SUMMARY
Bacteria display an abundance of cellular forms and can change shape during their life cycle. Many plausible models regarding the functional significance of cell morphology have emerged. A greater understanding of the genetic programs underpinning morphological variation in diverse bacterial groups, combined with assays of bacteria under conditions that mimic their varied natural environments, from flowing freshwater streams to diverse human body sites, provides new opportunities to probe the functional significance of cell shape. Here we explore shape diversity among bacteria, at the levels of cell geometry, size, and surface appendages (both placement and number), as it relates to survival in diverse environments. Cell shape in most bacteria is determined by the cell wall. A major challenge in this field has been deconvoluting the effects of differences in the chemical properties of the cell wall and the resulting cell shape perturbations on observed fitness changes. Still, such studies have begun to reveal the selective pressures that drive the diverse forms (or cell wall compositions) observed in mammalian pathogens and bacteria more generally, including efficient adherence to biotic and abiotic surfaces, survival under low-nutrient or stressful conditions, evasion of mammalian complement deposition, efficient dispersal through mucous barriers and tissues, and efficient nutrient acquisition.
INTRODUCTION
Van Leeuwenhoek's microscopes opened our eyes to the microbial world and its abundance of forms. Bacteria were initially characterized on the basis of morphology, but over time, phylogenetics has proven to be a more robust method for classifying bacteria. In the clinic, biochemical characteristics (recently including those observed by mass spectrometry) and growth on various indicator media robustly distinguish bacterial pathogens, and thus there is little incentive to directly observe bacteria. Even in the research lab, cell biology approaches often focus on morphological changes to the host cell during infection, not the morphology of the pathogen itself. Still, both pathogenic and nonpathogenic bacteria show considerable morphological diversity. In this review, we consider the functional consequences of differences in form on bacterial survival in native environments (as opposed to culture flasks of rich media). The work reviewed here builds on pioneering studies and hypotheses described in several excellent reviews on the possible selective consequences of bacterial cell shape diversity (1–5).
We explore several aspects of morphological diversity (Table 1). Bacteria have distinct cell body shapes, ranging from spheres (cocci) to rods (bacilli) of various curvatures and helicities and to more exotic shapes, such as stars, formed by elaboration of prosthecae through polar growth. Bacteria can also produce a variety of appendages, such as pili or flagella, which show diversity in overall shape, length, and width as well as placement with respect to the cell body. Finally, bacteria can change morphology during their life cycle or in response to environmental conditions. Much of our understanding of the mechanisms that produce different shapes comes from the study of model organisms under laboratory growth conditions. Increasingly, researchers studying pathogenic bacteria in the context of infection models have observed morphological diversity and/or observed that mutants with altered colonization or virulence properties have an altered shape. This has led to speculation that cell shape itself may be a virulence factor or that the mammalian host environment(s) imposes a selective pressure driving morphological diversity. In some cases, studies of shape in model organisms under conditions mimicking natural environments have revealed selective forces that likely are relevant to pathogenic lifestyles. This field has made significant progress recently in part due to advances in imaging that allow interrogation of individual bacterial cells on multiple time scales and resolution of bacterial subcellular structures. While this review focuses mostly on recent work on pathogens, we also highlight work on nonpathogenic organisms that illuminates functional principles and methodologies that will likely translate to the study of pathogens.
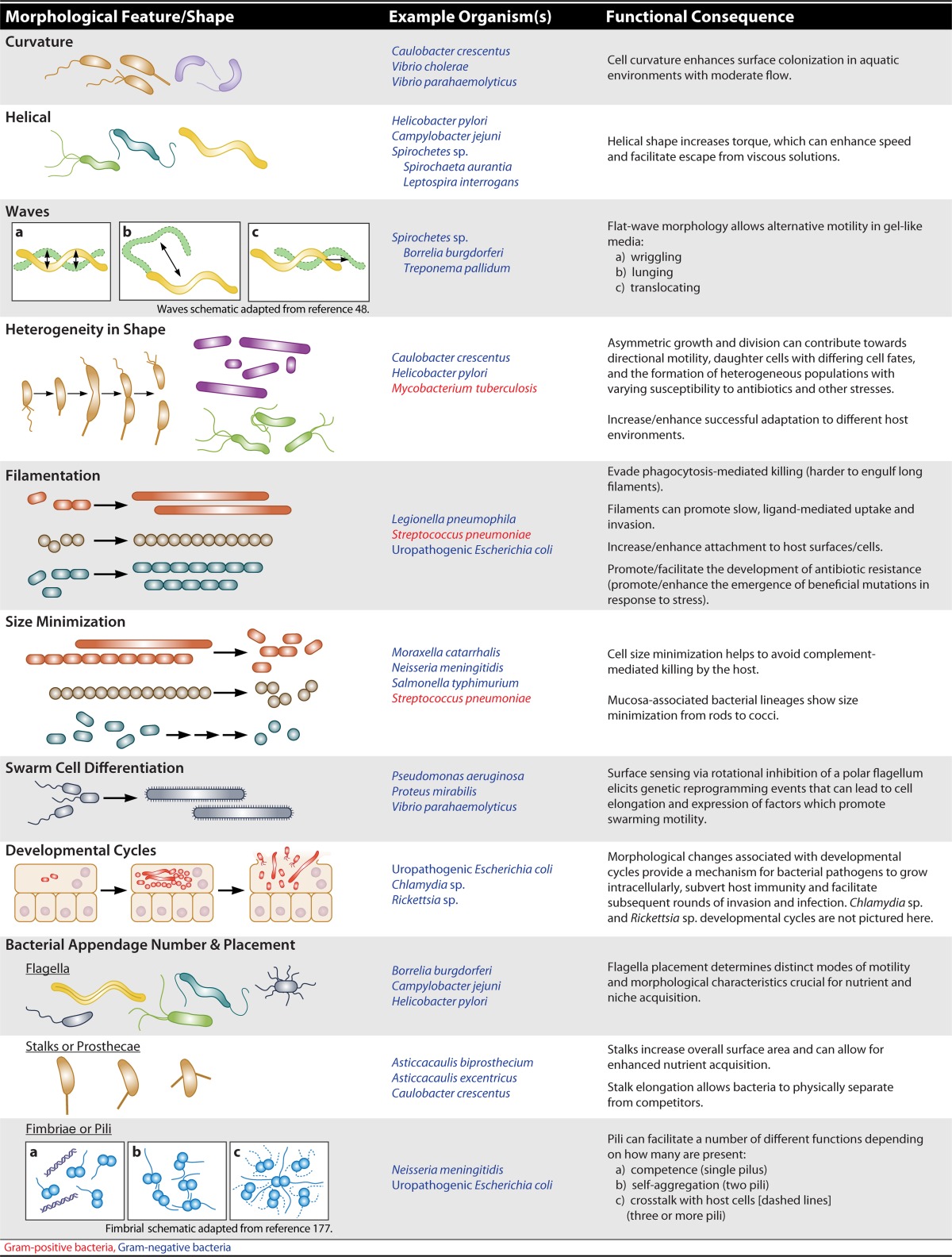
TABLE 1.
Examples of bacterial morphological variation and functional consequencesa
CELL BODY MORPHOLOGY
In most bacteria, the cell wall determines the shape of the cell. Peptidoglycan (PG) is the major structural component of the cell wall in both Gram-positive and Gram-negative bacteria. Thus, much research in the bacterial morphology field focuses on defining the mechanisms by which the cell wall is synthesized and modified to produce different cell body shapes. We refer the reader to several excellent reviews on cell wall synthesis and the evolution of bacterial morphogenesis for an in-depth consideration of this topic (6–9). To summarize, studies in a variety of organisms have revealed that cell shape can be derived from asymmetric positioning of new cell wall synthesis, alteration of cell wall thickness, and/or changes in the chemical composition of the cell wall polymer or its extent of cross-linking. Because the cell wall provides tensile strength and a diffusion barrier in addition to specifying cell shape, studies linking changes in cell morphology to particular phenotypes must carefully consider cell wall structural alterations as a possible confounder.
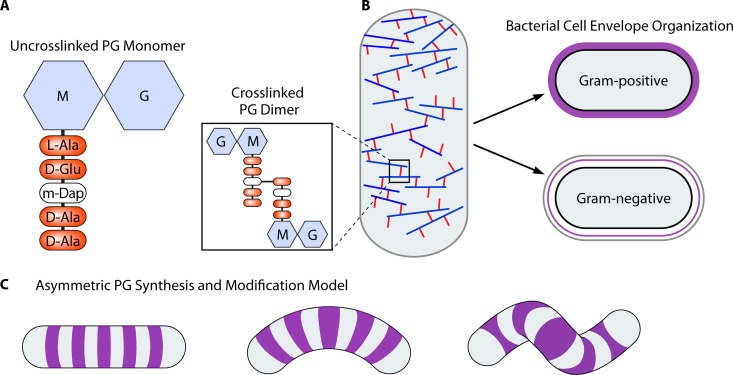
Because the structure of the PG cell wall is so intimately coupled with cell shape, we summarize its major features in Fig. 1. The PG polymer consists of a repeating disaccharide with a short peptide stem attached to one of the sugars (Fig. 1A). Transpeptidation between peptide stems can link the sugar strands together, forming a rigid meshwork that encases the cell and prevents lysis due to turgor pressure. The biosynthesis of this essential structure has been an important antibiotic target and an active area of research, which is the subject of several excellent reviews (6, 10, 11). In Gram-negative bacteria, the PG cell wall resides in the periplasm between the inner and outer membranes. Multiple lines of evidence suggest that the PG layer in Gram-negative bacteria is relatively thin, perhaps a single monolayer, with the glycan strands oriented primarily in a circumferential direction as a disordered lattice (Fig. 1B) (6, 10–14). The outer membrane of Gram-negative bacteria protects the PG from lytic enzymes produced by the host or other microorganisms. Gram-positive bacteria lack an outer membrane and have a thicker PG layer that is exposed to the external environment and includes additional structural constituents, such as wall teichoic acids and mycolic acids (15). Recent work suggests that even though the Gram-positive cell wall has multiple layers, it has a similar organization of circumferentially oriented glycan strands, with newly synthesized material incorporated on the inner face (16). PG is thought to be the structural determinant of cell shape because purified PG sacculi retain the shape of the cell from which they are isolated, be it coccoid or a straight, curved, or helical rod (17–19). The major theoretical model of shape generation for the cell wall posits asymmetric synthesis driven by cytoskeletal elements that direct the activity of PG synthesis enzymes to particular regions of the cell (7), though mathematical modeling has suggested that changes in cross-linking could also drive alternate shapes (Fig. 1C) (20). In this section, we explore the functional consequences of different cell shapes. We limit our discussion of the underlying mechanisms of generating cell shape to the possible selective forces that may have driven the convergent evolution of specific cell shapes in different bacterial lineages.
FIG 1.
Cell wall composition and architecture in different bacterial classes and shapes. (A) Monomeric and cross-linked peptidoglycan (PG) structure depicting the N-acetylglucosamine (G)–N-acetylmuramic acid (M) disaccharide with a cross-link mediating meso-diaminopimelic acid (m-Dap) in the third position of the pentapeptide stem. (B) Glycan strands (repeating PG disaccharide subunits) are generally arranged circumferentially around the cell in both Gram-positive and Gram-negative bacteria. Gram-positive bacteria have a thick layer of PG (shown in purple), while Gram-negative bacteria have a thin layer between the inner (black outline) and outer (gray outline) membranes. (C) Model for the generation of curvature and twist in curved and helical bacteria whereby asymmetric wedges or zones of PG synthesis and/or modification may promote increased or decreased PG incorporation on one side or region of the cell.
The Not-So-Simple Sphere
Perhaps the simplest bacterial cell shape that can be adopted is that of a sphere. Bacteria displaying this shape are classified as either coccoid (entirely round, such as staphylococci and micrococci) or ovococcoid (slightly ovoid, such as streptococci or lactococci) (Table 1). A number of bacterial pathogens fall into these morphological categories, including the Gram-positive bacteria Staphylococcus aureus (coccoid) and Streptococcus pneumoniae (ovococcoid) as well as the Gram-negative coccoid pathogens Neisseria gonorrhoeae and Neisseria meningitidis. Both S. aureus and S. pneumoniae have been studied in great depth by numerous groups over the years, and much of our understanding of bacterial processes in cocci has come from this body of work.
Studies looking at PG synthesis in both coccoid and ovococcoid bacteria have helped to inform our understanding of how these basic shapes can be generated. It has been proposed that at least two different PG machineries exist to generate the ovococcoid shape, whereas only one PG synthesis complex is required to form a coccoid cell. Coccoid bacteria, such as S. aureus, grow the cell wall exclusively by using the cell division machinery, which drives the synthesis of septal PG. However, S. pneumoniae and other ovococcoid bacteria synthesize new cell walls by using both the cell division machinery (septal PG) and an elongation complex (peripheral PG) (21, 22). Incomplete or delayed cell division of ovococci results in chains or filaments, as these cells divide in one plane perpendicular to the long axis of the cell. Coccoid cells, however, alternate their division axes along two or three planes, which can result in clusters of cells if division is inefficient (21, 22). Since coccoid cells synthesize new PG strictly via their division septa, these cells are often found as diplococci because they must divide in order to grow their cell wall (23).
The possible implications for bacteria possessing one or more PG synthesis machineries are intriguing. One might speculate that coccoid species are more streamlined in the number of factors required for PG synthesis, since these organisms use only the cell division machinery to make new cell walls. This implies that the total number of cell wall genes in coccoid bacteria is smaller and that these genes contain less redundancy than in organisms possessing complexes for both septal and peripheral PG incorporation. Since the cell wall is a target for many important antibiotics, including the β-lactams, this raises the possibility that these differences may affect the susceptibility of these organisms to treatments that specifically disrupt PG biosynthesis. Interestingly, a bioinformatic search found that many coccoid pathogens have an expanded number of PG synthesis enzymes (24). Systematic deletion of seven of the nine PG synthesis genes in S. aureus resulted in a mutant whose morphology and growth appeared to be wild type under standard laboratory conditions, indicating that only two PG synthesis enzymes are required for in vitro growth (24). However, this mutant showed increased susceptibility to lytic enzymes, such as lysozyme, and to cell wall-specific antibiotics and was attenuated for infection and killing in Drosophila flies (24). These findings demonstrate that many of the PG synthesis enzymes are functionally redundant in vitro (24) and suggest that bacteria are under selective pressure to maintain this redundancy in order to survive in more diverse environments.
Although the coccoid morphology is the simplest bacterial form to generate, it is not believed to be the shape of the first bacterium. Multiple lines of evidence support the model that the ancestral bacterial morphology was that of a rod, not a sphere. Phylogenetic analyses indicate that the coccal morphology evolved multiple times in very distantly related bacteria and that once a lineage displays this particular shape, subsequent branches are always coccoid, suggesting that this is the degenerate form (25–27). These observations imply that coccoid cells result from rods that have lost the ability to elongate. Moreover, examples of rods changing into spheres have been demonstrated both genetically and biochemically with the use of antibiotics, thus lending support to this model (28–32). Indeed, a recent paper describing the possible evolutionary steps that might have driven the bacillus-to-coccoid shape transition in Neisseria meningitidis provides evidence that the process happened in two distinct genetic steps. The authors further argue that convergent evolution in Moraxella catarrhalis resulted in a similar shape transition for a pathogen that occupies the same niche as N. meningitidis, suggesting that environmental selective pressure was the driving force behind this shape change (32).
The Default Shape
Escherichia coli (Gram negative) and Bacillus subtilis (Gram positive) are two of the most extensively investigated rod-shaped bacteria. Studies of these two model organisms have laid the groundwork for much of what we know about cell shape generation in both rods and nonbacillary bacteria. Most rods result from a combination of septal cell wall growth and peripheral sidewall elongation; however, some rods, such as mycobacteria, agrobacteria, and corynebacteria, elongate specifically at the poles (33–36). For Proteus mirabilis, a rod-shaped Gram-negative bacterium that can cause urinary tract infections, evidence suggests that the bacillary morphology may be important for multicellular swarming motility (refer to “Running with the Herd,” below) (37). A P. mirabilis mutant displaying a curved-rod morphology could differentiate into elongated swarmer cells but was defective in swarming motility (37). It is thought that these irregularly shaped cells cannot properly align to form multicellular rafts for productive swarming, indicating the importance of rod shape maintenance for this organism. Recent findings from work on CetZ1, an archaeal FtsZ/tubulin homolog in Haloferax volcanii, support this idea. It was found that upon disruption of CetZ1 function, H. volcanii cells lose their rod shape and turn into irregularly shaped cells with reduced swimming speed, suggesting that the archaeal rod shape has been maintained to promote robust swimming motility (38).
Crescents
Perhaps one of the best-known examples for how a particular bacterium achieves its unique cell shape can be found in the model organism Caulobacter crescentus, which resides in freshwater streams and lakes. As its name implies, C. crescentus displays a characteristic crescent shape or vibrioid morphology that is determined by a single protein, crescentin (39, 40). Crescentin is a bacterial intermediate filament-like protein that possesses multiple coiled-coil domains (39). Although crescentin homologs appear to be restricted to the Caulobacter genus, many bacterial genomes contain coiled-coil-rich proteins (Ccrps) (39, 41). Null mutations in creS, the gene that encodes crescentin, result in straight-rod cells that have no obvious growth defects under standard laboratory conditions (39). However, isolates of C. crescentus from the wild maintain the vibrioid morphology, strongly suggesting that this particular shape provides a selective benefit in nature (42). Intriguingly, many other aquatic microorganisms adopt a vibrioid morphotype, bolstering the idea that this shape is somehow selectively maintained (3).
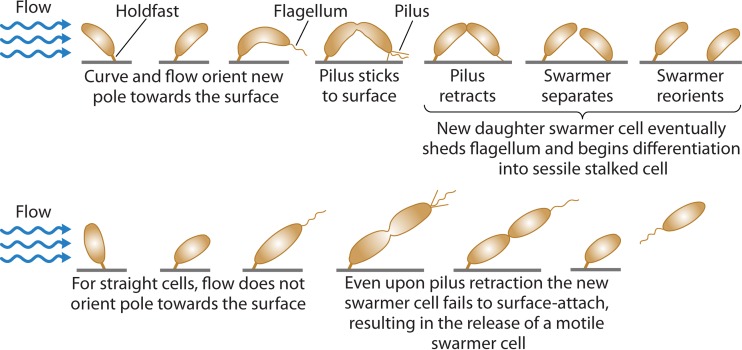
The first experimentally supported model for why C. crescentus displays its distinctive curved-cell shape was reported by Persat and colleagues (42). They demonstrated that the C. crescentus cell curvature enhances the surface colonization of this bacterium under conditions where there is moderate flow over the cells (Fig. 2) (42). Using time-lapse imaging to monitor microcolony formation of wild-type cells (crescent rods) in competition with a straight-rod creS mutant, Persat et al. found that the wild-type cells outcompeted their straight-rod counterparts on surfaces with flow. The data support this model, showing that once a wild-type cell is initially attached to a surface, it elongates in preparation for cell division. Under moderate amounts of flow, the curvature of the growing cell causes it to orient into an arc that eventually brings the piliated pole of the emerging daughter cell in closer proximity to the surface. Seconds before complete cell division, the surface-attached stalked cell undergoes a single pilus retraction event which increases the likelihood of daughter cell attachment. The new surface-attached sessile swarmer cell eventually sheds its flagellum and redifferentiates into a stalked cell (Fig. 2) (42, 43). Taken together, these findings provide a mechanistic model for how a given cell shape can be advantageous and begin to uncover the selective pressures that drive the maintenance and propagation of specific bacterial morphologies (42).
FIG 2.
Model for how the curved shape of Caulobacter crescentus enhances surface colonization under flow conditions. The curved shape of the bacterium allows the dividing cell to reorient the new daughter cell pole closer to the surface, thus increasing the likelihood of attachment. The dividing cell also increases the probability of daughter cell attachment by retracting the pilus right before complete division. Cells lacking curvature cannot orient the new daughter cell pole closer to the surface, resulting in the release of the swarmer daughter cell instead of surface attachment and colonization. (Adapted from reference 42 by permission from Macmillan Publishers Ltd.)
For other curved rods, including the human pathogens Vibrio cholerae and Vibrio parahaemolyticus, it is less clear how this particular shape is achieved (19). These bacteria do not possess an equivalent creS homolog, indicating that the vibrioid morphology is generated by an alternate mechanism. Interestingly, members of the Vibrionaceae family contain homologs of genes that promote helical shape in Helicobacter pylori and Campylobacter jejuni (described in the next section) (19, 44–47). Identification of the genes governing curved shape in the Vibrionaceae would allow exploration of the functional significance of the vibrioid shape of these organisms in the aquatic environment, the mammalian host, or both.
Corkscrews and Waves
Helical shapes have arisen multiple times during the evolution of bacteria and are represented by distinct groups of bacterial pathogens that include the spirochetes (e.g., Treponema, Spirochaeta, Leptospira, and Borrelia spp.) and the proteobacteria (e.g., gastric and nongastric Helicobacter spp. and the foodborne pathogen Campylobacter jejuni), all of which cause a wide variety of diseases in humans and animals. It has been shown for both helical proteobacteria and spirochetes that a loss of motility and/or helical cell shape causes major defects in the colonization potential of these organisms (19, 47, 48). For clarity, we define a helical or flat-wave-shaped cell as distinct from a spiral-shaped cell, although these descriptors are often used interchangeably in the literature to define truly helical cells with a fixed or constant helical radius along the length of the cell body. While it is true that, by definition, a spiral may be described as a helix, a spiral-shaped cell would have a continually expanding helical radius (i.e., a nautilus shell), and to our knowledge, there are no described bacteria with a truly spiral morphology.
Interestingly, helical and planar-wave cell morphologies appear to be found mainly in Gram-negative bacteria. A notable exception is the helical shape of Streptomyces aerial hyphae that result from polar growth (49). The relative restriction of helical shape to Gram-negative bacteria is likely a consequence of the biophysical properties of the PG cell wall which encases the cell. Because Gram-positive bacteria possess a thick, multilayered PG sacculus surrounding the cell, it is likely a more rigid structure than the single layer of PG found in Gram-negative bacteria (Fig. 1). In order to deviate from a straight-rod morphology, there must be a mechanism of asymmetric incorporation of new PG into the cell wall and/or localized changes in PG cross-linking (19). Consistent with this hypothesis, the majority of the genes involved in cell shape determination in the proteobacteria H. pylori and C. jejuni alter the cross-linking or peptide composition of the cell wall when absent or overexpressed (19, 45–47, 50, 51). There is evidence for genetic and functional conservation of helical shape-generating pathways. For example, pgp1, a homolog of the H. pylori csd4 gene, was identified in C. jejuni and shown to be important for proper cell shape (47, 50).
Spirochetes have evolved alternative mechanisms for generating helical shape as well as flat-wave morphology. Some spirochetes have coupled their motility functions with cell shape in the form of periplasmic flagella (PF). In Borrelia burgdorferi (which causes Lyme disease and is transmitted to mammals via the tick Ixodes scapularis), the inherent wave-form structure of the periplasmic flagella imparts the flat-wave or planar-wave morphology of the bacterium. Deletion of the periplasmic flagella results in the loss of both motility and helical shape in B. burgdorferi (52). When the flagella rotate within the periplasmic space, they deform the shape of the cell envelope and the PG layer, leading to a propagating wave that travels down the cell axis, displacing fluid to propel the bacterium forward. Biophysical modeling studies of B. burgdorferi PF indicate two distinct wave forms of the PF, with one of these wave forms imparting the flat or planar-wave shape of the bacterium (53). The number and positioning of these PF are quite diverse within the spirochetes and impart distinct modes of motility. In these studies, it was suggested that different species of Borrelia alter their morphology by adjusting the number and ratio of different flagellar wave forms, which could lead to increased or decreased stiffness of the periplasmic flagellar bundle relative to the stiffness of the cell body.
Interestingly, the spirochetes Treponema primitia, Spirochaeta aurantia, and Leptospira interrogans do not undergo cell wall deformation, and their morphology does not change when the flagella rotate (54, 55). In L. interrogans, the PF do not extend the entire length of the cell, and this organism retains a helical shape in mutants lacking flagella (56). Furthermore, purified PG sacculi from L. interrogans have a helical morphology, suggesting that the shape-determining function of PF is less important for this bacterium (57).
The functional significance of helical cell shape has been hypothesized to promote enhanced motility through viscous solutions or gels via a corkscrew or burrowing model of motility (56). Until recently, there was little experimental evidence to support this theory beyond the early and foundational experiments investigating the motility of spirochetes [Leptospira interrogans (biflexa), Spirochaeta aurantia, and Spirochaeta halophila P1] in high-viscosity solutions of polyvinylpyrrolidone or methylcellulose. Greenberg and Canale-Parola measured the minimum inhibitory viscosity (MIV) of these organisms and found that their translational motility was enhanced under conditions of high viscosity, unlike their motile and flagellated E. coli counterparts, in which motility was inhibited in high-viscosity solutions (56). Furthermore, cell shape mutants of S. aurantia with reduced coiling (i.e., reduced helicity) but otherwise normal motility had an MIV of 1/10 that of the normally coiled parental strain, suggesting a direct role for helical shape in promoting motility through a viscous environment (56).
Translational motility mediated by chemotaxis is a major contributor to the pathogenesis of diverse pathogens, including H. pylori, which regulates its chemotactic behavior to properly colonize the gastric glands in a mouse stomach model (58–60). For the spirochete B. burgdorferi, a major challenge is to separate the distinct contributions of cell shape and motility, as they are intimately linked. To address this, Harman et al. (48) developed an in vitro mouse dermis model to investigate the temporal dynamics of B. burgdorferi motility and demonstrated that B. burgdorferi exhibits multiple motility states (nonmotile, wriggling, lunging, and translocation) due to transient adherence to host dermis tissues (Table 1). They also showed that these bacteria are able to “wriggle” through pores smaller than their own diameter, suggesting a certain degree of elasticity or malleability to the cell wall, a property not often considered in thinking of shapes. It has become apparent that the helical and flat-wave shapes that are produced by unique flagellar arrangements likely promote motility through multiple mechanisms.
More recently, studies of the pathogens H. pylori and C. jejuni provided further support for the functional importance of helical cell shape in the natural environment of the host. For H. pylori, nonhelical cell shape mutants exhibited stomach colonization defects in a murine colonization model. In these experiments, mice were challenged with equal amounts of wild-type (helical) and mutant (curved-rod) H. pylori bacteria. After a 1-week infection, the helical bacteria outnumbered the nonhelical mutants by a magnitude of 1 log10, demonstrating that helical shape is an important pathogenesis factor (19, 44). Additionally pgp1, a homolog of the H. pylori csd4 gene, was identified in C. jejuni and shown to be important for proper cell shape (mutants show a straight-rod morphology) and colonization of chicks (poultry are a major reservoir of C. jejuni and a major source of human infection) (47).
Studies utilizing soft agar to investigate motility are commonly utilized to mimic gel-like environments or environments with increased viscosity in which the bacteria naturally reside. However, there are important differences between gels and solutions of high viscosity. For example, a study on the motility of B. burgdorferi revealed multiple motility states for this pathogen in both a gelatin matrix and a murine dermis model that had not previously been observed using viscous methylcellulose solutions (48). Straight-rod C. jejuni pgp1 and H. pylori csd4 mutants exhibit soft agar motility defects, again suggesting an important synergy between cell shape and motility toward the pathogenic program of these organisms (47, 50).
H. pylori resides in and colonizes the epithelial cell surface of the human stomach. To reach its niche, this bacterium must escape the highly acidic gastric lumen and traverse the gastric mucus layer. Gastric mucins display pH- and concentration-dependent viscoelastic properties, behaving as a viscoelastic gel at low pH or high mucin concentrations (>25 mg/ml) and as a viscoelastic solution at neutral pH and lower mucin concentrations (61–64). It has been proposed that H. pylori utilizes the corkscrew model to penetrate and traverse the mucus gel to escape the acidic lumen, which is consistent with the motility defects observed for straight and curved mutants in soft agar. However, the localized production of urease by H. pylori actually changes the viscoelastic properties of the surrounding mucus from a gel-like state to a viscous solution, thus challenging the corkscrew model (61). While initial studies suggested that like B. burgdorferi, H. pylori cell shape mutants showed swimming behavior similar to that of the wild type in methylcellulose viscous solutions (19), single-cell tracking of multiple H. pylori strains revealed large temporal fluctuations in the speed of individual bacteria that may have confounded prior studies (65). Analysis of 50 to 100 cells per genotype revealed contributions of both helical shape and increased flagellar number to swimming speed in viscous gastric mucin solutions, in addition to an enhanced ability to escape from a mucin gel (65). Further work on motility specifically at physiologic concentrations of gastric and intestinal mucins, combined with exploration of the localization of helical versus nonhelical mutants within the infected host, will be needed to definitively establish the mechanism(s) by which nonhelical mutants of H. pylori and C. jejuni fail to robustly colonize their hosts.
Nonidentical Twins
Bacteria divide by binary fission, usually producing daughter cells of equal size. However, these resulting cells are intrinsically asymmetric, possessing an old pole (originating from the mother cell) and a new pole (originating from the most recent cell division event). Mounting evidence suggests that bacteria take advantage of this inherent polarity for a number of important cellular processes, including directional motility, producing daughter cells with different cell fates, and generating population heterogeneity in cell sizes and growth rates (66). C. crescentus displays one of the more extreme examples of asymmetric cell division, producing daughter cells that differ in size, morphology, replicative potentials, and function (67, 68). The larger, sessile, “stalked” cell possesses a polar prostheca or stalk, which enables the cell to attach to surfaces via an exopolysaccharide-based holdfast (69, 70). The stalked cell is competent for DNA replication and initiates this program directly after division. The smaller, “swarmer” cell is motile and capable of chemotaxis by way of a single polar flagellum, but it cannot replicate its chromosome. However, under the right conditions, the swarmer cell can differentiate into a stalked cell and initiate DNA replication. Although more is known about C. crescentus asymmetric cell division than perhaps that of any other bacterium, many of the mechanistic details have yet to be elucidated fully. What is apparent, however, is that many bacteria, including several important human pathogens, exploit this asymmetry in cell division (Table 1).
Rod-shaped actinomycetes, such as mycobacteria, and corynebacteria grow in a distinct polar manner (33–35). In contrast to the case in most rod-shaped bacteria, such as E. coli and B. subtilis, where longitudinal growth results from insertion of new cell wall material into the sidewall, polar growth results from new cell wall material inserted only at the cell poles. Interestingly, in mycobacteria this polar growth results in unequally sized daughter cells that elongate at different rates, resulting in population heterogeneity (71–74). Several studies have reported that daughter cells who inherit the old pole are larger and grow faster than cells which inherit the new pole (71, 74). For Mycobacterium tuberculosis, the causative agent of tuberculosis, the variability in cell size resulting from asymmetric growth and division has been suggested to increase the fitness of this pathogen by producing cells with differing antibiotic susceptibilities within microcolonies (71). Aldridge and colleagues found that larger cells were more sensitive to cell wall synthesis inhibitors than smaller sibling cells (71); however, this observation may apply only to cells grown in microcolonies (74, 75). Nevertheless, the presence of asymmetrically sized progeny suggests that the resulting daughter cells are not equal and may be in different physiologic growth states. Additionally, since division is not symmetric, inheritance of new cell wall material will also be skewed to one daughter cell over the other. The consequences of this size heterogeneity may provide a built-in mechanism for M. tuberculosis to adapt to the host environment (76).
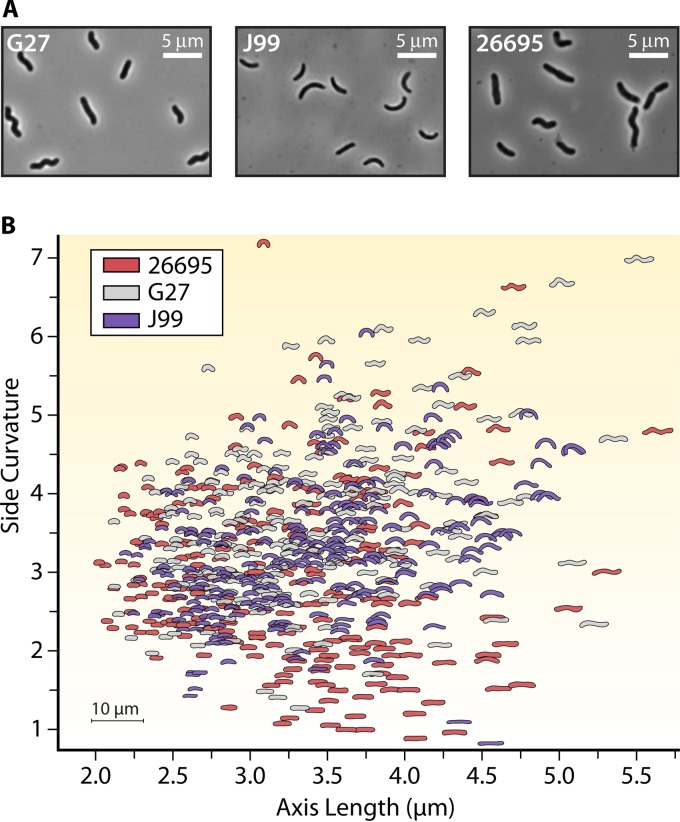
Morphological plasticity as a mechanism for pathogens to better adapt to different environments may also explain the enormous cell shape heterogeneity seen in clonal isolates of H. pylori (Fig. 3) (19). Although H. pylori is described as a helical rod, cultures grown from clonal isolates display a range of morphologies, including both curved and straight rods of various lengths and helicities (19). It has been postulated that this heterogeneity of shape allows this pathogen to infect and adapt to different stomach niches depending on the particular individual that is infected. Given the diverse shapes observed for each clonal isolate, a single H. pylori strain may be poised for more successful transmission between hosts because, presumably, there will always be cells with different morphologies that are better suited for different gastric environments.
FIG 3.
Helicobacter pylori intraspecies cell shape heterogeneity. (A) Phase-contrast images of three clinical strains of Helicobacter pylori (188–190). (B) Quantitative Celltool scatterplot analysis (19) showing cell side curvature versus axis length for clonal populations of the indicated strains of H. pylori (>100 cells/strain).
MORPHOLOGICAL TRANSITIONS
While bacterial species have been defined by specific morphologies, they can also adopt alternate shapes and sizes over the course of their life cycles. Diverse bacteria respond to starvation conditions by forming metabolically inert spores. While spore formation indeed represents a major morphological reprogramming, we refer the reader to several excellent reviews on this subject (49, 77–79) and instead focus on morphological transitions induced by environmental conditions that still support some level of metabolic activity. For pathogens, these include morphological transitions that accompany colonization of distinct tissues and cell types within a host, transmission between hosts, and transit through environmental reservoirs.
Bigger Is Better
Microbes employ a variety of strategies to evade and combat host defenses. One such strategy used by a number of different bacterial and fungal pathogens is the increasing of cell size and overall surface area to avoid phagocytosis and facilitate enhanced attachment to host cell surfaces (80–85). Bacteria can effectively increase their cell surface area by a number of different mechanisms, including producing appendages (i.e., prosthecae [86–88]), increasing the cellular diameter, or elongating into filaments. Monds et al. (89) described the isolation of E. coli cells that have evolved to be larger due to specific mutations in the cytoskeletal protein MreB. The observed increase in cell size scales linearly with fitness under conditions where cells are exiting stationary phase; larger cells reach their maximal growth rate more quickly than their smaller, ancestral counterparts (89). These findings provide a mechanistic framework to better understand the interplay between cell shape (i.e., getting big) and fitness (89).
Another way that cells can become larger is by forming long filaments. Filamentation is the result of continual longitudinal growth without cell division. For this review, filaments encompass both single-nucleus and multinucleated cells that can be separated by septa and are connected by a contiguous outer membrane (with or without evidence of outer membrane invagination in Gram-negative bacteria). Interestingly, multiple bacterial pathogens have been reported to undergo filamentation in vivo, suggesting that filamentation may be important for pathogenesis (81, 90).
Several studies have started to illuminate the mechanistic details of how the filamentous morphology is beneficial to different microbial pathogens. One common theme that has emerged is the use of filamentation to evade phagocytosis-mediated killing by the host. It was recently reported that filamentous Legionella pneumophila cells are harder for professional phagocytes to engulf (91). These filaments are slowly phagocytized along the long axis of the cell, resulting in prolonged phagocytic cups that fail to develop complete hydrolytic activity, thus allowing escape of the pathogen from phagosomal killing in a filament length-dependent manner (91). Similar findings have also been reported for uropathogenic E. coli (UPEC), where bacillary cells are preferentially killed over their filamentous counterparts (85).
A recent report found that filamentous L. pneumophila is able to invade lung epithelial cells and produce infectious progeny, demonstrating the invasive potential of this morphotype (92). Although it had been appreciated that L. pneumophila could infect lung epithelial cells, the invading bacteria were either coccoid or rod-like in shape (92, 93). Interestingly, internalization of L. pneumophila filaments occurs in a manner distinct from that for nonfilamentous cells, consisting of a more gradual mode of invasion and specific ligand-mediated binding to host cell surface receptors for phagocytosis (92).
In addition to preventing efficient phagocytosis by immune cells, filamentation has also been proposed to aid in the attachment of microbial pathogens to host cells, thereby reducing pathogen shedding as well as facilitating persistence within the host (80). S. pneumoniae, which colonizes the mucosal surface of the upper respiratory tract, grows in chains of various lengths. In culture, longer-chain S. pneumoniae variants adhere better to human respiratory epithelial cells than shorter chains (94). In accordance with this model, competition experiments in a murine nasopharyngeal colonization model showed that three different chain mutants outcompeted the parental wild-type strain (94). Thus, in both cell culture and whole-animal infections, filamentation promotes adherence.
Filamentation in response to different environmental stresses has been observed for numerous bacterial species, pathogenic or not (95–104). Insults that cause DNA damage generally induce the SOS response, which can cause cells to undergo filamentation due to induction and expression of the division inhibitor SulA (105, 106). In UPEC, filamentation appears to be a direct response to DNA damage induced by the host innate immune system (98). UPEC sulA mutants do not produce filaments and are severely attenuated in the murine cystitis model, suggesting that filamentation is required for optimal persistence during infection (98). In addition to sulA, over 30 other genes involved in DNA repair and mutagenesis are induced by the SOS response (96, 107, 108). Filamentation has also been proposed to facilitate the development of antibiotic resistance against genotoxic agents (109). By accumulating SOS response-induced mutations, multinucleated filaments may provide cells the opportunity to recombine and select for advantageous alleles between chromosomes (109). This process could not only facilitate the emergence of antibiotic resistance but also promote the generation of beneficial mutations in response to different environmental stresses that induce filamentation.
Mini Me
The ability of a cell to increase its size can be a double-edged sword: in some cases, becoming bigger can be an advantageous trait (as discussed above in “Bigger Is Better”), but in other situations, largeness can be a lethal liability. One intriguing observation is that several common invasive pathogens are relatively small, including S. pneumoniae, N. meningitidis, and Haemophilus influenzae (80, 110). Indeed, microbial cell size is an important pathogenesis factor, and work by Dalia and Weiser demonstrated that minimization of bacterial size is a mechanism used by S. pneumoniae to circumvent complement-mediated killing by the host (110). They found that during invasive systemic infection, S. pneumoniae cells with decreased chain length were more virulent than their corresponding long-chain variants (110). Longer bacterial chains collectively possess a larger surface area and thus serve as larger targets for complement deposition and opsonophagocytic killing (110). Nevertheless, although minimization of cell size may be an effective way to evade complement-dependent killing, it appears that the host has a response to this strategy. Antibodies can agglutinate smaller microbial targets to effectively increase their size for better targeting and clearance by the host (110). Previous studies have also reported a correlation with the protective effect of antibody-mediated clumping or agglutinating against systemic infections by S. pneumoniae and Salmonella enterica serovar Typhimurium (111–113). Taken together, these findings suggest a potentially important role that cell size minimization plays in bacterial pathogenesis.
Running with the Herd
Bacteria exhibit diverse mechanisms of motility to gain access to favorable microhabitats and for successful colonization of abiotic and biotic surfaces. Swarming motility is operationally defined as “a rapid multicellular movement of bacteria across a surface, powered by a rotating flagellum” (114). Swarming motility is characterized by dramatic changes in overall cell length and increases in the number of flagella that vary significantly depending on the species of bacterium (Table 1). Bacterial swarming is a hallmark characteristic and virulence determinant for numerous pathogens (e.g., Pseudomonas aeruginosa, Proteus mirabilis, and Vibrio parahaemolyticus) and nonpathogens (B. subtilis) alike. For more exhaustive information on swarming motility in these organisms, the reader is referred to two excellent reviews (114, 115). Swarm cell differentiation is a complex physiological response to environmental cues and signaling events. As bacteria transition from the motile swimmer state to the swarmer state, there is an elongation of the bacterial cell as well as widespread changes in gene expression, an increase in the number of flagella, and the production of a surfactant to reduce surface tension. It is hypothesized that this elongation serves to provide additional surface area on the bacterium to accommodate the increased number of flagella. This idea is supported by recent findings in Salmonella where cells that were not actively upregulating flagellum production but elongated to double the normal cell length possessed twice the number of flagella. This 2-fold increase in flagellation was sufficient to overcome surface friction and promote swarming motility (116). The signals that drive the switch from the motile cell to the swarmer state are varied and are the subject of active research; however, the functional consequences of flagellar placement are discussed further below (refer to Bacterial Cell Surface Appendages).
Swarming behavior is perhaps best understood for the Gram-positive model organism B. subtilis (117) and the Gram-negative bacterium P. mirabilis, which causes opportunistic urinary tract infections in immunocompromised individuals (115, 118). In the case of P. mirabilis, there is considerable debate in the field as to the absolute requirement of swarming motility for virulence, since it has been shown that a vast majority of the bacteria in ascending urinary tract infections are undifferentiated swimmer cells (119). Thus, going forward, a better understanding of the signaling events that lead to swarm cell differentiation and the temporal dynamics of swarm cell migration and group behavior is needed to better understand how the swarming state and virulence are temporally coordinated.
As Bacteria Grow Up
Morphological changes can be observed for bacteria that undergo developmental programs, including several important pathogens. Examples of such organisms include obligate intracellular pathogens in the Chlamydia and Rickettsia genera (120–125). These organisms display distinct morphological forms as they transition from various host environments and growth states. However, their complex life cycles and requirement for propagation within mammalian cells have made it challenging to study how these shape changes contribute to pathogenesis. Such challenges include the genetic intractability of these pathogens; however, recent advances, including whole-genome sequencing technologies, are enabling researchers to begin investigating these kinds of questions. Moreover, the identification of host-cell-free (axenic) media that permit growth outside the host is facilitating these studies. Axenic propagation of the obligate intracellular pathogen Coxiella burnetii, which is the causative agent of Q fever, has been reported (126). These new developments will allow for more in-depth studies on pathogens that were previously inaccessible.
One pathogen whose morphological plasticity through its complex developmental life cycle has been studied in greater depth is UPEC, the predominant causative agent of urinary tract infections (Table 1) (82, 127, 128). Upon initial invasion, UPEC begins to form early intracellular bacterial communities (IBCs) within the cytoplasm of bladder umbrella cells, and each IBC originates from infection by a single bacterium (129). These early IBCs consist of nonmotile, rod-shaped cells that are loosely associated into colonies. However, the cells eventually transition into slower-growing coccoids that form more organized biofilm-like communities (mid-IBCs). At this point, a small subset of cells begins to further differentiate into filaments within these mid-IBCs. As mentioned previously, filamentation of UPEC within these IBCs is due to induction of the SOS response as the cells cope with host immune insults that damage their DNA (98). Eventually, the coccoid UPEC cells become motile and bacillary (late IBCs), lysing the host cell and releasing both filaments and motile rods for further rounds of invasion into neighboring bladder cells (egress and second-generation IBCs) (90). Since each IBC represents a single invasion event, this suggests that the morphological changes observed within these communities are part of a developmental program (90). Each phase and its associated morphotype presumably function to facilitate intracellular growth and subsequent rounds of infection. As discussed earlier in this review, filamentation has been shown to decrease efficient phagocytosis as well as to provide a larger surface area for attachment to host cell surfaces, thus providing these cells with several key advantages for establishing second-generation IBCs. Additionally, the transition from the coccoid cells seen in mid-IBCs to the motile rods in late IBCs, prior to host cell lysis, results in released progeny that can rapidly spread and establish infection at more distal sites. Although more work is needed to elucidate the molecular mechanisms that underlie the UPEC developmental program, it is clear that shape plays an important role in the survival and propagation of this pathogen.
To Be or Not To Be
Many Gram-negative pathogens, including V. cholerae, Vibrio vulnificus, V. parahaemolyticus, E. coli, S. enterica, Shigella dysenteriae, L. pneumophila, C. jejuni, and H. pylori, transition from their characteristic rod-shaped forms to smaller coccoid forms after incubation for days to weeks in fresh or salt water, particularly under low-temperature conditions (130–133). These coccoid forms often show resistance to culture but have continued metabolic activity as measured by incorporation of radiolabeled thymidine or glutamic acid and can be induced to elongate in the presence of nutrients (134). While thousands to millions of organisms may be detected by direct microscopy, no or very few such bacteria can be cultured on standard laboratory media, leading to their designation as viable but nonculturable (VBNC). Some VBNC bacteria retain infectivity. For example, VBNC V. cholerae showed growth in ligated rabbit ileal loops (134), and VBNC L. pneumophila became fully virulent in a guinea pig model after passage through the freshwater amoeba Acanthamoeba castellanii (135). While these studies indicate that VBNC organisms can cause disease, they do not address whether their altered shape provides specific advantages for host infection given their altered metabolic and growth capabilities.
Not unexpectedly, both coccoid formation and resuscitation of VBNC forms are linked to changes in the cell wall. While signals that can promote resuscitation from the VBNC state remain elusive for many bacteria, a variety of Vibrio species can be resuscitated back to a cultivable state by a temperature upshift (136, 137), allowing further dissection of this developmental program. Early studies revealed progressive cell shortening during conversion of V. cholerae to the VBNC state until the bacteria became small cocci. During resuscitation, cells were observed to elongate and initially divide reductively, yielding shorter cells than those seen during exponential-phase growth (137). Subsequent ultrastructural studies revealed a mixed population of irregular and coccoid forms during VBNC conversion at room temperature, while the population became entirely coccoid when VBNC conversion occurred at 4°C (138). During recovery, growth appeared to occur via budding of extremely small coccoid cells that could themselves divide by binary fission, and in some cases, the budded daughters retained connections to the mother cell that resembled strands or fimbriae. Recovery of E. coli L forms (bacteria lacking cell wall material through growth in the presence of cell wall inhibitors) similarly showed abundant morphological diversity and apparent budding (139). Studies of V. parahaemolyticus revealed that the proportion of irregular shapes observed during VBNC conversion increases in low-salt media, which leads to increased cellular turgor pressure (140). Computational modeling of the Gram-negative cell wall combined with limited antibiotic perturbation in E. coli suggests that localized disruption of cell wall integrity produces similar cell bulges and bends (20). Quantitative PCR analysis of candidate cell wall synthesis and cell wall modification genes during the time window where irregular forms peaked revealed increased expression of DacB, a PG d-Ala-d-Ala carboxypeptidase. Disruption of dacB suppressed the accumulation of aberrant shapes during VBNC conversion, while complementation reversed this suppression (140). Comparison of the global PG compositions of H. pylori during exponential phase, during stationary phase, and in the coccoid VBNC form revealed a substantial increase in the dipeptide content within the sacculus, with a concomitant loss of tripeptides and tetrapeptides (141), in coccoid cells. Dipeptide-rich PG has been suggested to be a marker of old or inert cell wall and cannot serve as a cross-linking acceptor for linkage to new PG strands. These studies suggest that remodeling of the cell wall structure and composition accompanies VBNC conversion and reversion. However, it remains unclear whether these changes have been selected to promote the shape change or are merely a by-product of the change in cell wall structure that promotes survival under stressful conditions.
While VBNC states have primarily been described for Gram-negative proteobacteria, the Gram-positive enterococci have also been shown to enter a VBNC state (142). Interestingly, Enterococcus faecalis becomes slightly longer instead of shorter during VBNC formation, and this morphological transition correlates with a thickened cell wall. Analysis of the cell wall revealed increased cross-linking of the PG and a higher level of expression of PBP5, a d,d-transpeptidase (143). VBNC E. faecalis also showed increased autolytic activity and lipoteichoic acid compared to those of exponentially growing cells. Thus, while remodeling of the cell wall appears to be a shared feature of VBNC induction in diverse organisms, conversion to a coccoid shape is not conserved. The inherent differences in cell wall organization between Gram-positive and Gram-negative organisms could underlie the differences in morphology of VBNC forms among bacterial groups. Gram-positive organisms can elaborate a thicker wall with altered chemical properties as a barrier to noxious chemicals and conditions, while the thin and relatively simpler cell wall of Gram-negative organisms may necessitate minimizing the surface area as a primary strategy to avoid interactions with a hostile environment. Early experiments suggested the Gram-positive cell wall as a barrier to release of secreted α-amylase in Bacillus amyloliquefaciens (144), and the production of branched PG stem peptides in pneumococcus PG was recently found to limit pneumolysin release (145).
The functional impacts of VBNC conversion are varied. VBNC S. dysenteriae maintained Shiga toxin expression and a lower level of adherence to cultured cells but was unable to invade these cells, a phenotype essential for full virulence (146). H. pylori coccoid cells showed 50-fold reductions in adherence and induction of the proinflammatory cytokine interleukin-8 (IL-8) (147), both of which are phenotypes associated with increased pathogenicity. Studies with mutants of amiA, encoding the sole amidase in H. pylori required for septum cleavage, suggest that in addition to lower adherence, the lower IL-8 induction by coccoids may also result from the diminished levels of tripeptide in the sacculi of coccoids, since amiA mutants fail to show lower tripeptide levels upon extended culture (148). While VBNC cells may have lower virulence, they still can serve as reservoirs for infectious organisms and can maintain plasmids, in the case of E. coli (149), and vancomycin resistance, in the case of Enterococcus (142). Thus, a major open question remains as to the conditions and mechanisms of VBNC resuscitation. Recently, two studies suggested that in vibrios, quorum-sensing autoinducer molecules promote the efficiency and kinetics of resuscitation (150, 151). A further understanding of resuscitation will allow assessment of the specific properties of VBNC and resuscitated forms.
BACTERIAL CELL SURFACE APPENDAGES
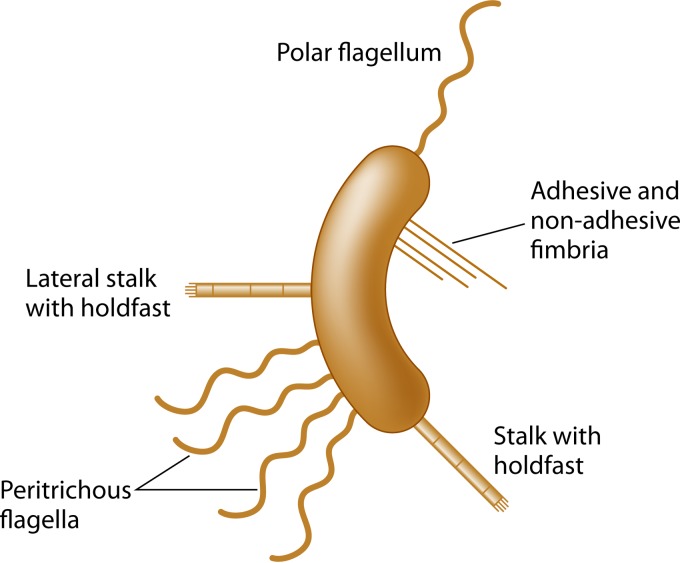
Thus far, we have primarily concerned ourselves with the overall form of a bacterial cell in the context of its main body. However, the vast majority of bacteria produce extracellular flagella and/or pili that are located on the outside of the cell and may be thought of as surface-associated appendages. These mostly extracellular structures are not morphological determinants, but their presence and physical placement on the cell surface do indeed affect the overall shape and physical reach of the organism in distinct ways that promote survival, niche acquisition, and/or pathogenesis. In particular, flagella, type I pili, and type IV pili have all been linked to pathogenesis in a variety of organisms (Fig. 4). For the purposes of this review, we are expanding the definition of bacterial morphology to include the contributions of the aforementioned extracellular appendages in addition to the morphology of the cell body.
FIG 4.
Schematic of bacterial cellular appendages discussed in this review.
Flagella function to allow both chemotaxis-directed motility and swarming motility but can also have adhesive properties toward host cells. Flagella also serve as a frequent target of the host innate and adaptive immune systems, and thus Salmonella, for example, downregulates flagellin expression during systemic host colonization (152–154). For the human pathogens V. cholerae and H. pylori, the flagellum is sheathed by an extension of the outer membrane that was once thought to function in preventing immune recognition of the flagellar filament by the host. However, a recent study by Brennan and colleagues challenges this idea and suggests an alternative role in promoting immune activation through the shedding of lipopolysaccharide (LPS) during flagellar rotation (155). Pili promote adhesion and a third type of motility, known as twitching motility. Pili can also play a role in DNA uptake, the induction of cytokines and immunogenic responses, hemagglutination, and biofilm formation (156–159). Other bacteria, such as C. crescentus, elaborate a polar stalk with an adhesive tip, a structure distinct from pili that may be important for nutrient uptake and attachment to surfaces (160). In this section, we explore how the number, length, and location of these appendages affect their functional properties.
Locomotion Defined by Location
The bacterial flagellum is one of the most well-studied and complex bacterial organelles (161, 162). For an excellent review on the incredible diversity and importance of this molecular motor, the reader is referred to a previous review (163). At a rudimentary level, the most basic function of the bacterial flagellum is to provide a means of locomotion via a process known as chemotaxis that allows the bacterium to change its direction by modulating motor function in response to environmental signals (i.e., nutrient gradients). Flagellar arrangements and numbers on the cell can have dramatic effects on the form and type of motility manifested by the bacterium.
The Vibrio and Pseudomonas flagella are of the monotrichous variety, consisting of a single polar flagellum used for swimming motility in the planktonic growth state of these bacteria, which functions basically like a propeller on a boat. Another form of motility, termed darting motility, is promoted by bacteria with an amphitrichous, or bipolar arrangement, with one flagellum on each of the poles. This type of motility involves straight runs followed by reorientation and then reversal of direction by engagement of the opposite polar flagellum. Campylobacter jejuni achieves this bipolar arrangement by using a protein called FlhF that simultaneously directs polar placement of flagella and prevents FtsZ assembly, and thus cell division, at the poles (164). Pseudomonas aeruginosa also utilizes an FlhF protein to direct polar localization and regulation of late-stage flagellar genes and flagellar numbers. Furthermore, loss of FlhF results in the misplacement of nonpolar flagella that results in impaired swimming and swarming phenotypes (165). Overexpression of FlhF in Pseudomonas putida results in a lophotrichous (multiple polar flagellar) phenotype. Interestingly, despite having more flagella, these lophotrichous P. putida mutants are unable to spread on 0.4% motility agar plates. In contrast, a mutation of a small RNA gene that leads to a larger median number of polar flagella in H. pylori leads to increased swimming speeds (65).
The peritrichous arrangement, in which many flagella are found along the entire surface of the cell, promotes a type of motility termed “runs and tumbles” whereby the bacterium changes swimming direction from the smooth “run” to a “tumble” when the flagella unbundle during reversal of motor rotation. In addition to their function in cell propulsion, flagella can also function as a surface sensing apparatus. The seafood-associated diarrheal pathogen Vibrio parahaemolyticus lives in marine environments and is motile in its planktonic state by means of a single polar flagellum. Upon inhibition of rotation of its flagellum, either by contact with a surface or from a highly viscous environment, a genetic reprogramming event occurs that triggers swarmer cell differentiation (166). The swarmer cells express an independent lateral flagellar gene system that results in peritrichous swarmer cells capable of swarming motility and is independent from the polar signal-transducing flagellum. Shewanella putrefaciens CN-32 also possesses a lateral flagellar gene system, and recent work by Bubendorfer et al. (167) demonstrated that the placement of the additional flagella alters directionality changes, resulting in improved directional persistence of swimming and spreading of this bacterium. Taken together, these results suggest that both the placement and number of flagella are important motility and virulence determinants.
Standing Tall
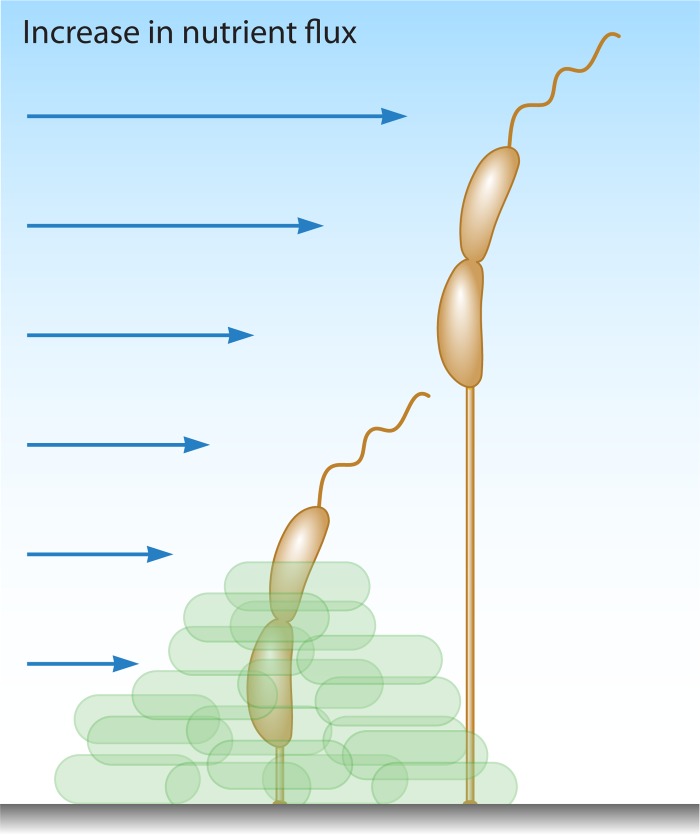
Prosthecae are true cell envelope extensions found in many nonpathogenic Gram-negative bacteria, possessing both PG and cell membranes, in contrast to flagellar or pilus appendages (168, 169). In a recent paper by Jiang et al. (170), the authors investigated the evolution of stalk positioning in several stalk-possessing bacteria, including C. crescentus, Asticcacaulis biprosthecium, and Asticcacaulis excentricus. They demonstrated that stalk positioning in Asticcacaulis spp. was driven by coopting the developmental regulator SpmX by evolution of protein function with modular protein domains to specify the location (polar versus nonpolar) of stalk synthesis. Nevertheless, the function of these stalks is not entirely clear, but one long-standing hypothesis is that they function to enhance nutrient acquisition from the environment by increasing the cell surface area with minimal change to the surface-to-volume ratio of the cell (3, 86, 171). Evidence to support this model came from numerous examples of stalk lengthening in response to phosphate starvation in different bacteria, including Caulobacter, Asticcacaulis, Hyphomicrobium, and Rhodomicrobium (171). The most well-studied prostheca-possessing bacterium is C. crescentus, which has a single polar prostheca, more often referred to as a stalk, with an adhesive holdfast at its end (172). Under nutrient-rich conditions, the stalk of C. crescentus measures approximately 1 μm in length; however, in phosphate-limiting media, the stalk can lengthen to up to 30 μm (173, 174). Wagner and colleagues reported that these stalks can take up phosphate by using the high-affinity phosphate-binding protein PstS, which was proposed to shuttle the bound phosphate to a transporter complex for entry into the cell body cytoplasm (168). However, the feasibility of this model has been challenged by more recent work demonstrating the diffusion limitation of PstS within stalks by protein barriers that prevent exchange between these two compartments (175). One previously proposed idea that has taken favor in recent years is that stalk lengthening may function to raise cells above attached surfaces, via the holdfast, into the path of increased nutrient flux (Fig. 5) (5, 160). This model, although not mutually exclusive, may best explain why C. crescentus employs stalk formation and lengthening to survive in oligotrophic (low nutrient) environments; however, this may not hold true for other organisms that possess prosthecae. Asticcacaulis biprosthecium, which contains stalks that also elongate under nutrient-poor conditions, lacks sticky holdfasts, suggesting that stalk lengthening may function in a manner distinct from that in C. crescentus (160). In this case, stalk elongation might serve to provide “nutrient whiskers” to facilitate nutrient acquisition (3) or as a mechanism to avoid protozoan predation by effectively increasing cell size (176).
FIG 5.
Model for the physiological role of stalk elongation in Caulobacter crescentus. Stalk elongation may serve to elevate cells away from attached surfaces to increase nutrient availability and to distance cells from other competitors. (Adapted from reference 160 by permission of Taylor & Francis LLC.)
The Long and Short of It
There are many different flavors of pili (also referred to as fimbriae), and they vary based on length, thickness, chemical decorations, tip adhesions, and the ability to retract (Fig. 4) (156). Differences in pilus length and number can directly affect binding to host factors and thus may affect pathogenesis. A study by Imhaus and Duménil (177) showed that in N. meningitidis, modulation of the degree or amount of type IV piliation alters functional properties of those pili (Table 1). Diplococci expressing only a single pilus could import DNA for natural transformation but could not undergo intrabacterial aggregation. As the number of pili increased to two, the bacteria were able to self-aggregate. A further increase to approximately five pili was sufficient to promote bacterial cross talk with host cells (Table 1) (177). A recent modeling and experimental study with N. meningitidis suggested that twitching motility utilizes both temporal retraction and physical placement of type IV pili for cell orientation and to optimize directionality during twitching motility (178).
UPEC utilizes both P and type I pili to promote successful pathogenesis. Melican et al. (179) used a live animal (murine) renal bacterial infection model to show that P-type fimbriae enhance initial colonization steps primarily by promoting bacterium-host cell interactions in synergy with the type I fimbriae, promoting interbacterial binding and biofilm formation. This is mediated by the FimH tip adhesin of the type I pilus and provides a fitness advantage for withstanding sheer forces induced by filtrate flow in vivo (179, 180). Furthermore, attachment and shortening of type I pili are required for bladder cell invasion by UPEC and progression through the IBC transmission cycle in a murine model of infection (181, 182). The fact that different pili are used for different functions likely relates to their differences in receptors and/or differences in their structural features, which have not been examined extensively to date, especially in the context of temporal effects during pathogenic processes.
Another interesting example of division of labor among distinct pili occurs in the oral cavity, where numerous microbes engage in niche acquisition and host interaction processes facilitated by their pili. Disease in the oral cavity is thought to arise through polymicrobial synergy and dysbiosis, whereby communities of different bacteria influence one another, leading to the onset of a polymicrobial disease state. In the mouth, the keystone pathogens Porphyromonas gingivalis, Streptococcus gordonii, Treponema denticola, and Tannerella forsythia are collectively known as the “red complex” bacteria (183). The red complex has been strongly associated with the periodontal disease state (184). Porphyromonas gingivalis expresses two types of fimbriae. Long (major) fimbriae of ∼3 μm in length that are comprised of the FimA protein bind to glyceraldehyde-3-phosphate dehydrogenase expressed on the surfaces of a variety of commensal Streptococcus and Actinomyces bacteria in the oral cavity (185). The shorter (minor), type 2 fimbriae, comprised of the Mfa1 protein, are only 100 to 500 nm long and bind to the SspA and SspB antigen I/II family of Streptococcus gordonii surface proteins (186). These are species-specific proteins that promote specific polymicrobial community interactions in the oral biofilm (187). This species specificity is an important characteristic because it has been noted that P. gingivalis does not interact with Streptococcus mutans (which promotes dental caries) because it does not express the SspA/B surface proteins, explaining why this organism is not found in polymicrobial biofilms with P. gingivalis. Initial stages of P. gingivalis colonization are mediated by the major type I pili to promote successful establishment of a polymicrobial interaction between P. gingivalis and oral Streptococcus bacteria, followed by community structure diversification and biofilm formation mediated by the short type II pili. Thus, these fimbrial appendages mediate specific protein-protein interactions that are important virulence determinants whose function is determined in part by the physical reach of pili and the specific protein-protein interactions mediated through pili of differing lengths.
CONCLUSIONS
Bacterial morphological diversity manifests at multiple levels, including cell body geometry and size and the elaboration and diverse placement of appendages. Human bacterial pathogens encompass much of the morphological diversity found in the bacterial kingdom (Table 1). Furthermore, extensive research on the physiology, cell biology, and immunology of the mammalian host provides an excellent opportunity to probe the functional significance of cell shape, as each of these host features likely exerts selective pressures on bacteria that transiently or chronically colonize humans to cause disease. The elucidation of genetic programs that drive distinct morphologies (e.g., csd genes that drive helical shape in H. pylori and the induction of cell filamentation by sulA expression during UPEC infection) has provided a means to explore the functional significance of cell morphology for pathogenesis through infection studies with mutants that have altered morphologies or cannot transition between distinct morphological states. As described in Table 1, we can say that bacterial morphology affects the ability of organisms to colonize distinct host niches, their susceptibility to host defenses, and disease progression for many, if not all, pathogens. While in many cases the cell morphology has been associated only with the pathogenic state, precise molecular mechanisms have begun to emerge. The enhanced susceptibility of cell chaining mutants of S. pneumoniae to complement-mediated killing provides an elegant example. Future work in this area will hopefully bring this level of molecular detail to more of the cases highlighted in this review.
We expect much progress in further elucidating the functional significance of shape for pathogenic and nonpathogenic bacteria in the coming years. Studies both in model bacterial systems and directly in pathogens should allow a deeper mechanistic understanding of the molecular processes that generate and regulate the morphology of bacteria and thus illuminate strategies to experimentally perturb these parameters. Furthermore, increasingly sophisticated methods to image bacteria in the host and to genetically manipulate the host will allow more detailed testing of hypotheses relating to the functional significance of bacterial morphology.
ACKNOWLEDGMENTS
We thank Jenny Taylor for assistance with the illustration in Fig. 1 and Lauren Saunders for providing the images of H. pylori used in Fig. 3.
This work was supported by the National Institutes of Health under award numbers RO1AI094839 (N.R.S.) and T32CA009657 (D.C.Y. and K.M.B.), by a National Science Foundation graduate research fellowship to K.M.B. (grant DGE-1256082), and by an American Cancer Society—Lakeshore Division postdoctoral fellowship to D.C.Y. (grant PF-13-391-01-MPC).
REFERENCES
- 1.Chang F, Huang KC. 2014. How and why cells grow as rods. BMC Biol 12:54. doi: 10.1186/s12915-014-0054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young KD. 2007. Bacterial morphology: why have different shapes? Curr Opin Microbiol 10:596–600. doi: 10.1016/j.mib.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young KD. 2006. The selective value of bacterial shape. Microbiol Mol Biol Rev 70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawler ML, Brun YV. 2007. Advantages and mechanisms of polarity and cell shape determination in Caulobacter crescentus. Curr Opin Microbiol 10:630–637. doi: 10.1016/j.mib.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner JK, Brun YV. 2007. Out on a limb: how the Caulobacter stalk can boost the study of bacterial cell shape. Mol Microbiol 64:28–33. doi: 10.1111/j.1365-2958.2007.05633.x. [DOI] [PubMed] [Google Scholar]
- 6.Typas A, Banzhaf M, Gross CA, Vollmer W. 2011. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheffers DJ, Pinho MG. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev 69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang C, Caccamo PD, Brun YV. 2015. Mechanisms of bacterial morphogenesis: evolutionary cell biology approaches provide new insights. Bioessays 37:413–425. doi: 10.1002/bies.201400098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Randich AM, Brun YV. 2015. Molecular mechanisms for the evolution of bacterial morphologies and growth modes. Front Microbiol 6:580. doi: 10.3389/fmicb.2015.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lovering AL, Safadi SS, Strynadka NCJ. 2012. Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem 81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 11.Desmarais SM, de Pedro MA, Cava F, Huang KC. 2013. Peptidoglycan at its peaks: how chromatographic analyses can reveal bacterial cell wall structure and assembly. Mol Microbiol 89:1–13. doi: 10.1111/mmi.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vollmer W, Höltje JV. 2004. The architecture of the murein (peptidoglycan) in gram-negative bacteria: vertical scaffold or horizontal layer(s)? J Bacteriol 186:5978–5987. doi: 10.1128/JB.186.18.5978-5987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gan L, Chen S, Jensen GJ. 2008. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci U S A 105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gumbart JC, Beeby M, Jensen GJ, Roux B. 2014. Escherichia coli peptidoglycan structure and mechanics as predicted by atomic-scale simulations. PLoS Comput Biol 10:e1003475. doi: 10.1371/journal.pcbi.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown S, Santa Maria JP, Walker S. 2013. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beeby M, Gumbart JC, Roux B, Jensen GJ. 2013. Architecture and assembly of the Gram-positive cell wall. Mol Microbiol 88:664–672. doi: 10.1111/mmi.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Pedro MA, Quintela JC, Höltje JV, Schwarz H. 1997. Murein segregation in Escherichia coli. J Bacteriol 179:2823–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodwin SD, Shedlarski JG. 1975. Purification of cell wall peptidoglycan of the dimorphic bacterium Caulobacter crescentus. Arch Biochem Biophys 170:23–36. doi: 10.1016/0003-9861(75)90094-6. [DOI] [PubMed] [Google Scholar]
- 19.Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. 2010. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori's helical shape and stomach colonization. Cell 141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. 2008. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci U S A 105:19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zapun A, Vernet T, Pinho MG. 2008. The different shapes of cocci. FEMS Microbiol Rev 32:345–360. doi: 10.1111/j.1574-6976.2007.00098.x. [DOI] [PubMed] [Google Scholar]
- 22.Pinho MG, Kjos M, Veening JW. 2013. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol 11:601–614. doi: 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- 23.Margolin W. 2009. Sculpting the bacterial cell. Curr Biol 19:R812–R822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed P, Atilano ML, Alves R, Hoiczyk E, Sher X, Reichmann NT, Pereira PM, Roemer T, Filipe SR, Pereira-Leal JB, Ligoxygakis P, Pinho MG. 2015. Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance. PLoS Pathog 11:e1004891. doi: 10.1371/journal.ppat.1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stackebrandt E. 1979. A phylogenetic dissection of the family Micrococcaceae. Curr Microbiol 2:317–322. doi: 10.1007/BF02602867. [DOI] [Google Scholar]
- 26.Woese CR, Blanz P, Hespell RB, Hahn CM. 1982. Phylogenetic relationships among various helical bacteria. Curr Microbiol 7:119–124. doi: 10.1007/BF01568426. [DOI] [Google Scholar]
- 27.Siefert JL, Fox GE. 1998. Phylogenetic mapping of bacterial morphology. Microbiology (Reading, Engl) 144:2803–2808. doi: 10.1099/00221287-144-10-2803. [DOI] [PubMed] [Google Scholar]
- 28.Aldea M, Hernández-Chico C, de la Campa AG, Kushner SR, Vicente M. 1988. Identification, cloning, and expression of bolA, an ftsZ-dependent morphogene of Escherichia coli. J Bacteriol 170:5169–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray T, Popham DL, Setlow P. 1997. Identification and characterization of pbpA encoding Bacillus subtilis penicillin-binding protein 2A. J Bacteriol 179:3021–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lleo MM, Canepari P, Fontana R, Satta G. 1997. Inhibition of bacterial cell surface extension by various means causes blocking of macromolecular synthesis. Res Microbiol 148:11–20. doi: 10.1016/S0923-2508(97)81895-5. [DOI] [PubMed] [Google Scholar]
- 31.Henriques AO, Glaser P, Piggot PJ, Moran CP. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol Microbiol 28:235–247. doi: 10.1046/j.1365-2958.1998.00766.x. [DOI] [PubMed] [Google Scholar]
- 32.Veyrier FJ, Biais N, Morales P, Belkacem N, Guilhen C, Ranjeva S, Sismeiro O, Pehau-Arnaudet G, Rocha EP, Werts C, Taha MK, Boneca IG. 2015. Common cell shape evolution of two nasopharyngeal pathogens. PLoS Genet 11:e1005338. doi: 10.1371/journal.pgen.1005338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thanky NR, Young DB, Robertson BD. 2007. Unusual features of the cell cycle in mycobacteria: polar-restricted growth and the snapping-model of cell division. Tuberculosis (Edinb) 87:231–236. doi: 10.1016/j.tube.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767–776. doi: 10.1016/S0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 35.Letek M, Fiuza M, Ordóñez E, Villadangos AF, Ramos A, Mateos LM, Gil JA. 2008. Cell growth and cell division in the rod-shaped actinomycete Corynebacterium glutamicum. Antonie Van Leeuwenhoek 94:99–109. doi: 10.1007/s10482-008-9224-4. [DOI] [PubMed] [Google Scholar]
- 36.Cameron TA, Anderson-Furgeson J, Zupan JR, Zik JJ, Zambryski PC. 2014. Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. mBio 5:e01219-14. doi: 10.1128/mBio.01219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hay NA, Tipper DJ, Gygi D, Hughes C. 1999. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol 181:2008–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duggin IG, Aylett CHS, Walsh JC, Michie KA, Wang Q, Turnbull L, Dawson EM, Harry EJ, Whitchurch CB, Amos LA, Löwe J. 2015. CetZ tubulin-like proteins control archaeal cell shape. Nature 519:362–365. doi: 10.1038/nature13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ausmees N, Kuhn JR, Jacobs-Wagner C. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705–713. doi: 10.1016/S0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 40.Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, Jacobs-Wagner C. 2009. Bacterial cell curvature through mechanical control of cell growth. EMBO J 28:1208–1219. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagchi S, Tomenius H, Belova LM, Ausmees N. 2008. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol Microbiol 70:1037–1050. doi: 10.1111/j.1365-2958.2008.06473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Persat A, Stone HA, Gitai Z. 2014. The curved shape of Caulobacter crescentus enhances surface colonization in flow. Nat Commun 5:3824. doi: 10.1038/ncomms4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li G, Brown PJB, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. 2012. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol 83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonis M, Ecobichon C, Guadagnini S, Prévost MC, Boneca IG. 2010. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol Microbiol 78:809–819. doi: 10.1111/j.1365-2958.2010.07383.x. [DOI] [PubMed] [Google Scholar]
- 45.Sycuro LK, Rule CS, Petersen TW, Wyckoff TJ, Sessler T, Nagarkar DB, Khalid F, Pincus Z, Biboy J, Vollmer W, Salama NR. 2013. Flow cytometry-based enrichment for cell shape mutants identifies multiple genes that influence Helicobacter pylori morphology. Mol Microbiol 90:869–883. doi: 10.1111/mmi.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frirdich E, Vermeulen J, Biboy J, Soares F, Taveirne ME, Johnson JG, Dirita VJ, Girardin SE, Vollmer W, Gaynor EC. 2014. Peptidoglycan ld-carboxypeptidase Pgp2 influences Campylobacter jejuni helical cell shape and pathogenic properties and provides the substrate for the dl-carboxypeptidase Pgp1. J Biol Chem 289:8007–8018. doi: 10.1074/jbc.M113.491829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frirdich E, Biboy J, Adams C, Lee J, Ellermeier J, Gielda LD, Dirita VJ, Girardin SE, Vollmer W, Gaynor EC. 2012. Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni. PLoS Pathog 8:e1002602. doi: 10.1371/journal.ppat.1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harman MW, Dunham-Ems SM, Caimano MJ, Belperron AA, Bockenstedt LK, Fu HC, Radolf JD, Wolgemuth CW. 2012. The heterogeneous motility of the Lyme disease spirochete in gelatin mimics dissemination through tissue. Proc Natl Acad Sci U S A 109:3059–3064. doi: 10.1073/pnas.1114362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flärdh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 50.Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, Salama NR. 2012. Multiple peptidoglycan modification networks modulate Helicobacter pylori's cell shape, motility, and colonization potential. PLoS Pathog 8:e1002603. doi: 10.1371/journal.ppat.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan ACK, Blair KM, Liu Y, Frirdich E, Gaynor EC, Tanner ME, Salama NR, Murphy MEP. 2015. Helical shape of Helicobacter pylori requires an atypical glutamine as a zinc ligand in the carboxypeptidase Csd4. J Biol Chem 290:3622–3638. doi: 10.1074/jbc.M114.624734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motaleb MA, Corum L, Bono JL, Elias AF, Rosa P, Samuels DS, Charon NW. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci U S A 97:10899–10904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dombrowski C, Kan W, Motaleb MA, Charon NW, Goldstein RE, Wolgemuth CW. 2009. The elastic basis for the shape of Borrelia burgdorferi. Biophys J 96:4409–4417. doi: 10.1016/j.bpj.2009.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berg HC, Turner L. 1979. Movement of microorganisms in viscous environments. Nature 278:349–351. doi: 10.1038/278349a0. [DOI] [PubMed] [Google Scholar]
- 55.Murphy GE, Matson EG, Leadbetter JR, Berg HC, Jensen GJ. 2008. Novel ultrastructures of Treponema primitia and their implications for motility. Mol Microbiol 67:1184–1195. doi: 10.1111/j.1365-2958.2008.06120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greenberg EP, Canale-Parola E. 1977. Relationship between cell coiling and motility of spirochetes in viscous environments. J Bacteriol 131:960–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slamti L, de Pedro MA, Guichet E, Picardeau M. 2011. Deciphering morphological determinants of the helix-shaped Leptospira. J Bacteriol 193:6266–6275. doi: 10.1128/JB.05695-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Howitt MR, Lee JY, Lertsethtakarn P, Vogelmann R, Joubert LM, Ottemann KM, Amieva MR. 2011. ChePep controls Helicobacter pylori infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. mBio 2:e00098-11. doi: 10.1128/mBio.00098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aihara E, Closson C, Matthis AL, Schumacher MA, Engevik AC, Zavros Y, Ottemann KM, Montrose MH. 2014. Motility and chemotaxis mediate the preferential colonization of gastric injury sites by Helicobacter pylori. PLoS Pathog 10:e1004275. doi: 10.1371/journal.ppat.1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lertsethtakarn P, Howitt MR, Castellon J, Amieva MR, Ottemann KM. 2015. Helicobacter pylori CheZ(HP) and ChePep form a novel chemotaxis-regulatory complex distinct from the core chemotaxis signaling proteins and the flagellar motor. Mol Microbiol 97:1063–1078. doi: 10.1111/mmi.13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, Ewoldt RH, McKinley GH, So P, Erramilli S, Bansil R. 2009. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci U S A 106:14321–14326. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bansil R, Celli JP, Hardcastle JM, Turner BS. 2013. The influence of mucus microstructure and rheology in Helicobacter pylori infection. Front Immunol 4:310. doi: 10.3389/fimmu.2013.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sellers LA, Allen A, Morris ER, Ross-Murphy SB. 1987. Mechanical characterization and properties of gastrointestinal mucus gel. Biorheology 24:615–623. [DOI] [PubMed] [Google Scholar]
- 64.Taylor C, Allen A, Dettmar PW, Pearson JP. 2004. Two rheologically different gastric mucus secretions with different putative functions. Biochim Biophys Acta 1674:131–138. [DOI] [PubMed] [Google Scholar]
- 65.Martínez LE, Hardcastle JM, Wang J, Pincus Z, Tsang J, Hoover TR, Bansil R, Salama NR. 14 September 2015. Helicobacter pylori strains vary cell shape and flagellum number to maintain robust motility in viscous environments. Mol Microbiol doi: 10.1111/mmi.13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsokos CG, Laub MT. 2012. Polarity and cell fate asymmetry in Caulobacter crescentus. Curr Opin Microbiol 15:744–750. doi: 10.1016/j.mib.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Curtis PD, Brun YV. 2010. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev 74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skerker JM, Laub MT. 2004. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat Rev Microbiol 2:325–337. doi: 10.1038/nrmicro864. [DOI] [PubMed] [Google Scholar]
- 69.Bodenmiller D, Toh E, Brun YV. 2004. Development of surface adhesion in Caulobacter crescentus. J Bacteriol 186:1438–1447. doi: 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levi A, Jenal U. 2006. Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J Bacteriol 188:5315–5318. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. 2012. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335:100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joyce G, Williams KJ, Robb M, Noens E, Tizzano B, Shahrezaei V, Robertson BD. 2012. Cell division site placement and asymmetric growth in mycobacteria. PLoS One 7:e44582. doi: 10.1371/journal.pone.0044582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singh B, Nitharwal RG, Ramesh M, Pettersson BMF, Kirsebom LA, Dasgupta S. 2013. Asymmetric growth and division in Mycobacterium spp.: compensatory mechanisms for non-medial septa. Mol Microbiol 88:64–76. doi: 10.1111/mmi.12169. [DOI] [PubMed] [Google Scholar]
- 74.Santi I, Dhar N, Bousbaine D, Wakamoto Y, McKinney JD. 2013. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat Commun 4:2470. doi: 10.1038/ncomms3470. [DOI] [PubMed] [Google Scholar]
- 75.Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. 2013. Dynamic persistence of antibiotic-stressed mycobacteria. Science 339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 76.Kieser KJ, Rubin EJ. 2014. How sisters grow apart: mycobacterial growth and division. Nat Rev Microbiol 12:550–562. doi: 10.1038/nrmicro3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kroos L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu Rev Genet 41:13–39. doi: 10.1146/annurev.genet.41.110306.130400. [DOI] [PubMed] [Google Scholar]
- 78.Setlow P. 2014. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol 196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McKenney PT, Driks A, Eichenberger P. 2013. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol 11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weiser JN. 2013. The battle with the host over microbial size. Curr Opin Microbiol 16:59–62. doi: 10.1016/j.mib.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Justice SS, Hunstad DA, Cegelski L, Hultgren SJ. 2008. Morphological plasticity as a bacterial survival strategy. Nat Rev Microbiol 6:162–168. doi: 10.1038/nrmicro1820. [DOI] [PubMed] [Google Scholar]
- 82.Justice SS, Hung C, Theriot JA, Fletcher DA, Anderson GG, Footer MJ, Hultgren SJ. 2004. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci U S A 101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zaragoza O, García-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodríguez-Tudela JL, Casadevall A. 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog 6:e1000945. doi: 10.1371/journal.ppat.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chrétien F, Heitman J, Dromer F, Nielsen K. 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog 6:e1000953. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Horvath DJ Jr, Li B, Casper T, Partida-Sanchez S, Hunstad DA, Hultgren SJ, Justice SS. 2011. Morphological plasticity promotes resistance to phagocyte killing of uropathogenic Escherichia coli. Microbes Infect 13:426–437. doi: 10.1016/j.micinf.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Poindexter JS. 1981. The caulobacters: ubiquitous unusual bacteria. Microbiol Rev 45:123–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morgan P, Dow CS. 1985. Environmental control of cell-type expression in prosthecate bacteria, p 131–169. In Fletcher M, Floodgate GD (ed), Bacteria in their natural environments, 1st ed Academic Press, London, United Kingdom. [Google Scholar]
- 88.Neidhardt FC, Ingraham JL, Schaechter M. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates Inc, Sunderland, MA. [Google Scholar]
- 89.Monds RD, Lee TK, Colavin A, Ursell T, Quan S, Cooper TF, Huang KC. 2014. Systematic perturbation of cytoskeletal function reveals a linear scaling relationship between cell geometry and fitness. Cell Rep 9:1528–1537. doi: 10.1016/j.celrep.2014.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Justice SS, Harrison A, Becknell B, Mason KM. 2014. Bacterial differentiation, development, and disease: mechanisms for survival. FEMS Microbiol Lett 360:1–8. doi: 10.1111/1574-6968.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Prashar A, Bhatia S, Gigliozzi D, Martin T, Duncan C, Guyard C, Terebiznik MR. 2013. Filamentous morphology of bacteria delays the timing of phagosome morphogenesis in macrophages. J Cell Biol 203:1081–1097. doi: 10.1083/jcb.201304095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Prashar A, Bhatia S, Tabatabaeiyazdi Z, Duncan C, Garduño RA, Tang P, Low DE, Guyard C, Terebiznik MR. 2012. Mechanism of invasion of lung epithelial cells by filamentous Legionella pneumophila. Cell Microbiol 14:1632–1655. doi: 10.1111/j.1462-5822.2012.01828.x. [DOI] [PubMed] [Google Scholar]
- 93.Mody CH, Paine R, Shahrabadi MS, Simon RH, Pearlman E, Eisenstein BI, Toews GB. 1993. Legionella pneumophila replicates within rat alveolar epithelial cells. J Infect Dis 167:1138–1145. doi: 10.1093/infdis/167.5.1138. [DOI] [PubMed] [Google Scholar]
- 94.Rodriguez JL, Dalia AB, Weiser JN. 2012. Increased chain length promotes pneumococcal adherence and colonization. Infect Immun 80:3454–3459. doi: 10.1128/IAI.00587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pine L, Boone CJ. 1967. Comparative cell wall analyses of morphological forms within the genus Actinomyces. J Bacteriol 94:875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Radman M. 1975. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci 5A:355–367. [DOI] [PubMed] [Google Scholar]
- 97.Bos J, Yakhnina AA, Gitai Z. 2012. BapE DNA endonuclease induces an apoptotic-like response to DNA damage in Caulobacter. Proc Natl Acad Sci U S A 109:18096–18101. doi: 10.1073/pnas.1213332109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Justice SS, Hunstad DA, Seed PC, Hultgren SJ. 2006. Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc Natl Acad Sci U S A 103:19884–19889. doi: 10.1073/pnas.0606329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stackhouse RR, Faith NG, Kaspar CW, Czuprynski CJ, Wong ACL. 2012. Survival and virulence of Salmonella enterica serovar Enteritidis filaments induced by reduced water activity. Appl Environ Microbiol 78:2213–2220. doi: 10.1128/AEM.06774-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zobell CE, Cobet AB. 1964. Filament formation by Escherichia coli at increased hydrostatic pressures. J Bacteriol 87:710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rolinson GN. 1980. Effect of beta-lactam antibiotics on bacterial cell growth rate. J Gen Microbiol 120:317–323. [DOI] [PubMed] [Google Scholar]
- 102.Ryan DM, Monsey D. 1981. Bacterial filamentation and in vivo efficacy: a comparison of several cephalosporins. J Antimicrob Chemother 7:57–63. doi: 10.1093/jac/7.1.57. [DOI] [PubMed] [Google Scholar]
- 103.Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. 2004. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science 305:1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- 104.Chauhan A, Madiraju MV, Fol M, Lofton H, Maloney E, Reynolds R, Rajagopalan M. 2006. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J Bacteriol 188:1856–1865. doi: 10.1128/JB.188.5.1856-1865.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huisman O, D'Ari R. 1981. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature 290:797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- 106.Bi E, Lutkenhaus J. 1993. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol 175:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Michel B. 2005. After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol 3:e255. doi: 10.1371/journal.pbio.0030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Al Mamun AA, Lombardo MJ, Shee C, Lisewski AM, Gonzalez C, Lin D, Nehring RB, Saint-Ruf C, Gibson JL, Frisch RL, Lichtarge O, Hastings PJ, Rosenberg SM. 2012. Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science 338:1344–1348. doi: 10.1126/science.1226683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bos J, Zhang Q, Vyawahare S, Rogers E, Rosenberg SM, Austin RH. 2015. Emergence of antibiotic resistance from multinucleated bacterial filaments. Proc Natl Acad Sci U S A 112:178–183. doi: 10.1073/pnas.1420702111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dalia AB, Weiser JN. 2011. Minimization of bacterial size allows for complement evasion and is overcome by the agglutinating effect of antibody. Cell Host Microbe 10:486–496. doi: 10.1016/j.chom.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bull CG. 1915. The agglutination of bacteria in vivo. J Exp Med 22:484–491. doi: 10.1084/jem.22.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bull CG. 1915. The mechanism of the curative action of antipneumococcus serum. J Exp Med 22:457–464. doi: 10.1084/jem.22.4.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Margni RA, Parma EA, Cerone S, Erpelding A, Perdigón G. 1983. Agglutinating and non-agglutinating antibodies in rabbits inoculated with a particulate antigen (Salmonella typhimurium). Immunology 48:351–359. [PMC free article] [PubMed] [Google Scholar]
- 114.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Armbruster CE, Mobley HLT. 2012. Merging mythology and morphology: the multifaceted lifestyle of Proteus mirabilis. Nat Rev Microbiol 10:743–754. doi: 10.1038/nrmicro2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Partridge JD, Harshey RM. 2013. More than motility: Salmonella flagella contribute to overriding friction and facilitating colony hydration during swarming. J Bacteriol 195:919–929. doi: 10.1128/JB.02064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kearns DB, Losick R. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol 49:581–590. [DOI] [PubMed] [Google Scholar]
- 118.Mobley HL, Belas R. 1995. Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol 3:280–284. doi: 10.1016/S0966-842X(00)88945-3. [DOI] [PubMed] [Google Scholar]
- 119.Jansen AM, Lockatell CV, Johnson DE, Mobley HLT. 2003. Visualization of Proteus mirabilis morphotypes in the urinary tract: the elongated swarmer cell is rarely observed in ascending urinary tract infection. Infect Immun 71:3607–3613. doi: 10.1128/IAI.71.6.3607-3613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Abdelrahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol Rev 29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 121.Sunyakumthorn P, Bourchookarn A, Pornwiroon W, David C, Barker SA, Macaluso KR. 2008. Characterization and growth of polymorphic Rickettsia felis in a tick cell line. Appl Environ Microbiol 74:3151–3158. doi: 10.1128/AEM.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Blouin EF, Kocan KM. 1998. Morphology and development of Anaplasma marginale (Rickettsiales: Anaplasmataceae) in cultured Ixodes scapularis (Acari: Ixodidae) cells. J Med Entomol 35:788–797. doi: 10.1093/jmedent/35.5.788. [DOI] [PubMed] [Google Scholar]
- 123.Boldis V, Strus J, Kocianová E, Tusek-Znidaric M, Stefanidesová KN, Schwarzová KN, Kúdelová M, Sekeyová Z, Spitalská E. 2009. Life cycle of Rickettsia slovacain L929 cell line studied by quantitative real-time PCR and transmission electron microscopy. FEMS Microbiol Lett 293:102–106. doi: 10.1111/j.1574-6968.2009.01510.x. [DOI] [PubMed] [Google Scholar]
- 124.Elfving K, Lukinius A, Nilsson K. 2012. Life cycle, growth characteristics and host cell response of Rickettsia helvetica in a Vero cell line. Exp Appl Acarol 56:179–187. doi: 10.1007/s10493-011-9508-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bastidas RJ, Elwell CA, Engel JN, Valdivia RH. 2013. Chlamydial intracellular survival strategies. Cold Spring Harb Perspect Med 3:a010256. doi: 10.1101/cshperspect.a010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Omsland A, Beare PA, Hill J, Cockrell DC, Howe D, Hansen B, Samuel JE, Heinzen RA. 2011. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol 77:3720–3725. doi: 10.1128/AEM.02826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hooton TM, Stamm WE. 1997. Diagnosis and treatment of uncomplicated urinary tract infection. Infect Dis Clin North Am 11:551–581. doi: 10.1016/S0891-5520(05)70373-1. [DOI] [PubMed] [Google Scholar]
- 128.Svanborg C, Godaly G. 1997. Bacterial virulence in urinary tract infection. Infect Dis Clin North Am 11:513–529. doi: 10.1016/S0891-5520(05)70371-8. [DOI] [PubMed] [Google Scholar]
- 129.Schwartz DJ, Chen SL, Hultgren SJ, Seed PC. 2011. Population dynamics and niche distribution of uropathogenic Escherichia coli during acute and chronic urinary tract infection. Infect Immun 79:4250–4259. doi: 10.1128/IAI.05339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barer MR, Gribbon LT, Harwood CR, Nwoguh CE. 1993. The viable but non-culturable hypothesis and medical bacteriology. Rev Med Microbiol 4:183–191. doi: 10.1097/00013542-199310000-00001. [DOI] [Google Scholar]
- 131.Roszak DB, Colwell RR. 1987. Survival strategies of bacteria in the natural environment. Microbiol Rev 51:365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rollins DM, Colwell RR. 1986. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl Environ Microbiol 52:531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shahamat M, Mai U, Paszko-Kolva C, Kessel M, Colwell RR. 1993. Use of autoradiography to assess viability of Helicobacter pylori in water. Appl Environ Microbiol 59:1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Roszak DB, Colwell RR. 1987. Metabolic activity of bacterial cells enumerated by direct viable count. Appl Environ Microbiol 53:2889–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Steinert M, Emödy L, Amann R, Hacker J. 1997. Resuscitation of viable but nonculturable Legionella pneumophila Philadelphia JR32 by Acanthamoeba castellanii. Appl Environ Microbiol 63:2047–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nilsson L, Oliver JD, Kjelleberg S. 1991. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. J Bacteriol 173:5054–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ravel J, Knight IT, Monahan CE, Hill RT, Colwell RR. 1995. Temperature-induced recovery of Vibrio cholerae from the viable but nonculturable state: growth or resuscitation? Microbiology (Reading, Engl) 141:377–383. doi: 10.1099/13500872-141-2-377. [DOI] [PubMed] [Google Scholar]
- 138.Chaiyanan S, Chaiyanan S, Grim C, Maugel T, Huq A, Colwell RR. 2007. Ultrastructure of coccoid viable but non-culturable Vibrio cholerae. Environ Microbiol 9:393–402. doi: 10.1111/j.1462-2920.2006.01150.x. [DOI] [PubMed] [Google Scholar]
- 139.Billings G, Ouzounov N, Ursell T, Desmarais SM, Shaevitz J, Gitai Z, Huang KC. 2014. De novo morphogenesis in L-forms via geometric control of cell growth. Mol Microbiol 93:883–896. doi: 10.1111/mmi.12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hung WC, Jane WN, Wong HC. 2013. Association of a d-alanyl-d-alanine carboxypeptidase gene with the formation of aberrantly shaped cells during the induction of viable but nonculturable Vibrio parahaemolyticus. Appl Environ Microbiol 79:7305–7312. doi: 10.1128/AEM.01723-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Costa K, Bacher G, Allmaier G, Dominguez-Bello MG, Engstrand L, Falk P, de Pedro MA, García del Portillo F. 1999. The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J Bacteriol 181:3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lleò MM, Bonato B, Signoretto C, Canepari P. 2003. Vancomycin resistance is maintained in enterococci in the viable but nonculturable state and after division is resumed. Antimicrob Agents Chemother 47:1154–1156. doi: 10.1128/AAC.47.3.1154-1156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Signoretto C, Lleo MM, Tafi MC, Canepari P. 2000. Cell wall chemical composition of Enterococcus faecalis in the viable but nonculturable state. Appl Environ Microbiol 66:1953–1959. doi: 10.1128/AEM.66.5.1953-1959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gould AR, May BK, Elliott WH. 1975. Release of extracellular enzymes from Bacillus amyloliquefaciens. J Bacteriol 122:34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Greene NG, Narciso AR, Filipe SR, Camilli A. 2015. Peptidoglycan branched stem peptides contribute to Streptococcus pneumoniae virulence by inhibiting pneumolysin release. PLoS Pathog 11:e1004996. doi: 10.1371/journal.ppat.1004996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rahman I, Shahamat M, Chowdhury MA, Colwell RR. 1996. Potential virulence of viable but nonculturable Shigella dysenteriae type 1. Appl Environ Microbiol 62:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cole SP, Cirillo D, Kagnoff MF, Guiney DG, Eckmann L. 1997. Coccoid and spiral Helicobacter pylori differ in their abilities to adhere to gastric epithelial cells and induce interleukin-8 secretion. Infect Immun 65:843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chaput C, Ecobichon C, Cayet N, Girardin SE, Werts C, Guadagnini S, Prévost M-C, Mengin-Lecreulx D, Labigne A, Boneca IG. 2006. Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog 2:e97. doi: 10.1371/journal.ppat.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Byrd JJ, Colwell RR. 1990. Maintenance of plasmids pBR322 and pUC8 in nonculturable Escherichia coli in the marine environment. Appl Environ Microbiol 56:2104–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bari SMN, Roky MK, Mohiuddin M, Kamruzzaman M, Mekalanos JJ, Faruque SM. 2013. Quorum-sensing autoinducers resuscitate dormant Vibrio cholerae in environmental water samples. Proc Natl Acad Sci U S A 110:9926–9931. doi: 10.1073/pnas.1307697110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ayrapetyan M, Williams TC, Oliver JD. 2014. Interspecific quorum sensing mediates the resuscitation of viable but nonculturable vibrios. Appl Environ Microbiol 80:2478–2483. doi: 10.1128/AEM.00080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Stewart MK, Cookson BT. 2014. Mutually repressing repressor functions and multi-layered cellular heterogeneity regulate the bistable Salmonella fliC census. Mol Microbiol 94:1272–1284. doi: 10.1111/mmi.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Miao EA, Andersen-Nissen E, Warren SE, Aderem A. 2007. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin Immunopathol 29:275–288. doi: 10.1007/s00281-007-0078-z. [DOI] [PubMed] [Google Scholar]
- 154.Salazar-Gonzalez RM, McSorley SJ. 2005. Salmonella flagellin, a microbial target of the innate and adaptive immune system. Immunol Lett 101:117–122. doi: 10.1016/j.imlet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 155.Brennan CA, Hunt JR, Kremer N, Krasity BC, Apicella MA, McFall-Ngai MJ, Ruby EG. 2014. A model symbiosis reveals a role for sheathed-flagellum rotation in the release of immunogenic lipopolysaccharide. eLife 3:e01579. doi: 10.7554/eLife.01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Proft T, Baker EN. 2009. Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell Mol Life Sci 66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Durand E, Bernadac A, Ball G, Lazdunski A, Sturgis JN, Filloux A. 2003. Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J Bacteriol 185:2749–2758. doi: 10.1128/JB.185.9.2749-2758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Herrington DA, Hall RH, Losonsky G, Mekalanos JJ, Taylor RK, Levine MM. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med 168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bradley DE. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol 26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 160.Klein EA, Schlimpert S, Hughes V, Brun YV, Thanbichler M, Gitai Z. 2013. Physiological role of stalk lengthening in Caulobacter crescentus. Commun Integr Biol 6:e24561. doi: 10.4161/cib.24561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 162.Berg HC. 2004. E. coli in motion. Springer, New York, NY. [Google Scholar]
- 163.Pallen MJ, Matzke NJ. 2006. From the origin of species to the origin of bacterial flagella. Nat Rev Microbiol 4:784–790. doi: 10.1038/nrmicro1493. [DOI] [PubMed] [Google Scholar]
- 164.Balaban M, Hendrixson DR. 2011. Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni. PLoS Pathog 7:e1002420. doi: 10.1371/journal.ppat.1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Murray TS, Kazmierczak BI. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J Bacteriol 188:6995–7004. doi: 10.1128/JB.00790-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gode-Potratz CJ, Kustusch RJ, Breheny PJ, Weiss DS, McCarter LL. 2011. Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol Microbiol 79:240–263. doi: 10.1111/j.1365-2958.2010.07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bubendorfer S, Koltai M, Rossmann F, Sourjik V, Thormann KM. 2014. Secondary bacterial flagellar system improves bacterial spreading by increasing the directional persistence of swimming. Proc Natl Acad Sci U S A 111:11485–11490. doi: 10.1073/pnas.1405820111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wagner JK, Setayeshgar S, Sharon LA, Reilly JP, Brun YV. 2006. A nutrient uptake role for bacterial cell envelope extensions. Proc Natl Acad Sci U S A 103:11772–11777. doi: 10.1073/pnas.0602047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Poindexter JS. 1992. Dimorphic prosthecate bacteria: the genera Caulobacter, Asticcacaulis, Hyphomicrobium, Pedomicrobium, Hyphomonas, and Thiodendron, p 2176–2196. In Ballows A, Tr̈uper HG, Dworkin M, Harder W, Schleifer KH (ed), The prokaryotes, vol 3 Springer, New York, NY. [Google Scholar]
- 170.Jiang C, Brown PJB, Ducret A, Brun YV. 2014. Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature 506:489–493. doi: 10.1038/nature12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Poindexter JS. 1984. Role of prostheca development in oligotrophic aquatic bacteria, p 33–40. In Klug MJ, Reddy CA (ed), Current perspectives in microbial ecology: proceedings of the Third International Symposium on Microbial Ecology, Michigan State University, 7–12 August 1983 American Society for Microbiology, Washington, DC. [Google Scholar]
- 172.Bustamante VH, Martínez-Flores I, Vlamakis HC, Zusman DR. 2004. Analysis of the Frz signal transduction system of Myxococcus xanthus shows the importance of the conserved C-terminal region of the cytoplasmic chemoreceptor FrzCD in sensing signals. Mol Microbiol 53:1501–1513. doi: 10.1111/j.1365-2958.2004.04221.x. [DOI] [PubMed] [Google Scholar]
- 173.Schmidt JM, Stanier RY. 1966. The development of cellular stalks in bacteria. J Cell Biol 28:423–436. doi: 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gonin M, Quardokus EM, O'Donnol D, Maddock J, Brun YV. 2000. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J Bacteriol 182:337–347. doi: 10.1128/JB.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schlimpert S, Klein EA, Briegel A, Hughes V, Kahnt J, Bolte K, Maier UG, Brun YV, Jensen GJ, Gitai Z, Thanbichler M. 2012. General protein diffusion barriers create compartments within bacterial cells. Cell 151:1270–1282. doi: 10.1016/j.cell.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bianchi M. 1989. Unusual bloom of star-like prosthecate bacteria and filaments as a consequence of grazing pressure. Microb Ecol 17:137–141. doi: 10.1007/BF02011848. [DOI] [PubMed] [Google Scholar]
- 177.Imhaus AF, Duménil G. 2014. The number of Neisseria meningitidis type IV pili determines host cell interaction. EMBO J 33:1767–1783. doi: 10.15252/embj.201488031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zaburdaev V, Biais N, Schmiedeberg M, Eriksson J, Jonsson A-B, Sheetz MP, Weitz DA. 2014. Uncovering the mechanism of trapping and cell orientation during Neisseria gonorrhoeae twitching motility. Biophys J 107:1523–1531. doi: 10.1016/j.bpj.2014.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Melican K, Sandoval RM, Kader A, Josefsson L, Tanner GA, Molitoris BA, Richter-Dahlfors A. 2011. Uropathogenic Escherichia coli P and type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog 7:e1001298. doi: 10.1371/journal.ppat.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. 2002. Bacterial adhesion to target cells enhanced by shear force. Cell 109:913–923. doi: 10.1016/S0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 181.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J 19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 183.Hajishengallis G, Lamont RJ. 2012. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol 27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lamont RJ, Hajishengallis G. 2015. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med 21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Maeda K, Nagata H, Kuboniwa M, Kataoka K, Nishida N, Tanaka M, Shizukuishi S. 2004. Characterization of binding of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase to Porphyromonas gingivalis major fimbriae. Infect Immun 72:5475–5477. doi: 10.1128/IAI.72.9.5475-5477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Park Y, Simionato MR, Sekiya K, Murakami Y, James D, Chen W, Hackett M, Yoshimura F, Demuth DR, Lamont RJ. 2005. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun 73:3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, Demuth DR. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology (Reading, Engl) 148:1627–1636. doi: 10.1099/00221287-148-6-1627. [DOI] [PubMed] [Google Scholar]
- 188.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 189.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 190.Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. 2009. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol 191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]