Abstract
Collagen IV is the main structural protein that provides a scaffold for assembly of basement membrane proteins. Posttranslational modifications such as hydroxylation of proline and lysine and glycosylation of lysine are essential for the functioning of collagen IV triple-helical molecules. These modifications are highly abundant posing a difficult challenge for in-depth characterization of collagen IV using conventional proteomics approaches. Herein, we implemented an integrated pipeline combining high-resolution mass spectrometry with different fragmentation techniques and an optimized bioinformatics workflow to study posttranslational modifications in mouse collagen IV. We achieved 82% sequence coverage for the α1 chain, mapping 39 glycosylated hydroxylysine, 148 4-hydroxyproline, and seven 3-hydroxyproline residues. Further, we employed our pipeline to map the modifications on human collagen IV and achieved 85% sequence coverage for the α1 chain, mapping 35 glycosylated hydroxylysine, 163 4-hydroxyproline, and 14 3-hydroxyproline residues. Although lysine glycosylation heterogeneity was observed in both mouse and human, 21 conserved sites were identified. Likewise, five 3-hydroxyproline residues were conserved between mouse and human, suggesting that these modification sites are important for collagen IV function. Collectively, these are the first comprehensive maps of hydroxylation and glycosylation sites in collagen IV, which lay the foundation for dissecting the key role of these modifications in health and disease.
Keywords: basement membrane, collagen IV, posttranslational modifications, glycosylation, hydroxylysine, hydroxyproline, mass spectrometry, MyriMatch
Graphical abstract
INTRODUCTION
Collagen IV is the most abundant structural protein in basement membranes (BMs), a specialized form of extracellular matrix underlying all epithelia, where it assembles into an independent network that provides structural support essential for tissue development and maintenance.1 Collagen IV also interacts with cells playing a critical role for a variety of biological processes including cell adhesion, migration, survival, proliferation, and differentiation.2–4 Collagen IV is a family of six genetically distinct α-chains designated col4a1–col4a6.5 Out of many potential combinations, the chains assemble to form only three distinct heterotrimers, also known as protomers, α121, α345, and α556. The α121 protomers are formed by two α1 and one α2 chains and are present in the BMs of all tissues. The α345 and α556 protomers are characterized by their well-defined tissue distribution and differential expression during different developmental stages.6,7 Triple-helical protomers are secreted into the extracellular space where they form supramolecular networks by end-to-end interactions.6,8 At the carboxyl terminal end, two protomers are linked head-to-head by interactions of their trimeric noncollagenous (NC1) domains and enzymatically reinforced by peroxidasin-catalyzed sulfilimine cross-link bonds forming a structure known as the NC1 hexamer.9–12 At the amino terminal, four protomers aggregate, forming a structure known as the 7S dodecamer. Each chain of collagen IV is composed of a long triple helical collagenous region of more than one-thousand residues characterized by the repetitive Gly-Xaa-Yaa motif flanked by the amino terminal 7S and carboxyl terminal noncollagenous (NC1) domains.13,14 The 7S domain is predominantly collagenous in nature and shares many features of the triple helical region including a large number of posttranslationally modified amino acid residues such as hydroxylation of proline and lysine residues as well as O-glycosylation of hydroxylysine (HyK) residues. The 7S domain also contains a single N-glycosylated asparagine residue, which is thought to be important for assembly and registration of collagen IV chains.8,13
Proline is the most common amino acid residue in collagenous sequences usually found in the Xaa and Yaa positions of the distinct triple-helical Gly-Xaa-Yaa motif. Previous amino acid analyses demonstrated that about 50–60% of the proline content is found as hydroxyproline (HyP) in type IV collagen.15,16 Proline residues are modified by specific hydroxylases that can either hydroxylate the nitrogen-containing ring of proline in positions 4 to form 4-hydroxyproline (4-HyP) and less frequently in position 3 to form 3-hydroxyproline (3-HyP).17 4-HyP occurs in the Yaa position of the Gly-Xaa-Yaa motif, which is critical for the stabilization of the triple helix conformation by forming intramolecular hydrogen bonding.17–19 The posttranslational formation of 4-HyP in collagens is catalyzed by prolyl 4-hydroxylase (P4H). Among the three prolyl 4-hydroxylates known in vertebrates, prolyl 4-hydroxylase 1 has been demonstrated to be important for collagen IV assembly.20 Although the content of 3-HyP in collagen IV is distinctly higher than in other collagens, significant variation in prolyl 3-hydroxylation across different tissues has been documented.15,21 These 3-HyP sites are most commonly found in the Xaa position when 4-Hyp occupies the Yaa position. Out of the three known prolyl 3-hydroxylase isoenzymes present in vertebrates, prolyl 3-hydroxylase 2 (P3H2) has been identified as responsible for the prolyl 3-hydroxylation of collagen IV.22 More recently, genetic deletion of P3H2 in mice has recently demonstrated that 3-HyP in collagen IV may be important for regulating platelet aggregation and normal development of eye tissues.23,24
Most lysine residues in collagen IV are posttranslationally modified by hydroxylation and glycosylation. Classical amino acid analysis showed that about 90% of all lysine residues are hydroxylated, and depending on which BMs are analyzed, between 70 and 90% are further glycosylated.25,26 These numbers are significantly higher than those observed for fibrillar collagens.27–29 Lysine residues found within the Gly-Xaa-Lys motif are likely to be hydroxylated by members of the lysyl hydrolase family; lysyl hydroxylase 1 (LH1), LH2, and LH3.30,31 In addition, HyK residues are the substrate for galactosyl and glucosyl transferases.32–35 These enzymes catalyze O-glycosylation of HyK residues by the sequential addition of galactose and glucose. During the first step, a galactose (Gal) molecule is attached to the hydroxyl group of the HyK by a β-glycosidic bond. This galactosyl-hydroxylysine can be further modified by the addition of glucose (Glu) to the C-2 of Gal previously attached to the HyK via a α-glycosidic bond. O-glycosylation and hydroxylation of lysine residues in collagen IV is primarily catalyzed by a multifunctional LH3 enzyme. Gene ablation experiments demonstrated its essential role in collagen IV biosynthesis during early development as the loss of glycosylations results in premature collagen IV aggregation in mouse.36 Regulation of lysine hydroxylation and glycosylation is not only important for stabilization of collagen supramolecular networks, but also may regulate cell–matrix interactions through integrin membrane receptors important for cellular adhesion and signaling.37,38 Further, it has been also suggested that these modifications may control the formation of stable intermolecular cross-linking in collagens.39–41 However, the spread of hydroxylation and glycosylation modifications depends on the type of collagen, the tissue of origin,25,42 functional region within the tissue,43 maturation,44 and pathological conditions.45–48
Although collagen IV has been studied for many years and some of the basic structural features are well-understood, the distribution and composition of hydroxylation and glycosylation sites within protomers have not been studied in detail. Liquid chromatography–tandem mass spectrometry (LC–MS/MS) has become the favorite technique to routinely identify posttranslational modifications (PTMs) of proteins of biological interest; however, a number of practical issues remain to be scrutinized for the analyses of highly modified large proteins such as collagens.49–51 Indeed, although collagen IV is very abundant, a surprising underrepresentation of sequence coverage has been observed in proteomics inventories of highly enriched BM preparations.52–56 This may be explained by the inherent properties of collagen IV such as repetitive nature of primary sequence, high insolubility, cross-linking, and resistance to cleavage due to extensive PTM, which could lead to low peptide abundance, poor primary sequence coverage, and poor identification of the location of PTMs. Alternative approaches such as use of complementary ionization techniques,57–59 glycosylated peptide enrichment,60 and multiple enzyme digestions61 have been used for MS characterization of other collagens but not tried with the BM collagen IV. In this paper, we aimed to characterize hydroxylation and glycosylation of mouse collagen IV using MS and bioinformatics. We present an integrated optimized pipeline to maximize the coverage and identification of PTMs of mouse collagen IV. Further we employed this workflow to map the PTMs on human collagen IV by analyzing publicly available MS data obtained from human lens capsule. Significant improvement in the generation, detection, and identification of posttranslationally modified peptides will not only positively increase collagen IV sequence coverage in proteomics experiments, but also lay the foundation for dissecting the key role of these modifications in health and disease.
EXPERIMENTAL SECTION
Materials
Commercial Murine Engelbreth-Holm-Swarm (EHS) collagen IV was purchased from BD Biosciences (MA, USA). Ammonium bicarbonate, sodium phosphate, and MS grade formic acid and dithiothreitol (DTT) were purchased from Sigma-Aldrich (St. Louis, MO). Iodoacetamide was acquired from GE Healthcare (Piscataway, NJ). HPLC-grade solvents, including water, methanol, and acetonitrile, were procured from Sigma-Aldrich (St. Louis, MO). MS-grade Trypsin (V511A) was obtained from Promega (Madison, WI), and PNGase-F was purchased from New England Biolabs (Ipswich, MA). LysC was obtained from Waka Chemicals (Richmond, VA). Endoproteinase GluC was obtained from Roche (USA). OMIX C18 tips were procured from Agilent technologies (USA).
Methods
In Silico Analysis
Different PTMs like hydroxylation of proline and lysine residues and O-glycosylation of lysine residues were considered based on specific motifs and sites in mouse col4a1. It has been reported that tryptic cleavage does not occur if hydroxylysine residues are glycosylated.62 The extent of glycosylation of col4a1 reduces the efficiency of tryptic cleavage. To mimic this enzymatic behavior, the col4a1 sequence (without signal peptide) was preprocessed replacing the potential hydroxylysine residues (in “Gly-Xaa-Lys” motif) with the symbol “X” employed to identify unspecified amino acid residues. After this manipulation, the modified theoretical col4a1 chain was then subjected to fully specific in silico tryptic and Glu-C digestions using the “cleave” function as it is included in MATLAB Bioinformatics Toolbox.63 Default cleavage rules were set for the in silico enzymatic digestion. Once the lists of potential peptides were obtained, the “X” residue was replaced with “K”. After the generation of total number of potential tryptic and Glu-C peptides, both lists were stratified using bins corresponding to the number of modifications per peptide. The peptide median size based on amino acid length and peptide size range for each specific number of modifications were calculated and plotted using the left axis, while the theoretical peptides in each bin were used as an input to calculate the sequence coverage curve for col4a1 and plotted using the right axis.
Collagen IV 7S Dodecamer Purification
PFHR-9 mouse endodermal cells (ATCC CLR-2423) were cultured in DMEM medium supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin, and incubated at 37 °C in 5% and 10% of CO2, respectively. PFHR-9 cells were grown past confluence between 5 and 7 days with medium supplemented with 50 μg/mL of ascorbic acid to accumulate BM proteins as previously described.11 PFHR-9 cells were homogenized in 1% (w/v) deoxycholate with sonication, and the insoluble material was isolated after centrifugation at 20 000g for 15 min. The pellet was first extracted with 50 mM Tris-Cl pH 7.5 containing 1 M NaCl, and then it was digested with 0.1 mg/mL of bacterial collagenase (Worthington) in 50 mM Tris-Cl pH 7.5, 5 mM CaCl2, 5 mM benzamidine, 25 mM 6-aminocaproic acid, and 0.4 mM phenylmethylsulfonyl fluoride (PMSF). Collagenase-solubilized proteins were dialyzed overnight against 50 mM Tris-Cl, pH 7.5, at 4 °C. The 7S dodecamer was purified using DEAE-52 column followed by S-200 gel filtration chromatography using an AKTA purifier chromatography system. Protein concentrations were determined with the Pierce BCA protein assay kit.
Enzymatic Digestions
A solution of purified PFHR-9 7S dodecamer was mixed with an equal volume of 0.1 M Tris-HCl, pH 7.5, buffer containing 8 M Guanidine-HCl, and 50 mM dithiothreitol to denature proteins and reduce disulfide bonds. After samples were heated in a boiling water bath for 10 min, the samples were allowed to reach room temperature before proteins were alkylated with 50mM iodoacetamide in the dark for 45 min. Following ethanol precipitation at −20 °C for 2 h, the pellets were resuspended in 0.1 M ammonium bicarbonate pH 7.5 and digested with trypsin overnight using an enzyme-to-substrate ratio of 1:20 (w/w). Peptide samples were deglycosylated with PNGAse F at 37 °C overnight and cleaned using OMIX C18 tips according to manufacturer instructions.
For EHS collagen IV, the samples were spiked with laminin, denatured, reduced, alkylated, and ethanol precipitated as described for 7S dodecamer samples. The proteins were dissolved in 100 mM ammonium bicarbonate solution before being subjected to either trypsin or GluC digestion at an enzyme/substrate ratio of 1:20 w/w or incubating at 37 °C for 16 h. A solution of benzamidine was added to a 1 mM final concentration to deactivate GluC before performing PNGAseF treatment at 37 °C overnight. An additional sample was first digested with LysC for 4 h at 37 °C in 25 mM ammonium bicarbonate, pH 7.8 containing 2 M Gnd, 1 mM EDTA, and 1 mM CaCl2. The sample was then diluted to reduce guanidine concentration below 0.75 M with 25 mM ammonium bicarbonate before further digestion with trypsin for 16 h at room temperature. The peptide mixtures were cleaned using OMIX C18 tips following the manufacturer recommendations.
LC–MS/MS
Collagen IV peptides were loaded onto a capillary reversed-phase analytical column (360 μm o.d. × 100 μm i.d.) using an Eksigent NanoLC Ultra HPLC and autosampler. The analytical column was packed with 20 cm of C18 reversed-phase material (Jupiter, 3 μmbeads, Phenomenex) directly into a laser-pulled emitter tip. Peptides were gradient-eluted at a flow rate of 500 nL/min, and the mobile phase solvents consisted of water containing 0.1% formic acid (solvent A) and acetonitrile containing 0.1% formic acid (solvent B). A 90 min gradient was performed, consisting of the following: 0–15 min, 2% B (during sample loading); 15–70 min, 2–40% B; 70–75 min, 40–90% B; 75–77 min, 90% B; 77–78 min 90–2% B; and 78–90 min, 2% B (column re-equilibration). Upon gradient elution, peptides were mass analyzed on an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific) equipped with a nanoelectrospray ionization source. The instrument was operated using a data-dependent method with dynamic exclusion enabled. Full-scan spectra were acquired with the Orbitrap (resolution 60 000), and the top 16 and 12 most abundant ions were selected for fragmentation when collision induced dissociation (CID) and electron transfer dissociation (ETD) fragmentation were respectively employed. An isolation width of 2 m/z, activation time of 10 ms, and 35% normalized collision energy were used to generate CID MS2 spectra. For ETD, the isolation width was set to 2 m/z, and the default charge state was set to 4. The reaction time was set to 90 ms with a supplemental activation. The charge state screening was not enabled. The LTQ Orbitrap Velos mass spectrometer was externally calibrated, permitting <2 ppm mass accuracy. Dynamic exclusion settings were allowed for a repeat count for 15 s.
Collagen IV peptides were also analyzed on a Q Exactive mass spectrometer equipped with an Easy nLC-1000 system (Thermo Scientific). A 2 uL injection volume of peptides, representing approximately a total of 1 ug, was separated on a PicoFrit (New Objective, Woburn, MA) column (75 μm ID × 110 mm, 10 μm ID tip) packed with ReproSil-Pur C18-AQ resin (3 μm particle size and 120 Å pore size). Peptides were eluted using a flow rate of 300 nL/min, and the mobile phase solvents consisted of water containing 0.1% formic acid (solvent A) and acetonitrile containing 0.1% formic acid (solvent B). A 70 min gradient was performed, consisting of the following: 0–5 min, increase to 2% B; 5–55 min, 5–35% B; 55–60 min, 90% B and held at 90% B for 10 min before returning to the initial conditions of 2% B. Data were collected using a data-dependent method with dynamic exclusion enabled with a scanning window of 300–1800 m/z. Full scans were acquired at a resolution of 70 000, an AGC target of 3 × 106, and a 64 ms max injection time. The top 20 most abundant ions were selected for fragmentation with higher energy C-trap dissociation (HCD) at a resolution of 17 500, an AGC target of 2 × 105, 100 ms max injection time, 2 m/z isolation width, and 27% normalized collision energy. The data were collected with unassigned and +1 charge excluded in the charge state settings. Dynamic exclusion was set to 20 s. A summary of enzymatic digestions and LC–MS/MS experiments for 7S dodecamer and EHS collagen IV samples is provided in Supporting Table S1 in detail.
Lens Capsule MS/MS Data from ProteomeXchange
Three MS data sets from lens capsule BM isolated from human eyes submitted by Uechi et al. with identifier PXD001025 were downloaded from ProteomeXchange. In this study, lens capsule BM proteins were fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and in-gel digested with trypsin before LC–MS/MS analyses using CID fragmentation in a LTQ Orbitrap Velos Pro mass spectrometer. A total of 14, 44, and 48 .raw fles were downloaded for the three lens capsule samples.52
Database Searching
Thermo .raw files were independently searched by MyriMatch algorithm run on windows server maintained by the Vanderbilt University Center for Structural Biology.64 Files were initially searched against full proteome-level mouse FASTA databases (NCBI) and human Uniprot database (downloaded on July 14, 2015), which also included the reverse sequences for each protein to compute the false discovery rate (FDR) with a maximum of two missed cleavages and one missed termini cleavage (semitryptic digest), with a minimum peptide length of five amino acids and a maximum length of 50 amino acids. An initial simple search was configured to only identify peptides with the following amino acid modifications: variable oxidation of methionine (+15.9949) as well as fixed carbamidomethylation of cysteine (+57.021). For .raw files obtained from LTQ Orbitrap, Velos precursor and fragment mass tolerance were set to 10 ppm and 0.5 Da, respectively, whereas for .raw files obtained from Q Exactive, tolerances were set to 10 and 20 ppm, respectively. The pepXML files obtained in this first search were imported into IDPicker (version 3.1) for the parsimonious grouping of identified proteins with a maximum Q value of 2% for peptide spectrum match (PSM). About 150 proteins in trypsin or trypsin and LysC digested samples and about 300 proteins in GluC digested samples were exported as subset FASTA protein database using an utility in IDPicker 3.1.65,66 For downloaded lens capsule data, 210, 520, and 148 protein sequences were retained in the subset FASTA databases for three different samples pertaining 14, 44, and 48 .raw files, respectively. The .raw files were further searched against this subset FASTA database allowing for a maximum of four missed cleavages and expanding the search space with the addition of dynamic modifications (proline hydroxylation, lysine glycosylation, etc.) up to 10 per peptide (Table 1). We employed a very unique and useful feature of MyriMatch, which allows the use of a limited set of regular expressions to consider dynamic PTMs linked to primary structure peptide motifs such as the ones found in collagen IV. Table 1 lists the regular expressions and monoisotopic masses to mimic the collagen motifs where the exclamation signs (e.g., “!”) point to the residue with the modification. The ability of defining these motifs as dynamic modifications increases the sensitivity and specificity of the analysis considerably since not all the amino acid residues affected by the modification are considered to have the dynamic modification. For more details about the use of this feature, one can go to the documentation page located at https://svn.code.sf.net/p/proteowizard/code/trunk/pwiz/pwiz_tools/Bumbershoot/myrimatch/doc/index.html. To estimate FDRs, each sequence of the database was reversed and concatenated to the database. Candidate peptides were required to feature trypsin cleavages or protein termini at least at one end. Further, peptide identification, filtering, and protein assembly were done with the IDPicker 3.1 algorithm with a maximum Q value of 2% for PSM.66 Resulting PSMs were manually inspected, and MS2 spectra were examined to confirm correct assignment of PTMs. In addition, known peptides containing 3-HyP were searched for manually by searching possible ion masses in the raw data using Thermo Xcalibur software.24
Table 1.
List of Dynamic Modifications and Their Respective Regular Expressions Employed by MyriMatch during Database Search
| PTM name | regular expressiona | monoisotopic mass |
|---|---|---|
| methionine oxidation | M | +15.994915 |
| hydroxylation of lysine | K!G | +15.994915 |
| galactosyl-hydroxylation of lysine | K!{)}G | +178.047738 |
| glucosyl-galactosyl hydroxylation of lysine | K!{)}G | +340.100562 |
| hydroxylation of proline in Xaa position of “Gly-Xaa-Yaa” motif | GP![AVQP!] | +15.994915 |
| hydroxylation of proline in Yaa position of “Gly-HyP-Yaa” motif | GP!P! | +15.994915 |
| deamidation of asparagine after PNGAse F treatment | N!{P}[ST] | +0.984016 |
The exclamation sign “!” points to the residue with the modification.
RESULTS
In Silico Analysis of α1 Chain of Collagen IV
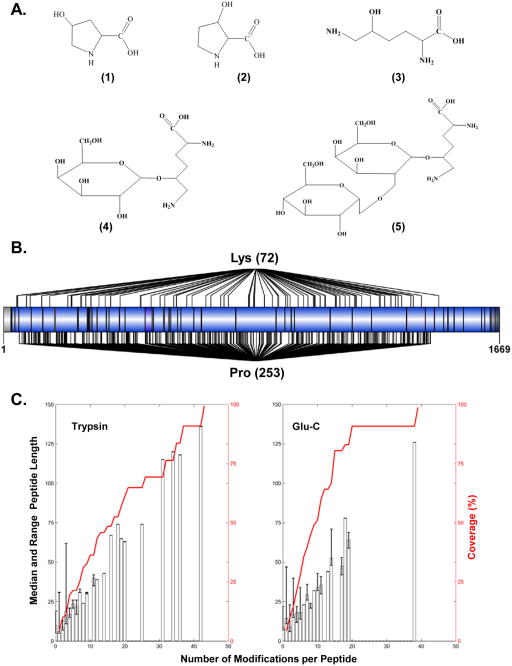
Collagen IV is highly modified during biosynthesis to form stable triple helical protomers. The chemical structures of lysine and proline related known PTMs are represented in Figure 1, panel A. Since our goal is to map and characterize hydroxylation and glycosylation of mouse collagen IV, we concentrated our efforts to develop and implement a comprehensive strategy to maximize identification of peptides containing modified lysine and proline residues. Figure 1, panel B shows that out of the 322 proline residues contained within the col4a1 sequence, 54 are in the Xaa position of Gly-Xaa-Yaa (where Y = HyP, Val, Ala, Gln) motif, and 213 are in the Yaa position of the Gly-Xaa-Yaa motif, which makes them potential candidates for hydroxylation in the 3 and 4 positions of five-membered nitrogen-containing ring, respectively. Additionally, from a total of 89 lysine residues, 72 are found in the Yaa position within the Gly-Xaa-Yaa motif, which is needed for hydroxylation and glycosylation.
Figure 1.
Theoretical sites of hydroxylation and glycosylation in the col4a1 sequence. (A) Chemical structures of 3-hydroxyproline (1), 4-hydroxyproline (2), hydroxylysine (3), galactosyl-hydroxylysine (4), and glucosylgalactosyl-hydroxylysine (5). (B) Schematic representation of col4a1 sequence showing fully tryptic peptides (separated by a vertical black line “|”). Lys (74) and Pro (253) designate the total number of potential lysine modification (hydroxylation and glycosylation) and proline hydroxylation sites derived from in silico analyses of mouse col4a1 sequence, respectively. Pro (253) is distributed as 40 3-HyP and 213 4-HyP sites. (C) Plots representing the distribution of trypsin and GluC peptides generated by in silico digestion of col4a1. Proline residues in the Yaa position within “Gly-Xaa-Yaa” motif and also in the Xaa position of the “Gly-Xaa-HyP” motif were considered hydroxylated in the in silico analyses. Lysine residues within the “Gly-Xaa-Lys” motif were considered as miscleaved sites in in silico trypsin digestion as they are likely to be glycosylated. The resulting pools of theoretical tryptic and Glu-C peptides were grouped by number of modifications. For each group, the median length (white bars) and range (black lines) were plotted against the number of modifications per peptide. Red lines represent the cumulative contributions to sequence coverage for the col4a1 chain.
Because glycosylated lysine residues do not undergo tryptic cleavage,62 an in silico digestion of full-length collagen IV α1 chain (col4a1) sequence with trypsin was performed to evaluate its influence on peptide length and number of potential PTMs per peptide. As can be observed in Figure 1, panel C, trypsin digestion generated a significant number of long peptides with a high number of PTMs, revealing a potential unfavorable situation for MS analysis. Because peptides of more than ~40 residues in length pose a challenge for MS identification, our in silico analysis anticipated a low sequence coverage for col4a1 sequence identification. Also, peptides carrying five or fewer potential PTMs only account for less than 30% of the col4a1 sequence, while a number of much longer peptides featuring up to 40 PTMs cover significantly greater portion of the col4a1 sequence. The uniform distribution of these potentially modified lysine residues may negatively impact the detectability of tryptic peptides by MS, and thus a large number of amino acid residues within the col4a1 sequence would not be accounted for. Among several commonly used proteolytic enzymes tested with collagen IV in silico, GluC produced the best results in terms of shorter peptide length and number of PTMs per peptide (Figure 1C).
PTM Characterization of the 7S Domain by Optimization of Proteomics and Bioinformatics Workflow
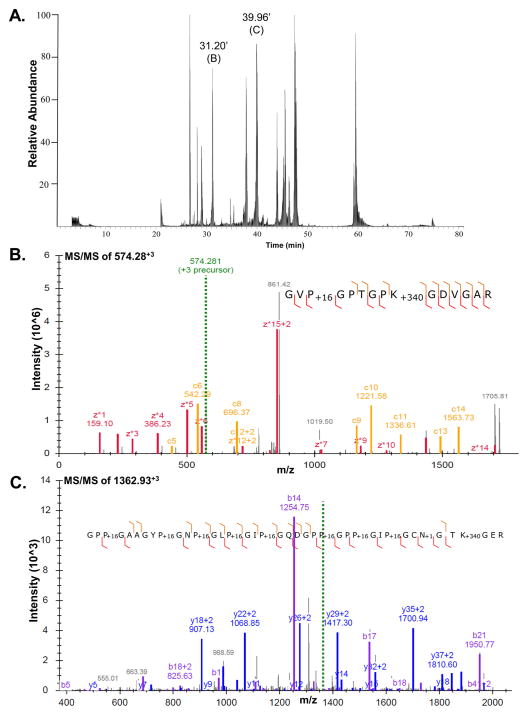
To achieve the comprehensive characterization of collagen IV, optimization of protein digestion, MS, and bioinformatics was needed. We chose to start by analyzing the 7S dodecamer because it is a soluble domain that can be easily purified in large quantities from mouse PHFR-9 cells, and a miniaturized version of the entire type IV collagen molecule containing many potential sites of hydroxylation and O-glycosylation of lysine residues as well as hydroxylation of proline residues. A multilevel strategy was implemented for achieving better sequence coverage of the 7S domain of col4a1 and col4a2, which are composed of 145 and 158 amino acids, respectively.67 Trypsin-digested peptides were subjected to analyses using CID and ETD fragmentation methods. Figure 2 depicts the typical base peak chromatogram for a 7S domain tryptic digest showing two examples that illustrate the benefits of using different peptide fragmentations strategies and increasing the search space during database searching. The chromatographic peak observed at 31.2 min peak is dominated by an ion m/z 574.28 (+3) produced a very uninformative CID MS/MS dominated by only two fragment ions at m/z values of 783.19 and 522.50 (not shown). These two fragment ions correspond to the +2 and +3 ions, respectively, derived from the neutral loss of Glu from the parent ion. However, no further information regarding the primary structure of the parent ion can be inferred. On the other hand, Figure 2, panel B shows a very informative tandem mass spectrum obtained for the same parent ion analyzed by ETD fragmentation. MyriMatch assigned the primary structure to the tryptic peptide 94GVp(OH)GPTGPk(GluGal)GDVGAR108. The PSM is excellent with c- and z*-ion ladders represented by most of the ions present in the tandem mass spectrum and providing supporting evidence for both hydroxylation of P96 and glycosylation of K102.
Figure 2.
Mass spectrometric analyses of glycosylated peptides from 7S dodecamer of collagen IV purified from mouse PHFR9 cells. (A) Base peak chromatogram of a tryptic digests of 7S dodecamer deglycosylated with PNGase F. (B) MS/MS spectrum of m/z 574.28+3 ion by ETD fragmentation represents the O-glycosylated peptide sequence in which P96 is hydroxylated (+16) and K102 is glucosylgalactosylated (+340). The c- and z*-ion series are colored orange and red, respectively. (C) CID MS/MS spectrum of m/z 1362.93+3 showing a PSM for an N- and O-glycosylated tryptic peptide containing a total of 10 PTMs identified after expanding the search space used by MyriMatch. The modifications include eight sites of prolyl 4-hydroxylation (+16), one site of N-glycosylation (+1), and one site of O-glucosylgalactosylation (+340).
Figure 2, panel C shows a CID tandem mass spectrum with a very complex and information rich fragmentation pattern that was not identified by a standard database search (i.e., three dynamic modifications). ETD tandem mass spectrum of this peptide was also complex and informative, but also was not identified by MyriMatch (not shown). Additional deglycosylation experiments with PNGAse F revealed that this peptide was initially N-glycosylated. Knowing that both col4a1 and col4a2 have a single N-Glycosylation site each in the 7S region, we identified tryptic peptide9 3GPPGAAGYPGNPGLPGIPGQDGPPGPPGIPGCNGTKGER132 as a potential candidate. Because this peptide sequence features up to 13 potential modifications when proline, lysine, and asparagine residues were taken into account, the number of dynamic modifications allowed in MyriMatch search was changed accordingly. After importing and filtering out the results in IDPicker using a very stringent FDR cutoff value of 1%, we were able to see that this tandem mass spectrum was assigned to the same peptide sequence, but with the following modifications: 93GPp(OH)GAAGYp(OH)GNp(OH)GLp(OH)GIp(OH) GQDGPp(OH)GPp(OH)GIp(OH)GCn(-NH2)GTk-(GluGal)GER132. Out of the 11 proline residues present in Gly-Pro-Pro motif, eight of them were found to be hydroxylated. In addition, Asn126 and Lys129 were both modified with N- and O-glycosylations moieties, respectively. The high quality of the PSM shown in Figure 2, panel C as well as the perfect match of the PTMs with literature reports based on Edman degradation of placenta collagen IV lends credence to the correctness of the identification.13 These optimizations were successfully achieved to probe the PTMs in the 7S domain of collagen IV.
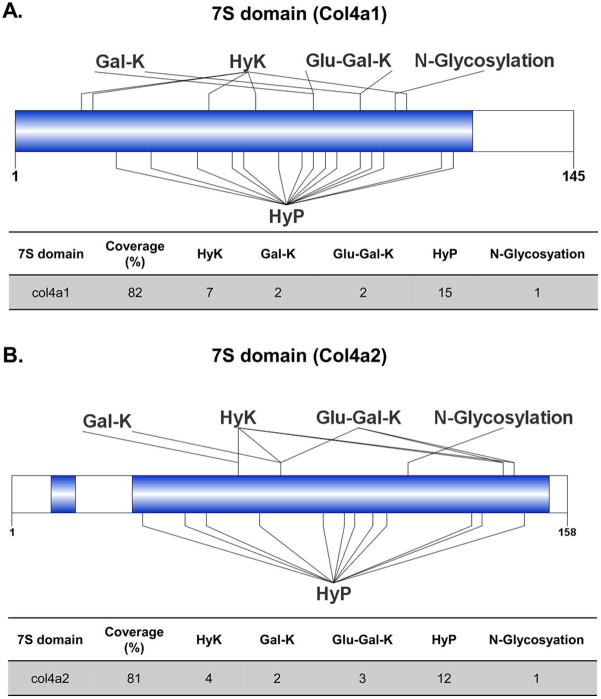
By using our improved method, 82% of sequence coverage was achieved for the 7S domain of col4a1 including 15 HyP, 7 HyK sites, and 2 O-linked glycosylation sites (Figure 3A). Further, the data revealed that these two glycosylation sites are microheterogeneous in nature as both galactosyl- and glucosylgalactosyl moieties were mapped to the sites. Similarly, 81% of sequence coverage was achieved for the 7S domain of col4a2 including 12 prolines and four lysine hydroxylation sites. Two unique sites with glucosylgalactosyl hydroxylysines and one unique site with galactosyl hydroxylysine were identified (Figure 3B). Lys78 showed heterogeneous distribution with the presence of either galactosyl or glucosylgalactosyl glycosylation.
Figure 3.
Optimization of MS and bioinformatics workflow improves identification of hydroxylation and O-linked glycosylation sites in the 7S domain of (A) col4a1 and (B) col4a2 purified from mouse PHFR9 cells. The first 27 and 25 amino acids corresponding to the signal peptides of col4a1 and col4a2 chains were removed, respectively. The blue color represents sequence coverage. The following abbreviations Glu-Gal-K, Gal-K, HyK, and HyP denote glucosylgalactosyl-hydroxylysine, galactosyl-hydroxylysine, hydroxylysine, and hydroxyproline, respectively.
Maximizing Coverage and PTM Characterization of Mouse col4a1
To test whether the significant improvements achieved in the purified 7S dodecamer could translate to better coverage and PTM identification in full-length collagen IV alpha chains, we analyzed collagen IV from EHS tumors. Analysis of a tryptic digest of EHS collagen IV by using a standard database search, which includes allowing for only two dynamic modifications per peptide, achieved very poor sequence coverage with most detected peptides matching the NC1 domain, similar to what was observed in proteomics studies of enriched BM. However, optimization of bioinformatics parameters by increasing the number of dynamic modifications significantly enhanced the sequence coverage as depicted in Supporting Figure S1.
To improve the results even further (in addition to trypsin), we also prepared GluC digests of EHS collagen IV to generate a new set of shorter peptides more amenable for MS identification as suggested by our in silico analyses (Figure 1C). In addition, peptide mixtures were analyzed by ETD and CID fragmentations using a Thermo LTQ Orbitrap Velos mass spectrometer and by HCD fragmentation using a Q-Exactive instrument. The .raw data sets were searched with the strategy described in the Methods section using MyriMatch and the dynamic modification motifs indicated in Table 1. The search results of different enzymes with multiple fragmentation types were assembled in IDPicker. Figure 4 shows the comprehensive mapping of hydroxylation and O-glycosylation sites in col4a1. By concatenation of these searches, we achieved about 81.5% sequence coverage of the entire col4a1 molecule (excluding signal peptide) (Table 2). A total of 156 HyP sites were identified (Figure 5A). Seven of which are likely to be 3-hydroxyprolines because they are found in the Xaa position of the Gly-Xaa-HyP motif (Figure 4 and Supporting Figure S2). An additional site of prolyl hydroxylation was identified within the Gly-HyP-Gln motif, which has been recently described in types II and V collagens.59,68 However, at this time it is not know whether hydroxylation occurs in the 3- or 4- position of the proline ring in this motif. Of note, no HyP sites were identified using Gly-Xaa-Ala or Gly-Xaa-Val motifs. In Supporting Figure S2, we show PSM match for all 3-HyP sites in mouse col4a1 found in this study. In addition, a total of 39 O-glycosylation sites were identified (Table 2). Out of these, seven sites (K78, K90, K573, K825, K1043, K1046, and K1081) were heterogeneous containing either galactosyl or glucosylgalactosyl moieties. Ten HyK sites were identified without any glycosylation moieties attached to it.
Figure 4.
Comprehensive mapping of hydroxylation and O-linked glycosylation sites in mouse col4a1. Collagen IV from mouse EHS tumor and 7S domain of PFHR9 cells were used to generate maps. Peptide sequences identified by MS are shown in black and represent an overall sequence coverage of 81.5%. Amino acid residues 1–27 corresponding to the signal peptide and sequences not identified in this study are colored gray. Red “P” indicates 4-hydroxyproline in the Yaa position of Gly-Xaa-Yaa motif, and a bold, red “P” indicates 3-hydroxyproline in the Xaa position of Gly-Xaa-HyP. Additionally, red “P*” indicates a hydroxyproline site in the Xaa position of “Gly-HyP-Gln” motif. Green and orange diamonds denote glucosylgalactosyl-hydroxylysine, and orange diamond denotes galactosyl-hydroxylysine residues. The bold “K” indicates hydroxylation of lysine (HyK). For added completeness, Lys1651 is denoted as K as shown in our previous studies.10,11 Red “N” indicates N-glycosylation of Asn126. A summary of the PTMs is presented in Table 2, and PSMs for O-glycosylated lysine and 3-hydroxyproline sites are provided in Supporting Figure S2.
Table 2.
Comparative Analyses between Mouse and Human col4a1 MS Results
| summary | mouse col4a1 | human col4a1 |
|---|---|---|
| sequence coverage | 81.5% | 84.73% |
| total O-glycosylation sites | 39 | 35 |
| galactosyl-hydroxylysine | 14 | 15 |
| glucosyl-galactosyl-hydroxylysine | 30 | 29 |
| hydroxylysine sites | 10 | 18 |
| hydroxyproline sites in Xaa position of “Gly-Xaa-Gln” motif | 1 | 3 |
| 4-hydroxyproline sites (Yaa position of “Gly-Xaa-Yaa” mottif) | 148 | 163 |
| 3-hydroxyproline sites (Xaa position of “Gly-Xaa-HyP” motif) | 7 | 14 |
Figure 5.
Venn diagrams summarizing the complementary contributions to sequence coverage, O-glycosylation, and hydroxylation in mouse EHS col4a1 by using (A) trypsin and GluC or (B) different fragmentation methods.
Venn diagrams in Figure 5 show that while tryptic peptides matched a greater portion of col4a1 sequence and identified more O-glycosylation and hydroxylation sites, peptides identified in the GluC digest also contributed significantly (Figure 5A). Even though there was a number of overlapping peptide sequences, CID, ETD, and HCD fragmentation methods contributed with unique coverage gains by 6%, 3%, and 32%, respectively. The identification of O-glycosylated and hydroxylated peptides also benefited from the use of different fragmentation strategies (Figure 5B). Identification of O-glycosylated and hydroxylated peptides by HCD contributed a significantly higher proportion compared to CID and ETD.
PTM Characterization of Human col4a1 from Public Repository Database
Collagen IV has been shown to be the most abundant component of human lens capsule BMs and hence could serve as a convenient resource for the PTM characterization. To test the capabilities of our improved bioinformatics workflow for the mapping of hydroxylation and O-glycosylation of lysine residues and hydroxylation of proline residues, we downloaded the lens capsule LC–MS/MS data generated by Uechi et al., which provided a proteomic overview of human eye BM.52 Although the data sets were generated by trypsin digestion using conventional CID fragmentation, an extensive 1D-gel LC fractionation strategy was undertaken for each sample analyzed. Analyses of the multiple .raw files generated for these sample sets with our bioinformatics approach identified 66, 68, and 58 distinct peptides corresponding to 1207, 1364, and 767 spectra, resulting in an increased sequence coverage of the entire col4a1 molecule (excluding signal peptide) from 29% to about 85% (Figure 6, Supporting Figure S3, and Table 2).
Figure 6.
Mapping of hydroxylation and O-linked glycosylation sites in col4a1 from human lens capsule. Three sets of MS data were analyzed from a publicly available data set from lens capsule BM isolated from human eyes submitted by Uechi et al. with identifier number PXD001025.52 Peptide sequences identified by MS are shown in black and represent an overall sequence coverage of 85%. Amino acid residues 1–27 corresponding to the signal peptide and sequences not identified in this study are colored gray. Red “P” indicates 4-hydroxyproline in the Yaa position of Gly-Xaa-Yaa motif, and a bold, red “P” indicates 3-hydroxyproline in the Xaa position of Gly-Xaa-HyP. Additionally, red “P*” indicates a hydroxyproline site in the Xaa position of “Gly-HyP-Gln” motif. Green and orange diamonds denote glucosylgalactosyl-hydroxylysine, and orange diamond denotes galactosyl-hydroxylysine residues. The bold “K” indicates hydroxylation of lysine (HyK). For added completeness, Lys1651 is denoted as K as shown in our previous studies.10 A summary of the PTMs is presented in Table 2, and PSMs for O-glycosylated lysine and 3-hydroxyproline sites are provided in Supporting Figure S3.
A total of 180 HyP sites were identified (Figure 6), 14 of which we report as potential 3-HyP in human col4a1 chain because they are found in the Xaa position of the Gly-Xaa-HyP motif. Although three additional HyP sites were identified within the Gly-HyP-Gln motif, they were not classified as 3- or 4-HyP because there is no prior chemical evidence to support either isomer (Table 2). No HyP sites were identified using Gly-Xaa-Ala or Gly-Xaa-Val motifs in MyriMatch searches of human lens capsule samples. In addition, a total of 35 O-glycosylation sites were identified (Figure 6, Table 2). Out of these, nine sites (K78, K90, K361, K1049, K1066, K1132, K1188, K1304, and K1340) were heterogeneous with either galactosyl or glucosylgalactosyl moieties attached to HyK residues. Eighteen lysine residues were identified in their hydroxylated form without any further glycosylation. Notably, 21 O-glycosylation sites were conserved between mouse and human col4a1 (Supporting Table S2). Significant enhancement of PTM characterization and greater sequence coverage of human collagen IV from publicly available data sets demonstrate the utility of our optimized bioinformatics workflow.
DISCUSSION
Collagen IV networks provide structural support to BM and ligands for cell membrane receptors regulating cellular function.2,4 The presence of a large number of PTMs, mainly on proline and lysine residues, contributes to the structural features relevant for different biological functions and disease of collagen IV.3 For instance, the importance of hydroxylation and O-glycosylation of lysine residues of collagen IV has been demonstrated by deleting lysyl hydroxylase-3 in mice, which resulted in the accumulation of collagen IV inside the cell leading to embryonic lethality.36 In this study, we report the first comprehensive PTM characterization of collagen IV mapping a large number of hydroxylation and glycosylation sites despite inherent challenges presented by this highly modified extracellular molecule. A unique distribution of 3- and 4-hydroxyproline residues as well as hydroxylysine residues was observed along the collagenous sequences. Although the majority of lysine residues were predicted to be glycosylated, they were also found in their unmodified state. Thus, our findings allow us to evaluate the extent of collagen IV modification and lay the foundation for dissecting the key role of these modifications in health and disease.
High-resolution MS has become the tool of choice for the characterization of PTMs in proteins. However, MS-based PTM characterization has remained a challenge for a molecule like collagen IV because of several reasons. In silico analyses suggested that trypsin, the most commonly proteolytic enzyme used in proteomic analyses, would generate very long and heavily modified collagen IV peptides, which are not easily detectable through conventional MS-based identification workflows. These problems are evident in many proteomic analyses of BM proteins, in which collagen IV is reported with low abundance and low sequence coverage.52–56 In fact, when we analyzed collagen IV by using a standardized proteomics approach, we obtained extremely poor coverage (Supporting Figure S1) likely due to the extensive presence of PTMs that hampered identification of modified peptides. Interestingly, the majority of peptides identified when searching with standard database strategies mapped to the NC1 domain sequence. This is consistent with our initial assessment because, unlike the 7S and triple helical domains, the NC1 domain does not contains HyP or glycosylated lysine residues, a feature that surely facilitated the identification of NC1 domain peptides. These findings prompted us to optimize our bioinformatics workflow to achieve significant improvements in peptide identification as well as sequence coverage.
We expanded the database search space by increasing the number of allowable modifications per peptide and number of miscleavages. Notably, the unique option available in MyriMatch that restricts the assignment of a modification to a specific sequence motif, not only reduced the search space and computing time, but also increased the specificity of output search results. This new strategy produced very significant improvements increasing both sequence coverage and the number of PTMs identifications. For instance, two distinct and highly modified 7S domain derived peptides, which had been previously missed, could only be identified after bioinformatics workflow was optimized (Figure 2). The use of our optimized strategy significantly improved coverage rising up to about 82% of col4a1 sequence. In addition, the overall number of identified PTMs significantly increased from only a few oxidations of methionine to a large number of hydroxylations of both proline and lysine residues as well as glycosylation of lysine residues.
The presence of glycosylated lysine residues prevents tryptic enzymatic cleavage potentially generating very long peptides with a large number of PTMs.26 Our analysis revealed that about 70% of the lysine residues present in the Yaa position of Gly-Xaa-Yaa motif are hydroxylated, consistent with classical amino acid analyses reports.26 These peptides produce very complex MS2 spectra that pose a challenge for its identification by any database search engine. Although in silico analysis suggested that GluC digestion of col4a1 would generate a significant number of peptides amenable for MS detection, the results showed that tryptic digestion still contributed with the largest number of collagen IV peptide identifications. However, GluC yielded a significant number of peptide identifications, many of which were unique and complemented trypsin results. Although glycosylated peptides may undergo neutral loss under collision-induced dissociation (CID) hampering MS/MS sequencing efforts, this unwanted event did not seem to impact the identification of glycosylated collagen IV peptides. This finding is consistent with previous studies, which demonstrated CID stability of glycopeptides from other collagens.69 However, HCD fragmentation was superior at detecting collagen IV glycopeptides as it identified 82% of the O-glycosylation sites while ETD and CID fragmentation techniques identified 41% of the sites. Similarly, HCD fragmentation yielded the majority (33%) of unique amino acid sequence compared to CID and ETD based fragmentation methods. ETD and CID contributed about 6% and 4% of nonoverlapping amino acid residues, respectively. Nonetheless, all three fragmentation strategies produced significant complementary information.
Proline hydroxylation is a key PTM that could either stabilize or destabilize the triple-helical domain of collagen molecules depending on the site of hydroxylation. Deletion of the prolyl 4-hydroxylase in mice highlighted the importance of 4-HyP for collagen IV function as knock out animals experienced frequent rupture of capillary walls as a result of defective BM.20 Our results demonstrated that about 70% of proline residues located in the Yaa position of the Gly-Xaa-Yaa motif (4-HyP) are hydroxylated in mouse collagen IV, a finding that is in agreement with previous amino acid analysis.15,16 In addition, we also identified seven potential 3-HyP sites (P444, P478, P602, P605, P647, P1424, P1436) located in the Xaa position of the Gly-Xaa-HyP motif, which is consistent with previous amino acid analysis of EHS collagen IV.15 We also searched for HyP residues in the Xaa position of “Gly-Xaa-[Ala/Gln/Val]” motifs, which have been recently identified in other collagens.59,68 These searches led to the identification of an additional HyP (P481) residue in the Xaa position of the Gly-Xaa-Gln motif. Although the Xaa position of HyP residue suggests that hydroxylation may occur in the third position of the proline ring, there is no previous chemical evidence to support this assignment. Interestingly, a previous study in type XI collagen reported a 4-HyP residue in the Xaa position of the Gly-Xaa-Ala motif,70 indicating that not all HyP residues found in the Xaa position are hydroxylated in the 3-position of the proline ring. To identify HyP residues in the Gly-HyP-Gln motif as either P3H or P4H substrates, it would be important to determine their isomeric nature by other experimental approaches such as Edman microsequencing or MS/MS analyses.71
To assess the robustness of our optimized bioinformatics workflow, MS data of human lens capsule BM were downloaded from the ProteomeXchange repository and analyzed to map glycosylation and hydroxylation of lysine as well as hydroxylation of proline residues in the human col4a1 chain. Although these files were generated using only trypsin digestion and CID fragmentation, sequence coverage for col4a1 was three-fold higher than originally reported52 after the data were analyzed with our bioinformatics pipeline. In addition, we detected 35 O-glycosylation sites, 21 of which are conserved with the mouse col4a1 (Figure 6 and Supporting Table S2). Following the same pattern, 76% of proline residues located in the Yaa position of the Gly-Xaa-Yaa motif were identified as 4-HyP, and a total of 14 Pro residues were classified as 3-HyP in human lens capsule col4a1 (Figure 6 and Supporting Figure S4).
A comparative analysis of prolyl 3-hydroxylation sites in col4a1 from EHS mouse tumor, human lens capsule, and previously reported bovine lens capsule23 was undertaken. The analysis shows that a total of 19 3-HyP sites have been mapped within col4a1 chain, with 10 of which (P74, P95, P372, P421, P444, P478, P484, P786, P803, and P1367) being newly identified in this study. More importantly, the analysis also revealed four sites (P602, P647, P1424, and P1436) that are invariably conserved, suggesting that these modified residues are important for collagen IV function in different tissues (Supporting Figure S4). The four conserved 3-HyP residues are located in two different GPP-rich clusters (601–606; 1423–1437) present in col4a1 sequence. However, a third GPP-rich cluster (197–214) in 7S domain harboring 3 3-HyP sites identified by Pokidysheva et al. by Edman microsequencing in bovine lens capsule was not observed in our MS results (Supporting Figure S4), identifying possible prolyl 3-hydroxylation differences between the mouse tumor and bovine lens capsule collagen IV samples. This notion is supported by previous biochemical analyses showing that 3-HyP levels in collagen IV from mouse EHS tumor are lower than in other animal tissues.14,21,72,73 In addition, these results suggest that the distribution of 3-HyP in collagen IV could change in different tissues, a concept that has been evidenced in fibrillar collagens.24 Notably, the physiological function of prolyl 3-hydroxylation by P3H2 in collagen IV has been investigated in two different mouse models. Although the evidence initially supported a molecular mechanism in which prolyl 3-hydroxylation is important to prevent collagen IV-mediated coagulation,23 more recent genetic and biochemical evidence has challenged this hypothesis and demonstrated that 3-HyP residees in different collagens are required for normal development of eye tissues.24 Thus, further studies will be required to define the function of 3-HyP in collagen IV.
Site-specific glycosylation is important for the assembly of collagen molecules and cell–matrix interaction. Although the number of O-glycosylation sites in type I collagen is significantly fewer than in collagen IV, they have been shown to be essential for the assembly of collagen fibers.29,30,74 Consistently, our results show that 39 out of 72 (50%) potential glycosylation sites were found as glycosylated hydroxylysines in mouse col4a1, which is significantly more than the six out of 23 sites (26%) reported for mouse col1a1 (Supporting Figure S5).24,28,75,76 Increased glycosylation may be explained by slower biosynthesis of collagen IV,77,78 which may permit the addition of more O-glycosylations than collagen I.79 Interestingly, the importance of the exquisite regulation of collagen glycosylation has been revealed in osteogenesis imperfecta where col1a1 mutations promote overglycosylation by slowing down triple helix folding rate and altering the structure of the collagen fiber, resulting in reduced bone strength.48,80
Glycosylation in collagen IV has been shown to be important for secretion,81 supramolecular assembly,13 endocytic collagen uptake,82 and modulation of cell adhesion.37 However, no quantitative information on site-specific modification heterogeneity has been pursued. Although our global analyses were not intended to look at this question, in human lens capsule, we estimated a surprisingly low amount of the glycosylated form of K1265 (approximately 9.89%), a glycosylation site known to modulate signal transduction and cell–matrix interactions in melanoma tumor cells.37,38,83 Furthermore, the type of glycosylation was revealed as glucosylgalactosyl-hydroxylysine, something that has not been previously distinguished by Edman degradation studies.84 This finding suggests that collagen IV synthesized by melanoma cells may increase glycosylation of this site to promote cell adhesion. Thus, it will be interesting to confirm the glycosylation heterogeneity at this site and others by developing new high-throughput MS workflows. This would also facilitate the quantitation of tissue-specific differences that may elucidate the role of site-specific PTMs in collagen IV network assembly and cell–matrix interactions.
Supplementary Material
Acknowledgments
We are grateful to Mohamed Rafi for the excellent technical assistance and Matt Chambers for the invaluable help with software use. We would like to thank Dr. Daniel Liebler for facilitating many laboratory instruments and reagents for MS analyses. We also thank Dr. Kristie Rose, from the Proteomics Laboratory-Mass Spectrometry Research Center at Vanderbilt University, for the assistance with the LTQ-Orbitrap Velos instrument. This work was supported in part by NIH R01 Grant Nos. DK099467 and DK065138. The LTQ-Orbitrap Velos mass spectrometer used in these studies was purchased with funds from the NIH S10 Grant No. RR027714 awarded to the Vanderbilt proteomics shared resource.
ABBREVIATIONS
- BM
basement membrane
- col4a1
collagen IV alpha 1 chain
- col4a2
collagen IV alpha 2 chain
- col1a1
type I collagen alpha 1 chain
- Gal
galactosyl
- GluGal
glucosylgalactosyl
- HyP
hydroxyproline
- HyK
hydroxylysine
- PTMs
posttranslational modifications
- CID
collision induced dissociation
- ETD
electron transfer dissociation
- HCD
higher energy C-trap dissociation
- FDR
false discovery rate
- PSM
peptide spectrum match
Footnotes
Author Contributions
R.M.V. and B.G.H. contributed to the overall conception of the study. R.M.V. and D.L.T. designed the experiments and methodology. R.M.V., T.B., L.V.-M. and L.J.Z. performed the experiments. R.M.V., D.L.T., L.V.-M., and T.B. analyzed the data. R.M.V. and T.B. wrote the manuscript.
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome. 5b00767. The data may be accessed at https://massive.ucsd.edu/ProteoSAFe/datasets.jsp#%7B%22title_input%22%3A%22collagen%22%7D in ProteomeXchange with identifier number PXD003237.
Representative diagram showing improved sequence coverage of mouse col4a1 by optimizing the number of allowed PTMs and miscleavages in the database search using MyriMatch; PSMs of O-glycosylation and 3-hydroxyproline sites in mouse col4a1 from EHS tumor; PSMs of O-glycosylation and 3-hydroxyproline sites in human col4a1 from lens capsule; comparison of 3-HyP sites identified in col4a1 from mouse, human, and bovine; summary of prolyl 3-hydroxylation and glycosylation sites in mouse col1a1 (PDF)
References
- 1.Pöschl E, Schlötzer-Schrehardt U, Brachvogel B, Saito K, Ninomiya Y, Mayer U. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development. 2004;131:1619–1628. doi: 10.1242/dev.01037. [DOI] [PubMed] [Google Scholar]
- 2.Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 3.Parkin JD, San Antonio JD, Pedchenko V, Hudson B, Jensen ST, Savige J. Mapping structural landmarks, ligand binding sites, and missense mutations to the collagen IV heterotrimers predicts major functional domains, novel interactions, and variation in phenotypes in inherited diseases affecting basement membranes. Hum Mutat. 2011;32:127–43. doi: 10.1002/humu.21401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harbor Perspect Biol. 2011;3:a004911. doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudson BG, Reeders ST, Tryggvason K. Type IV collagen: structure, gene organization, and role in human diseases. Molecular basis of Goodpasture and Alport syndromes and diffuse leiomyomatosis. J Biol Chem. 1993;268:26033–6. [PubMed] [Google Scholar]
- 6.Khoshnoodi J, Pedchenko V, Hudson BG. Mammalian collagen IV. Microsc Res Tech. 2008;71:357–370. doi: 10.1002/jemt.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson WE, Rose KL, Hudson BG, Vanacore RM. Supramolecular organization of the alpha121-alpha565 collagen IV network. J Biol Chem. 2014;289:25601–10. doi: 10.1074/jbc.M114.571844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timpl R, Wiedemann H, Delden V, Furthmayr H, Kuhn K. A network model for the organization of type IV collagen molecules in basement membranes. Eur J Biochem. 1981;120:203–11. doi: 10.1111/j.1432-1033.1981.tb05690.x. [DOI] [PubMed] [Google Scholar]
- 9.McCall AS, Cummings CF, Bhave G, Vanacore R, Page-McCaw A, Hudson BG. Bromine is an essential trace element for assembly of collagen IV scaffolds in tissue development and architecture. Cell. 2014;157:1380–92. doi: 10.1016/j.cell.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fidler AL, Vanacore RM, Chetyrkin SV, Pedchenko VK, Bhave G, Yin VP, Stothers CL, Rose KL, McDonald WH, Clark TA, Borza DB, Steele RE, Ivy MT, Aspirnauts T, Hudson JK, Hudson BG. A unique covalent bond in basement membrane is a primordial innovation for tissue evolution. Proc Natl Acad Sci U S A. 2014;111:331. doi: 10.1073/pnas.1318499111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhave G, Cummings CF, Vanacore RM, Kumagai-Cresse C, Ero-Tolliver IA, Rafi M, Kang JS, Pedchenko V, Fessler LI, Fessler JH, Hudson BG. Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nat Chem Biol. 2012;8:784–90. doi: 10.1038/nchembio.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanacore R, Ham AJ, Voehler M, Sanders CR, Conrads TP, Veenstra TD, Sharpless KB, Dawson PE, Hudson BG. A sulfilimine bond identified in collagen IV. Science. 2009;325:1230–4. doi: 10.1126/science.1176811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langeveld J, Noelken M, Hård K, Todd P, Vliegenthart J, Rouse J, Hudson B. Bovine glomerular basement membrane. Location and structure of the asparagine-linked oligosaccharide units and their potential role in the assembly of the 7 S collagen IV tetramer. J Biol Chem. 1991;266:2622–2631. [PubMed] [Google Scholar]
- 14.Risteli J, Bachinger HP, Engel J, Furthmayr H, Timpl R. 7-S collagen: characterization of an unusual basement membrane structure. Eur J Biochem. 1980;108:239–50. doi: 10.1111/j.1432-1033.1980.tb04717.x. [DOI] [PubMed] [Google Scholar]
- 15.Kleinman HK, McGarvey ML, Liotta LA, Robey PG, Tryggvason K, Martin GR. Isolation and characterization of type IV procollagen, laminin, and heparan sulfate proteoglycan from the EHS sarcoma. Biochemistry. 1982;21:6188–93. doi: 10.1021/bi00267a025. [DOI] [PubMed] [Google Scholar]
- 16.Kefalides NA. Structure and biosynthesis of basement membranes. Int Rev Connect Tissue Res. 1973;6:63–104. doi: 10.1016/b978-0-12-363706-2.50008-8. [DOI] [PubMed] [Google Scholar]
- 17.Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg RA, Prockop DJ. The thermal transition of a non-hydroxylated form of collagen. Evidence for a role for hydroxyproline in stabilizing the triple-helix of collagen. Biochem Biophys Res Commun. 1973;52:115. doi: 10.1016/0006-291x(73)90961-3. [DOI] [PubMed] [Google Scholar]
- 19.Sakakibara S, Inouye K, Shudo K, Kishida Y, Kobayashi Y, Prockop DJ. Synthesis of (Pro-Hyp-Gly) n of defined molecular weights. Evidence for the stabilization of collagen triple helix by hydroxypyroline. Biochim Biophys Acta, Protein Struct. 1973;303:198. doi: 10.1016/0005-2795(73)90164-5. [DOI] [PubMed] [Google Scholar]
- 20.Holster T, Pakkanen O, Soininen R, Sormunen R, Nokelainen M, Kivirikko KI, Myllyharju J. Loss of assembly of the main basement membrane collagen, type IV, but not fibril-forming collagens and embryonic death in collagen prolyl 4-hydroxylase I null mice. J Biol Chem. 2007;282:2512–9. doi: 10.1074/jbc.M606608200. [DOI] [PubMed] [Google Scholar]
- 21.Dean DC, Barr JF, Freytag JW, Hudson BG. Isolation of type IV procollagen-like polypeptides from glomerular basement membrane. Characterization of pro-alpha 1(IV) J Biol Chem. 1983;258:590–6. [PubMed] [Google Scholar]
- 22.Tiainen P, Pasanen A, Sormunen R, Myllyharju J. Characterization of recombinant human prolyl 3-hydroxylase isoenzyme 2, an enzyme modifying the basement membrane collagen IV. J Biol Chem. 2008;283:19432–9. doi: 10.1074/jbc.M802973200. [DOI] [PubMed] [Google Scholar]
- 23.Pokidysheva E, Boudko S, Vranka J, Zientek K, Maddox K, Moser M, Fassler R, Ware J, Bachinger HP. Biological role of prolyl 3-hydroxylation in type IV collagen. Proc Natl Acad Sci U S A. 2014;111:161–6. doi: 10.1073/pnas.1307597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson DM, Joeng KS, Werther R, Rajagopal A, Weis M, Lee BH, Eyre DR. Post-translationally abnormal collagens of prolyl 3-hydroxylase-2 null mice offer a pathobiological mechanism for the high myopia linked to human LEPREL1 mutations. J Biol Chem. 2015;290:8613–22. doi: 10.1074/jbc.M114.634915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddi A, Piez KA. Extracellular Matrix Biochemistry. Elsevier; 1984. [Google Scholar]
- 26.Spiro RG, Fukushi S. The lens capsule. Studies on the carbohydrate units. J Biol Chem. 1969;244:2049–58. [PubMed] [Google Scholar]
- 27.Terajima M, Perdivara I, Sricholpech M, Deguchi Y, Pleshko N, Tomer KB, Yamauchi M. Glycosylation and cross-linking in bone type I collagen. J Biol Chem. 2014;289:22636–47. doi: 10.1074/jbc.M113.528513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perdivara I, Yamauchi M, Tomer KB. Molecular Characterization of Collagen Hydroxylysine -Glycosylation by Mass Spectrometry: Current Status. Aust J Chem. 2013;66:760–769. doi: 10.1071/CH13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamauchi M, Sricholpech M. Lysine post-translational modifications of collagen. Essays Biochem. 2012;52:113–33. doi: 10.1042/bse0520113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sricholpech M, Perdivara I, Yokoyama M, Nagaoka H, Terajima M, Tomer KB, Yamauchi M. Lysyl hydroxylase 3-mediated glucosylation in type I collagen: molecular loci and biological significance. J Biol Chem. 2012;287:22998–3009. doi: 10.1074/jbc.M112.343954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kivirikko KI, Ryhanen L, Anttinen H, Bornstein P, Prockop DJ. Further hydroxylation of lysyl residues in collagen by protocollagen lysyl hydroxylase in vitro. Biochemistry. 1973;12:4966–71. doi: 10.1021/bi00748a023. [DOI] [PubMed] [Google Scholar]
- 32.Risteli J, Kivirikko KI. Activities of prolyl hydroxylase, lysyl hydroxylase, collagen galactosyltransferase and collagen glucosyltransferase in the liver of rats with hepatic injury. Biochem J. 1974;144:115–22. doi: 10.1042/bj1440115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spiro RG, Spiro MJ. Studies on the biosynthesis of the hydroxylysine-linked disaccharide unit of basement membranes and collagens. 3. Tissue and subcellular distribution of glycosyltransferases and the effect of various conditions on the enzyme levels. J Biol Chem. 1971;246:4919–25. [PubMed] [Google Scholar]
- 34.Liefhebber JM, Punt S, Spaan WJ, van Leeuwen HC. The human collagen beta(1-O)galactosyltransferase, GLT25D1, is a soluble endoplasmic reticulum localized protein. BMC Cell Biol. 2010;11:33. doi: 10.1186/1471-2121-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schegg B, Hulsmeier AJ, Rutschmann C, Maag C, Hennet T. Core glycosylation of collagen is initiated by two beta(1-O)galactosyltransferases. Molecular and cellular biology. 2009;29:943–52. doi: 10.1128/MCB.02085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rautavuoma K, Takaluoma K, Sormunen R, Myllyharju J, Kivirikko KI, Soininen R. Premature aggregation of type IV collagen and early lethality in lysyl hydroxylase 3 null mice. Proc Natl Acad Sci U S A. 2004;101:14120–5. doi: 10.1073/pnas.0404966101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stawikowski MJ, Aukszi B, Stawikowska R, Cudic M, Fields GB. Glycosylation modulates melanoma cell alpha2beta1 and alpha3beta1 integrin interactions with type IV collagen. J Biol Chem. 2014;289:21591–604. doi: 10.1074/jbc.M114.572073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lauer-Fields JL, Malkar NB, Richet G, Drauz K, Fields GB. Melanoma cell CD44 interaction with the alpha 1(IV)1263–1277 region from basement membrane collagen is modulated by ligand glycosylation. J Biol Chem. 2003;278:14321–30. doi: 10.1074/jbc.M212246200. [DOI] [PubMed] [Google Scholar]
- 39.Robins SP. Cross-linking of collagen. Isolation, structural characterization and glycosylation of pyridinoline. Biochem J. 1983;215:167–173. doi: 10.1042/bj2150167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamauchi M, Katz EP, Mechanic GL. Intermolecular crosslinking and stereospecific molecular packing in type I collagen fibrils of the periodontal ligament. Biochemistry. 1986;25:4907–4913. doi: 10.1021/bi00365a027. [DOI] [PubMed] [Google Scholar]
- 41.Yamauchi M, Noyes C, Kuboki Y, Mechanic GL. Collagen structural microheterogeneity and a possible role for glycosylated hydroxylysine in type I collagen. Proc Natl Acad Sci U S A. 1982;79:7684–7688. doi: 10.1073/pnas.79.24.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uzawa K, Yeowell HN, Yamamoto K, Mochida Y, Tanzawa H, Yamauchi M. Lysine hydroxylation of collagen in a fibroblast cell culture system. Biochem Biophys Res Commun. 2003;305:484–487. doi: 10.1016/s0006-291x(03)00799-x. [DOI] [PubMed] [Google Scholar]
- 43.Moro L, Romanello M, Favia A, Lamanna M, Lozupone E. Posttranslational modifications of bone collagen type I are related to the function of rat femoral regions. Calcif Tissue Int. 2000;66:151–156. doi: 10.1007/s002230010030. [DOI] [PubMed] [Google Scholar]
- 44.Bailey AJ, Paul RG, Knott L. Mechanisms of maturation and ageing of collagen. Mech Ageing Dev. 1998;106:1–56. doi: 10.1016/s0047-6374(98)00119-5. [DOI] [PubMed] [Google Scholar]
- 45.Brinckmann J, Notbohm H, Tronnier M, Açil Y, Fietzek PP, Schmeller W, Müller PK, Bätge B. Overhydroxylation of lysyl residues is the initial step for altered collagen cross-links and fibril architecture in fibrotic skin. J Invest Dermatol. 1999;113:617–621. doi: 10.1046/j.1523-1747.1999.00735.x. [DOI] [PubMed] [Google Scholar]
- 46.Lehmann HW, Wolf E, Röser K, Bodo M, Delling G, Müller PK. Composition and posttranslational modification of individual collagen chains from osteosarcomas and osteofibrous dysplasias. J Cancer Res Clin Oncol. 1995;121:413–418. doi: 10.1007/BF01212948. [DOI] [PubMed] [Google Scholar]
- 47.Michalsky M, Norrissuarez K, Bettica P, Pecile A, Moro L. Rat cortical and trabecular bone collagen glycosylation are differently influenced by ovariectomy. Biochem Biophys Res Commun. 1993;192:1281–1288. doi: 10.1006/bbrc.1993.1555. [DOI] [PubMed] [Google Scholar]
- 48.Tenni R, Valli M, Rossi A, Cetta G. Possible role of overglycosylation in the type I collagen triple helical domain in the molecular pathogenesis of osteogenesis imperfecta. American journal of medical genetics. 1993;45:252–256. doi: 10.1002/ajmg.1320450219. [DOI] [PubMed] [Google Scholar]
- 49.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 50.Holman JD, Dasari S, Tabb DL. Informatics of protein and posttranslational modification detection via shotgun proteomics. Methods Mol Biol. 2013;1002:167–79. doi: 10.1007/978-1-62703-360-2_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hill RC, Wither MJ, Nemkov T, Barrett A, D’Alessandro A, Dzieciatkowska M, Hansen KC. Preserved Proteins from Extinct Bison latifrons Identified by Tandem Mass Spectrometry; Hydroxylysine Glycosides are a Common Feature of Ancient Collagen. Mol Cell Proteomics. 2015;14:1946–58. doi: 10.1074/mcp.M114.047787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uechi G, Sun Z, Schreiber EM, Halfter W, Balasubramani M. Proteomic View of Basement Membranes from Human Retinal Blood Vessels, Inner Limiting Membranes, and Lens Capsules. J Proteome Res. 2014;13:3693. doi: 10.1021/pr5002065. [DOI] [PubMed] [Google Scholar]
- 53.Balasubramani M, Schreiber EM, Candiello J, Balasubramani GK, Kurtz J, Halfter W. Molecular interactions in the retinal basement membrane system: a proteomic approach. Matrix Biol. 2010;29:471–83. doi: 10.1016/j.matbio.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 54.Lennon R, Byron A, Humphries JD, Randles MJ, Carisey A, Murphy S, Knight D, Brenchley PE, Zent R, Humphries MJ. Global analysis reveals the complexity of the human glomerular extracellular matrix. J Am Soc Nephrol. 2014;25:939–51. doi: 10.1681/ASN.2013030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pierchala BA, Munoz MR, Tsui CC. Proteomic analysis of the slit diaphragm complex: CLIC5 is a protein critical for podocyte morphology and function. Kidney Int. 2010;78:868–82. doi: 10.1038/ki.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO. The Extracellular Matrix: Tools and Insights for the “Omics” Era. Matrix Biol. 2015 doi: 10.1016/j.matbio.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen Y, Tolic N, Xie F, Zhao R, Purvine SO, Schepmoes AA, Moore RJ, Anderson GA, Smith RD. Effectiveness of CID, HCD, and ETD with FT MS/MS for degradomic-peptidomic analysis: comparison of peptide identification methods. J Proteome Res. 2011;10:3929–43. doi: 10.1021/pr200052c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mechref Y. Use of CID/ETD mass spectrometry to analyze glycopeptides. Curr Protoc Protein Sci. 2012:1111. doi: 10.1002/0471140864.ps1211s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song E, Mechref Y. LC–MS/MS identification of the O-glycosylation and hydroxylation of amino acid residues of collagen alpha-1 (II) chain from bovine cartilage. J Proteome Res. 2013;12:3599–609. doi: 10.1021/pr400101t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taga Y, Kusubata M, Ogawa-Goto K, Hattori S. Development of a novel method for analyzing collagen O-glycosylations by hydrazide chemistry. Mol Cell Proteomics. 2012;11:M111.010397. doi: 10.1074/mcp.M111.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choudhary G, Wu SL, Shieh P, Hancock WS. Multiple enzymatic digestion for enhanced sequence coverage of proteins in complex proteomic mixtures using capillary LC with ion trap MS/MS. J Proteome Res. 2003;2:59–67. doi: 10.1021/pr025557n. [DOI] [PubMed] [Google Scholar]
- 62.Lapolla A, Fedele D, Reitano R, Arico NC, Seraglia R, Traldi P, Marotta E, Tonani R. Enzymatic digestion and mass spectrometry in the study of advanced glycation end products/peptides. J Am Soc Mass Spectrom. 2004;15:496–509. doi: 10.1016/j.jasms.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 63.Henson R, Cetto L. The MATLAB bioinformatics toolbox. Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics. 2005 doi: 10.1002/047001153X.g409308. [DOI] [Google Scholar]
- 64.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6:654–61. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma ZQ, Dasari S, Chambers MC, Litton MD, Sobecki SM, Zimmerman LJ, Halvey PJ, Schilling B, Drake PM, Gibson BW, Tabb DL. IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J Proteome Res. 2009;8:3872–81. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.French WR, Zimmerman LJ, Schilling B, Gibson BW, Miller CA, Townsend RR, Sherrod SD, Goodwin CR, McLean JA, Tabb DL. Wavelet-based peak detection and a new charge inference procedure for MS/MS implemented in ProteoWizard’s msConvert. J Proteome Res. 2015;14:1299–307. doi: 10.1021/pr500886y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qian RG, Glanville RW. Separation and characterization of two polypeptide chains from the 7S cross-linking domain of basement-membrane (type IV) collagen. Biochem J. 1984;222:447–52. doi: 10.1042/bj2220447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang C, Park AC, Davis NA, Russell JD, Kim B, Brand DD, Lawrence MJ, Ge Y, Westphall MS, Coon JJ, Greenspan DS. Comprehensive mass spectrometric mapping of the hydroxylated amino acid residues of the alpha1(V) collagen chain. J Biol Chem. 2012;287:40598–610. doi: 10.1074/jbc.M112.406850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perdivara I, Perera L, Sricholpech M, Terajima M, Pleshko N, Yamauchi M, Tomer KB. Unusual fragmentation pathways in collagen glycopeptides. J Am Soc Mass Spectrom. 2013;24:1072–81. doi: 10.1007/s13361-013-0624-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kimura T, Cheah KS, Chan SD, Lui VC, Mattei MG, van der Rest M, Ono K, Solomon E, Ninomiya Y, Olsen BR. The human alpha 2(XI) collagen (COL11A2) chain. Molecular cloning of cDNA and genomic DNA reveals characteristics of a fibrillar collagen with differences in genomic organization. J Biol Chem. 1989;264:13910–6. [PubMed] [Google Scholar]
- 71.Kassel DB, Biemann K. Differentiation of hydroxyproline isomers and isobars in peptides by tandem mass spectrometry. Anal Chem. 1990;62:1691–5. doi: 10.1021/ac00214a032. [DOI] [PubMed] [Google Scholar]
- 72.Glanville RW, Qian RQ, Siebold B, Risteli J, Kuhn K. Amino acid sequence of the N-terminal aggregation and cross-linking region (7S domain) of the alpha 1 (IV) chain of human basement membrane collagen. Eur J Biochem. 1985;152:213–9. doi: 10.1111/j.1432-1033.1985.tb09186.x. [DOI] [PubMed] [Google Scholar]
- 73.Timpl R, Martin GR, Bruckner P, Wick G, Wiedemann H. Nature of the collagenous protein in a tumor basement membrane. Eur J Biochem. 1978;84:43–52. doi: 10.1111/j.1432-1033.1978.tb12139.x. [DOI] [PubMed] [Google Scholar]
- 74.Bornstein P, Sage H. Structurally distinct collagen types. Annu Rev Biochem. 1980;49:957–1003. doi: 10.1146/annurev.bi.49.070180.004521. [DOI] [PubMed] [Google Scholar]
- 75.Cabral WA, Perdivara I, Weis M, Terajima M, Blissett AR, Chang W, Perosky JE, Makareeva EN, Mertz EL, Leikin S, Tomer KB, Kozloff KM, Eyre DR, Yamauchi M, Marini JC. Abnormal type I collagen post-translational modification and cross-linking in a cyclophilin B KO mouse model of recessive osteogenesis imperfecta. PLoS Genet. 2014;10:e1004465. doi: 10.1371/journal.pgen.1004465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pokidysheva E, Zientek KD, Ishikawa Y, Mizuno K, Vranka JA, Montgomery NT, Keene DR, Kawaguchi T, Okuyama K, Bachinger HP. Posttranslational modifications in type I collagen from different tissues extracted from wild type and prolyl 3-hydroxylase 1 null mice. J Biol Chem. 2013;288:24742–52. doi: 10.1074/jbc.M113.464156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grant ME, Kefalides NA, Prockop DJ. The biosynthesis of basement membrane collagen in embryonic chick lens. I. Delay between the synthesis of polypeptide chains and the secretion of collagen by matrix-free cells. J Biol Chem. 1972;247:3539–44. [PubMed] [Google Scholar]
- 78.Juva K, Prockop DJ. Formation of enzyme-substrate complexes with protocollagen proline hydroxylase and large polypeptide substrates. J Biol Chem. 1969;244:6486–92. [PubMed] [Google Scholar]
- 79.Vuust J, Piez KA. A kinetic study of collagen biosynthesis. J Biol Chem. 1972;247:856–62. [PubMed] [Google Scholar]
- 80.Dominguez LJ, Barbagallo M, Moro L. Collagen overglycosylation: a biochemical feature that may contribute to bone quality. Biochem Biophys Res Commun. 2005;330:1–4. doi: 10.1016/j.bbrc.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 81.Sipila L, Ruotsalainen H, Sormunen R, Baker NL, Lamande SR, Vapola M, Wang C, Sado Y, Aszodi A, Myllyla R. Secretion and assembly of type IV and VI collagens depend on glycosylation of hydroxylysines. J Biol Chem. 2007;282:33381–8. doi: 10.1074/jbc.M704198200. [DOI] [PubMed] [Google Scholar]
- 82.Jurgensen HJ, Madsen DH, Ingvarsen S, Melander MC, Gardsvoll H, Patthy L, Engelholm LH, Behrendt N. A novel functional role of collagen glycosylation: interaction with the endocytic collagen receptor uparap/ENDO180. J Biol Chem. 2011;286:32736–48. doi: 10.1074/jbc.M111.266692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Malkar NB, Lauer-Fields JL, Fields GB. Convenient synthesis of glycosylated hydroxylysine derivatives for use in solid-phase peptide synthesis. Tetrahedron Lett. 2000;41:1137–1140. [Google Scholar]
- 84.Babel W, Glanville RW. Structure of human-basement-membrane (type IV) collagen. Complete amino-acid sequence of a 914-residue-long pepsin fragment from the alpha 1(IV) chain. Eur J Biochem. 1984;143:545–56. doi: 10.1111/j.1432-1033.1984.tb08404.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.