHI/NotI) and ligated into pBIISK (digested with PstI/NotI), resulting in pBIISK_ata_up/down. This plasmid was transformed into A. baumannii ATCC 19606 by electroporation. The resulting clones were selected on LB agar supplemented with 50 μg/ml kanamycin. Transformed bacteria were grown overnight in LB medium containing 10% sucrose to select cells that lost the plasmid after homologous recombination. Integration of homologous regions was proven by colony PCR using ata_ctrl_fwd and ata_ctrl_rev. The next day, single colonies were picked and transferred to LB agar and LB agar supplemented with 50 μg/ml kanamycin to finally identify the colonies that had lost the plasmid (no growth on LB agar supplemented with kanamycin). Successful deletion of ata was verified by sequencing using ata_seq_fwd and ata_seq_rev (data not shown).
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristics | Resistance | Source |
|---|---|---|---|
| TOPO_TA_2.1 | Standard cloning vector for amplicons with TA-overhangs, lacZα for blue/white screening | Kmr Ampr | Thermo Fisher Scientific |
| TOPO_TA_2.1_glyA | TOPO_TA.2.1 including glyA fragment | Kmr Ampr | This study |
| TOPO_TA_2.1_rpoB | TOPO_TA.2.1 including rpoB fragment | Kmr Ampr | This study |
| TOPO_TA_2.1_hmbs | TOPO_TA.2.1 including hmbs fragment | Kmr Ampr | This study |
| pBluescript II SK(+) (pBIISK) | Cloning vector for A. baumannii ATCC 19606, sacB (levansucrase) for segregation | Kmr | Stratagene |
| pBIISK_ata_up/down | pBIISK including the 1.5-kbp ata_up and 1.5-kbp ata_down fragments | Kmr | This study |
TABLE 3.
Primers used in this study
| Primer | 5′→ 3′ sequence | Source or reference |
|---|---|---|
| glyA_fwd | GAC AGG AAA ATG TGC CGA AT | This study |
| glyA_rev | GCA GGT GAA CCA AGA CGA AT | This study |
| rpob_fwd | GAG TCT AAT GGC GGT GGT TC | 41 |
| rpob_rev | ATT GCT TCA TCT GCT GGT TG | 41 |
| hmbs_fwd | TTC CTT CCC TGA AGG GAT TCA CTC AG | Galaxy Diagnostics, Durham, NC, USA |
| hmbs_rev | TTA AGC CCA GCA GCC TAT CTG ACA CCC | Galaxy Diagnostics, Durham, NC, USA |
| ata_up_PstI_fwd | GAC ACT GCA GCA TTT AGA AAG TAT TCA CGT TGG TTC | This study |
| ata_up_BamHI_rev | GAT CGG ATC CCT TTA TTC ATA AAA CTT CTC AGA C | This study |
| ata_down_BamHI_fwd | GAG AGG ATC CGA AAC TGG TTA GGA GGG CAA TGC TC | This study |
| ata_down_NotI_rev | GAC AGC GGC CGC GAT TTC TCC ATG AAT GAA TTA AG | This study |
| ata_ctrl_fwd | GTC CAA CCA AGC AAA GGA TAG GGC CTT TGA G | This study |
| ata_ctrl_rev | CTA ACT GAT CTA AAA ATT CAC CAA TTT TTC GG | This study |
| ata_seq_fwd | GTC GTT GAG TTC GGT ATT TGT CTG AGA AG | This study |
| ata_seq_rev | TAA TCT CTT TAA TCT GTC AGC AAG GAG AGC | This study |
Infection of endothelial cells and confocal laser scanning microscopy.
Human umbilical vein endothelial cells (HUVECs) were cultured in EC growth medium (PromoCell, Mannheim, Germany) as described previously (24). For static adherence assays, 1.0 × 105 HUVECs were seeded onto collagen G (Biochrom, Hamburg, Germany)-coated coverslips in 24-well plates, grown overnight without antibiotics, and infected with a multiplicity of infection (MOI) of 100 for B. henselae or 200 for A. baumannii by centrifugation (300 × g, 5 min). Cells were incubated for 60 min in a humidified atmosphere at 37°C and 5% CO2. After a gentle washing with prewarmed cell culture medium, the infection was stopped by adding 3.75% paraformaldehyde (PFA). For quantification of adherent bacteria by fluorometry and qRT-PCR, 5 × 105 cells were seeded (in six-well plates) and infection was done as described above.
For dynamic adherence assays, HUVECs (100 μl of a 1.0 × 106 cell suspension) were seeded overnight in growth medium without antibiotics on collagen G-coated multichannel slides (Ibidi, Martinsried, Germany). Bacteria were resuspended in 12 ml EC growth medium (MOI of 100 for B. henselae or 200 for A. baumannii), and HUVECs were infected with a constant flow rate of 0.2 ml (∼0.26 dyne/cm2 [1 dyne is 10−5 N], according to the manufacturer's indications)/min for 60 min. After a gentle washing with prewarmed cell culture medium for 5 min, the infection was stopped by adding 3.75% PFA and microscopic slides were prepared. When adherent bacteria were quantified, cells were dissolved by adding trypsin-EDTA (Gibco BRL-Invitrogen, Karlsruhe, Germany), flushed with EC medium, and collected in 15-ml Falcon tubes.
Bacterial adherence was quantified using confocal laser scanning microscopy (CLSM) as described previously (24). Cells were permeabilized by incubation with 0.1% Triton X-100 (Sigma, Steinheim, Germany) in phosphate-buffered saline (PBS) for 15 min. The actin cytoskeleton was stained with tetramethylrhodamine isothiocyanate (TRITC)-labeled phalloidin (Sigma, Steinheim, Germany) for 1 h, and EC and bacterial DNA was stained with 4′,6-diamidin-2′-phenylindole dihydrochloride (DAPI) for 10 min. Finally, slides were mounted with Fluoprep medium (bioMérieux, Marcy l'Etoile, France). Cellular fluorescence was evaluated using a Leica DM IRE 2 confocal laser scanning microscope. Images were digitally processed with Photoshop 7.0 (Adobe Systems, Mountain View, CA).
Dynamic adhesion assay using human umbilical cords.
All experiments were done in accordance with ethical permission (4/12; University Hospital Frankfurt am Main). Human umbilical cords were collected in the midwifery of the Klinikum Sachsenhausen (Frankfurt am Main, Germany). Autolytic or otherwise affected samples were omitted from further experiments. For infection, fresh human umbilical cord veins were cut into two pieces of ∼5 cm in length each for comparison of wild-type and mutant bacteria. Veins were connected with buttoned cannulas, fixed with cable binders, and washed with prewarmed EC culture medium (0.2 ml/min, 15 min). Bacterial suspensions were prepared as described above and adjusted to 1.0 × 108 CFU/ml in prewarmed EC culture medium. The infection was started by switching a three-way valve and injecting the bacteria into the umbilical cord vein by a pump. Samples were continuously infected for 60 min in a recirculating system (Fig. 1). After this, nonadherent bacteria were flushed by washing the veins with EC cell culture medium. For determination of adherent bacteria, ECs were removed by injecting prewarmed 0.05% trypsin–EDTA into the vein and incubating the sample for 10 min. Dissolved cells were washed out of the veins and collected in 15-ml Falcon tubes for further processing.
FIG 1.
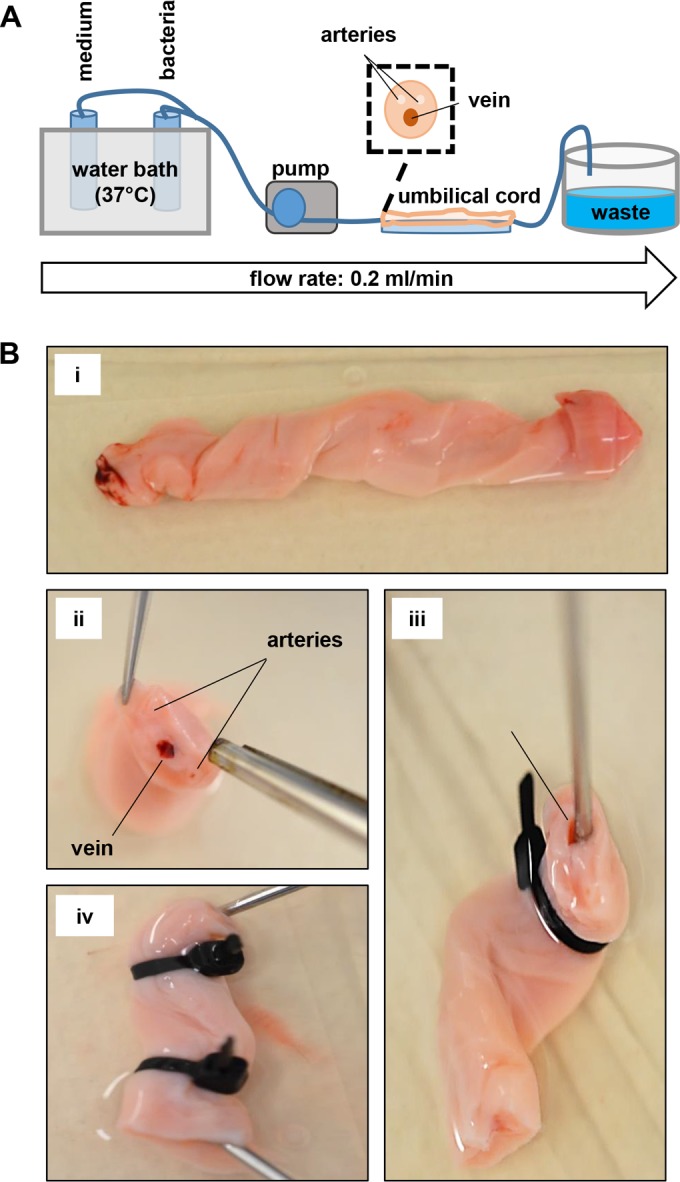
Experimental setup of the ex vivo umbilical cord infection assays. (A) Schematic draft of the experimental setup. A pump is connected with a fresh human umbilical cord vein. Blood clots and debris are flushed by pumping prewarmed medium through the vein for 15 min. Bacteria were pumped in prewarmed medium for 60 min with a constant defined flow (0.2 ml/min). After this, nonadherent bacteria were removed by flushing the vein for an additional 15 min with prewarmed medium. (B) Photography of processing of human umbilical cords for ex vivo infections: (i) fresh umbilical cord of ∼10 cm in length; (ii) situs of a cross-section of the umbilical cord (note the umbilical vein); (iii and iv) connection of buttoned cannulas (arrow) (iii), which were fixed (iv) by using cable binders.
Microcomputed tomography, shear stress calculation, and Doppler analysis.
For ex vivo microcomputed tomography (CT) imaging, the umbilical cord vein was perfused with PBS and a three-way valve connected to polyethylene tubing was introduced. The vein was perfused with the radiopaque contrast solution MicroFil (Flow Tech, Carver, MA, USA) with a pressure of 10 to 15 mm Hg until the silicon polymer cured. The umbilical cords were subsequently scanned in a micro-CT (Skyscan 1176; Bruker micro-CT, Kontich, Belgium) with the following settings: 50 kV; 0.5-mm aluminum filter; 500-μA source current; exposure time, 210 ms; 18-μm isotopic resolution; 3 projection images per 0.5° rotation step; rotation range, 180°. Volumetric data were reconstructed with the NRecon/InstaRecon CBR Server—Premium software (Skyscan, Kontich, Belgium/InstaRecon, Champaign, IL, USA). Image analysis, segmentation of micro-CT data, and quantification of vein lumen area were performed with the Imalytics Preclinical Software (Gremse-IT, Aachen, Germany). Discrimination of contrast agent and soft tissue was achieved by applying a threshold defined as the mean of contrast agent and soft tissue intensity. The lumen area of the vein was measured with the virtual elastic sphere tool (25) (Fig. 2A). Shear stress (τ) was calculated according to the following formula: τ = (4 × μ × v)/(π × r3), where μ is the viscosity (1.011 mPa × s), v is the flow rate (3.3 × 10−9 m3/s), and r is the diameter (26).
FIG 2.
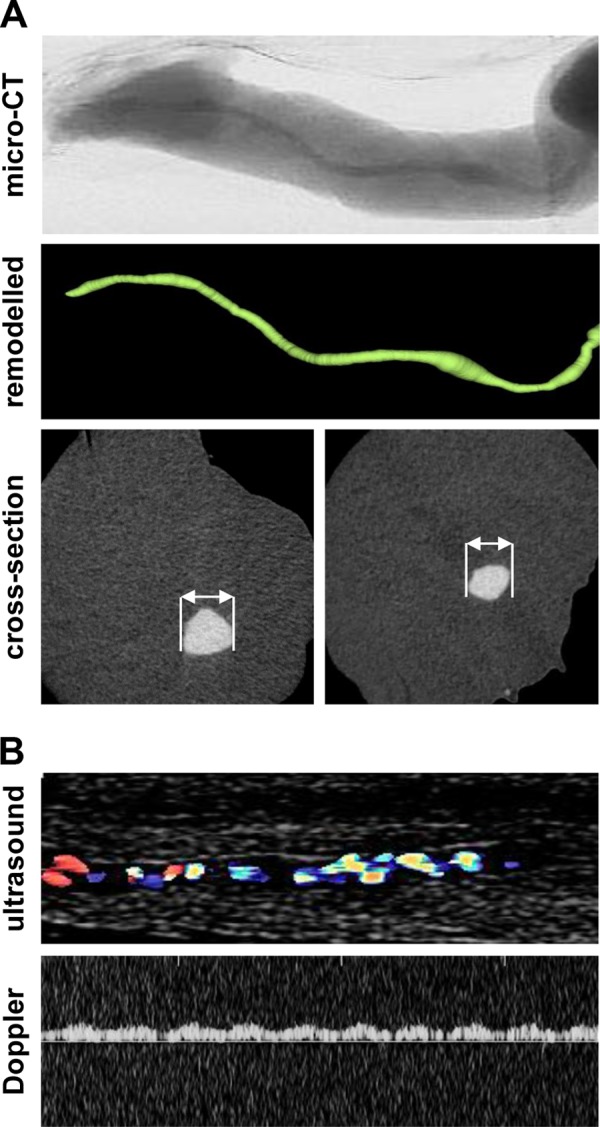
Microcomputed tomography and Doppler ultrasound analysis of umbilical cord veins. (A) Umbilical cord veins were perfused with MicroFil contrast solution, recorded by microcomputed tomography, and digitally processed; total length analyzed, 1.8 cm; diameter, 1.3 (left) or 0.9 (right) mm. (B) For ultrasound analysis, umbilical cord veins were washed and infected with barium sulfate (10%, resuspended in PBS).
To confirm laminar flow conditions in the umbilical cord vein during infection procedures, ultrasound Doppler analysis was performed. The flow pattern was recorded using a linear ultrasound head (5 to 12 MHz; here 9 MHz) connected with a Phillips Envisor ultrasound device (Hamburg, Germany) while the vein was flushed with 10% barium sulfate (resuspended in PBS). A monodirectional laminar flow with a flow velocity directly corresponding to the infusion pump velocity for 10% barium sulfate (0.2 ml/min, resuspended in PBS) was detectable (Fig. 2B).
Quantification of endothelial cell-adherent bacteria by fluorometry.
For fluorometric quantification of bacterial adherence to ECs, bacteria were stained with the life-staining dye carboxyfluorescein succinimidyl ester (CFSE; Santa Cruz biotechnology, Heidelberg, Germany) prior to infection. Bacteria were grown as described above, centrifuged for 10 min at 10,000 × g, washed with PBS (Gibco), and resuspended in 10 μM CFSE (dissolved in PBS) for 15 min at 37°C. CFSE-stained bacteria were washed with PBS. Potential bactericidal effects were excluded by comparing the amounts of CFU prior to and after the labeling procedures, which revealed no significant reduction of bacterial viability (quantified by determination of CFU; data not shown).
To correlate the total number of CFSE-labeled adherent bacteria to the total number of ECs, cells were labeled with anti-CD31-Alexa 647-conjugated antibodies. PFA-fixed ECs were incubated with 0.2% bovine serum albumin (BSA; dissolved in PBS) to block unspecific antibody binding. ECs were washed with PBS and incubated with 10 μg of anti-CD31-Alexa Fluor 647 (dissolved in PBS) for 60 min at room temperature and protected from light. Nonbound antibodies were removed by washing the cells with PBS.
For quantification of the EC-adherent bacteria, total cell samples from infection experiments (including CFSE bacteria and CD31-Alexa Fluor 647 HUVECs) were dispersed in 200 μl PBS, and the total volume was added to a black 96-well plate (Greiner BioOne GmbH, Frickenhausen, Germany). Additionally, standards were prepared: for CFSE-labeled bacteria, 1.0 × 105 to 1.0 × 107 CFU/well; for CD31-Alexa Fluor 647-labeled ECs, 1.0 × 104 to 1.0 × 106 cells/well. Calibration was performed by standard methods: bacterial CFU were calculated from cultivating serial dilutions on agar plates, and ECs were quantified using a Thoma cell counting chamber. Quantification of the relative fluorescence units (RFU) was performed using a multiple plate reader (Infinite 200 PRO; Tecan, Männedorf, Switzerland). The adjusted emission and excitation wavelengths for CFSE were 482 nm and 517 nm, and for Alexa Fluor 674, 647 nm and 678 nm, respectively. A standard curve was calculated by the internal standard in each experiment, and absolute bacterial and EC amounts were determined.
Quantification of endothelial cell-adherent bacteria by quantitative real-time PCR.
To quantify the number of adherent bacteria and HUVECs by using numbers of gene copy equivalents, qRT-PCRs were established. Samples (not PFA fixed) were prepared as described above, centrifuged (10,000 × g, 10 min), and dissolved in 30 μl distilled water. DNA was isolated by incubating the cells for 15 min at 95°C. After this, samples were centrifuged again at 10,000 × g for 10 min to remove cellular debris. DNA concentration was determined via 260 nm/280 nm photometry. The determination of bacterial and EC numbers was performed by determining gene copy numbers. The isolated genomic DNA of the samples was used to amplify species-specific genes. For quantifying adherent B. henselae isolates, a glyA (serine hydroxymethyltransferase, 120-bp) gene fragment was used, and for quantifying adherent A. baumannii, an rpoB (RNA polymerase beta-subunit, 110 bp) gene fragment was used. To determine the amount of ECs, the hmbs (hydroxymethylbilane synthase, 209 bp) gene was amplified. Primers are given in Table 3. DNA samples were added in a qRT-PCR mixture (5 μl qRT-PCR-SYBR green master mix [Qiagen, Hilden, Germany] together with 0.125 μl of forward and reverse primers [each 100 pmol] and 2.75 μl distilled water). For each qRT-PCR, an internal standard with known gene copy numbers was amplified. These PCR standards were produced by amplifying the respective gene fragments of glyA, rpoB, and hmbs and ligation into a TOPO_TA_2.1 vector according to the manufacturer's instructions (Thermo Fisher Scientific, Waltham, MA, USA). Total copy numbers of the resulting plasmids were calculated using the molecular mass of the single plasmids, the plasmid concentration, and the Avogadro constant (NA = 6 × 1023 mol−1) according to the following formula: plasmid copy number = NA × m/M, where m is the plasmid mass after isolation in grams and M is the molecular mass of plasmids in grams per mole. For calculation of adherent bacteria per EC, the following formula was used: binding ratio (bacteria/EC) = bacterial gene equivalent (rpoB, glyA)/0.5 EC gene equivalent (hmbs).
Statistical analysis and bioinformatics.
All experiments were performed at least three times. Differences between mean values of experimental and control groups were analyzed by Student's t test (Graph Pad Prism 4.03; Graph Pad Software, San Diego, CA, USA). A P value of <0.05 was considered to be statistically significant.
RESULTS
The aim of this study was to establish a new ex vivo organ infection model for analyzing bacterial adherence and to investigate whether Ata-dependent mechanisms of A. baumannii interactions with ECs are principally existing. For this approach, human umbilical cord veins were used, and adhesion of TAA-expressing and -deficient B. henselae and A. baumannii was analyzed. Quantification of adherent bacteria was done by fluorometry and qRT-PCR.
Quantification of bacterial adherence to ECs using fluorometry and quantitative qRT-PCR.
Adhesion of B. henselae to ECs is usually determined by counting cell-adherent bacteria, e.g., via fluorescence microscopy (2). It is obvious that such procedures—although they might be of use when working with slow-growing bacteria—are time-consuming and suffer from interobserver variability. Therefore, methods independent from microscopy or cultivation techniques might be useful for determining bacterial adherence. We established two independent technologies to overcome these limitations by using fluorometry and qRT-PCR analysis to quantify the adherent bacteria.
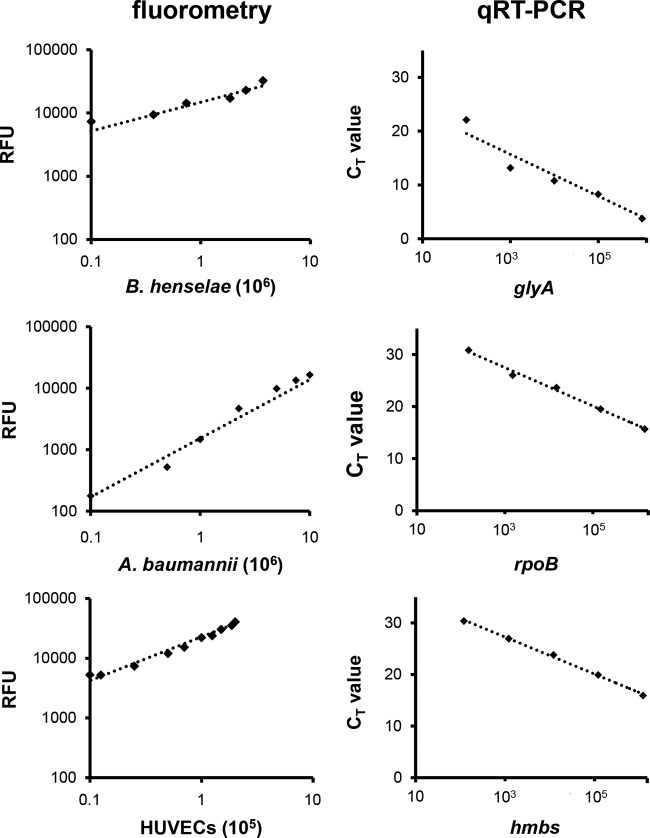
For fluorometric analysis, bacteria were stained with CFSE, and ECs were labeled with an anti-CD31-Alexa Fluor 647 antibody (see Materials and Methods), resulting in a bright green fluorescence of the stained bacteria and a bright orange fluorescence of the labeled ECs in fluorescence microscopy (data not shown). As emission and excitation spectra do not overlap, fluorescence was clearly distinguishable by fluorometry. The numbers of labeled bacteria (B. henselae and A. baumannii) and ECs showed a strict linear correlation with relative fluorescence units (RFU); the correlation coefficients were 0.978 (B. henselae), 0.989 (A. baumannii), and 0.993 (ECs) (Fig. 3).
FIG 3.
Quantification of bacteria and ECs using fluorometry or qRT-PCR. (Left) Correlation between bacterial CFSE- or EC-anti-CD31 fluorescence intensity quantified by fluorometry. y axis, relative fluorescence units (RFU). (Right) Correlation between CT values (y axis) of the respective amplified (log copy number, x axis) bacterial (B. henselae glyA; A. baumannii rpoB) or endothelial (hmbs) genes.
For the determination of bacterial adherence by the quantification of genome equivalents, a qRT-PCR method was developed. Therefore, specific primers for bacteria were designed (for B. henselae, targeting glyA; for A. baumannii, targeting rpoB) resulting in specific amplicons of 120 bp and 110 bp, respectively. For EC-specific amplicons, an hmbs PCR was used (209 bp) (Table 3). All primers were tested and found not to result in unspecific amplicons within the described setting (data not shown). The generation of internal standards was done by ligation of the respective amplicons into TOPO_TA vectors and transformation into Escherichia coli DH5α, and the correct insertion of the fragments was verified by PCR and restriction fragment length polymorphism (RFLP) (data not shown). Plasmids were propagated in E. coli DH5α, and the respective plasmid copy number was determined as described above (see Materials and Methods). Using these internal standards (from 10 to 106 copies), a linear correlation between threshold cycle (CT) values and log copy number was observed (Fig. 3). Correlation coefficients were 0.946 (B. henselae), 0.993 (A. baumannii), and 0.997 (ECs).
TAA-dependent adherence of B. henselae and A. baumannii to ECs under static infection conditions.
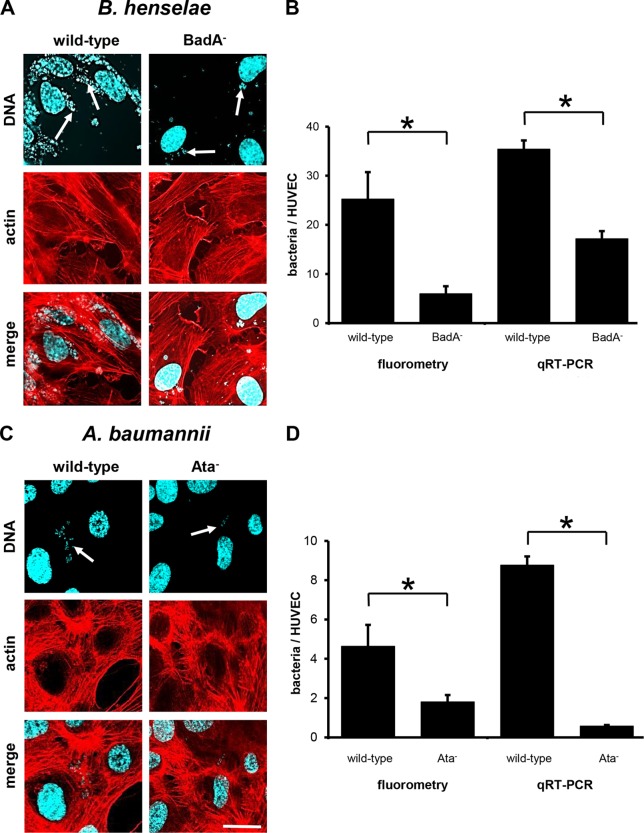
To analyze the TAA-dependent attachment of B. henselae and A. baumannii to ECs, HUVECs were infected as described above for 60 min. Adherence was determined via fluorescence microscopy, and as expected, BadA-expressing B. henselae adhered to ECs whereas B. henselae BadA− showed a clearly reduced cell adherence (Fig. 4A). Fluorescence microscopy findings correlated with those obtained by fluorometry or qRT-PCR (Fig. 4B). For wild-type B. henselae, ∼25.3 ± 5.5 adherent bacteria (by fluorometry) or ∼35.4 ± 1.8 bacteria per HUVEC (by qRT-PCR) were calculated. Binding rates were significantly reduced when HUVECs were infected with B. henselae BadA− (∼6.0 ± 1.5 bacteria [fluorometry] or ∼17.2 ± 1.6 bacteria per HUVEC [qRT-PCR]). To address the role of Ata in binding to ECs, an ata deletion mutant was constructed as described in Materials and Methods. It turned out that the presence or absence of Ata correlated with bacterial binding to ECs. As observed by fluorescence microscopy, total numbers of adherent bacteria were lower than numbers of B. henselae (Fig. 4C). These data were confirmed by fluorometry (wild type, ∼4.6 ± 1.1 bacteria per HUVEC) and qRT PCR (∼8.7 ± 0.4 bacteria per HUVEC) (Fig. 4D). EC adherence of Ata-deficient A. baumannii was significantly reduced (∼1.8 ± 0.4 bacteria [fluorometry] and ∼0.6 ± 0.1 bacteria [qRT PCR] per HUVEC). Taken together, loss of BadA or Ata expression resulted in an ∼3-fold (B. henselae) or ∼6-fold (A. baumannii) reduction in adherence to ECs.
FIG 4.
TAA-dependent adhesion of B. henselae and A. baumannii to HUVECs (static infection). HUVECs (1.0 × 105) were seeded on coverslips and infected with the respective bacteria (for B. henselae at an MOI of 100; for A. baumannii at an MOI of 200) for 60 min. For fluorescence microscopy (A, C), actin cytoskeleton of ECs was stained with TRITC-phalloidin (red), bacteria and nuclei were stained with DAPI (blue), and images were digitally overlaid (merge). Scale bar, 20 μm. (B, D) Adherence to ECs was determined in parallel by using fluorometry and qRT-PCRs. *, P < 0.05.
TAA-dependent adherence of B. henselae and A. baumannii to ECs under dynamic infection conditions.
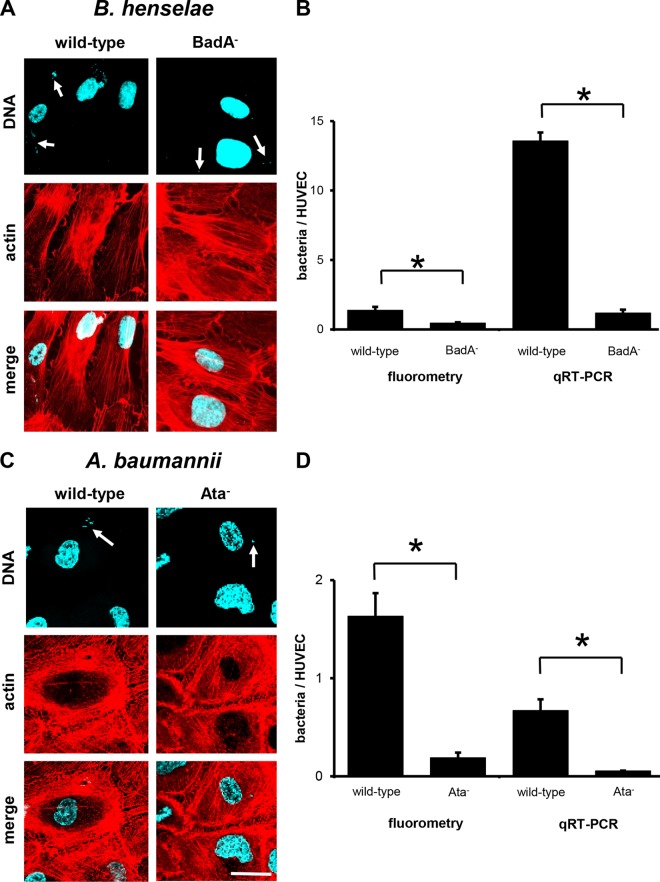
Next, we used the dynamic infection models to analyze the role of TAA-mediated EC binding under shear stress conditions (0.26 dyne/cm2). Here, only very few adherent bacteria (both B. henselae and A. baumannii) were detectable by fluorescence microscopy, indicating strongly reduced bacterial adherence under shear stress conditions (Fig. 5A and C). These observations were confirmed by fluorometry and qRT-PCRs: ∼1.4 ± 0.2 B. henselae wild-type isolates (fluorometry) and ∼13.6 ± 1.2 bacteria (qRT-PCR) adhered per HUVEC (Fig. 5B and D). For B. henselae BadA−, only ∼0.5 ± 0.1 bacteria (fluorometry) and ∼1.2 ± 0.5 bacteria (qRT-PCR) adhered to HUVECs. Similar results were obtained when analyzing dynamic EC binding of A. baumannii (Fig. 5D): for the wild type, ∼1.6 ± 0.2 bacteria (fluorometry) and ∼0.7 ± 0.1 bacteria (qRT-PCR) per HUVEC; for Ata-deficient bacteria, ∼0.2 ± 0.1 bacteria (fluorometry) and ∼0.1 ± 0.02 bacteria (qRT-PCR) per HUVEC. In summary, under dynamic infection conditions, TAA-deficient bacteria showed a more reduced EC adherence than under static infection conditions (B. henselae, ∼9-fold reduction; A. baumannii, ∼8-fold reduction).
FIG 5.
TAA-dependent adhesion of B. henselae and A. baumannii to HUVECs (dynamic conditions). HUVECs (1.0 × 104) were seeded on multichannel slides and infected with 1.0 × 108 bacteria per ml under constant flow conditions (flow rate, 0.2 ml per min) for 15 min (B. henselae) or 60 min (A. baumannii). (A, C) For fluorescence microscopy, actin cytoskeleton of ECs was stained with TRITC-phalloidin (red), bacteria and nuclei were stained with DAPI (blue), and images were digitally overlaid (merge). Scale bar, 20 μm. (B, D) Adherence to ECs was determined in parallel by using fluorometry and qRT-PCR. *, P < 0.05.
TAA-dependent adherence of B. henselae and A. baumannii to human umbilical cord endothelial cells ex vivo.
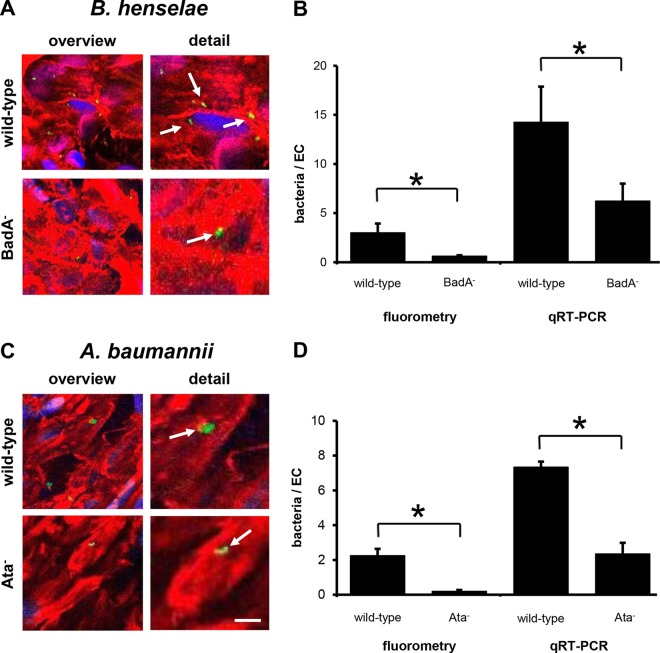
Finally, we transferred the infection procedures in an ex vivo organ infection model using human umbilical cords. Fresh human whole-organ grafts were prepared and dynamically infected with B. henselae and A. baumannii strains as described above. As the micro-CT revealed an umbilical cord vein with a diameter of 4.0 ± 0.6 mm (n = 20) (Fig. 2A), the mean shear stress was calculated at 0.5 ± 0.2 dyne/cm2. Moreover, flow pattern analysis via ultrasound and Doppler revealed a unidirectional and laminar flow (Fig. 2B). Fluorescence microscopy analysis of vessel samples (taken after 60 min of infection) (Fig. 6A and C) demonstrated TAA-dependent binding of B. henselae and A. baumannii to human endothelium to a certain degree. However, counting of EC-adherent bacteria was not feasible because of limitations in fluorescence microscopy due to the thickness of the microscopic samples. In contrast, fluorometry and qRT-PCR technology allowed to calculate the number of adherent bacteria in this ex vivo organ infection model. Here, total binding rates for B. henselae wild-type strain were ∼2.4 ± 0.9 bacteria (fluorometry) and ∼14.3 ± 3.6 bacteria (qRT-PCR) per EC. B. henselae BadA− showed a ∼3-fold-lower adhesion: ∼0.50 ± 0.08 bacteria (fluorometry) and ∼6.3 ± 1.8 bacteria (qRT-PCR) attached per human EC (Fig. 6B). For wild-type A. baumannii, ∼2.3 ± 0.4 bacteria (fluorometry) and ∼7.3 ± 0.3 bacteria (qRT-PCR) adhered per EC, whereas, in contrast, A. baumannii Ata− showed a ∼4-fold-lower adhesion (∼0.20 ± 0.08 bacteria [fluorometry] and ∼2.4 ± 0.6 bacteria [qRT-PCR] per EC) (Fig. 6D). Notably, total binding rates of B. henselae were similar to those obtained in the dynamic in vitro approach and those of A. baumannii were ∼4-fold higher.
FIG 6.
TAA-dependent adhesion of B. henselae and A. baumannii to human umbilical cord endothelial cells ex vivo. Human umbilical cord veins were infected for 60 min as described in Materials and Methods. (A, C) For fluorescence microscopy, actin cytoskeleton of ECs was stained with TRITC-phalloidin (red), bacteria were stained with CFSE (green) prior to infection, and nuclei were stained with DAPI (blue). Scale bar, 10 μm. (B, D) Adherent bacteria were quantified by using fluorometry (B) and qRT-PCRs (D). *, P < 0.05.
DISCUSSION
Adhesion of pathogenic bacteria to host cells is an early step for establishing an infection. Elucidating the details of how the bacteria attach to host cells is of great interest in infection biology research because it allows to understand this crucial pathogenicity mechanism. Moreover, such analyses might help to prevent infections by, e.g., identifying promising vaccination targets or via inhibiting adhesion to host cells by “antiligands.” These two strategies are getting more and more important as many pathogenic and multidrug-resistant Gram-negative bacteria (e.g., A. baumannii) have emerged in the last decade (27, 28).
The aim of our study was to develop an infection model that mimics infection conditions closer to those encountered in nature as previously used in in vitro approaches. For this purpose, we set up a dynamic human organ infection model using human umbilical cord veins. This model was proven to render reliable results in bacterial adherence for the two emerging pathogens B. henselae and A. baumannii.
Determination of bacterial adherence by fluorometry and qRT-PCR.
For the quantification of adherent bacteria, we set up two independent approaches based on fluorometry and qRT-PCR. Principally, our results revealed that fluorometry is an easy and valuable tool in analyzing bacterial adherence rates. We found that this method is highly reproducible, showing a linear correlation between the number of CFSE-stained bacteria or antibody-labeled ECs and the respective relative fluorescence units (Fig. 3). Using internal standards for fluorometry, the number of bacteria and cells was determinable, allowing to calculate the absolute number of adherent bacteria per endothelial cell. Further advantages are that no specific antibodies are needed for bacterial labeling and that adhesion of slow-growing bacteria like B. henselae to ECs can be analyzed within hours instead of weeks by omitting long-lasting bacterial subcultivation. CD31 (also known as platelet/endothelial cell adhesion molecule-1 [PECAM-1]) was used as a marker for ECs (29). It can be suggested that our fluorometry approach might be expanded to other cell types, since many specific cell antibodies are available (e.g., to lymphatic vessels specifically expressing the lymphatic vessel marker 1 [LYVE-1]) (30). Furthermore, bacterial adherence to host cells could be quantified using a qRT-PCR approach by determining the number of bacterial and host cell genome equivalents. This method was highly reproducible and also showed a linear correlation between the number of bacteria or ECs and the CT values of the specific amplified fragments. Using internal plasmid standards for qRT-PCR, the absolute number of adherent bacteria per EC can be determined.
In our study, the detection limits of fluorometry (upcoming background signals of PBS and unstained cells) are 50,000 bacteria and 5,000 ECs (Fig. 3). Therefore, this method is helpful in experiments in which such bacterial or host cell numbers can be expected. In contrast, qRT-PCR-based determination of adherence is more sensitive by nature (the limit of detection was one bacterium or one host cell). This method is more useful for experiments in which low numbers of bound bacteria are expected. Notably, the two methods revealed comparable results. In the case of B. henselae, e.g., under static infection conditions, ∼25 bacteria per EC were detected via fluorometry whereas ∼35 bacteria were calculated from qRT-PCR. These data are congruent to those from our earlier observations (2, 6). In the case of A. baumannii, ∼5 bacteria were found to be adherent to ECs by fluorometry and ∼9 bacteria via qRT-PCR. We conclude that both methods might expand the spectrum of methods in adherence research in future, as they might be adapted to many adherence assays with some minor modifications.
Notably, fluorescence microscopy of tissue samples (e.g., human umbilical cord tissue) is difficult by nature: on the one hand, the sample is relatively thick (>1 mm), and on the other hand, the so-called “Wharton's jelly” (substantia gelatina funiculi umbilicalis), which is the major structure component of the umbilical cord, prevents reliable fluorescence microscopy. Here, fluorometry and qRT-PCR-based bacterial quantification are clearly superior to microscopic techniques when analyzing bacterial adherence. The use of “multiphoton microscopy” might overcome such limitations, as thicker tissue samples (>1 mm) can be analyzed (31); however, such microscopes are highly expensive and of limited access.
TAA-dependent adherence of B. henselae and A. baumannii.
Since the role of BadA in mediating bacterial adherence to ECs has been analyzed in detail (2, 5, 6, 32, 33), B. henselae was used in our study as a control to calibrate the experiments with A. baumannii. Previous works described mainly Ata-dependent adhesion abilities of A. baumannii to ECMs under static infection conditions (3). In contrast, not much is known about the role of Ata in mediating the binding of A. baumannii to host cells, especially to ECs, or in a dynamic infection model. It is highly likely that at least in systemic infections (e.g., bloodstream infections) A. baumannii bacteria will interact with human endothelium (10, 11); however, this process has not been analyzed in detail. Therefore, our data show for the first time that Ata mediates in fact the binding of A. baumannii to human ECs. Our study revealed a ∼5-fold-higher binding of A. baumannii wild type to ECs than of isogenic Ata deletion mutants under static infection conditions. When ECs were infected under shear stress conditions, the binding rate decreased significantly. In static infection assays, ∼7 wild-type bacteria and ∼1 Ata-deficient bacterium attached to the ECs, whereas under shear stress the binding rates decreased to ∼1 wild-type bacterium and ∼0.2 Ata-deficient A. baumannii isolate. Interestingly, the binding rates to ECs of A. baumannii are much lower than those of B. henselae (see above). It can be assumed that the binding to ECs is much more pronounced in B. henselae, as this pathogen is an endotheliotropic bacterium (33). In contrast, ECs might not represent the typical target cells in A. baumannii infections, since this pathogen targets mainly respiratory and skin epithelia (34). Consequently, the exact molecular basis for endothelial or epithelial tropism of A. baumannii and the binding affinities still remain to be elucidated.
Infection of human tissue grafts: a step toward “organ microbiology.”
Cell culture-based static infection models are widely accepted in cellular microbiology and pathogenicity research. Recently, dynamic cellular and live murine infection models have been applied for analyzing the interaction of bacteria with ECs in vitro or using intravital microscopy, e.g., for B. henselae, Bartonella quintana, Yersinia enterocolitica, Neisseria meningitidis, Borrelia burgdorferi, and Staphylococcus aureus (6, 35–38). Although these methods are of high importance in the field of pathogenicity research, they suffer from some shortcomings by nature: (i) dynamic in vitro adherence assays are restricted to one particular cell line in an artificial experimental environment (lacking the complexity of natural human tissues; e.g., cell-cell-adhesions, extracellular matrices, mixed cell types, elasticity of human blood vessels) and (ii) cell growth conditions (addition of growth factors, fetal calf serum, etc.) or passaging of the cells might lead to unexpected cellular differentiation patterns influencing the specific characteristics of human cell lines (39); moreover, (iii) intravital microscopy is laborious and also limited to animal, mostly murine infection models. In the case of B. henselae, no appropriate murine infection model exists. Thus, alternative experimental settings are needed (33). Moreover, murine infection models (e.g., pneumonia models [3, 17]) might be limited in transferability to humans (4).
To overcome these restrictions, we developed a new infection model for analyzing bacterial adherence using human umbilical cords and shear stress conditions to mimic the human bloodstream conditions. Umbilical cord veins are suitable tools for this for many reasons: high disposability, the constant flow and shear stress allowed by the laminar structure of the veins, ethical acceptance, and the avoidance of animal infection experiments. We demonstrate that our infection model mimics the physiological shear stress of ∼0.5 dyne/cm2 and unidirectional laminar fluid flow in venous vessels. Similar conditions are found in the human blood circulation system (26), and thus our model is close to nature. Using this model, we demonstrated as a “proof of principle” that TAA-dependent adherence to ECs can be analyzed under dynamic flow infection conditions. Our experiments revealed for wild-type B. henselae total binding rates of ∼9.0 bacteria and for B. henselae BadA− up to ∼3.0 bacteria per EC (Fig. 6A and B). For A. baumannii, total binding rates were lower than for B. henselae; however, the Ata-deficient mutant showed ∼4-fold-lower adhesion rates than wild-type bacteria (Fig. 6C and D). Compared to dynamic in vitro infection settings, endothelial ex vivo bacterial adhesion was generally ∼2-fold higher. This might be caused by more-physiological additional binding sites in the human umbilical cord veins.
The work described here demonstrates that dynamic ex vivo infection models using human umbilical cords might represent a new, valuable tool in infection biology. Together with the calculation of adherent bacteria via fluorometry and qRT-PCR, this method has already allowed to analyze the role of the TAAs BadA and Ata in the infection process of human endothelial cells. Further studies with this newly developed infection model could allow detailed investigations of, e.g., host cell responses during dynamic infection procedures under more-physiological conditions. These findings might have the potential to expand the technologies present in “cellular microbiology” to a seminal new field in infection biology (“organ microbiology”).
Supplementary Material
ACKNOWLEDGMENTS
We thank Horst Buxmann and Rolf L. Schlößer (Department of Neonatology, University Hospital Frankfurt am Main), the midwife team from the Hospital Frankfurt Sachsenhausen for ensuring the support with fresh human umbilical cords, and Evelyn Skiebe for excellent technical assistance.
Funding Statement
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG-FOR 2251 to V. Kempf, S. Göttig, B. Averhoff, G. Wilharm, and V. Müller and DFG-SFB 815 to V. Kempf and R. Brandes).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01502-15.
REFERENCES
- 1.Schülein R, Seubert A, Gille C, Lanz C, Hansmann Y, Piemont Y, Dehio C. 2001. Invasion and persistent intracellular colonization of erythrocytes: a unique parasitic strategy of the emerging pathogen Bartonella. J Exp Med 193:1077–1086. doi: 10.1084/jem.193.9.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riess T, Andersson SG, Lupas A, Schaller M, Schäfer A, Kyme P, Martin J, Wälzlein J-H, Ehehalt U, Lindroos H, Schirle M, Nordheim A, Autenrieth IB, Kempf VA. 2004. Bartonella adhesin A mediates a proangiogenic host cell response. J Exp Med 200:1267–1278. doi: 10.1084/jem.20040500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentancor LV, Camacho-Peiro A, Bozkurt-Guzel C, Pier GB, Maira-Litran T. 2012. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J Bacteriol 194:3950–3960. doi: 10.1128/JB.06769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG. 2013. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kempf VA, Lebiedziejewski M, Alitalo K, Walzlein JH, Ehehalt U, Fiebig J, Huber S, Schütt B, Sander CA, Müller S, Grassl G, Yazdi AS, Brehm B, Autenrieth IB. 2005. Activation of hypoxia-inducible factor-1 in bacillary angiomatosis: evidence for a role of hypoxia-inducible factor-1 in bacterial infections. Circulation 111:1054–1062. doi: 10.1161/01.CIR.0000155608.07691.B7. [DOI] [PubMed] [Google Scholar]
- 6.Müller NF, Kaiser PO, Linke D, Schwarz H, Riess T, Schäfer A, Eble JA, Kempf VA. 2011. Trimeric autotransporter adhesin-dependent adherence of Bartonella henselae, Bartonella quintana, and Yersinia enterocolitica to matrix components and endothelial cells under static and dynamic flow conditions. Infect Immun 79:2544–2553. doi: 10.1128/IAI.01309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Rourke F, Mändle T, Urbich C, Dimmeler S, Michaelis UR, Brandes RP, Flotenmeyer M, Döring C, Hansmann ML, Lauber K, Ballhorn W, Kempf VA. 2015. Reprogramming of myeloid angiogenic cells by Bartonella henselae leads to microenvironmental regulation of pathological angiogenesis. Cell Microbiol 17:1447–1463. doi: 10.1111/cmi.12447. [DOI] [PubMed] [Google Scholar]
- 8.Göttig S, Gruber TM, Higgins PG, Wachsmuth M, Seifert H, Kempf VA. 2014. Detection of pan drug-resistant Acinetobacter baumannii in Germany. J Antimicrob Chemother 69:2578–2579. doi: 10.1093/jac/dku170. [DOI] [PubMed] [Google Scholar]
- 9.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 10.Seifert H, Strate A, Pulverer G. 1995. Nosocomial bacteremia due to Acinetobacter baumannii: clinical features, epidemiology, and predictors of mortality. Medicine 74:340–349. doi: 10.1097/00005792-199511000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Sebeny PJ, Riddle MS, Petersen K. 2008. Acinetobacter baumannii skin and soft-tissue infection associated with war trauma. Clin Infect Dis 47:444–449. doi: 10.1086/590568. [DOI] [PubMed] [Google Scholar]
- 12.Brossard KA, Campagnari AA. 2012. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect Immun 80:228–233. doi: 10.1128/IAI.05913-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77:3150–3160. doi: 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smani Y, McConnell MJ, Pachón J. 2012. Role of fibronectin in the adhesion of Acinetobacter baumannii to host cells. PLoS One 7:e33073. doi: 10.1371/journal.pone.0033073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eijkelkamp BA, Stroeher UH, Hassan KA, Paulsen IT, Brown MH. 2014. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics 15:1020. doi: 10.1186/1471-2164-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishikawa M, Nakatani H, Hori K. 2012. AtaA, a new member of the trimeric autotransporter adhesins from Acinetobacter sp. Tol 5 mediating high adhesiveness to various abiotic surfaces. PLoS One 7:e48830. doi: 10.1371/journal.pone.0048830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentancor LV, Routray A, Bozkurt-Guzel C, Camacho-Peiro A, Pier GB, Maira-Litran T. 2012. Evaluation of the trimeric autotransporter Ata as a vaccine candidate against Acinetobacter baumannii infections. Infect Immun 80:3381–3388. doi: 10.1128/IAI.06096-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VA. 2006. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol 14:264–270. doi: 10.1016/j.tim.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Leo JC, Grin I, Linke D. 2012. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philos Trans R Soc Lond B Biol Sci 367:1088–1101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyskowski A, Leo JC, Goldman A. 2011. Structure and biology of trimeric autotransporter adhesins. Adv Exp Med Biol 715:143–158. doi: 10.1007/978-94-007-0940-9_9. [DOI] [PubMed] [Google Scholar]
- 21.Riess T, Dietrich F, Schmidt KV, Kaiser PO, Schwarz H, Schäfer A, Kempf VA. 2008. Analysis of a novel insect cell culture medium-based growth medium for Bartonella species. Appl Environ Microbiol 74:5224–5227. doi: 10.1128/AEM.00621-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser PO, Linke D, Schwarz H, Leo JC, Kempf VA. 2012. Analysis of the BadA stalk from Bartonella henselae reveals domain-specific and domain-overlapping functions in the host cell infection process. Cell Microbiol 14:198–209. doi: 10.1111/j.1462-5822.2011.01711.x. [DOI] [PubMed] [Google Scholar]
- 23.Stahl J, Bergmann H, Göttig S, Ebersberger I, Averhoff B. 2015. Acinetobacter baumannii virulence is mediated by the concerted action of three phospholipases D. PLoS One 10:e0138360. doi: 10.1371/journal.pone.0138360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kempf VA, Schaller M, Behrendt S, Volkmann B, Aepfelbacher M, Cakman I, Autenrieth IB. 2000. Interaction of Bartonella henselae with endothelial cells results in rapid bacterial rRNA synthesis and replication. Cell Microbiol 2:431–441. doi: 10.1046/j.1462-5822.2000.00072.x. [DOI] [PubMed] [Google Scholar]
- 25.Gremse F, Grouls C, Palmowski M, Lammers T, de Vries A, Grull H, Das M, Muhlenbruch G, Akhtar S, Schober A, Kiessling F. 2011. Virtual elastic sphere processing enables reproducible quantification of vessel stenosis at CT and MR angiography. Radiology 260:709–717. doi: 10.1148/radiol.11110069. [DOI] [PubMed] [Google Scholar]
- 26.Malek AM, Alper SL, Izumo S. 1999. Hemodynamic shear stress and its role in atherosclerosis. JAMA 282:2035–2042. doi: 10.1001/jama.282.21.2035. [DOI] [PubMed] [Google Scholar]
- 27.Gruber TM, Göttig S, Mark L, Christ S, Kempf VA, Wichelhaus TA, Hamprecht A. 2015. Pathogenicity of pan-drug-resistant Serratia marcescens harbouring blaNDM-1. J Antimicrob Chemother 70:1026–1030. doi: 10.1093/jac/dku482. [DOI] [PubMed] [Google Scholar]
- 28.Göttig S, Gruber TM, Stecher B, Wichelhaus TA, Kempf VA. 2015. In vivo horizontal gene transfer of the carbapenemase OXA-48 during a nosocomial outbreak. Clin Infect Dis 60:1808–1815. doi: 10.1093/cid/civ191. [DOI] [PubMed] [Google Scholar]
- 29.Miettinen M, Lindenmayer AE, Chaubal A. 1994. Endothelial cell markers CD31, CD34, and BNH9 antibody to H- and Y-antigens: evaluation of their specificity and sensitivity in the diagnosis of vascular tumors and comparison with von Willebrand factor. Mod Pathol 7:82–90. [PubMed] [Google Scholar]
- 30.Jackson DG. 2003. The lymphatics revisited: new perspectives from the hyaluronan receptor LYVE-1. Trends Cardiovasc Med 13:1–7. doi: 10.1016/S1050-1738(02)00189-5. [DOI] [PubMed] [Google Scholar]
- 31.Werkmeister E, Kerdjoudj H, Marchal L, Stoltz JF, Dumas D. 2007. Multiphoton microscopy for blood vessel imaging: new non-invasive tools (Spectral, SHG, FLIM). Clin Hemorheol Microcirc 37:77–88. [PubMed] [Google Scholar]
- 32.Lu YY, Franz B, Truttmann MC, Riess T, Gay-Fraret J, Faustmann M, Kempf VA, Dehio C. 2013. Bartonella henselae trimeric autotransporter adhesin BadA expression interferes with effector translocation by the VirB/D4 type IV secretion system. Cell Microbiol 15:759–778. doi: 10.1111/cmi.12070. [DOI] [PubMed] [Google Scholar]
- 33.Kaiser PO, Riess T, O'Rourke F, Linke D, Kempf VA. 2011. Bartonella spp.: throwing light on uncommon human infections. Int J Med Microbiol 301:7–15. doi: 10.1016/j.ijmm.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Visca P, Seifert H, Towner KJ. 2011. Acinetobacter infection: an emerging threat to human health. IUBMB Life 63:1048–1054. doi: 10.1002/iub.534. [DOI] [PubMed] [Google Scholar]
- 35.Soyer M, Dumenil G. 2012. A laminar-flow chamber assay for measuring bacterial adhesion under shear stress. Methods Mol Biol 799:185–195. doi: 10.1007/978-1-61779-346-2_12. [DOI] [PubMed] [Google Scholar]
- 36.Claes J, Liesenborghs L, Lox M, Verhamme P, Vanassche T, Peetermans M. 2015. In vitro and in vivo model to study bacterial adhesion to the vessel wall under flow conditions. J Vis Exp 100:e52862. doi: 10.3791/52862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laschke MW, Kerdudou S, Herrmann M, Menger MD. 2005. Intravital fluorescence microscopy: a novel tool for the study of the interaction of Staphylococcus aureus with the microvascular endothelium in vivo. J Infect Dis 191:435–443. doi: 10.1086/427193. [DOI] [PubMed] [Google Scholar]
- 38.Moriarty TJ, Norman MU, Colarusso P, Bankhead T, Kubes P, Chaconas G. 2008. Real-time high resolution 3D imaging of the Lyme disease spirochete adhering to and escaping from the vasculature of a living host. PLoS Pathog 4:e1000090. doi: 10.1371/journal.ppat.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartung T. 2013. Food for thought look back in anger—what clinical studies tell us about preclinical work. ALTEX 30:275–291. doi: 10.14573/altex.2013.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drancourt M, Birtles R, Chaumentin G, Vandenesch F, Etienne J, Raoult D. 1996. New serotype of Bartonella henselae in endocarditis and cat-scratch disease. Lancet 347:441–443. doi: 10.1016/S0140-6736(96)90012-4. [DOI] [PubMed] [Google Scholar]
- 41.Hornsey M, Ellington MJ, Doumith M, Thomas CP, Gordon NC, Wareham DW, Quinn J, Lolans K, Livermore DM, Woodford N. 2010. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemother 65:1589–1593. doi: 10.1093/jac/dkq218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.