Abstract
To characterize T cell epitopes in monkeypox virus (MPXV) infected rhesus macaques, we utilized IFNγ Elispot assay to screen 400 predicted peptides from 20 MPXV proteins. Two peptides from the F8L protein, an analog of E9L protein in vaccinia, were found to elicit CD8+ T cell responses. Prediction and in vitro MHC binding analyses suggest that one is restricted by Mamu-A1⁎001 and another by Mamu-A1⁎002. The Mamu-A1⁎002 epitope is completely identical in all reported sequences for variola, vaccinia, cowpox and MPXV. The Mamu-A1⁎001 epitope is conserved in MPXV and vaccinia, and has one residue substitution (V6>I) in some cowpox sequences and all variola sequences. Given CD8+ T-cell epitopes from E9L were also identified in humans and mice, our data suggested that F8L/E9L may be a dominant pox viral protein for CD8+ T cell responses, and may be considered as a target when designing vaccines that target pox-specific T cell responses.
Keywords: Monkeypox virus, Rhesus, T cell epitopes, CD8+ T cells, Bioinformatics analysis, Peptides, Elispot
Introduction
Monekypox virus (MPXV) is a member of the Orthopoxvirus family that also includes variola virus (VARV), the causative agent of human smallpox, and vaccinia virus (VACV). Since cessation of broad VACV immunization, MPXV infections are now emerging as a public health concern. MPXV infection can cause up to 10% lethality in humans, and poses a risk to human health as an infectious agent and a potential biological weapon. MPXV infection resembles in many aspects the human clinical symptoms of smallpox, including fever, weight loss, lesion development, and death (Huhn et al., 2005, Jezek et al., 1987). To better understand the pathogenesis of MPXV and related orthopoxviruses, such as variola virus, and to provide a model to evaluate counter measures against these poxviruses, non-human primate (NHP) models of MPXV infection have been developed using aerosol (Zaucha et al., 2001), intranasal (Saijo et al., 2006), and intratracheal (Stittelaar et al., 2005, Stittelaar et al., 2006), as well as intravenous (IV) (Johnson et al., 2011, Hooper et al., 2004) routes of exposure. The respiratory pathogen delivery routes are generally technically demanding and may also require specialized facilities and equipment, while the IV inoculation is technically easier and has been used successfully to evaluate the immunogeneicity and protective role of various forms of poxvirus vaccines against MPX infection in NHPs (Hooper et al., 2004, Edghill-Smith et al., 2005, Earl et al., 2004, Heraud et al., 2006).
MPXV-infected NHPs have symptoms resembling those of humans infected with monkeypox and smallpox. Macaques infected with MPXV generate both neutralizing antibodies (Johnson et al., 2011, Keasey et al., 2010) and T cell responses (Estep et al., 2011). Upon proteomic array analysis, while antibodies from MPXV-infected macaques and VACV-vaccinated humans recognized a few common proteins, antibodies from MPXV-infected macaques also recognized proteins that were not recognized by vaccinated human sera (Keasey et al., 2010). It is interesting to determine if this also applies to T cell responses. T cell responses, as the other major arm of adaptive immune responses, have also been shown to play a critical role in protection against poxvirus infection (Redfield et al., 1987, Howell et al., 2006, Xu et al., 2004, Snyder et al., 2004). The precise definition of T cell epitopes allows rigorous assessment of antigen-specific T cell responses in viral pathogenesis and in vaccine studies. Specifically in the case of poxviruses, a relatively large number of CD8+ and CD4+ T cell epitopes have been characterized in human and murine systems (Kennedy and Poland, 2007, Moutaftsi et al., 2010). However, only a few T cell epitopes have been reported in rhesus macaques infected with VACV WR (Walsh et al., 2009), and no specific epitopes have been reported for variola or monkeypox. Given the pivotal role of non-human primates for the study of poxvirus pathogenesis and counteracting agents, including vaccine studies, it is important to have specific reagents to study pox-specific T cell function in rhesus macaques. In the current study, we utilized an approach combining bioinformatic epitope predictions and Elispot immunological assays to identify the first CD8+ T cell epitopes for monkeypox virus recognized in rhesus macaques. Our results illustrate the feasibility of this approach to identify epitopes from biosafety level 3 (BSL-3) poxviruses.
Results
Selection of antigens for study
To identify candidate antigens for analysis, we compiled T cell recognition data from three sources. These included an analysis undertaken by Moutaftsi et al. (2010) to identify prevalently recognized VACV WR ORFs, a compilation and analysis of donor recognition data available in the immune epitope data base (IEDB) (Vita et al., 2010), and a study of CD8 T cell responses to VACV WR in Mamu-A1 ⁎ 001 positive macaques (Walsh et al., 2009). While several orthopoxvirus antigens have been shown to elicit antibodies that are protective in challenge models, and linkage between CD4+ T cell recognition and antibody production following VACV has also been reported, for the present study we have selected only from ORFs for which CD8 T cell epitopes have been described.
From the Moutaftsi et al. (2010) analysis, a list of the 23 most prevalently recognized VACV WR ORFs was compiled, where prevalence was defined as an ORF being recognized in the context of 3 or more different HLA or H-2 alleles. Next, we examined human CD8 T cell recognition data available in the IEDB. For this analysis we compiled the number of positive donor responses for each of the 218 VACV WR ORFs versus the total number of donors tested from all studies curated by the IEDB as of June 1, 2012. From these data a frequency of positive responses was calculated. Overall, 49 ORFs were identified that had response rates of 25% or greater (138 had at least one positive response). Finally, we tabulated a set of VACV WR ORFs described in a previous study that were recognized by CD8 T cells in vaccinated Mamu-A1 ⁎ 001 macaques (Walsh et al., 2009). This identified 16 ORFs that were recognized in one or both of two animals studied.
Taken together, 70 ORFS were identified that were selected by one or another analysis. To limit the analysis to a more manageable set, we selected a set of 20 ORFs to include all ORFs identified in at least 2 of the 3 analyses described above, or that were recognized in both of the macaques studied in Walsh et al. (2009). This set of ORFs is listed in Supplemental Table 1.
Selection of peptides
Among the NHPs, we chose to work with those expressing Mamu-A1 ⁎ 001 and Mamu-A1 ⁎ 002. Both of these alleles are prevalent in our cohort, and have been extensively characterized in previous studies of simian immunodeficiency virus infection in rhesus macaques (Loffredo et al., 2004, Allen et al., 2001, Allen et al., 1998). Furthermore, multiple efficient approaches for predicting peptide binders have been developed for both of these alleles (Peters et al., 2005, Peters et al., 2006).
To select peptides for Mamu-A1 ⁎ 001 and Mamu-A1 ⁎ 002, we first analyzed the previously published CD8 T cell epitope data (Walsh et al., 2009, Loffredo et al., 2004, Allen et al., 2001, Allen et al., 1998) to determine the optimal peptide length(s) for consideration. We tabulated the number of epitopes identified for each allele, and determined their distribution as a function of peptide length. For both alleles it was found that the vast majority of epitopes defined previously were 9 residues in length (78 and 74%, respectively, for Mamu-A1 ⁎ 001 and Mamu-A1 ⁎ 002). Because 9-mers represent the canonical epitope length for most class I alleles, we checked that the predominance of 9-mers in the cases Mamu-A1 ⁎ 001 and Mamu-A1 ⁎ 002 did reflect a selection bias. Accordingly, for each of the studies, we also tabulated the number of peptides synthesized of various lengths and then calculated the fraction of peptides examined that were eventually identified as T cell epitopes. As shown in Supplemental Table 2, 9-mers were more likely to be identified as epitopes than peptides of other sizes. We also note that the differences observed are likely to be conservative estimates, as each of the studies evaluated included only the top predicted or most canonical motifs for peptides of 8-, 10- and 11-residues, while for 9-mers broader selection criteria were considered to include peptides with poorer predicted affinities. Accordingly, for the present study, we have focused solely on 9-mer peptides.
Finally, to identify potential epitopes derived from the set of 20 antigens selected above, we scanned the corresponding MPXV Zaire 79 and Sierra Leone sequences utilizing the Mamu-A1 ⁎ 001 and Mamu-A1 ⁎ 002 consensus algorithms hosted by IEDB (www.iedb.org). Unique peptides were selected as described in the Materials and Methods. The final set selected represented the top 2.25%-scoring, 9-mer peptides amongst the 20 antigens in both MPXV isolates, further supplemented by ensuring that for each antigen at least 5 peptides or, if larger, 2.25% of the total peptides in the antigen, were included for study. Following this approach, 400 peptides (194 for Mamu-A ⁎ 001, and 206 for Mamu-A1 ⁎ 002) were selected for further analysis (Supplemental Table 3).
Mapping epitope specific responses using Elispot and intracellular staining
We assayed PBMCs from two different MPXV-infected NHPs for each Mamu-A haplotype ( Table 1). In the case of Mamu-A1 ⁎001, we assayed PBMCs from animal A5E073, which was infected with the MPXV Sierra Leone strain, and DC11, which was infected with the MPXV Zaire 79. In the case of the Mamu-A1 ⁎ 002 we utilized PBMCs from the DC22 and FLR macaques, both NHPs were infected with the MPXV Sierra Leone.
Table 1.
MPXV-infected NHPs and IFNγ Elispot results.
| NHPs | Virus inoculation and dose (PFU) | Mamu haplotype | Day of necropsy | IFNγ Elispot no./1×106 PBMCs stimulated by peptides | IFNγ Elispot no./1×106 PBMCs stimulated by inactivated VACV | Peptides triggering IFNγ secretion |
|---|---|---|---|---|---|---|
| DC11 | Zaire 79 1.5×107 | A001 | 14 | 44 | 72 | ISPDGCYSL |
| A5E073 | Sierra Leone 2.5×106 | A001 | 30 | 171 | 494 | ISPDGCYSL |
| DC22 | Sierra Leone 2.5×106 | A002 | 30 | 180 | 317 | LTFDYVVTF |
| FLR | Sierra Leone 2.5×108 | A002 | 30 | 158 | 257 | LTFDYVVTF |
While DC11 succumbed to MPXV infection at day 14 post-infection, the remaining three NHPs survived to the end of the study (day 30). To identify specific epitopes, predicted peptide pools (containing an average of 20 peptides/pool) were tested in IFNγ Elispot assays. Positive peptide pools were deconvoluted to identify the individual peptide associated with the positive IFNγ signal. Each experiment was repeated at least 3 times with similar results. From A5E073 and DC 11 (A001 haplotype), we detected a positive response for pool 15. Upon deconvolution the response was found to be associated with the E7 peptide (sequence ISPDGCYSL), which is derived from the F8L protein (aa 119–127). In the case of both DC22 and FLR (A002 haplotype), peptide D2 (sequence LTFDYVVTF), also derived from the F8L protein (aa 254–262), was positive in the Elispot assay. While only F8L-derived epitopes were identified in the animals studied here, it is important to note that F8L peptides represented only 22/194 (11.3%) of the A1⁎001 peptides screened, and 24/206 (11.7%) of the A1⁎002 peptides. The number of IFNγ- positive PBMCs (SFC/1×106 cells) from each animal is shown in Table 1. It was notable that inactivated VACV stimulated more IFNγ-secreting cells than peptides in the Elispot test. It is worth noting that these numbers may underestimate the actual number of positive cells, since PBMCs had been frozen at −80 °C for over two years, and the viability of the cells upon thawing was 50–90%. However, based on the results above we are able to conclude that the E7 and D2 epitopes of the F8L protein are consistently recognized in Mamu-A1 ⁎ 001 and Mamu-A1 ⁎ 002 animals, respectively.
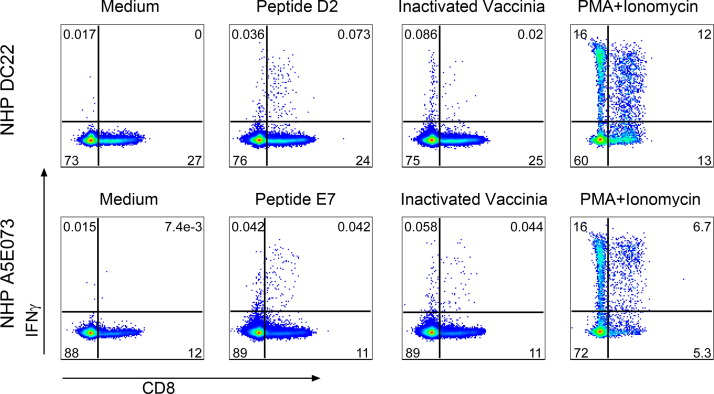
To further characterize these responses, we also performed IFNγ intracellular staining. As shown in Fig. 1, IFNγ production in response to inactivated VACV or a PMA+Ionomycin positive control was mediated by both CD8+ and CD8− cells, while stimulation with F8L peptides D2 or E7 skewed the response to CD8+ T cells. However, cultures with inactivated VACV stimulation did not gave a higher frequency of IFNγ-secreting cells than the peptide-stimulated cultures.
Fig. 1.
Peptides D2 and E7 triggered IFNγ-secreting CD8+ T cell responses. Frozen PBMCs were thawed and stimulated with either peptide or inactivated vaccinia virus, or with PMA plus ionomycin as a positive control, or the culture with medium alone as a negative control. The culture was incubated for overnight in the presence of BD Golgi plug/Golgi stop in the last 5 h of culture. Cultured cells were stained with mixture of surface markers, fixed with BD cytofix/perm, followed by intracellular staining of IFNγ. Samples were acquired using a BD LSR Fortessa cytometer and analyzed with Flowjo software. This figure illustrates the IFNγ-expression on CD3+CD8+ T cells.
Finally, MHC-peptide binding assays utilizing purified Mamu-A1⁎001 and Mamu-A1 ⁎ 002 molecules were performed to determine the affinity of each epitope for its predicted restricting allele. Both peptides were found to bind their corresponding allele with very high affinities, with IC50s <0.5 nM. Specifically, E7 (sequence ISPDGCYSL) bound Mamu-A1 ⁎ 001 with an affinity of 0.20 nM (+/− 0.077), while D2 (sequence LTFDYVVTF) bound Mamu-A1 ⁎ 002 with an affinity of 0.42 nM (+/− 0.11). Notably, these affinities are higher than those reported for previously identified simian- immunodeficiency-virus-derived epitopes restricted by Mamu-A1 ⁎ 001 and Mamu-A1 ⁎ 002 (Loffredo et al., 2004, Allen et al., 2001).
D2 and E7 epitope sequences are largely conserved in different poxviruses
We next examined the conservation of these F8L epitope sequences in different poxviruses, and in different MPXV strains. MPXV F8L is the analog protein of E9L in VACV. Utilizing the National Center for Biotechnology Information resource, we compared the epitope sequences with all sequences available for various monkeypox strains, as well as vaccinia, variola, and cowpox viruses. The E7 peptide (sequence ISPDGCYSL) is completely conserved in all viruses. The D2 peptide (sequence LTFDYVVTF) is also completely conserved amongst all monkeypox and vaccinia strains. In the case of cowpox virus however, this E9L sequence is identical in some strains, while other cowpox virus sequences represent a single highly conservative V6>I variation (LTFDYIVTF). Interestingly, all reported variola E9L sequences carry the V6>I variant. The residue in position 6 is not expected to function as a primary anchor residue influencing Mamu A1⁎002 binding and, perhaps not surprisingly given the conservative nature of the variation, the two variants are predicted to have essentially identical Mamu A1⁎002 binding affinities (13 versus 15 nM according to the ANN algorithm, and 27 versus 28 nM according to the SMM algorithm; see tools.immuneepitope.org/mhci).
Discussion
The precise identification of T cell epitopes allows accurate quantification of T cell immune responses. This in turn enables effective studies of host-pathogen interactions and virus pathogenesis, and is a key element for the evaluation of different vaccine candidates (Sette and Rappuoli, 2010). Many T cell epitopes have been characterized for poxviruses in humans and mice (Kennedy and Poland, 2007, Moutaftsi et al., 2010), with most of the epitopes being specific for MHC-I CD8+ T cells (total 246 peptides, 145-MHC-I in humans, and 103-MHC-I in mice) and fewer (61peptides) restricted by MHC-II on CD4+ T cells (Moutaftsi et al., 2010). Interestingly, only a handful of T cell epitopes from VACV proteins have been reported for NHPs (Walsh et al., 2009). The unique niche of NHP models in studying the select agents MPXV and VARV makes it of great interest to define T cell epitopes in these models.
The main goal of our study was to illustrate that selection of predicted epitopes from antigens previously shown to be targeted by CD8 responses in other species is an effective means to identify potential epitopes in a model system such as the select agent monkeypox. In this study, we identified for the first time two CD8+ T cell epitopes derived from MPXV that are recognized by rhesus macaques. Based on binding predictions and in vitro binding assays, it has been inferred that one of the epitopes is restricted by Mamu-A1 ⁎ 001 and the other epitope by Mamu-A1 ⁎ 002. It is important to note that all of the monkeys studied expressed several other Mamu A and B class I alleles, and it is thus possible that the responses may be restricted by any of these other specificities.
Interestingly, both peptides are derived from the monkeypox F8L protein. Previous results from humans and mice have identified epitopes derived from E9L, including one human CD8+ epitope (aa 107–115) and two murine CD8+ T cell epitopes (aa 716–724 and aa 858–866). In addition, two peptides from E9L (aa 562–570 and aa 883–891) were also identified as CD8+ T cell epitopes in rhesus monkeys infected with vaccinia virus (Walsh et al., 2009). These data support the assertion that F8L/E9L is a major poxvirus target of CD8+ T cell responses. Previous studies have concluded that, unlike some other viruses that are associated with a narrow pattern of immunodominance, poxvirus-specific immune responses are directed against a broad array of antigenic proteins. Indeed, of the more than 200 potential poxvirus proteins, at least 114 proteins have been reported as being recognized by CD8+ T cell responses (Moutaftsi et al., 2010). Based on the numbers of epitopes identified for each of these poxvirus proteins, F8L/E9L is amongst the top 23 antigens that contains CD8+ epitopes recognized by both humans and mice (Moutaftsi et al., 2010).
Our results are interesting as the F8L/E9L protein is the only one amongst the 20 proteins tested to be recognized in different strains of monkeypox, and in the context of both Mamu class I molecules studied (Mamu-A1 ⁎ 001 and Mamu-A1 ⁎ 002). Whether this reflects a narrow immunodominance associated with monkeypox infection in rhesus macaques and potentially also reflects its high pathogenicity, remains to be ascertained by more comprehensive studies. On the other hand, this data could also be the result of the limited amount of animals screened in our study. Furthermore, previous studies with VACV and other viruses in mouse models showed that T cell immunodominance can be modulated by the route of virus inoculation (Tscharke et al., 2005). This level of immunodominance regulation may also exist in NHPs with MPXV infection. In addition, PBMCs from three of four NHPs were derived from day 30 post-inoculation when the peak primary response could be over and the remaining MPXV-specific T cells have undergone clonal affinity selection. Therefore, it is possible that new or more peptides from other proteins or restricted by other MHC molecules will be identified as immunogenic following more extensive screening.
Because of the rather large size of the MPXV proteome, the present study focused on only a subset of ORFs found in previous studies to be recognized in both monkeys and humans. While some previous efforts in orthopoxvirus models have been biased towards early expressed ORFs that are not necessarily either immunodominant or relevant for protection, the selection of ORFs for the present study was based on published data from a set of studies analyzing responses on a complete genome-wide basis, to include early through late expressed ORFs. Only 2 out of 400 peptides screened in the present study were identified as T cell epitopes. In this context, it is important to emphasize that the algorithms utilized in this study predict MHC binding, not T cell recognition. The capacity to bind MHC is a necessary, but not sufficient, requirement for recognition by T cells. Most certainly other factors are involved, such that only a few peptides with the capacity to bind MHC end up being actual T cell epitopes. Potential factors involved include, but are likely not limited to, the rate of expression of the protein of provenance, efficiency of generation of the epitope sequence by cellular processing mechanisms, resistance of the peptide to intracellular enzymatic degradation, and the presence of a T cell repertoire with the capacity to recognize the peptide in the context of the corresponding MHC molecule. This issue has been reported upon and reviewed elsewhere (Assarsson et al., 2007, Yewdell, 2006, Yewdell and Del Val, 2004, Yewdell and Bennink, 1999), where it has been estimated that about 1 in 100 high-binding peptides and less than 0.01% of all peptides end up being T cell epitopes. The “low” success rate observed in the present study is thus not much different from what has been reported in other contexts. It is unknown whether the CD8+ T cell response triggered by F8L/E9L protein or by the specific peptides in F8L/E9L is protective. A previous study in the mouse model demonstrated that F8L/E9L epitopes displayed 66.7% protective efficacy (Moutaftsi et al., 2009). Similar studies may need to be done in rhesus monkeys to determine whether peptides from F8L/E9L can induce protective T cell responses against MPXV or other poxvirus infections.
F8L/E9L is a DNA polymerase and is expressed at the early stage of virus infection. It shares 98 and 99% homology with VARV and vaccinia virus, respectively. Given this high degree of conservation, one could imagine that the CD8+ T cell responses triggered by F8L/E9L are likely to be cross-reactive across all the poxviruses. In addition, we found that one of the two epitopes identified in this study is completely identical among all the poxviruses in question. The other one is also completely conserved amongst in the vast majority of poxvirus sequences and is associated with only one amino acid substitution in some cowpox virus isolates and all the variola viruses. We speculate that the CD8+ response triggered by these two peptides should be conserved in poxviruses, such as monkeypox, cowpox virus, vaccinia and variola viruses. Thus, the two peptides may serve as possible reagents to monitor CD8+ T cell responses in rhesus macaques.
It could be interesting to test if the two epitopes identified in rhesus macaques are also immunogenic in humans. If it can be shown that humans and rhesus macaques do not share the same epitopes, it underscores the importance to take caution when interpreting T cell response data from rhesus macaques during vaccine or viral pathogenesis studies. For example, a vaccine containing human T cell epitopes may not be detected in rhesus macaques. On the other hand, a strong CD8+ T cell response to a specific peptide in rhesus macaques may not be a reflection of a human scenario. Thus, it is important to know if a protein contains T cell epitopes for both humans and rhesus macaques. In the case of F8L/E9L, a positive CD8+ response detected by these peptides in rhesus macaques probably suggests T cell responses in humans.
Previous efforts to characterize poxvirus-specific T cell epitopes have mostly used vaccinia virus (using Dryvax for human and vaccinia WR strain for mice). To our knowledge, our effort is different from previous studies in that we have tried to characterize poxvirus-specific T cell epitopes in rhesus macaques, and also in that we used MPXV-infected rhesus PBMCs. Although poxviruses share a high degree homology, it is not guaranteed that the epitopes identified from one virus such as VACV are also immunogenic during infection with another virus. This is because different viruses may use different mechanisms to execute their infection process and to interact with host cells, such as dendritic cells. This in turn may affect antigen processing and presentation. Thus, our effort to characterize monkeypox-specific T cell epitopes in rhesus macaques is significant in that it provides reagents for future monkeypox studies in rhesus macaques, but also in that it provides evidence to show that the F8L/E9L protein is immunogenic in humans, mice and NHPs.
Our future efforts will be to extend the current study by testing the peptides in additional donors, and to also analyze peptides predicted to be specific for other Mamu class I haplotypes from different time points of MPXV infection.
Funding
The study was supported under the Battelle Memorial contract with the National Institute of Allergy and Infectious Diseases (NIAID) (Contract No. HHS N272200700016I) and was also supported in part by NIAID Division of Intramural Research funds. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests
The authors have declared that no competing interests exist.
Materials and methods
Ethics statement
All animal experiments were approved by the National Institute of Allergy and Infectious Diseases, Division of Intramural Research, Animal Care and Use Committee (ACUC) and adhered to National Institutes of Health (NIH) policies. The entire procedure to handle NHPs, including injections or inoculations of virus, blood withdrawals, biopsy and necropsy, strictly adhered to NIH NHP regulations and guidelines. Investigators and animal care personnel provided additional comfort and care, such as supplemental heat and IV, subcutaneous, or oral fluids, consistent with the scientific integrity of the protocol. Animals that were not eating were tube-fed under sedation, and given metoclopromide 0.3 mg/kg intramuscular (IM) 15 min prior to tube feeding to prevent vomiting.
Infection of rhesus monkeys with monkeypox virus
MPXV Zaire 79 and Sierra Leone stocks were propagated in BSC-1 cells at a multiplicity of infection (MOI) of 0.1 for 3 days. Virus inoculum was crude virus prepared by 3 freeze/thaw cycles followed by centrifugation and ultrasonic treatment, then pelleted over a 36% sucrose cushion. Rhesus macaques of both sexes, ranging from 4 to 8 kg, were housed in BSL-3 bio-containment cages and all animals were acclimated to the study facility for a minimum of 2 weeks prior to the start of the study. Prior to enrollment, NHPs were screened and found to be seronegative for simian retrovirus, simian T cell leukemia, VACV, cowpox virus, and MPXV. Since intravenous inoculation (IV) was a well-established skill in our team, thus was used in this experiment. MPXV at desired virus doses in 1 ml phosphate buffered saline (PBS) was IV administrated into NHPs. At different time points before and after virus inoculation, blood was collected aseptically in ethylenediamine tetra acetic acid (EDTA)-treated tubes or heparin tubes.
All animal handling procedures were performed under anesthesia by intramuscular injection of 10–25 mg/kg of ketamine hydrochloride. Animals were euthanized in accordance with the 2007 American Veterinary Medical Association (AVMA) guidelines on euthanasia utilizing exsanguination via direct cardiac puncture following induction of deep anesthesia by IV injection of 100 mg/kg of sodium pentobarbital (Fatal Plus®, Vortech Pharmaceuticals, Dearborn, MI). Complete necropsies at the end of the experiment were performed following standard surgical procedures.
Peripheral blood mononuclear cell (PBMC) preparation and major histocompatibility complex (MHC) class I sequence-based genotyping
Heparinized blood was diluted 2-fold in PBS, layered over Ficoll solution (Sigma, WI), and centrifuged at 1000×g at room temperature for 30 min. Then, the interface cell layer was collected and washed twice with PBS+2% Fetal Bovine Serum (FBS). The PBMCs were frozen in −80 °C for future analysis.
Total RNA was isolated from PBMCs and used as a template for the first-round cDNA synthesis. PCR was performed on cDNA samples with universal primers containing the multiplex identifier sequence tags that amplify the highly polymorphic 568bp region of rhesus macaque MHC-I exons 2–4 that encodes the peptide binding domain. These cDNA-PCR amplicons were then purified, quantitated, and pooled prior to pyrosequencing on a Roche/454 GS Junior instrument. The resulting nucleotide sequences were compared to known rhesus macaque MHC class I alleles in a reference database of Mamu-A, Mamu-B allele sequences to identity genotypes for each NHP.
Prediction of candidate peptides
Candidate peptides for Mamu-A1⁎001 and Mamu-A1 ⁎002 binding were selected using the immune epitope database (IEDB) consensus algorithm specific for each allele (www.iedb.org) (Vita et al., 2010). As described below, only 9-mer peptides for 20 selected antigens were considered. Starting with the Sierra Leone sequence, peptides for the 20 antigens (collectively about 7100 peptides) were scored together, and for each allele the top 2.25% scoring peptides were selected. This set was then supplemented with additional peptides to ensure that for each antigen we included at least 5 peptides or, if larger, 2.25% of the total peptides in the antigen. Next, peptides from the corresponding MPXV Zaire 79 sequences were scored, and those that scored in the top 2.25% and that were not in the Sierra Leone sequences, were added to the set. As a result, 194 peptides were selected for Mamu-A1 ⁎ 001, and 206 for Mamu-A1 ⁎ 002. As constructed, the set of peptides selected represents the top 2.25% scoring peptides in both strains considered independently; 372 are present in both MPXV strains, 17 are present only in Sierra Leone, and 11 are present only in Zaire. This final set of 400 peptides was synthesized as crude material on a 1 mg scale by A & A Labs LLC (San Diego, CA). Here, the protein of provenance for the selected peptides has been designated based on VACV Copenhagen sequence orthologs.
In vitro assays utilizing purified MHC molecules to measure peptide binding to Mamu-A1 ⁎ 001 and Mamu-A1 ⁎ 002 were performed as previously described (Sidney et al., 2013, Loffredo et al., 2004, Allen et al., 2001).
Detection of interferon γ secreting T cells by Elispot assay and intracellular flow cytometric staining
Frozen PBMCs from necropsy blood were used for all Elispot and flow cytometric staining assays. IFNγ Elispot assays were performed using a monkey IFNγ Elispot kit (MabTech, Mariemont, OH) according to the manufacturer's recommended procedure. Briefly, anti-IFNγ antibody was diluted to 15 µg/ml in PBS and was used to coat Elispot plates (Millipore,) at 100 µl/well. Plates were then incubated at 4 °C overnight. The Elispot plates were washed 4 times with PBS, and then blocked with RPMI 1640 medium plus 5% FBS for 1-2 h at room temperature. PBMCs were thawed, washed and counted, and then re-suspended in RPMI 1640 medium supplemented with 5% FBS at a cell concentration of 2.5×106–3.5×106/ml. Each culture well contained 100 µl of PBMCs and total culture volume was 200 µl/well. The final concentration of culture for peptide pools and individual peptides was 2 µg/ml for each peptide; irradiated vaccinia virus, 0.5 PFU/cell; and positive control with PMA (50 ng/ml) plus ionomycin (200 ng/ml). Each stimulation condition was carried out in triplicate wells. Assay plates were incubated overnight at 37 °C in a CO2 incubator, and the signals were developed based on manufacturer's instructions.
To enumerate the IFNγ responses, plate wells were pictured with a Nikon D200 camera, and spot numbers were manually counted by three individuals. MPXV-specific spot number was obtained from the subtraction of spot number in media control wells from the number in peptide-stimulated wells. The average number of the three counts respectively from triplicate wells is presented.
To perform intracellular staining for IFNγ, PBMCs were cultured in a 96-well U-bottom plate with 1×106 cells per well. The dose of stimuli was the same as used in the Elispot culture. The plate was incubated at 37 °C overnight in the presence of BD Golgi plug and Golgi stop. The cells were washed twice with PBS+2% FBS, then incubated for 20 min at room temperature with a mixture of antibodies for various surface markers, including CD3, CD4, CD8, CD20 (BD Pharmingen, CA), and NKG2A (Beckman-Coulter). After two washes with PBS+2% FBS, the cells were fixed with Cytofix/Cytoperm (BD Biosciences) for 30–40 min at room temperature, followed by two washes with BDperm/wash (BD Biosciences). The cells were then incubated with antibody to IFNγ at 4 °C overnight. On the second day, the cells were washed and resuspended in 1× BDperm/wash buffer and acquired using a LSR-Fortessa flow cytometer (Becton Dickinson, San Jose, CA). The data were analyzed with Flowjo software (TreeStar Inc., Ashland, OR).
Acknowledgment
We would like to thank the staff at the Integrated Research Facility and the NIAID Emerging Viral Pathogens Section for their efforts in the execution of this study, in particular to Marisa St. Claire, Russell Byrum, Reed Johnson, Amy Papaneri, Louis Huzella, Catherine Jett, Krisztina Janosko, Cindy Allan, Dan Ragland, Kurt Copper, Isis Alexander, Nicholas Oberlander, and Bernardo Rosa for animal operation, sample collection, and technical support, and also to Dr. Jerry Jennings for his effort in coordination during the animal study.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2013.09.003.
Appendix A. Supplementary materials
Supplementary data
Supplementary data
Supplementary data
References
- Allen TM, Sidney J, del Guercio MF, Glickman RL, Lensmeyer GL, et al. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A⁎01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J. Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- Allen TM, Mothe BR, Sidney J, Jing P, Dzuris JL, et al. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A⁎01: implications for vaccine design and testing. J. Virol. 2001;75:738–749. doi: 10.1128/JVI.75.2.738-749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui HH, et al. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J. Immunol. 2007;178:7890–7901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- Earl PL, Americo JL, Wyatt LS, Eller LA, Whitbeck JC, et al. Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature. 2004;428:182–185. doi: 10.1038/nature02331. [DOI] [PubMed] [Google Scholar]
- Edghill-Smith Y, Bray M, Whitehouse CA, Miller D, Mucker E, et al. Smallpox vaccine does not protect macaques with AIDS from a lethal monkeypox virus challenge. J. Infect. Dis. 2005;191:372–381. doi: 10.1086/427265. (Epub 2005 Jan 2004) [DOI] [PubMed] [Google Scholar]
- Estep RD, Messaoudi I, O'Connor MA, Li H, Sprague J, et al. Deletion of the monkeypox virus inhibitor of complement enzymes locus impacts the adaptive immune response to monkeypox virus in a nonhuman primate model of infection. J. Virol. 2011;85:9527–9542. doi: 10.1128/JVI.00199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heraud JM, Edghill-Smith Y, Ayala V, Kalisz I., Parrino J., et al. Subunit recombinant vaccine protects against monkeypox. J. Immunol. 2006;177:2552–2564. doi: 10.4049/jimmunol.177.4.2552. [DOI] [PubMed] [Google Scholar]
- Hooper JW, Thompson E, Wilhelmsen C, Zimmerman M, Ichou MA, et al. Smallpox DNA vaccine protects nonhuman primates against lethal monkeypox. J. Virol. 2004;78:4433–4443. doi: 10.1128/JVI.78.9.4433-4443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MD, Gallo RL, Boguniewicz M, Jones JF, Wong C, et al. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity. 2006;24:341–348. doi: 10.1016/j.immuni.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Huhn GD, Bauer AM, Yorita K, Graham MB, Sejvar J, et al. Clinical characteristics of human monkeypox, and risk factors for severe disease. Clin. Infect. Dis. 2005;41:1742–1751. doi: 10.1086/498115. [DOI] [PubMed] [Google Scholar]
- Jezek Z, Szczeniowski M, Paluku KM, Mutombo M. Human monkeypox: clinical features of 282 patients. J. Infect. Dis. 1987;156:293–298. doi: 10.1093/infdis/156.2.293. [DOI] [PubMed] [Google Scholar]
- Johnson RF, Dyall J, Ragland DR, Huzella L, Byrum R, et al. Comparative analysis of monkeypox virus infection of cynomolgus macaques by the intravenous or intrabronchial inoculation route. J. Virol. 2011;85:2112–2125. doi: 10.1128/JVI.01931-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keasey S, Pugh C, Tikhonov A, Chen G, Schweitzer B, et al. Proteomic basis of the antibody response to monkeypox virus infection examined in cynomolgus macaques and a comparison to human smallpox vaccination. PLoS One. 2010;5:e15547. doi: 10.1371/journal.pone.0015547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy R, Poland GA. T-cell epitope discovery for variola and vaccinia viruses. Rev. Med. Virol. 2007;17:93–113. doi: 10.1002/rmv.527. [DOI] [PubMed] [Google Scholar]
- Loffredo JT, Sidney J, Wojewoda C, Dodds E, Reynolds MR, et al. Identification of seventeen new simian immunodeficiency virus-derived CD8+ T cell epitopes restricted by the high frequency molecule, Mamu-A⁎02, and potential escape from CTL recognition. J. Immunol. 2004;173:5064–5076. doi: 10.4049/jimmunol.173.8.5064. [DOI] [PubMed] [Google Scholar]
- Moutaftsi M, Salek-Ardakani S, Croft M, Peters B, Sidney J, et al. Correlates of protection efficacy induced by vaccinia virus-specific CD8+ T-cell epitopes in the murine intranasal challenge model. Eur. J. Immunol. 2009;39:717–722. doi: 10.1002/eji.200838815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutaftsi M, Tscharke DC, Vaughan K, Koelle DM, Stern L, et al. Uncovering the interplay between CD8, CD4 and antibody responses to complex pathogens. Future Microbiol. 2010;5:221–239. doi: 10.2217/fmb.09.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B, Bui HH, Sidney J, Weng Z, Loffredo JT, et al. A computational resource for the prediction of peptide binding to Indian rhesus macaque MHC class I molecules. Vaccine. 2005 doi: 10.1016/j.vaccine.2005.07.086. [DOI] [PubMed] [Google Scholar]
- Peters B, Bui H-H, Frankild S, Nielsen M, Lundegaard C, et al. A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLoS Comput. Biol. 2006;2:e65. doi: 10.1371/journal.pcbi.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield RR, Wright DC, James WD, Jones TS, Brown C, et al. Disseminated vaccinia in a military recruit with human immunodeficiency virus (HIV) disease. N. Engl. J. Med. 1987;316:673–676. doi: 10.1056/NEJM198703123161106. [DOI] [PubMed] [Google Scholar]
- Saijo M, Ami Y, Suzaki Y, Nagata N, Iwata N, et al. LC16m8, a highly attenuated vaccinia virus vaccine lacking expression of the membrane protein B5R, protects monkeys from monkeypox. J. Virol. 2006;80:5179–5188. doi: 10.1128/JVI.02642-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33:530–541. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Southwood S, Moore C, Oseroff C, Pinilla C, et al. Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. Curr. Protoc. Immunol. Unit. 2013;2013;1818:13. 13. doi: 10.1002/0471142735.im1803s100. (Chapter 18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JT, Belyakov IM, Dzutsev A, Lemonnier F, Berzofsky JA. Protection against lethal vaccinia virus challenge in HLA-A2 transgenic mice by immunization with a single CD8+ T-cell peptide epitope of vaccinia and variola viruses. J. Virol. 2004;78:7052–7060. doi: 10.1128/JVI.78.13.7052-7060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stittelaar KJ, van Amerongen G, Kondova I, Kuiken T, van Lavieren RF, et al. Modified vaccinia virus Ankara protects macaques against respiratory challenge with monkeypox virus. J. Virol. 2005;79:7845–7851. doi: 10.1128/JVI.79.12.7845-7851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stittelaar KJ, Neyts J, Naesens L, van Amerongen G, van Lavieren RF, et al. Antiviral treatment is more effective than smallpox vaccination upon lethal monkeypox virus infection. Nature. 2006;439:745–748. doi: 10.1038/nature04295. [DOI] [PubMed] [Google Scholar]
- Tscharke DC, Karupiah G, Zhou J, Palmore T, Irvine KR, et al. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J. Exp. Med. 2005;201:95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, et al. The immune epitope database 2.0. Nucleic Acids Res. 2010;38:D854–862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh SR, Gillis J, Peters B, Mothe BR, Sidney J, et al. Diverse recognition of conserved orthopoxvirus CD8+ T cell epitopes in vaccinated rhesus macaques. Vaccine. 2009;27:4990–5000. doi: 10.1016/j.vaccine.2009.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Johnson AJ, Liggitt D, Bevan MJ. Cellular and humoral immunity against vaccinia virus infection of mice. J. Immunol. 2004;172:6265–6271. doi: 10.4049/jimmunol.172.10.6265. [DOI] [PubMed] [Google Scholar]
- Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- Yewdell JW, Del Val M. Immunodominance in TCD8+ responses to viruses: cell biology, cellular immunology, and mathematical models. Immunity. 2004;21:149–153. doi: 10.1016/j.immuni.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Zaucha GM, Jahrling PB, Geisbert TW, Swearengen JR, Hensley L. The pathology of experimental aerosolized monkeypox virus infection in cynomolgus monkeys (Macaca fascicularis) Lab. Invest. 2001;81:1581–1600. doi: 10.1038/labinvest.3780373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data