Abstract
A number of transcriptional control elements are activated when Saccharomyces cerevisiae cells are submitted to various stress conditions, including high hydrostatic pressure (HHP). Exposure of Saccharomyces cerevisiae cells to HHP results in global transcriptional reprogramming, similar to that observed under other industrial stresses, such as temperature, ethanol and oxidative stresses. Moreover, treatment with a mild hydrostatic pressure renders yeast cells multi-stress tolerant. In order to identify transcriptional factors involved in coordinating response to high hydrostatic pressure, we performed a time series microarray expression analysis on a wild S. cerevisiae strain exposed to 50 MPa for 30 min followed by recovery at atmospheric pressure (0.1 MPa) for 5, 10 and 15 min. We identified transcription factors and corresponding DNA and RNA motifs targeted in response to hydrostatic pressure. Moreover, we observed that different motif elements are present in the promoters of induced or repressed genes during HHP treatment. Overall, as we have already published, mild HHP treatment to wild yeast cells provides multiple protection mechanisms, and this study suggests that the TFs and motifs identified as responding to HHP may be informative for a wide range of other biotechnological and industrial applications, such as fermentation, that may utilize HHP treatment.
Keywords: Biochemical reactions, Cell protection, DNA motifs, Environmental stresses, Microarray, RNA motifs, Yeast
INTRODUCTION
High hydrostatic pressure (HHP) is employed in a variety of biotechnological applications, including modulation of enzyme activities and food functionality, disaggregation of proteins, preparation of viral vaccines, and engineering of plant or animal tissues [1]. Pressure, similar to other environmental stresses, elicits a cohort of cellular responses. For instance, treatment of yeast cells to HHP results in structural and spatial compaction of biomolecules and cellular organelles, thereby leading to inhibition or activation of biochemical reactions responsive to changes in cellular volume [2], Moreover, HHP induces alteration of cell membrane and activation of antioxidant defences in yeast cells, which are also observed under other industrial stresses such as cold, heat, oxidative stresses, and high ethanol concentration [3]. These observations together with wide applications of HHP underscore the utility of HHP as a tool to model stress response in yeast and determine optimal Saccharomyces cerevisiae strains for specific industrial applications [4].
Saccharomyces cerevisiae has long been used as a model eukaryotic organism. In response to changes in environmental conditions, yeast cells must rapidly adjust their transcriptional output in order to adapt to the new conditions. This robust transcriptional reprogramming of hundreds of genes is essential for maintaining viability under diverse range of environmental conditions and stresses. Sensing external cues, transducing signals, and adjusting myriad number of gene and protein levels largely contribute to the cell’s defence mechanisms triggered by stress conditions. As transcriptional factors (TF) play central roles in cellular activity, understanding their genomic targets and corresponding outputs are undoubtedly crucial [5]. For example, microarray analyses have shown that TFs Msn2p and Msn4p control a broad subset of gene expressions responsive to environmental transitions, termed environmental stress response (ESR) [6, 7], Concomitantly, designated response mechanisms regulate the expression of genes specific to given stresses. Heat shock results in binding of the Hsf1p TF to heat shock elements (HSEs) present in the promoters of a large number of genes, whereas oxidative stress activates Yap family of TFs, mainly Yap1p, which translocates to the nucleus and directs gene expression, and osmotic stress induces the HOG pathway and nuclear accumulation of Hog1p TF [8].
Although microarray analyses of S. cerevisiae subjected to HHP have been previously performed [9, 10,11], the identities of transcriptional factors responsible for the observed transcriptional reprogramming remain unclear. Therefore, we examined temporal expression profiles of wild yeast cells exposed to pressure of 50 MPa for 30 min followed by recovery at atmospheric pressure of 0.1 MPa for 5, 10, and 15 min, and investigated transcriptional factors involved in the regulation of gene expression induced by HHP (Tabel 1). We showed previously that whereas pressure pretreatment alone (50 MPa for 30 min) does not induce tolerance to high pressures [12], brief incubation at atmospheric pressure following pressure treatment renders the cells tolerant to high pressure (220 MPa), to high (54°C) and low (−196°C) temperature [13]. To expand our understanding of global transcriptional response induced by HHP, we investigated temporal expression profiles of wild yeast cells exposed to 50 MPa for 30 min followed by recovery at atmospheric pressure for 5, 10 and 15 min, and explored transcriptional factors involved regulating the observed gene expression changes. In this report, we present identification of transcription factors and corresponding motifs responsive to pressure treatment, and demonstrate that whereas transcription of genes associated with adaptation to growth under pressure are regulated immediately after HHP, genes involved in cell cycle arrest and energy metabolism continue to be induced up to 15 min post-pressurization. Moreover, the described results should contribute to facilitating design of strategies to increase or decrease stress resistance in organisms of industrial interest.
Table 1.
Transcriptional factors involved in the regulation of gene expression induced by HHP
| Gene Name |
Description |
|---|---|
| MSN2 | Transcriptional activator related to Msn4p, activated in stress conditions, which results in translocation from the cytoplasm to the nucleus. Binds DNA at stress response elements of responsive genes, inducing gene expression. |
| MSN4 | Transcriptional activator related to Msn2p, activated in stress conditions, which results in translocation from the cytoplasm to the nucleus; Binds DNA at stress response elements of responsive genes, inducing gene expression |
| CBF1 | Helix-loop-helix protein. Binds the motif CACRTG present at several sites including MET gene promoters and centromere DNA element I (CDEI). Affects nucleosome positioning at this motif, associates with other transcription factors such as Met4p and Isw1p to mediate transcriptional activation or repression. |
| NRG1 | Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response. |
| RGM1 | Putative zinc finger DNA binding transcription factor; contains two N-terminal C2H2 zinc fingers and C-terminal proline rich domain; overproduction impairs cell growth and induces expression of genes involved in monosaccharide catabolism and aldehyde metabolism. |
| CIN5 | Basic leucine zipper (bZIP) transcription factor of the yAP-1 family. Physically interacts with the Tup1-Cyc8 complex and recruits Tup1p to its targets, Mediates pleiotropic drug resistance and salt tolerance. Nuclearly localized under oxidative stress and sequestered in the cytoplasm by Lot6p under reducing conditions. |
| OAF1 | Oleate-activated transcription factor, acts alone and as a heterodimer with Pip2p. Activates genes involved in beta-oxidation of fatty acids and peroxisome organization and biogenesis. |
| ABF1 | DNA binding protein with possible chromatin-reorganizing activity involved in transcriptional activation, gene silencing, and DNA replication and repair. |
| HMS2 | Chromatin associated high mobility group (HMG) family member involved in genome maintenance. rDNA-binding component of the Pol I transcription system. Associates with a 5'-3' DNA helicase and Fpr1p, a prolyl isomerase. |
| PHD1 | Transcriptional activator that enhances pseudohyphal growth. Physically interacts with the Tup1-Cyc8 complex and recruits Tup1p to its targets. Regulates expression of FLO11, an adhesin required for pseudohyphal filament formation; similar to StuA, an A. nidulans developmental regulator; potential Cdc28p substrate. |
| YER130C | Protein of unknown function. Transcription is regulated by Haa1p, Sok2p and Zap1p transcriptional activators. Computational analysis suggests a role as a transcription factor. C. albicans homolog (MNL1) plays a role in adaptation to stress. |
| STB5 | Transcription factor, involved in regulating multidrug resistance and oxidative stress response. Forms a heterodimer with Pdr1p; contains a Zn(II)2Cys6 zinc finger domain that interacts with a pleiotropic drug resistance element in vitro. |
| STP2 | Transcription factor, activated by proteolytic processing in response to signals from the SPS sensor system for external amino acids. Activates transcription of ammo acid permease genes. |
| GCN4 | Basic leucine zipper (bZIP) transcriptional activator of amino acid biosynthetic genes in response to amino acid starvation. Expression is tightly regulated at both the transcriptional and translational levels. |
| UPC2 | Sterol regulatory element binding protein, induces transcription of sterol biosynthetic genes and of DAN/TIR gene products. Ecm22p homolog; relocates from intracellular membranes to perinuclear foci on sterol depletion. |
| YRR1 | Zn2-Cys6 zinc-finger transcription factor that activates genes involved in multidrug resistance; paralog of Yrm1p, acting on an overlapping set of target genes. |
MATERIAL AND METHODS
Yeast strain and growth condition
The wild Saccharomyces cerevisiae yeast strain BT0605 was previously isolated from cachaça (Brazilian sugar cane spirit) distillery, as described in Bravim et al. [4] and is stored at −80 °C at the Agribusiness Applied Biotechnology Laboratory, UFES. This strain was selected for its flocculation ability, tolerance to ethanol, osmotic and heat shock stresses and for its high fermentation rate. Yeast cells were grown at 28 °C with aeration in liquid YEPD medium (1% yeast extract, 2% peptone, 2% glucose) to exponential growth phase (OD600nm = 1.0).
High hydrostatic pressure treatment (HHP)
Yeast cells were pressurized in the absence of air bubbles at room temperature as previously described [4]. Samples were subjected to four treatments: (1) 50 MPa for 30 min; 50 MPa for 30 min followed by incubation at atmospheric pressure (0.1 MPa) with aeration for (2) 5, (3) 10 and (4) 15 min. Experiments was performed twice in duplicate.
Microarray analyses
RNA preparation, amplification, microarray hybridization, and analysis were performed as described previously [14]. Total RNA was extracted using the Qiagen RNeasy Mini Kit (Valencia, CA), and cRNA was synthesized following standard protocol of Agilent Low RNA Input Linear Amplification Kit including additional DNase I purification step (Agilent Technologies, Palo Alto, CA). Briefly, 100 ng of total RNA was used as a template for first and second strand cDNA synthesis with reverse transcriptase using a primer containing poly dT and T7 polymerase promoter. Labeled cRNA was synthesized from cDNA using T7 RNA polymerase and cyanine3- (Cy3-) or Cy5-Iabeled CTP (PerkinEImer Life and Analytical Sciences, Boston, MA). The amount of cRNA synthesized and incorporation of Cy3- and Cy5-CTP into cRNA were measured using a NanoDrop (NanoDrop Technologies, Wilmington, DE). Equal amounts of Cy3- and Cy5-labeled cRNA were combined, mixed with the control target and fragmented for 30 min. Each sample was then hybridized to an Agilent yeast oligo microarray (VI, 4×44K, G2519F) or (V2, 8×l5K G4813A) for 17 h at 60 °C. The arrays were washed and scanned using Agilent Microarray Scanner (Agilent Technologies) at 100% PMT for red and green channels and at 5 µm resolution. The feature information was extracted from the microarrays using Agilent Feature Extraction Software version 9.5 with Linear Lowess dye normalization and no background subtraction and submitted to the Princeton University Microarray database for storage and analysis. Dye normalization for each array was determined by the rank consistency method and then spot intensities were calculated by the LOWESS method. Spots were retained for further analysis only if both the Cy3 and Cy5 channels were greater than 2.6σ of mean background intensity and were uniform in intensity. Only those genes for which 80% of the arrays yielded good data were retained for analysis. R2 values between experimental duplicate were greater than 0.99. All data described in this study can be publicly viewed and downloaded from the PUMAdb website: http://puma.princeton.edu/cgi-bin/publication/viewPublication.pl?pub_no=543
Gene expression confirmation
Total RNA was extracted from yeast cells using phenol/chloroform and precipitated with 3 M sodium acetate/absolute ethanol. Nucleic acids pellets were washed in 70% ethanol and resuspended in DEPC treated water. Extracted RNA samples were treated for 10 min with 0.5 U of RNAse-free DNAse I/µg RNA at 37 °C to remove any residual genomic DNA. cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biossystems, California, USA). Real-time RT-PCR experiments were carried out to determine the changes in the mRNA levels of several genes. The list of the validated genes and the primers used for amplification is shown in Table 2. Reactions were carried out in an Applied Biosystems Fast Real-Time PCR System (7.500, Applied Biosystems, California, USA). For each gene, calibration curve with 10-fold serial dilutions of cDNAs from the selected strain was obtained to determine the correlation coefficient (r2) which served as an indication of amplification efficiency, using the software supplied with the machine (Table 2). All analyses were performed in duplicate. Relative expression levels was obtained through the calculation of 2−ΔΔCt, where ΔΔCt = ΔCt treatment−ΔCt control [15]. The acquired data were normalized to ALG9 or TAF10 expression levels [16]. As the results for both genes were similar, just ALG9 data were used to the expression levels calculation (Supplementary material Figure SI).
Table 2.
Oligonucleotides used as primers for qRT-PCR analysis
| Target mRNA |
Primer sequence 5'-3' | Amplicon size (bp) |
PCR efficiency (%) |
|---|---|---|---|
| ADH1 | Forward, 5'ACTACGCCGGTATCAAATGG3' | ||
| ADH1 | Reverse, 5'TCAGCGGTAGCGTATTGTTG3' | 138 | 89 |
| ADH3 | Forward, 5'TATTCAAGCCGCCAAAATTC3' | ||
| ADH3 | Reverse, 5'TAACCCATCGCAGTTGCATA3' | 185 | 90 |
| HSP26 | Foward, 5'ATGCTGGCGCTCTTTATGAT3' | ||
| HSP26 | Reverse, 5'TTCTAGGGAAACCGAAACCA3' | 95 | 98 |
| PHM7 | Forward, 5'TTGGGGAATTGAACGAAGAG3' | ||
| PHM7 | Reverse, 5'TCTTCTGGCGAGTAGCCAAT3' | 180 | 88 |
| ROM1 | Forward, 5'AAACAAGTGGCACCAACACA3' | ||
| ROM1 | Reverse, 5'CATTCTTGGGATTGCTCGTT3' | 166 | 93 |
| RTN2 | Forward, 5'CGTGCTATCGACAGGATGAA3' | ||
| RTN2 | Reverse, 5'GGTTTGGGGTGGGATAACT3' | 110 | 106 |
| STF2 | Foward, 5'CGGTGAATCTCCAAATCACA3' | ||
| STF2 | Reverse, 5'CACTGGGGGTATTTCACCAT3' | 108 | 96 |
| TAF10 | Forward, 5'GCTAGGCAGCTATTGCAAGG3' | ||
| TAF10 | Reverse, 5'CAACAGCGCTACTGAGATCG3' | 129 | 98 |
| TFC1 | Forward, 5'TGGATGACGTTGATGCAGAT3' | ||
| TFC1 | Reverse, 5'GCTCGCTTTTCATTGTTTCC3' | 125 | 87 |
| USV1 | Forward, 5'AACGACAGCAACAACACCAA3' | ||
| USV1 | Reverse, 5'CGGAGGAAAGGACGATATGA3' | 214 | 80 |
| YGP1 | Forward, 5'TGACGGTGGTTACTCTTCCA3' | ||
| YGP1 | Reverse, 5'GAACGGCAGAACTCAAGGAG3' | 49 | 87 |
| YPS6 | Forward, 5'TGGGAGATGCTTTCCTTGTC3' | ||
| YPS6 | Reverse, 5'TCCTGTTCCGATGGGACTAC3' | 193 | 91 |
Regulatory motifs finding in DNA and proteins
The genes with expression changes of ≥2 or ≤−2-fold in the pressure treatment were selected for pre-clustering analysis using the Bingo 2.44 plugin [17], and then visualized using Cytoscape platform. After generation of clusters, the FIRE (Finding Informative Regulatory Elements) tool, part of the IGET platform (available on https://iget.c2b2.columbia.edu/), was used to find regulatory DNA and mRNA motifs shared between genes of the same cluster [18]. After that, a matrix was exported to the Cytoscape software to create the regulatory network. The preview layout was edited with “Spring Embedded Layout”.
For analysis of the rate of mRNA decay, genes induced ≥2-fold immediately after HHP with decay in the subsequent 5, 10 or 15 min of atmospheric pressure incubation were selected. Three groups were formed: (1) genes induced immediately after HHP with decay initiated in 5 min; (2) genes induced immediately after HHP with decay initiated in 10 min; (3) genes induced immediately after HHP with decay initiated in 15 min. The FIRE (Finding Informative Regulatory Elements) tool was used to find regulatory DNA and mRNA motifs shared between genes of the same group [18]. In parallel, the FIRE-pro tool was used to identify regulatory protein motifs shared between genes of the same group [19]. Functional motifs overrepresented and underrepresented were used for both analyses.
Dynamic Regulatory Events Miner analysis
DREM (Dynamic Regulatory Events Miner) software was used to determine bifurcation points by using hidden-input/hidden-output Markov model, as described in Ernst et al. [20], The software integrates TF-gene regulatory relationships derived from motif data with expression profiles from multiple time points. Significance was defined as P < 0.05 as assigned from Gene Ontology Term Finder and multiple hypotheses testing using randomization procedure for correction.
RESULTS AND DISCUSSION
Global microarray analysis of wild Saccharomyces cerevisiae strain BT0605 exposed to high hydrostatic pressure (HHP) of 50MPa revealed transcriptional changes in a broad range of genes. Among 6200 known or predicted genes in yeast, mRNA levels for approximately 2.7% genes were altered greater than 2-fold at 30 min of pressurization when compared to untreated cells, with 123 genes being induced and 47 genes repressed. After 15 min incubation at atmospheric pressure following HHP treatment, 12.9% of genes were affected, with 408 genes being upregulated and 392 genes being downregulated greater than 2-fold (Supplementary material Table S2).
Regulatory network of motifs
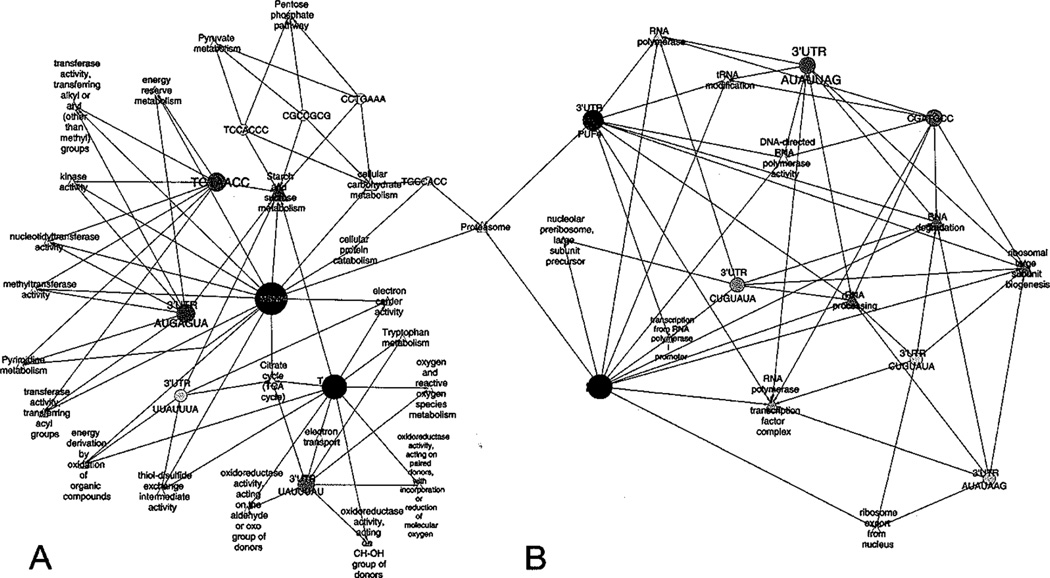
To corroborate DNA and mRNA motifs to observed transcriptional response induced by pressure treatment, we applied regulatory network analysis to the microarray data, and uncovered two modules of regulatory networks (Fig. 1). Briefly, a module in a network is composed of a set of nodes grouped together by strong interactions based on common cellular functions [21, 22, 23]. Genes which are part of a highly connected module have been shown to be relatively important in biological processes [24]. As shown in Fig. (1), module A is enriched in motifs present in genes associated with energy metabolism and stress response, which were mainly activated in response to pressurization. Module B is enriched with motifs of genes whose products are required for DNA, RNA and protein synthesis, with majority of them being downregulated in response to pressurization.
Fig. (1). Regulatory network of KNA and mRNA motifs after FIRE analysis.
Circles represent the regulatory motifs shared by a group of differentially expressed genes (> 2-fold induced or < −2-fold downregulated) after pressure treatment, with high (black) or low (gray) connectivity between motifs and genes groups. Groups of genes that shared the same ontology terms are also integrated in the network (triangle). A and B represent two modules of regulatory networks.
Genes involved in the proteasomic complex formation is the only node that shared the same motifs in both modules. Application of FIRE in silico program, which explores expression patterns and corresponding regulatory regions to discover informative motifs, identified MSN2/4, PAC, (3’UTR) PUF4 and TGCCACC motifs as regulatory elements in this group. After HHP treatment, we observed a negative correlation between PUF4 and a number of genes associated with proteasomic complex. Although Puf proteins have shown to be involved in mRNA degradation [25], our results suggest that decrease in PUF4 expression may lead to upregulation of genes required for proteasomic complex formation. In addition, our studies identified MSN2/4 DNA motifs in the promoters of genes encoding proteasomic complex, and we propose that Msn2/4 activate transcription of genes involved in protein degradation via proteasome in response to HHP-induced protein unfolding [26].
Module A also contains TATA and 3’UTR UAUUUAU motifs present in promoters of genes involved in tryptophan metabolism, electron transportable chain, ROS metabolism and oxidoreductase activity. TATA-containing genes are usually associated with environmental stress responses and are variably expressed, while most TATA-less genes represent housekeeping genes and are constitutively expressed [27, 28, 29]. Although 3’ UTR UAUUUAU element in mammalians cells are associated with mRNA stability [30], our results suggest that this motif in yeast may play a regulatory role on mRNA decay induced by TATA promoter during pressurization stress.
Furthermore, our results demonstrate that HHP-responsive genes encoding transferases, kinases, methyltransferase, and nucleotidyltransferase contain transcriptional regulatory elements TGTAACC and 3’UTR AUGAGUA. In addition, 3’UTR AUGAGUA is also involved in mRNA decay. However, the role of these elements and their corresponding transcriptional factors remain to be elucidated.
mRNA decay
Rates of mRNA decay were analyzed and the results were assembled in gene ontology categories. Table 3 shows the decay of genes groups differentially expressed at 5, 10 and 15 min of atmospheric pressure incubation following pressure treatment. Interestingly, whereas these genes were induced > 2-fold by HHP treatment, they were repressed at atmospheric pressure. Gene ontology (GO) analysis revealed that the categories affected 5 and 10 min post-pressurization were involved in regulation of sulfur metabolism. As shown in Table 3, after 15 min incubation at atmospheric pressure, affected categories were enriched in amine transporter activity and cell cycle. Three motifs are known to regulate the expression of these genes: whereas all 3 motifs were identified at 5 min post-piezotreatment, 1 and 2 of the motifs were identified at 10 and 15 min after treatment, respectively (Table 3).
Table 3.
Genes and motifs related to decay of relative expression as a function of time. Only genes with induced expression (≥ 2 fold) at time 0 relative to the control, but consequently down regulated are included. Results are presented for the genes for the time period in which expression is first downregulated after return to atmospheric pressure following HHP treatment
| Time (min) |
Gene name | Gene Ontology overrepresented terms |
p-value | Motifs overrepresented |
|---|---|---|---|---|
| 5 | MHT1, DAN1, METI4, AGP1, AYT1, MET3, MET2, MET6, MET17, SEO1 | Sulfur metabolism | 5.8 × 10−6 | CBF1 |
| 2.1 × 10−3 | (3’UTR) CGGAGC | |||
| 1.1 × 10−3 | (3'UTR) ACAUUCG | |||
| 10 | CIT2, YLL058w, MET16, NRG1 | Sulfur metabolism | 2.9 × 10−4 | (3’UTR) CGGAGC |
| 15 | YOR378w, YNL120C, FAR1, PRM5, HO, SPR28, MEP1, RTS3, REG2,MET10,MUP3, PRM1, YOR062c, PUT1, DIP5, YET2 | Amine transporter activity and Cell cycle | 1.2 × 10−3 | CBF1 |
| 2.8 × 10−3 | (3’UTR)ACAUUCG |
Sulfur metabolism genes (MET14, MET3, MET2, MET6, MET17, MET16 and MHT1) are involved in activation of methionine biosynthesis and/or regeneration. Although SHHP treatment resulted in upregulation of this group of genes, a high decay rate (at least fold-change < 0.03) was observed at 5 min and 10 min after pressure release (Table 3). The induction of genes associated with nitrogen and sulfur metabolism has previously been reported in yeast cells exposed to 40 MPa for 16 h [11]. Moreover, Murata et al. [31] reported that methionine biosynthesis might be correlated with lipid biosynthesis. Since pressure modifies phospholipid bilayers in part through compaction of fatty acyl chains thereby reducing membrane fluidity [3], and cells optimize membrane fluidity within the lipid matrix by modulating membrane composition [2], our results suggest that yeast cells utilize methionine metabolism and consequently lipid biosynthesis to provide membrane protection against HHP stress.
Interestingly, only three other treatments are known to induce genes involved in methionine biosynthesis: nitrogen and amino acids starvation, and treatment with diamide, a sulfhydryl oxidizing agent [6]. In nature, organisms often encounter conditions in which nutrients are limiting, and they use an array of metabolic changes to survive through these periods. Under nitrogen starvation, cells mobilize stored nitrogen sources such as vacuolar amino acid pool, express high affinity transporters to facilitate nitrogen uptake [32], and diamide stress elicits similar cellular response as that of oxidative stress. A variety of reactive oxygen species react readily with methionine residues in proteins to form methionine sulfoxide, thereby scavenging the reactive species. Most cells contain methionine sulfoxide reductases, which catalyze a thioredoxin-dependent reduction of methionine sulfoxide back to methionine. Thus, methionine residues may act as catalytic antioxidants, protecting both the protein where they are located and other macromolecules [33].
The transcriptional factor CBF1 is required for methionine prototrophy and chromatin-remodeling [34]. CBF1 negatively regulates genes by increasing production of S-adenosylmethionine (AdoMet), which is synthesized by the isoenzymes SAM1 and SAM2 [35]. AdoMet is involved in methylation of proteins, RNAs, biotin, polyamines and lipids [36, 37, 38]. The major phospholipids found in S. cerevisiae membranes are phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), and phosphatidyiserine (PS); and AdoMet is required for the synthesis of PC from PE [39]. Our results showed a decay of genes regulated by CBF1 (Table 3) and a high repression of SAM1 and SAM2, probably by its product formation (AdoMet) via retroinhibition [40] suggesting that activation of AdoMet may lead to cell membrane protection. Although our results show a basal level of expression of SAM1 and SAM2 (−0.29 and 1.19, respectively) immediately after HHP treatment, the MET6 gene, which is involved in AdoMet production [41], showed a high expression level (2.59).
Similarly to MET6, the gene DAN1, also related to sulfur metabolism, was highly expressed immediately after pressure treatment (2.81) and repressed 5 min after the piezotreatment (−0.01) (Table S1), whose gene product is a cell wall mannoprotein and associated with cell wall permeability, and is induced in response to environmental stress [42], These results further support our model described above that the induction of genes promoting phospholipid biosynthesis and cell membrane protection contributes to cellular mechanism responsive to hydrostatic pressure stress.
Genes related to cell cycle control were also affected after pressure treatment as shown in Table 3. For instance, we observed decrease in expression of genes involved in cell cycle progression at 15 min post-pressurization. Accordingly, the proportion of budded cells has shown to be decreased under pressure stress conditions [13].
DREM analyses
We applied Dynamic Regulatory Event Miner (DREM), a computational method, to investigate the temporal organization of dynamic regulatory events in the transcriptional response of S. cerevisiae under HHP stress. This approach integrates temporal dynamic transcriptome data with protein-DNA interactions, detects points in time when the expression pattern of a subset of genes deviates from the rest of the genes (bifurcation points), and predicts transcription factors regulating these transcriptional events [20].
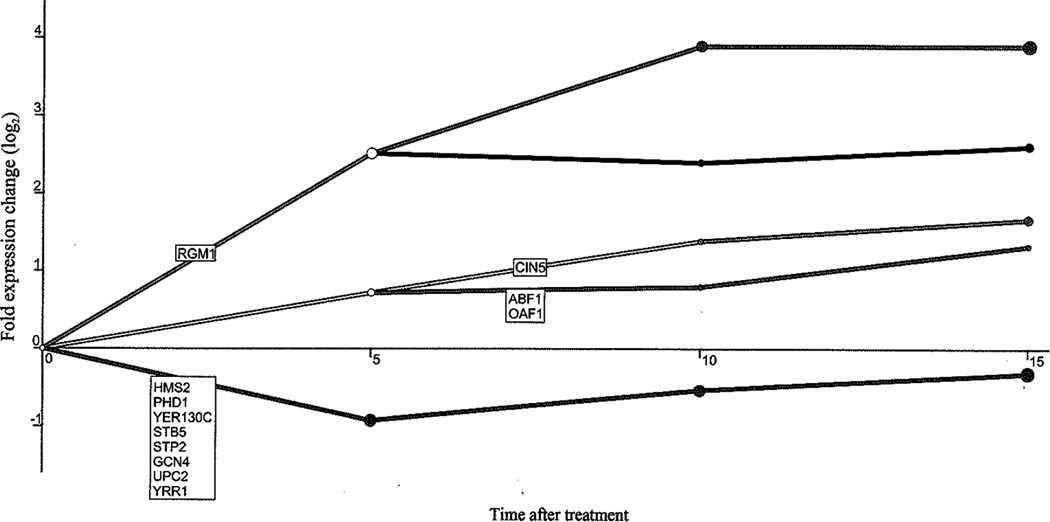
The dynamic transcriptional response to HHP treatment shows a complex pattern. Shown in fig. 2 is a temporal map from DREM analysis resulting in 5 unique paths controlled by a total of 12 TFs (using a TF split cutoff score of 0.005). The branch of upregulated transcripts (Table 4) includes regulators involved in repression of processes necessary for normal cell growth (Rgm1p), stress response (Cin5p), cell growth control and intracellular transport during stress response (Abf1p) and activation of genes involved in beta-oxidation of fatty acid as well as peroxisome organization and biogenesis (Oaf1p). Moreover, the transcripts in the lower upregulated branch are significantly associated with proteasome complex, regulated by ABF1 and OAF, and with mitochondria, regulated by CIN5, while genes in the upper upregulated branch are associated with plasma membrane enriched fraction and organelle envelope, regulated by RGM1 (Table 4).
Fig. (2). Identification of bifurcation points after pressure treatment.
Dynamic regulatory map based on time-series gene expression data and interaction data that associates TFs with the genes they regulate, highlighting bifurcation events in the time series. Each time point on the x-axis corresponds to the time immediately after pressure treatment (0) and 5, 10 and 15 min post-pressurization. Y-axis corresponds to relative expression of genes. The major paths and splits in the time series data were constructed for the genes that are assigned to these paths through the model. Each node is associated with a Gaussian distribution determining its y-axis location on the map. The area of a node is proportional to the standard deviation from the Gaussian distribution. A relatively small node implied the expression of the genes going through that node will be tightly centered on the node.
Table 4.
Gene Ontology Enrichment of transcripts following bifurcation.
| Branch of downregulated transcripts | Branch of upregulated transcripts | ||||
|---|---|---|---|---|---|
| Bifurcation controlling TFs |
Process gene ontology terms |
p-value | Bifurcation controlling TFs |
Process gene ontology terms |
p-value |
| HMS2, PHD1, YER130C, STB5, STP2, GCN4, UPC2, YRR1 | Monovalent inorganic cation transmembrane transporter activity | 5.2 × 10−8 | RGM1 | Organelle envelope | 2.4 × 10−4 |
| Plasma membrane enriched fraction | 4.3 × 10−4 | ||||
| Ion transmembrane transporter activity | 1.1 × 10−6 | CIN5 | Mitochondrion | 8.7 × 10−10 | |
| ATP biosynthetic process | 3.1 × 10−5 | Oxidation-reduction process | 2.2 × 10−9 | ||
| Cellular nitrogen compound biosynthetic process | 6.4 × 10−5 | Cellular glucan metabolic process | 2.5 × 10−6 | ||
| ABF1, OAF1 | Proteasome complex | 3.3 × 10−24 | |||
Previous reports have shown that the proteasome complex is essential to efficient modulation of genes involved in stress response [43, 44]. Pressure treatment results in abnormal distribution of mitochondria and their damage in yeast cells [45] that likely impair proper energy flux within the cell. Mitochondrial agglomeration is also associated with disruption of cytoskeleton proteins by pressure, which interferes with other cellular organelles [45]. Therefore, cells likely need to adjust transcriptional program of genes associated with repairing cellular organelles and proteolysis of damaged proteins from organelles during recovery from HHP [10].
The branch of downregulated transcripts includes monovalent inorganic cation transmembrane transporter activity and ion transmembrane transporter activity regulators (Table 4), which add to the cell’s protective mechanism from ionic toxicity [46]. Nevertheless, DREM analyses suggest that these transcriptional factors play an important role on downregulated genes during cellular protection to HHP stress (Table 4).
CONCLUSION
High hydrostatic pressure has shown to be responsible for the activation of stress response elements similarly observed under other environmental stresses, but the modulation of this response has not been conducted in a stochastic manner. The temporal transcriptional response profile to HHP presented in this report suggests that the regulation of gene groups follows a priority line: while genes corresponding to repair or modification of membranes, mitochondria, vacuole, as well as genes related to aggregation protection were immediately regulated, other groups of stress genes (for instance, genes that encode membrane proteins, and proteins involved in protein folding, cell respiration, spore formation) were regulated latter on. Several genes involved in adaptation to growth under HHP were induced during pressurization but the mRNA decayed rapidly during the post-pressurization period, such as genes induced by the TF CBF. Moreover, the temporal transcriptional profile analysis indicate that piezostress activates general stress response, for example cell cycle arrest and energy metabolism, which maintained at 15 min after HHP release. Comparison of motifs between these groups demonstrated that promoters of up- or downregutated genes responsive to HHP harbor different motifs governing transcriptional control. Several of the motifs described in the present work remain uncharacterized and their corresponding transcription factors are also unknown. Undoubtedly, elucidation of these factors and functions is necessary for complete understanding of cellular response to HHP.
Transcription factors Msn2 and Msn4 that participate in the activation of a broad class of stress-responsive genes did not show significant transcriptional changes. This is not surprising since transcription factors are primarily regulated at the level of cellular localization instead of expression level in order for cells to rapidly and robustly respond to environmental fluctuations. Namely, Msn2/4p migrate from the cytoplasm to the nucleus in response to stress [47].
Finally, studies focused on transcriptional and post-transcriptional regulation should provide important information on yeast cellular response to HHP stress. Moreover, these studies will catalyze a shift from a purely empirical approach towards scientific evidence-based methods in developing novel industrial products and processes that rely on utilization of pressurization.
Acknowledgments
This work was supported by grants from FINEP (Financiadora de Estudos e Projetos), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and FAPES (Fundação de Amparo à Pesquisa do Estado do Espfríto Santo).
LIST OF ABBREVIATIONS
- DREM
Dynamic Regulatory Events Miner
- DEPC
Diethylpyrocarbonate
- ESR
Environmental Stress Response
- FIRE
Finding Informative Regulatory Elements
- HOG1
Mitogen-activated protein kinase involved in osmoregulation
- HHP
High Hydrostatic Pressure
- HSE
Heat Shock element
- HSF1
heat shock transcription factor
- MPa
Mega Pascal
- Msn2p
Transcriptional activator related to Msn4p; activated in stress conditions, which results in translocation from the cytoplasm to the nucleus
- Msn4p
Transcriptional activator related to Msn2p
- ROS
Reactive oxygen species
- TF
Transcrptional factor
- YAP1
Transcription factor required for oxidative stress tolerance
Footnotes
SUPPLEMENTARY MATERIAL
Fig. (S1) Correlation plot of microarray (MA) and qRT-PCR fold value data for 11 genes (ADH1, ADH3, PHM7, HSP26, ROM1, RTN2, STF2, TFC1, USV1, ZEO1, YGP1 and YPS6) used in the validation of the microarray results on a wild S. cerevisiae strain exposed to 50 MPa for 30 min (A) followed by recovery at atmospheric pressure (0.1 MPa) for 5 (B), 10 (C) and 15 min (D). The correlation coefficient is indicated inside of each figure.
Table (S2) Transcriptional level of Saccharomyces cerevisiae BT0605 genes altered by HHP treatment of 50 MPa for 30 min followed by incubation at atmospheric pressure for up to 15 min. Induced genes have been given a positive value, suppressed genes a negative values.
REFERENCES
- 1.Aertsen A, Meersman F, Hendrickx ME, Vogel RF, Michiels CW. Biotechnology under high pressure: applications and implications. Trends Biotechnol. 2009;27(7):434–441. doi: 10.1016/j.tibtech.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Mentré P, Hui Bon Hoa G. Effects of high hydrostatic pressures on living cells: a consequence of the properties of macromolecules and macromolecule – associated water. Int. Ver. of Cytol. 2001;201:1–84. doi: 10.1016/s0074-7696(01)01001-4. [DOI] [PubMed] [Google Scholar]
- 3.Fernandes PMB. In: High Pressure Microbiology. Michiels Chris, Bartlett Douglas H, Aertsen Abram., editors. Vol. 1. Washington, DC: American Society for Microbiology; 2008. pp. 145–166. [Google Scholar]
- 4.Bravim F, Palhano FL, Fernandes AA, Fernandes PM. Biotechnological properties of distillery and laboratory yeasts in response to industrial stresses. J. Ind Microbiol. Biotechnol. 2010;37(10):1071–1079. doi: 10.1007/s10295-010-0755-0. [DOI] [PubMed] [Google Scholar]
- 5.Hahn S, Young ET. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189(3):705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11(12):4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12(2):323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hohmann S, Mager WH. Yeast stress responses. 2. Alemanha: Springer; 2003. [Google Scholar]
- 9.Fernandes PMB, Domitrovic T, Kao CM, Kurtenbach E. Genome expression pattern in Saccharomyces cerevisiae cells in response to high hydrostatic pressure. FEBS Lett. 2004;556(1–3):153–160. doi: 10.1016/s0014-5793(03)01396-6. [DOI] [PubMed] [Google Scholar]
- 10.Iwahashi H, Shimizu H, Odani M, Komatsu Y. Piezophysiology of genome wide gene expression levels in the yeast Saccharomyces cerevisiae. Extremophiles. 2003;7(4):291–298. doi: 10.1007/s00792-003-0322-y. [DOI] [PubMed] [Google Scholar]
- 11.Iwahashi H, Odani M, Ishidou E, Kitagawa E. Adaptation of Saccharomyces cerevisiae to high hydrostatic pressure causing growth inhibition. FEBS Lett. 2005;579(13):2847–2852. doi: 10.1016/j.febslet.2005.03.100. [DOI] [PubMed] [Google Scholar]
- 12.Fernandes PM, Panek AD, Kurtenbach E. Effect of hydrostatic pressure of a mutant of Saccharomyces cerevisiae deleted in the trehalose-6-phosphate synthase gene. FEMS Microbiol. Lett. 1997;152(1):17–21. doi: 10.1111/j.1574-6968.1997.tb10403.x. [DOI] [PubMed] [Google Scholar]
- 13.Palhano FL, Gomes HL, Orlando MT, Kurtenbach E, Fernandes PM. Pressure response in the yeast Saccharomyces cerevisiae: from cellular to molecular approaches. Cell. Mol. Biol. 2004;50(4):447–457. [PubMed] [Google Scholar]
- 14.Lippman SI, Broach JR. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc. Natl, Acad. Set. USA. 2009;106(47):19928–19933. doi: 10.1073/pnas.0907027106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Teste MA, Duquenne M, Françis JM, Parrou JL. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol. 2009;10:99–114. doi: 10.1186/1471-2199-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 18.Elemento O, Slonim N, Tavazoie S. A Universal Framework for Regulatory Element Discovery across All Genomes and Data Types. Mol. Cell. 2007;28(2):337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieber DS, Elemento O, Tavazoie S. Large-Scale Discovery and Characterization of Protein Regulatory Motifs in Eukaryotes. PLoS ONE. 2010;51(12):e14444, 1–12. doi: 10.1371/journal.pone.0014444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst J, Vainas O, Harbison CT, Simon I, Bar-Joseph Z. Reconstructing Dynamic Regulatory Maps. Mol. Syst. Biol. 2007;3(74):1–13. doi: 10.1038/msb4100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(6761 Suppl):C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 22.Alon U. Biological networks: the tinkerer as an engineer. Science. 2003;301(5641):1866–1867. doi: 10.1126/science.1089072. [DOI] [PubMed] [Google Scholar]
- 23.Palla G, Derenyi I, Farkas T, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435(7043):814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 24.Carlson MR, Zhang B, Fang Z, Mischel PS, Horvath S, Nelson SF. Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics. 2006;7(40):1–15. doi: 10.1186/1471-2164-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 2000;19(23):6602–6611. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorovits BM, Horowitz PM. High hydrostatic pressure can reverse aggregation of protein folding intermediates and facilitate acquisition of native structure. Biochemistry. 1998;37(17):6132–6135. doi: 10.1021/bi9730137. [DOI] [PubMed] [Google Scholar]
- 27.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116(5):699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 28.Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nat Genet. 2006;38(7):830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 29.Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. Genetic properties influencing the evolvability of gene expression. Science. 2007;317(5834):118–121. doi: 10.1126/science.1140247. [DOI] [PubMed] [Google Scholar]
- 30.Zubiaga AMJ, Belasco G, Greenberg ME. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell. Biol. 1995;15(4):2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murata Y, Watanabe T, Sato M, Momose Y, Nakahara T, Oka S, Iwahashi H. Dimethyl sulfoxide exposure facilitates phospholipid biosynthesis and cellular membrane proliferation in yeast cells. J. Biol. Chem. 2003;275(35):33185–33193. doi: 10.1074/jbc.M300450200. [DOI] [PubMed] [Google Scholar]
- 32.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol Chem. 2005;280(36):31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 33.Luo S, Levine RL. Methionine in proteins defends against oxidative stress. FASEB J. 2009;25(2):464–472. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai M, Davis RW. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;67(3):437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- 35.Thomas D, Cherest H, Surdin-Kerjan Y. Elements involved in S-adenosylmethionine-mediated regulation of the Saccharomyces cerevisiae MET25 gene. Mol. Cell. Biol. 1989;9(8):3292–3298. doi: 10.1128/mcb.9.8.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1997;67(4):503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phalip V, Kuhn I, Lemoine Y, Jeltsch JM. Characterization of the biotin biosynthesis pathway in Saccharomyces cerevisiae and evidence for a cluster containing BIO5, a novel gene involved in vitamer uptake. Gene. 1999;232(1):43–51. doi: 10.1016/s0378-1119(99)00117-1. [DOI] [PubMed] [Google Scholar]
- 38.Chattopadhyay MK, Tabor CW, Tabor H. Methylthioadenosine and polyamine biosynthesis in a Saccharomyces cerevisiae meuldelta mutant. Biochem. Biophys. Res. Commun. 2006;343(1):203–237. doi: 10.1016/j.bbrc.2006.02.144. [DOI] [PubMed] [Google Scholar]
- 39.Carman GM, Henry SA. Phospholipid biosynthesis in yeast. Annu. Rev. Biochem. 1989;58:635–669. doi: 10.1146/annurev.bi.58.070189.003223. [DOI] [PubMed] [Google Scholar]
- 40.Thomas D, Surdin-Kerjan Y. The synthesis of the two S-adenosyl-methionine synthetases is differently regulated in Saccharomyces cerevisiae. Mol Gen. Genet. 1991;226(1–2):224–233. doi: 10.1007/BF00273607. [DOI] [PubMed] [Google Scholar]
- 41.Pejchal R, Ludwig ML. Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication. PLoS Biol. 2005;3(2):e31. doi: 10.1371/journal.pbio.0030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abramova NE, Cohen BD, Sertil O, Kapoor R, Davies KJ, Lowry CV. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics. 2001;157(3):1169–1177. doi: 10.1093/genetics/157.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulahian R, Sikder D, Johnston SA, Kodadek T. The proteasomal ATPase complex is required for stress-induced transcription in yeast. Nucleic Acids Res. 2006;34(5):1351–1357. doi: 10.1093/nar/gkl012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sikder D, Johnston SA, Kodadek T. Widespread, but non-identical, association of proteasomal 19 and 20 S proteins with yeast chromatin. J Biol Chem. 2006;281(37):27346–27355. doi: 10.1074/jbc.M604706200. [DOI] [PubMed] [Google Scholar]
- 45.Fernandes PM, Farina M, Kurtenbach E. Effect of hydrostatic pressure on the morphology and ultrastructure of wild-type and trehalose synthase mutant cells of Saccharomyces cerevisiae. Lett. Appl. Microbiol. 2001;32(1):42–46. doi: 10.1046/j.1472-765x.2001.00853.x. [DOI] [PubMed] [Google Scholar]
- 46.Serrano R, Marquez JA, Rios G. In: Yeast stress responses. Hohmann S, Mager WH, editors. Vol. 1. New York: Springer; pp. 147–169. 21997. [Google Scholar]
- 47.Jacquet M, Renault G, Lallet S, De Mey J, Goidbeter A. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J. Cell. Biol. 2003;161(3):497–505. doi: 10.1083/jcb.200303030. [DOI] [PMC free article] [PubMed] [Google Scholar]