Abstract
Pluripotent stem cells often adopt a unique developmental program while retaining certain flexibility. The molecular basis of such properties remains unclear. Using differentiation of pluripotent Drosophila imaginal tissues as assays, we examined the contribution of epigenetic factors in ectopic activation of Hox genes. We found that over-expression of Trithorax H3K4 methyltransferase can induce ectopic adult appendages by selectively activating the Hox genes Ultrabithorax and Sex comb reduced in wing and leg discs, respectively. This tissue-specific inducibility correlates with the presence of paused RNA polymerase II in the promoter-proximal region of these genes. Although the Antennapedia promoter is paused in eye-antenna discs, it cannot be induced by Trx without a reduction in histone variants or their chaperones, suggesting additional control by the nucleosomal architecture. Lineage tracing and pulse-chase experiments revealed that the active state of Hox genes is maintained substantially longer in mutants deficient for HIRA, a chaperone for the H3.3 variant. In addition, both HIRA and H3.3 appeared to act cooperatively with the Polycomb group of epigenetic repressors. These results support the involvement of H3.3-mediated nucleosome turnover in restoring the repressed state. We propose a regulatory framework integrating transcriptional pausing, histone modification, nucleosome architecture and turnover for cell lineage maintenance.
Author Summary
During animal development, the primordia of different body parts undergo a series of transitions in which their developmental potency becomes more restricted. Hox genes encode a family of evolutionarily conserved transcriptional factors that are crucial for choosing different paths during transitions. Thus, the transcriptional status of Hox genes is directly linked to the maintenance and developmental direction of pluripotent tissues. As post-translational methylation of histone H3 is pivotal for transcriptional control, we could activate Hox genes and alter the subsequent development of some pluripotent Drosophila imaginal tissues by increasing the level of Trithorax that catalyzes activation-related methylation. However, other imaginal tissues remain refractory unless histone variants or their chaperones that directly affect nucleosome dynamics are simultaneously depleted. By monitoring the duration of Hox expression under these conditions, we found that the active state of Hox genes is substantially prolonged, resulting from effective conversion of promoter-associated paused RNA polymerase II into active transcription. Further analyses indicate that these factors are functionally linked to the Polycomb group of epigenetic factors that bestow long-term repression. Our studies demonstrate that developmental constraints are modulated by factors acting at multiple levels, offering a useful approach to tissue re-programming in regeneration medicine and stem cell research.
Introduction
During metazoan development, totipotent embryonic cells undergo a series of changes to adopt fixed adult features. Accordingly, the potency of these cells becomes progressively restricted. Intense interest has been focused on deciphering the regulatory molecules and mechanisms responsible for developmental restriction and the practical means of lifting such restrictions [1–5].
Drosophila larvae consist of a dozen disc-shaped tissues called imaginal discs that represent progenitors of adult appendages. Transplantation experiments show that they often endure passages of transfer in surrogate hosts without changing developmental fates. However in rare cases, discs may adopt alternative developmental paths and transform into corresponding appendages, revealing their pluripotent nature [6]. Their developmental plasticity is further corroborated by developmental switches resulting from mis-expression of transcriptional factors encoded by Hox genes that act as master developmental regulators [7–10].
Hox genes are mainly regulated by two groups of epigenetic factors categorically called Polycomb group (PcG) of repressors and Trithorax group (trxG) of activators after early embryonic stages. One salient function of PcG and trxG is the ability to trimethylate histone H3 on K27 and K4 (H3K27me3 and H3K4me3) to bestow repressive and active chromatin marks on Hox genes, respectively [11–13]. Although these factors often co-exist in the same cells, the on-off state of Hox genes and underlying chromatin marks are not perturbed through mitotic divisions. How such epigenetic inheritance is stably maintained is not fully understood [13–15].
The Hox genes are tightly regulated at the level of transcription. Recent studies have shown that repressed Hox promoters may contain paused RNA polymerase II (RNA-Pol II) in a region about 50 bp downstream of the transcriptional start site (TSS), with phosphorylation at Ser-5 of the C-terminal domain (CTD) of Rpb1 [16–19]. Genome-wide analyses have revealed a strong correlation between paused RNA-Pol II and PcG repressors and/or H3K27me3 marks [4, 20, 21]. In addition to canonical histones, histone variants have been implicated in transcriptional pausing of Hox genes. Enrichment of H2A.Z, an H2A variant, is widely found in nucleosomes immediately downstream of paused RNA-Pol II [22, 23]. Further, H3.3, a replication-independent H3 variant, is enriched at PcG-binding sites of Hox genes [24–26]. In mouse embryonic cells, depletion of H3.3 results in reduced levels of H3K27me3 at promoters of developmentally regulated genes [27]. However, deposition of H3.3 is also known to establish epigenetic memory of the transcriptionally active state [28].
HIRA, an H3.3 chaperone, is critical for the assembly of the paternal genome after fertilization in Drosophila [29] and is required for deposition of histone H3.3 at enhancers of active and poised genes [26, 30–32]. Recently, physical interaction between HIRA and PcG proteins has also been shown [27]. However, HIRA-deficient animals appear to be viable [33, 34], and H3.3 enrichment at telomeric regions and some transcription factor binding sites is HIRA-independent, indicating rather complex functions [35, 36].
Conceptually, maintenance of cell lineages must rely on an intricate interplay between factors acting at multiple levels. The identification of relevant factors and their contributions are imperative to gaining a deep understanding of the nature and potential manipulation of cell lineage maintenance [37, 38]. Here, we used a targeted Trithorax (Trx) expression system to alter developmental fates of several imaginal discs and examined the molecular mechanism employed by cooperating epigenetic factors in this process. We found disc-specific poised states are critical for activation of different Hox genes. In addition, the transiently induced active state may be rapidly obliterated by PcG and HIRA-dependent histone turnover, providing a molecular basis for maintenance of cell lineages.
Results
Altered tissue development and Hox expression by Trx
Using the Gal4/UAS induction system [39], we examined several Gal4 lines that can drive either ubiquitous (actin-Gal4) or selective expression in imaginal discs (e.g. A9-, engrailed- or dpp-Gal4 lines) (S1A Fig). The dpp-Gal4 line provides a versatile tool to correlate induction of Hox genes in larval stages with multiple phenotypical changes in adults. This line contains a ~4kb 3’ enhancer (i.e. blk) of dpp that drives expression primarily in larval imaginal discs [40]. In most imaginal discs, dpp-Gal4 is expressed in a strip of cells located along the anterior/posterior (A/P) compartmental boundary. Despite limited induction, Trx driven by dpp-Gal4 (henceforth dpp>Trx) resulted in profound adult phenotypes, including severely reduced wing size, absence of posterior notum, extra sex combs (ESC) on second and third legs (L2 and L3), and loss-of-arista (Fig 1A). Because these phenotypes resembled homeotic transformations resulting from ectopic expression of Hox genes [41, 42], we began to examine Hox protein expression in imaginal discs.
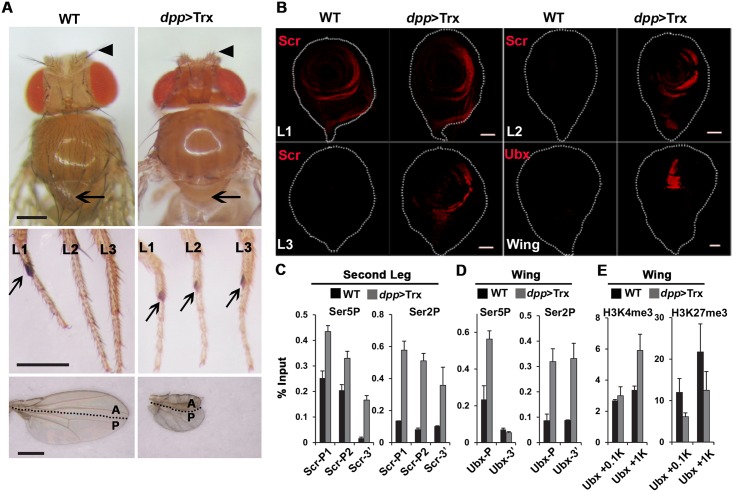
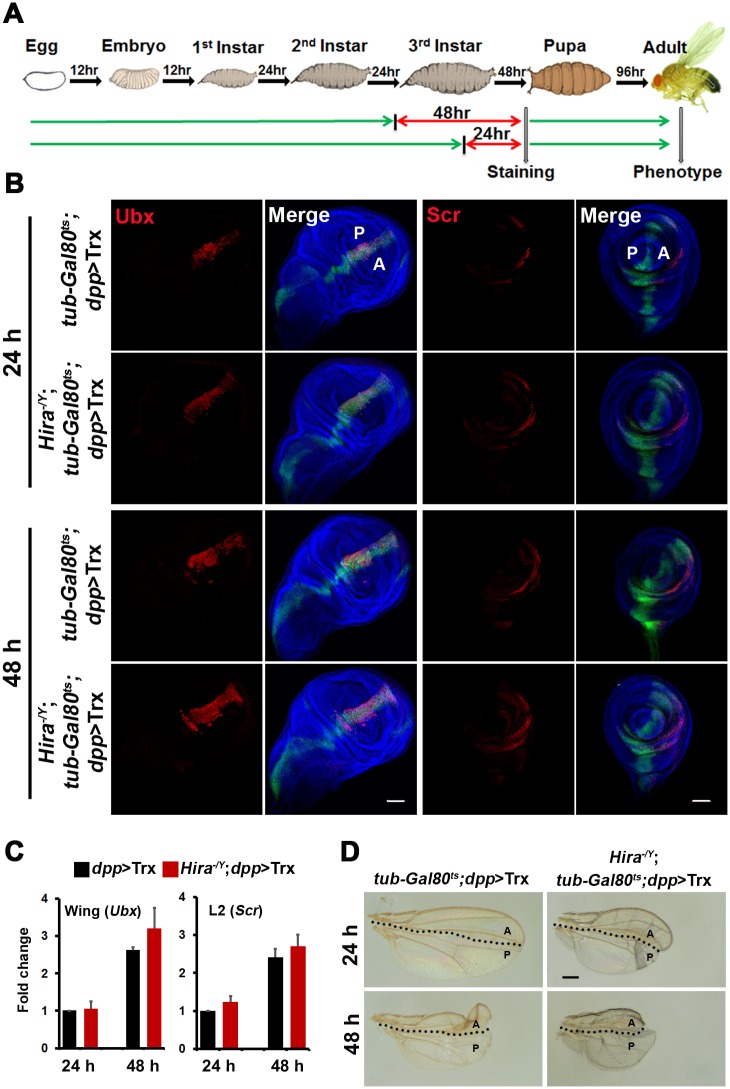
Fig 1. Tissue-specific activation of Hox genes by targeted Trx expression.
(A) Aberrant body parts induced by dpp-driven Trx (dpp>Trx). Top panel, wild-type (WT) adult with normal aristae (arrow head) and notum (arrow), dpp>Trx adult lacking aristae and notum. Middle panel, WT adult with sex comb on first leg only (L1, arrow), dpp>Trx adult with sex combs on all three legs (L1-L3). Bottom panel, WT adult with normal wings, dpp>Trx adult with severely reduced wings. Anterior (A) and posterior (P) compartments of wings and their boundaries are marked. Scale bars in all adult phenotype images represent 0.3 mm. (B) Induction of ectopic Hox proteins by dpp>Trx in imaginal discs. Leg and wing discs from third instar larvae were stained with anti-Scr and anti-Ubx antibodies (red), respectively. Ectopic Scr signals in L2/L3 discs and Ubx signals in wing discs are seen in dpp>Trx but not in WT samples. Discs are outlined by white dots. Scale bars in all confocal images represent 50 μm. (C-E) Occupancy of RNA polymerase II and tri-methylated histone H3 in Hox genes. Cross-linked chromatin from the second leg (C) and wing discs (D,E) of WT and dpp>Trx larvae were immunoprecipitated by different antibodies and quantified by qPCR for promoter-proximal and 3’ regions of different Hox genes. (C,D) Ser5P and Ser2P indicate specific phosphorylated isoforms of RNA-Pol II. (E) H3K4me3 and H3K27me3 indicate specific tri-methylation on histone H3. Bar graphs represent the averages ± SD of two separate ChIP experiments with two independent qPCR reactions. The y-axes represent the percentages of 1% input chromatin. P, promoter region; P1, first promoter; P2, second promoter; 3’, 3’ coding region; +0.1K, 100 bp downstream from TSS; +1K, 1 kb downstream from TSS.
The Hox gene Sex comb reduced (Scr) is normally expressed in the anterior compartment of the first leg (L1), but not the L2 or L3 discs, to give rise to the sex comb—a unique morphological marker of L1 in male adults. Consistent with the appearance of ESC on adult L2 and L3, dpp>Trx induced strong ectopic Scr signals (Fig 1B). In wing discs where the Ultrabithorax (Ubx) signal is normally absent, strong Ubx signals were induced in the central pouch region by dpp>Trx. Thus, dpp>Trx can alter the developmental fate of these discs via selective induction of key Hox genes. However, no ectopic Scr was detected in wing discs by dpp>Trx, nor was Ubx detected in L1/L2 discs. Moreover, we did not detect ectopic Scr or Ubx signals in eye-antenna discs. Thus, Trx is not capable of inducing Hox genes uniformly in these discs, indicating unique developmental restriction in different discs. We also noticed that not all Trx-expressing cells within the same disc can induce relevant Hox genes. For example, ubiquitous drivers such as actin>Trx could only induce Ubx in the wing pouch region and Scr in small regions of anterior L2/L3 discs (S1A and S1B Fig). Clearly, different parts of a given disc are not equally competent for activation of Hox genes.
Previous studies have shown that RNA-Pol II is paused at the proximal Ubx promoter in wing discs [18], raising the possibility of a general link between paused Hox promoters and developmental plasticity in different discs. To explore this possibility, we first examined the phosphorylation states of RNA-Pol II for Scr and Ubx promoters in leg and wing discs, respectively. Paused RNA-Pol II and actively elongating RNA-Pol II are preferentially phosphorylated at the CTD Ser-5 (Ser5P) and Ser-2 (Ser2P) residues, respectively. Using chromatin immuno-precipitation experiments (ChIP) for isolated discs, we found that the levels of Ser5P are much greater in proximal regions of two separate promoters (P1 and P2) of Scr than at 3’ distal region in L2 discs (Fig 1C and gene map in S2 Fig). Similarly, elevated Ser5P signals were observed for P1 and P2 promoters in L3 discs, albeit to a lesser extent (S3A Fig). Further analyses of L2 discs revealed low levels of Ser2P from P1 and P2 to 3’ distal regions, indicating the lack of active elongation. These results support that Scr promoters are in a paused state in L2/L3 discs. Under dpp>Trx induction, both Ser5P and Ser2P showed significant increases across these regions (Fig 1C). Clearly, paused RNA-Pol II in Scr promoters is readily stimulated into active elongation by dpp>Trx in L2/L3 discs.
Similar to Scr, stronger Ser5P, but not Ser2P, signals were found in the proximal region of the Ubx promoter than in the 3’ region in wing discs (Fig 1D). Again, dpp>Trx resulted in conversion from the paused state into active elongation with large increases of Ser2P in promoter and 3’ regions. We noted that Ser5P was not increased at the 3’ region of Ubx as at the 3’ region of Scr in L2 discs, presumably reflecting complete removal of Ser5P for large genes (>70 kb, Ubx; ~20 kb, Scr; see S2 Fig). We also measured the levels of Ser5P in these regions in L2 discs (where Ubx is inactive) and in L3 discs (where Ubx is constitutively expressed). Compared to L3 discs, low levels of Ser5P were found in both the promoter and 3’ regions of Ubx in L2 discs, suggesting that it is not in the paused state (S3A Fig). Interestingly, no ectopic Ubx signals were induced by dpp>Trx in L2 discs. Although both wings and L2 are derived from second thoracic segments, the inducibility of Hox genes by dpp>Trx apparently correlates with the paused state of Hox promoters rather than the tissue lineage.
We further examined the state of chromatin modification on Ubx. Interestingly, dpp>Trx did not cause a significant increase of H3K4me3 in the region immediately downstream (+0.1 kb) of the transcription start site (TSS) (Fig 1E). Instead, a substantial increase of H3K4me3 was observed in the region about 1 kb downstream (+1 kb) of the TSS. In addition, the levels of H3K27me3, a repressive modification, showed substantial decreases in both +0.1 kb and +1 kb regions. We noted that Trx was induced by dpp>Trx in small portions (< 20%) of wing and leg discs. Thus, our measurements based on whole discs undoubtedly represent underestimates for various changes. Taking this into consideration, we infer that dpp>Trx can induce substantial changes in the relative levels of H3K4me3 and H3K27me3 to facilitate conversion of the paused state to active transcription of Hox genes.
Hox activation is enhanced by mutations of histone chaperones
We next asked whether other factors contribute to conversion between the paused and active elongation states. Using the wing phenotype induced by dpp>Trx or A9>Trx, we examined the effect of knockdown of factors involved in transcription, chromatin remodeling and histone chaperones (S1 Table and S4 Fig). As expected for factors that promote transcriptional elongation, co-expression of RNAi for PAF1 or ASF showed dominant suppression of the wing phenotype caused by dpp>Trx. Unexpectedly, co-expression of RNAi for negative elongation factors such as components of the NELF (NELF-B, D, E) or DSIF complexes (Spt5) [43, 44] also resulted in rescue of the wing defect. Given that NELF knock-down results in reduction of paused RNA-Pol II and elevated nucleosome occupancy in the promoter-proximal region [45], it is conceivable that these effects undermine the efficacy of induction by dpp>Trx. In addition, the wing defect was partially rescued in nejp mutants, a hypomorphic allele of dCBP coding for a histone acetyltransferase. However, RNAi for ash1, a trxG, several positive elongation factors and chromatin remodeling factors failed to show any effect, suggesting that they might be functionally redundant or not critical under such conditions.
Canonical H3 is replaced by H3.3 in actively transcribed genes [46, 47], while H2A.Z is enriched in nucleosomes located immediately downstream of paused RNA-Pol II [23]. To examine whether they are involved in Hox regulation, we tested their interactions with Polycomb (Pc), an epigenetic repressor of Hox genes. Surprisingly, strong genetic interactions were observed (Table 1 and S5 Fig). The average number of extra sex comb teeth (ESCT) on L2/L3 of Pc4 heterozygotes was substantially enhanced from 0.8 to 3.7 by an H3.3B0 mutation and from 1.4 to 5.4 by an H2Av810 mutation. These results suggest that both H3.3 and H2A.Z are negatively involved in Hox gene regulation.
Table 1. Enhanced ESC phenotype by histone variant and chaperone mutations.
| Mutant | Pc-/Bal | Pc-/mutant |
|---|---|---|
| Hirassm | 0.6 ± 0.1 (120) | 4.4 ± 0.2 (120) |
| HiraHR1 | 0.7 ± 0.1 (120) | 3.4 ± 0.2 (120) |
| His3.3B0 | 0.8 ± 0.1 (120) | 3.7 ± 0.2 (120) |
| His3.3A2X1 | 1.2 ± 0.2 (120) | 1.4 ± 0.1 (120) |
| dom3 | 2.4 ± 0.2 (120) | 7.6 ± 0.2 (120) |
| dom9 | 2.7 ± 0.2 (120) | 4.3 ± 0.2 (120) |
| H2Av810 | 1.4 ± 0.2 (120) | 5.4 ± 0.2 (120) |
Females carrying mutations for histone variants or chaperones were crossed to Pc4 males. The numbers of extra sex comb teeth (ESCT) on both L2 and L3 of their progenies were scored. The averages and standard error of the mean (SEM) of ESCT per leg are shown. Total numbers of legs examined are indicated in parentheses. See S5 Fig for the range of % ESCT. Note that none of these histone chaperone or variant mutants show ESCT phenotype alone. Bal, Balancer.
We further examined the roles of HIRA, a H3.3 chaperone, and Domino (dom), a H2A.Z chaperone. Although Hira or dom mutants alone did not show ESCT phenotype in L2/L3 (S6A Fig), they significantly enhanced the ESCT phenotype of Pc4 mutants [48]. The average ESCT increased from ~0.6 to ~4.4 in the Hira background, and from ~2.4 to ~7.6 in the dom background (Table 1). Therefore, like the corresponding histone variants, these chaperones act as negative regulators of Hox genes. Reciprocal crosses also showed enhanced ESCT when Pc4 and other mutants were combined (S2 Table). Due to their stronger effects, we chose Hirassm and dom3 for further studies.
As expected, dpp>Trx-induced phenotypes became more pronounced in Hira or dom mutant backgrounds (Fig 2A). Adult wings from both Hira-/Y;dpp>Trx and dom-/+;dpp>Trx flies showed more severe size reductions than dpp>Trx alone. It is noteworthy that the anterior parts of wings were more severely affected than the posterior parts. Similar to the lack of ESCT phenotype, Hira and dom mutants did not show any wing defect (Fig 2A). However, when combined with overexpression of Trx, Hira and dom mutants increased the average ESCT for L2/L3 from 2.0/1.6 to 8.2/7.4 and to 6.1/5.2 in Hira and dom mutants, respectively (Table 2 and S6B Fig). More strikingly, about 90% of Hira or dom mutants showed strong arista-to-leg transformations, whereas only ~3% weak transformation with thin leg-like tissues was seen in the dpp>Trx line alone (Fig 2B).
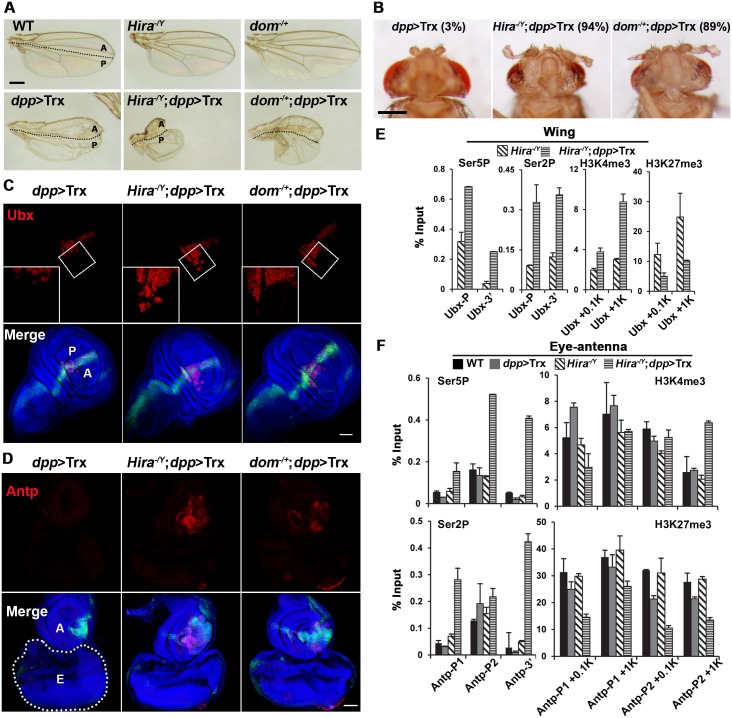
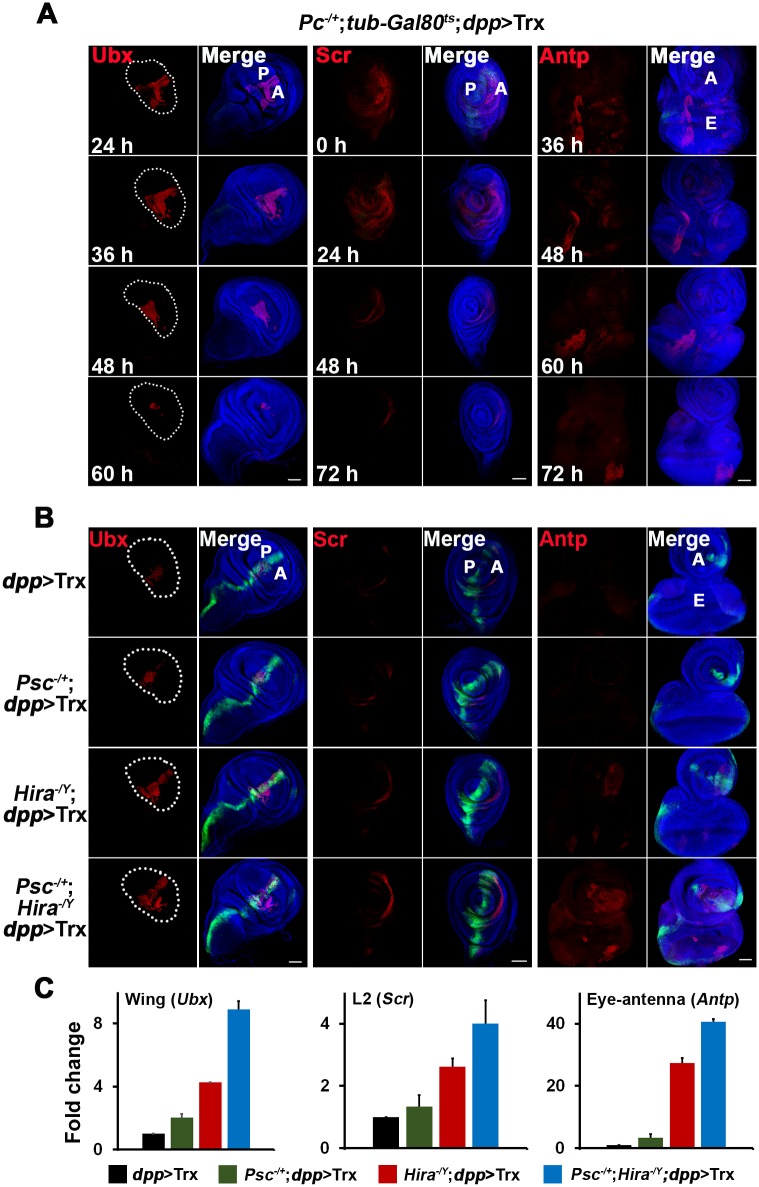
Fig 2. Enhancement of Trx-induced phenotypes in Hira and dom mutants.
(A) Wing phenotypes induced by dpp>Trx are enhanced in Hira hemizygotes or dom heterozygotes. (B) Arista phenotypes induced by dpp>Trx are enhanced in Hira hemizygotes or dom heterozygotes. The percentages of arista-to-leg transformations are indicated in parentheses. Only thin leg-like tissues are induced by dpp>Trx alone, while authentic leg tissues are seen in Hira or dom mutants. (C) Ectopic Ubx expression induced by dpp>Trx in wing discs is enhanced in Hira or dom mutants. Top panel, Ubx staining in wing discs (red). Based on GFP signals, the same disc regions are boxed. Enlarged (2.1X) images of boxed regions are shown as inserts. Lower panel, merged images of DNA (blue), Ubx staining (red) with A/P boundaries marked by dpp>GFP signals (green). (D) Ectopic Antp expression induced by dpp>Trx in eye-antenna discs is enhanced in Hira or dom mutants. Top panel, Antp staining in discs (red). No signal is detectable in the WT sample. Lower panel, merged images of DNA staining (blue), Antp staining (red) and dpp>GFP signals (green). Antenna and eye discs are marked by A and E, respectively. The outline of the eye disc is marked by white dots. (A-D) Experiments were carried out at 21°C. (E, F) Occupancy of RNA polymerase II and tri-methylated histone H3 in Hox genes. ChIP analyses were carried out as described in Fig 1C for chromatin samples prepared from wing (E) or eye-antenna (F) discs of different genotypes.
Table 2. Genetic interactions between dpp>Trx and chaperones.
| 2nd Leg | 3rd Leg | |
|---|---|---|
| dpp>Trx | 2.0 ± 0.1 (135) | 1.6 ± 0.1 (131) |
| Hira-/Y;dpp>Trx | 8.2 ± 0.1 (109) | 7.4 ± 0.1 (102) |
| dom-/+;dpp>Trx | 6.1 ± 0.1 (113) | 5.2 ± 0.1 (98) |
UAS-Trx females with or without chaperone mutations were crossed to dpp-Gal4 males. The ESCT on L2 and L3 were separately scored. The average and SEM are shown as described previously. Experiments were carried out at 21°C, which generally produced lower ESCT. See S6D Fig for distributions of % ESCT.
Consistent with more severe phenotypes in mutants, strong Ubx and Scr signals were detected in wider regions in wing and leg discs, respectively (Figs 2C and S6C). Using dpp>GFP to mark the A/P boundary, we found that ectopic Ubx signals extended beyond the A/P boundary in mutant discs, with more signals in the anterior compartment. These asymmetric patterns correlated with more severe deformation of anterior wings in these mutants. We also examined Hox gene expression in eye-antenna discs. Similar to dpp>Trx, there was little if any Ubx or Scr signals in mutant discs. However, strong Antennapedia (Antp) signals were detected in the region of antennal tissues corresponding to adult arista, providing a direct molecular basis for the arista-to-leg transformation in mutant adults (Fig 2D). Therefore, these chaperones and histone variants both appear to restrict the ability of Trx to activate Hox genes in tissue-specific manners.
We next examined the status of RNA-Pol II occupancy and histone H3 tri-methylation in Ubx and Antp genes in Hira mutant backgrounds. Both wing and eye-antenna discs were collected independently from Hira-/Y and Hira-/Y;dpp>Trx larvae and subjected to ChIP analyses. For the Ubx gene in wing discs, significant increases of Ser5P and Ser2P were induced by dpp>Trx in both promoter and 3’ regions (Fig 2E). These increases were accompanied by increases in H3K4me3 and decreases in H3K27me3 at the +0.1 kb and +1 kb regions. By and large, dpp>Trx induces similar changes in both WT and Hira backgrounds. However, we noted that either increases in H3K4me3 or decreases in H3K27me3 were more pronounced in Hira mutants, particularly in the downstream region.
In WT eye-antenna discs, the P2, but not P1, promoter region of the Antp gene showed relatively high levels of Ser5P and Ser2P (Fig 2F). However, only low levels of Ser5P and Ser2P were detected in the 3’ region, indicating a lack of active transcription. Thus, the P2 of Antp is poised in the paused state. Surprisingly, dpp>Trx failed to induce substantial increases of Ser5P or Ser2P in P1, P2 and downstream regions unless HIRA was inactivated. Thus, unlike Ubx in wing discs and Scr in L2/L3 discs, Antp induction by dpp>Trx is strictly controlled by HIRA in eye-antenna discs. Interestingly, Hira mutation alone was insufficient to cause any significant change in Ser5P or Ser2P of Antp promoters, suggesting that HIRA is not directly involved in modulation of RNA-Pol II. In addition, only low levels of Ser5P and Ser2P were detected in Scr and Ubx promoters under various genetic backgrounds (S3B Fig). These results further strengthen our earlier observation of a close relationship between inducibility and the paused state for Hox genes.
We further examined H3 tri-methylation on Antp in eye-antenna discs. In agreement with the lack of effects on RNA-Pol II occupancy, dpp>Trx alone failed to stimulate the level of H3K4me3 in +0.1 kb and +1 kb regions of both P1 and P2 promoters. In Hira mutants, however, a selective increase of H3K4me3 was detected in the +1 kb region of P2, but not other regions. In contrast, more uniform reductions were found for H3K27me3, albeit to different extents, in these regions. Thus the ability of dpp>Trx to alter the status of H3 tri-methylation is also restricted by HIRA activity in eye-antenna discs.
Lineage tracing of Hox activation
In addition to the Antp signals in antennal discs of Hira mutants, we noticed strong Antp signals in eye discs that are distant from the region that is under dpp-Gal4 induction, as marked by GFP signals (Fig 3A). Similarly, the ectopic Scr signals in L2/L3 discs were located in anterior regions that are distant from dpp>GFP in both WT and Hira backgrounds. Incompatible GFP and Hox patterns were also observed, albeit to a lesser extent, in anterior wing discs for Ubx. These Hox signals are hereafter referred to as Hira-dependent signals. Given the synergistic interactions between Hira and Pc—a key factor involved in epigenetic memory—we suspected that Hira-dependent signals might represent sustained Hox expression from earlier developmental stages. To test this possibility, we first traced the lineage of cells with active dpp enhancer at different stages. We exploited a lineage tracing strategy (G-TRACE) to mark cells with Ubi>GFP by removing FRT-flanked transcriptional termination signals residing in the 5’-UTR of Ubi>GFP via inducible UAS-FLP (S7A Fig) [49]. All descendent cells from these marked cells thus become continuously marked by Ubi>GFP. Additionally, cells with active dpp enhancer near the time of dissection were marked with RFP. Superimposition of these images enabled us to identify the region with active dpp enhancer at previous stages. Comparison of the location of Hox signals with superimposed images revealed that Hira-dependent signals clearly reside in cells with earlier lineages (Fig 3B).
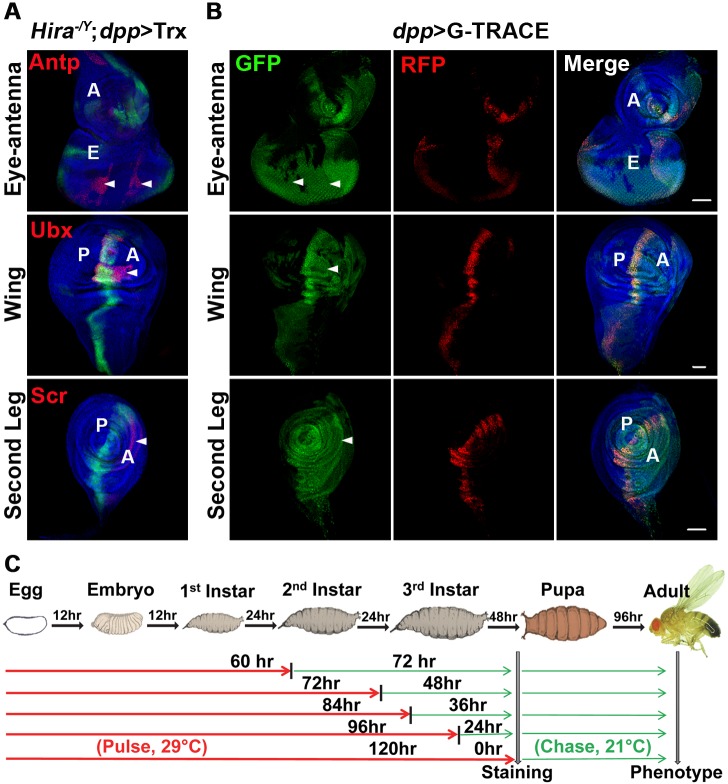
Fig 3. Lineage analyses of Hox-expressing cells that lack dpp>GFP.
(A) Incompatible Hox and GFP patterns in Hira mutants. Staining of Antp, Ubx and Scr in eye-antenna, wing and L2 leg discs, respectively, is shown in merged images with Hox staining (red), DNA staining (blue) and dpp>GFP signals (green). Regions showing Hox signals that do not match with dpp>GFP signals are indicated by arrowheads (Hira-dependent signal). Note that deviant Scr signals are also seen in the dpp>Trx background. (B) Comparison of cells with dpp-Gal4 activities at different stages. A G-TRACE strategy [49] was used to simultaneously mark cells with dpp-Gal4 activities throughout development by a FLP-dependent Ubi-GFP and cells with concurrent dpp-Gal4 activities by UAS-RFP in different discs (details in S7A Fig). Merged images of GFP (green), RFP (red) and DNA (blue) are shown. Arrowheads indicate regions with incompatible Hox expression in Fig 3A. (C) Protocol for pulse-chase experiments. A G-TRACE strategy was used for animals carrying a temperature-sensitive Gal80 (tub-Gal80ts). Embryos were raised at non-permissive temperature (29°C) for indicated periods (red line) until larval stages to allow cell marking by both Ubi-GFP and UAS-RFP. Subsequently, larvae were returned to permissive temperature (21°C, green line) to block Gal4 activity. The duration for 21°C was roughly estimated based on that for 29°C. The approximate developmental stages are indicated.
To further trace the developmental origin of these cells, we performed chase experiments with the introduction of a temperature sensitive Gal80 (Gal80ts) into G-TRACE lines to block Gal4 activity at defined stages. Animals were first raised at a non-permissive temperature of Gal80ts (29°C) to allow simultaneous induction of RFP- and FLP-dependent GFP, then shifted to a permissive temperature (21°C) at different time-points to block further expression of RFP and FLP until dissection at the third instar (Fig 3C). Comparison of the GFP- and RFP-marked regions from different time-points enabled us to map the region where dpp is temporarily active during different periods. As exemplified for leg discs (S7B Fig), RFP signals induced with either no chase or a 24-h chase were mostly located in regions that corresponded to the A/P boundary. Very little RFP signal was seen in the anterior part of the disc where Hira-dependent signals were detected. Conversely, strong GFP signals were observed in this region 48 h before dissection, strongly suggesting that Hira-dependent signals were induced by dpp activities from earlier stages. Indeed, dpp activities from these stages were sufficient to induce Scr signals and leg transformation (see below). Similarly, dpp activities were also evident from early stages in regions corresponding to Hira-dependent Ubx and Antp signals in wing and eye-antenna discs, respectively (S7B Fig).
Sustainable Hox expression in Hira mutants
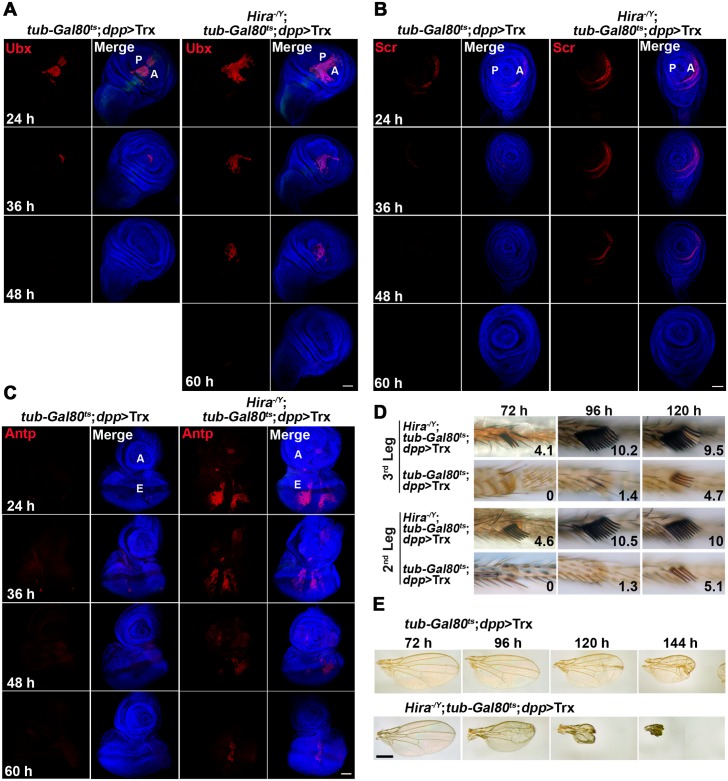
To ascertain the efficacy of dpp>Trx activity at early developmental stages, we examined Hox expression in animals treated under similar pulse-chase conditions (Fig 4A and 4B). Substantial amounts of Ubx and Scr signals could still be detected in WT discs after a 24-h chase, whereas very weak or no signals were seen after longer chases (36 h), indicating clearly that the effect of dpp>Trx activity during embryonic and early larval stages can’t sustain. However, Ubx and Scr signals were seen even after a 48-h chase in Hira mutants. Importantly, these signals appeared to match spatially with the Hira-dependent signals described earlier. Thus, both Ubx and Scr could be induced at earlier stages in Hira mutants. Remarkably, such induction was sustained for at least 48 h without continuous supply of Trx. A more striking contrast was observed in eye-antenna discs. In the WT background, dpp>Trx failed to induce any Antp signals. In contrast, a substantial amount of Antp signal was induced in mutant discs and they remained detectable after a 60-h chase (Fig 4C).
Fig 4. Maintenance of Hox expression in Hira mutants.
(A-C) Sustained Hox expression in Hira mutants. Pulse-chase experiments were performed with different chase periods as indicated (see Fig 3C). Individual staining of Ubx (A), Scr (B) and Antp (C) is shown for wing, L2 leg and eye-antenna discs, respectively. Merged images of Hox (red), DNA (blue) and GFP (green) are also shown. Note that only weak GFP signals can be seen after a 24-h chase. Compared to WT, Hox signals can be detected after longer chases in Hira mutants. (D, E) Enhanced adult phenotypes in Hira mutants. A similar pulse-chase protocol was adopted except that the chase was extended to eclosion. Hours indicate the duration of pulses. (D) Enhanced ESC phenotypes. The average numbers of sex comb teeth on L2 and L3 are indicated on the top. (E) Enhanced wing phenotypes in Hira mutants.
Next, we examined the adult phenotypes caused by these treatments. Although no obvious effect was found in adults treated with a 48-h pulse, extended treatment for 72 h resulted in appearance of ESCT on both L2 and L3 in mutants (avg. ESCT 4.6 and 4.1 for L2 and L3, respectively) (Table 3 and Fig 4D), but not in WT flies. Remarkably, ESCT on L2 and L3 of mutant flies produced by a 96-h pulse were more pronounced than WT flies under the same treatment (10.5 and 10.2 vs 1.3 and 1.4 for L2 and L3, respectively) or flies with continuous dpp>Trx throughout development (avg. ESCT ~8.1), and almost approached sex comb tooth number for L1 (10~12). The induction of almost complete transformation of posterior legs by the pulse clearly illustrates the developmental impact of sustainable Scr expression.
Table 3. ESC phenotypes induced by dpp>Trx pulses.
| Trx Induced | 2nd leg | 3rd leg | ||
|---|---|---|---|---|
| dpp>Trx | Hira-/Y; dpp>Trx | dpp>Trx | Hira-/Y; dpp>Trx | |
| 48 h | 0 (80) | 0.8 (79) | 0 (82) | 0 (83) |
| 72 h | 0 (89) | 4.6 ± 0.1 (87) | 0 (81) | 4.1 ± 0.1 (82) |
| 96 h | 1.3 ± 0.1 (85) | 10.5 ± 0.1 (82) | 1.4 ± 0.1 (84) | 10.2 ± 0.1 (81) |
| 120 h | 5.1 ± 0.1 (84) | 10 ± 0.1 (80) | 4.7 ± 0.1 (86) | 9.5 ± 0.2 (83) |
| 148 h | 6.1 ± 0.2 (71) | 9.1 ± 0.2 (48) | 6.2 ± 0.1 (76) | 8.6 ± 0.2 (47) |
Reductions in wing size began to appear in mutant flies treated with a 96-h pulse. Unlike the ESCT phenotype, wing phenotypes continued to deteriorate with longer pulses (Fig 4E). Similarly, a more pronounced arista-to-leg phenotype was also found in mutants with extended pulse period (S3 Table). The frequency of arista-to-leg transformations increased from ~3% to ~40% by prolonging induction from 96 to 168-h. Clearly, the effective period of phenotype induction for arista and wing discs is different from that of leg discs. Such differences probably reflect unique developmental properties of these discs.
Possible causes for sustainable Hox expression
The stronger effects observed in Hira mutants might result from several possibilities. For instance, much more Hox signals might be induced in Hira mutants. To examine this possibility, we compared the levels of Ubx and Scr induced by short pulses of dpp>Trx in WT and mutant discs. Larvae at early or late second instar were shifted from 21°C to 29°C to inactivate Gal80ts for 24 or 48 h until dissection (Fig 5A). In both cases, we observed comparable levels of Ubx and Scr signals in WT and mutant discs by immunostaining and quantitative RNA measurement (Fig 5B and 5C). When these larvae were shifted back to 21°C to complete morphogenesis, emerging adults showed marked differences in wing and leg phenotypes. Mutant larvae receiving either 24 h or 48 h pulses showed more severe reductions in wing size (Fig 5D). Similarly, ESCT numbers on L2 and L3 of mutants was significantly greater than those of WT adults (Table 4). These results strongly argue against the possibility that Hira mutation might directly affect the level of Hox induction. Instead, they suggest that higher levels of Hox signals and stronger phenotypes in Hira mutants may be attributed to cumulative effects of sustainable Hox expression during development.
Fig 5. Efficacy of short dpp>Trx pulses.
(A) Protocol for pulse experiments. Animals carrying tub-Gal80ts;dpp>Trx were raised at 21°C until late second instar or early third instar (green line) and shifted to 29°C for 24 h or 48 h (red line). These larvae were either dissected or returned to 21°C until eclosion. (B) Induction of Hox expression by short dpp>Trx pulses. After 24 or 48 h of dpp>Trx pulses, wing and L2 leg discs were stained for Ubx and Scr (red), respectively. For each pulse, comparable levels of Hox expression were induced in WT and Hira mutants. (C) Quantitative RNA measurement. Reverse transcription and qPCR were performed for RNA samples isolated from wing and L2 discs. Relative mRNA levels are shown after being normalized to corresponding Rpl32 mRNA expression. Means ± SD from two separate experiments with triplicate are shown. (D) Enhanced wing phenotypes by short dpp>Trx pulses in Hira mutants. Malformed adult wings induced by 24 h or 48 h dpp>Trx pulses are shown for WT and Hira mutants.
Table 4. ESC phenotypes induced by short dpp>Trx pulses.
| Trx Induced | 2nd leg | 3rd leg | ||
|---|---|---|---|---|
| dpp>Trx | Hira-/Y; dpp>Trx | dpp>Trx | Hira-/Y; dpp>Trx | |
| 24 h | 0.5 ± 0.1 (50) | 2.0 ± 0.2 (50) | 0.2 ± 0.1 (50) | 1.0 ± 0.1 (50) |
| 48 h | 4.2 ± 0.1 (50) | 6.0 ± 0.3 (50) | 3.2 ± 0.2 (50) | 6.0 ± 0.2 (50) |
It is noteworthy that although there were only modest increases in Scr signals from the 24 to 48-h treatment (Fig 5B), ESCT showed much greater increases. For example, ESCT number for L2 increased from 2.0 to 6.0 in Hira mutants and from 0.5 to 4.2 in WT animals (Table 4). These abrupt increases strongly suggest that the period between 24 and 48 h prior to dissection is critical for the developmental switch of posterior legs, further supporting our earlier observation that continuous dpp>Trx is not necessary for extensive leg transformation (Fig 4D).
Next, we examined whether sustainable Hox expression could result from the persistence of Hox or Trx proteins. The rapid decay of simultaneously-induced GFP signals argues against this possibility. To seek direct evidence, we used pulse-chase experiments to examine the persistence of Antp and Scr, which showed more prominent Hira-dependent signals, and Trx. These proteins were induced directly from UAS lines by dpp>Gal4 in WT or Hira mutant backgrounds at 29°C and then chased for various periods at 21°C (Fig 3C). In the case of Scr, induced signals disappeared almost completely in L2 discs within a 12-h chase (S10A Fig). Little, if any, Antp signal was detected in eye-antenna discs following an 18-h chase (S10B Fig). In both disc types, induced Trx signals became almost indistinguishable from endogenous Trx following a 6-h chase (S11 Fig). Clearly, the Hox signals observed after a 48-h chase cannot be attributed to the persistence of these proteins.
Interaction between Hira and PcG
Our observation that Hira mutation could enhance ESCT phenotypes of the Pc mutation suggested that the sustainable Hox expression in Hira mutants might be mechanistically related to the epigenetic memory conferred by PcG repressors (Table 1). To explore this possibility, we examined whether Pc mutation could also enhance Hox induction by dpp>Trx in a similar manner. Compared to levels in the WT background, much more Scr, Ubx and Antp signals were induced by dpp>Trx in Pc4 mutant discs (S12A Fig). Moreover, the overall patterns of Hox signals were similar to those observed in Hira mutants. Consistently, severe phenotypes such as arista-to-leg and ESC were also seen (S12B Fig). We then monitored the sustainability of Hox expression in Pc4 mutants in pulse-chase experiments. Ubx signals remained detectable after a 60-h chase in Pc4 mutants, while Scr and Antp signals could last for at least 72 h in L2 and eye-antenna discs, respectively (Fig 6A). Similar to Hira mutants, a significant portion of Hox signals were located outside the region where dpp>GFP was normally seen. These results indicate that Hox genes induced from earlier stages can also be sustained in Pc4 mutants.
Fig 6. Cooperativity between HIRA and PcG.
(A) Sustained Hox expression in Pc mutants. The protocol described in Fig 3C was used for pulse-chase experiments in Pc4 mutants. Hours indicate the duration of chases before dissection. The staining of Ubx, Scr and Antp in wing, L2 leg and eye-antenna is shown, respectively. Ubx signals are detectable after a 60-h chase, whereas Scr and Antp signals can still be seen after 72 h chases. Wing pouch is outlined by dots. (B) Enhanced Hox expression in Hira and Psc double mutants. Merged images of GFP (green), Hox (red) and DNA (blue) are shown. Hox expression induced by dpp>Trx in the following genotypes is compared: WT, Psc1, Hirassm, and Hirassm;Psc1. (C) Enhanced RNA levels in Hira and Psc double mutants. RT and qPCR analyses were carried out as described in Fig 5C for Ubx, Scr and Antp in wing, L2 and eye-antenna discs, respectively. (B, C) Experiments were carried out at 21°C.
We further asked whether the genetic interaction between Hira and Psc—a member of the PcG repressors—could be manifested at larval stages. As expected, introduction of a Psc mutation resulted in more Ubx, Scr and Antp signals than dpp>Trx alone. Importantly, a combination of both Psc and Hira mutations resulted in even more Ubx, Scr and Antp signals in broader areas and higher RNA levels (Fig 6B and 6C), supporting a common role in Hox repression.
H3.3 is required for restoration of the inactive state
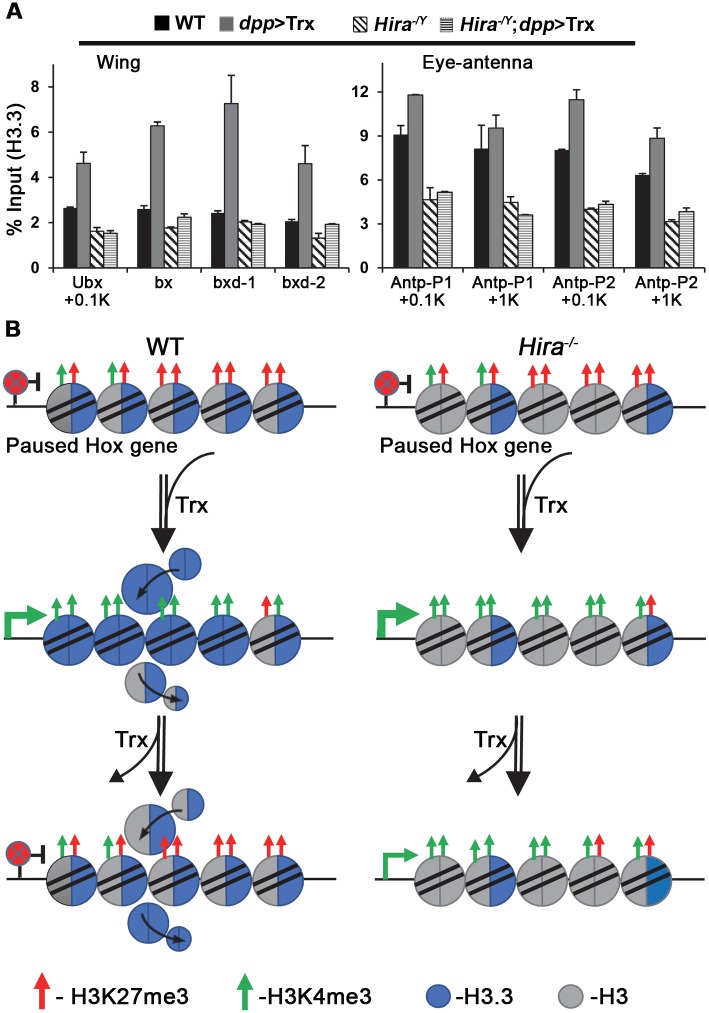
As a H3.3 chaperone, the main effect of HIRA on chromatin should be reflected by modulation of the level of H3.3. Using a constitutively active Hsp83-H3.3 (Flag) transgene introduced into different genetic backgrounds, we measured the levels of H3.3 on Ubx and Antp in wing and eye-antenna discs, respectively (Fig 7A). In wing discs, we monitored four regions, including the promoter-proximal region (+0.1 kb), an intragenic regulatory region at ~+25 kb (bx) [50, 51], and two upstream regulatory regions (bxd-1, bxd-2) at ~-28 kb and ~-29 kb [52, 53]. In the WT background, the level of H3.3 was substantially increased by dpp>Trx in all regions. As expected, relatively lower levels of H3.3 were detected in the promoter-proximal region of Hira mutants. However, no significant increase of H3.3 was induced by dpp>Trx in mutant discs, despite higher levels of RNA-Pol II and Ubx expression (Fig 2C and 2E). Compared to wing discs, relatively minor increases of H3.3 were induced by dpp>Trx in the +0.1 kb and +1 kb regions of the two Antp promoters in WT eye-antenna discs where no ectopic Antp signals were found. Similar to the Ubx promoter, the low levels of H3.3 detected in Antp promoters were not affected by dpp>Trx in mutant discs. Again, this was in sharp contrast to the induction of stronger Antp signals (Fig 2D). Although our results show that both H3.3 and HIRA are negatively involved in Hox gene regulation, the lack of a simple relationship between the level of H3.3 and Hox expression is inconsistent with a direct role in repression. Alternatively, H3.3-enriched regions have been recently reported to undergo faster turnover of chromatin marks [54, 55]. Therefore, our observations suggest that the low level of H3.3 in Hira mutants may defer the dynamic switch from active chromatin state to repressed state (see Discussion).
Fig 7. The involvement of H3.3 in maintenance of active Hox genes.
(A) Reduced H3.3 on Hox genes of Hira mutants. ChIP assays were performed using anti-flag antibody for wing (left) and eye-antenna (right) discs expressing H3.3-Flag in different genotypes. In addition to promoter-proximal regions of Ubx and Antp (Figs 1E and 2F), the levels of H3.3 in three Ubx regulatory elements (bx, bxd-1, bxd-2) were measured (S2 Fig). Bar graphs represent the average ± SD of two separate ChIP experiments with two independent qPCR reactions. (B) A model for the dynamic conversion between paused and active states of Hox genes. Hox promoters are poised in the paused state in imaginal discs with flexible developmental potential. Relatively high levels of H3K27me3 over H3K4me3 is maintained in these genes. Introduction of extra doses of Trx by dpp>Trx shifts the equilibrium towards relatively higher levels of H3K4me3, resulting in conversion of the paused state to the active state. Concomitantly, the level of H3.3 is increased. The high level of H3.3 facilitates the replacement of active H3K4me3 marks after Trx withdrawal. Consequently, the active Hox promoters are rapidly reverted to the paused state. With less H3.3 or HIRA, the replacement of active H3K4me3 marks becomes less efficient. Accordingly, the active state of Hox genes can be maintained for longer periods. The relative abundances of H3K27me3 and H3K4me3 are indicated by the numbers of red and green arrows, respectively. Nucleosomes containing H3.3 or H3 are indicated by blue or grey circles, respectively. Hybrid nucleosomes containing both H3.3 and H3 are also indicated.
Discussion
In this study, we show that the developmental restriction imposed on pluripotent imaginal tissues can be partially lifted through conversion of pre-existing transcriptional paused state of Hox genes into active elongation by simply increasing the level of a H3K4 methyltransferase. Furthermore, these developmental and molecular switches are regulated in tissue-specific manners by dynamic modulation of chromatin.
Mounting evidence suggests that the developmental potential of pluripotent progenitor cells is largely restricted by developmental regulators that are poised at a transcriptionally inactive paused state [1, 3, 4]. Our results support this view and extend it further with additional insights. We found that Ubx, Scr and Antp are poised at different transcriptional states in pluripotent imaginal discs. For example, Ubx possesses abundant RNA-Pol II Ser5P in the promoter-proximal region in wing discs, but little, if any, in L2 discs or eye-antenna discs. Although all these discs are formally considered inactive for Ubx expression, only wing discs exhibit features characteristic of the paused state. Similarly, Scr is poised at the paused state in L2/L3 discs, but not in eye-antenna or wing discs (S3B Fig). In contrast, Antp is kept at the paused state in eye-antenna discs. Thus, other than the paused state, an additional inactive state may exist. We refer to it as the “void state” for the lack of paused RNA-Pol II. Apparently, inactive Hox genes are in either paused or void states.
The paused state, rather than the void state, of Hox genes appears to correlate with the tendency of a given disc to adopt alternative developmental paths in animals with ectopic Hox expression [41, 42]. This relationship is further strengthened by our finding that dpp>Trx can induce similar developmental changes by converting the paused state of a specific Hox gene into the active state. In all cases examined, however, the void state remains refractory. Therefore, we suggest that developmental restriction can be effected by two mechanisms distinguishable by the presence or absence of paused RNA-Pol II of Hox genes. A given disc may adopt an alternative developmental path when the paused state of relevant Hox genes can be converted into the active state. In contrast, disc development becomes more restricted when Hox genes are in the void state.
It is generally believed that the paused state of Hox genes is stably maintained by PcG repressors through high levels of H3K27me3, a repressive histone mark [12, 56]. Our results clearly show that, for many discs, this state can be readily destabilized by an extra dose of Trx that gives rise to increases of active methylation marks such as H3K4me3. Concomitantly, the level of H3K27me3 is reduced. Although the direct cause of H3K27me3 reduction is unclear, the combined effects apparently results in greater increases of the relative ratio between active marks and repressive mark. Consequently, the paused state is converted into the active state. Conceptually, the paused state can be stably maintained by a constant ratio between functionally antagonistic marks. In support of this view, our results show that dpp>Trx fails to convert the paused state of Antp in eye-antenna discs in the absence of significant changes to these marks.
In addition to canonical histones, our results further show that non-canonical histones play important roles in maintaining the paused state of Hox genes, most evidently in eye-antenna discs. This role was initially revealed by the observation that mutations of two histone variants, H2A.Z and H3.3, and corresponding chaperones can cause substantial enhancement of various phenotypes mediated by ectopic Hox expression. Detailed analyses of Hira that encodes a H3.3 chaperone provided strong support to suggest that the enhanced Hox expression largely results from a prolonged active state via compromised nucleosome dynamics in Hira mutants. First, the extra Hox signals induced in the Hira background do not coincide with GFP signals induced by dpp>Gal4 (Fig 3A). Since GFP has a short half-life, its signals should match closely with the current activity of dpp>Gal4. Since the half-life of Trx is also short (~6 h), these deviant Hox signals, particularly for Antp, cannot be attributed to residual Gal4 or Trx in GFP-negative cells. Second, lineage tracing experiments revealed that cells showing deviant Hox signals may arise from those with active dpp>Gal4 as early as 48 h before the appearance of GFP-positive cells during the third instar. Since Hox signals can only last for about 12 h, these signals do not correspond to signals induced at earlier stages. Instead, they should result from continuous gene expression. Third, pulse-chase experiments showed that Hox genes, more extensively for Scr and Antp, are expressed for longer periods in mutant discs. For example, strong Antp signals were induced in mutant eye-antenna discs and remained obvious after a 48-h chase, whereas no signal was detected in WT discs (Fig 4C). Furthermore, we showed that comparable amounts of signals were induced in WT and mutant discs immediately after short pulses of dpp>Gal4 (Fig 5B). These results enabled us to deduce that the active state of Hox genes induced temporally by dpp>Gal4 can be maintained for longer periods in Hira mutants and that continuous Hox expression results in stronger cumulative effects.
Furthermore, our studies provide evidence to support a mechanistic link between the maintenance of the active state and the epigenetic memory conferred by trxG and PcG regulators [11, 57]. First, Hox expression was enhanced and maintained for longer periods in Pc4 mutants in a manner similar to that observed in Hira mutants. Second, strong genetic interactions were observed between Hira and PcG mutants (Pc4 and Psc1). Third, accompanied by Antp induction in mutant eye-antenna discs, the relative levels of H3K4me3 and H3K27me3 showed dynamic modulation. In light of the recent finding of physical association between HIRA and PcG [27], HIRA may be recruited by PcG. Previous studies have shown that H3.3 is enriched in chromatin regions that undergo rapid turnover to facilitate remodeling of chromatin landscapes [25, 58, 59]. Thus, the reduced levels of H3.3 observed in Hira mutants may slow down the turnover of pre-existing histone marks. Accordingly, H3K4me3 marks induced by dpp>Trx can be maintained much longer to allow continuous Hox expression.
For the conversion of the paused state to the active state, we propose a simple model for different situations (Fig 7B). At first, a Trx pulse can induce sufficient histone modification to destabilize the pre-existing balance between H3K4me3 and H3K27me3, and consequently promote active elongation of Hox genes through incorporation of more H3.3. In the absence of constant supply of Trx, the rapid turnover of active marks mediated by H3.3 results in reversion of the active state to the inactive paused state. However, with reduced levels of H3.3 or HIRA chaperone, the turnover of active marks becomes less efficient. Therefore, the active state can be maintained even after dpp>Trx is no longer expressed in those cells. We note that the unique property of each disc may render different responses to various regulatory factors. In addition, the presence of multiple cell lineages may result in variable responses in different parts of the same disc [6]. These complexities must be taken into consideration when cell-based models are being compared.
Recent findings that Trx is more active in catalyzing H3K4 mono-methylation than di- or tri-methylation suggest that our observations of increased H3K4me3 might not result solely from Trx activities [60–62]. It is possible that dSet1, the major Drosophila H3K4 trimethyltransferase [60, 61], is recruited to Hox genes and contributes significantly to the H3K4me3 increase. In addition, the ability of Trx to promote acetylation of H3K27 might also help antagonize the pre-existing balance between different methylation marks [13, 62]. It will be interesting to test these possibilities directly.
Materials and Methods
Fly stocks
P(GSV6)GS12194/TM3 (UAS-Trx) was obtained from DGRC, Kyoto. yw;dom3/SM6a, yw;dom9/CyO, yw;Spt5MGE-3/SM1, w;nejP/FM7c, w;ash122/TM6C, Psc1/CyO, Pc4/TM3, w;His2Av810/TM3, P{Gal4}A9, w;dppblk-Gal4/TM3 [40], en-Gal4/CyO, act-Gal4/TM3, UAS-Antp/TM3, UAS-Scr/CyO, and tub-Gal80ts/CyO were obtained from BDSC, Indiana. RNAi lines (listed in S1 Table) were obtained from VDRC, Vienna. Hirassm/FM7i [29], HiraHR1/FM7c [33], ywHis3.3B0, His3.3A2X1/CyO [34], Hsp83-H3.3(Flag) [63] and G-TRACE lines consisting of UAS-RedStinger (RFP) UAS-FLP Ubi-p63E(FRT.STOP)-Stinger (GFP)/CyO [49] were obtained from individual labs as indicated.
Genetic crosses
Flies were raised at 25°C on standard food unless otherwise mentioned. Homozygous UAS-Trx virgin females were crossed to dpp-Gal4/TM6B,Tb males at 25°C unless otherwise specified. Adult phenotypes of dpp-Gal4/UAS-Trx were scored three days after eclosion. In pulse-chase experiments, virgin females carrying UAS transgenes with or without mutations were crossed to tub-Gal80ts;dpp-Gal4 males and kept at either 29°C or 21°C for indicated periods to inactivate or activate Gal80ts, respectively. Flies were transferred every 12 h. The phenotypes wing-to-haltere, arista-to-antenna and sex comb number on L2 and L3 were assessed three days after flies hatched. For genetic interactions, virgin females heterozygous for Hirassm, HiraHR1, H2Av810, dom9, dom3, His3.3A2X1, His3.3B0 mutants were crossed to Pc4 heterozygous males. The number of sex comb teeth on L2 and L3 of their male progeny were counted as described previously [64]. The statistical analyses were performed by R language (http://www.r-project.org/). One-sided Wilcoxon rank sum test was used to compute the statistical significance for the increased sex comb number.
Immunostaining of discs
Imaginal discs isolated from third instar larvae carrying different genotypes were fixed by 3.7% formaldehyde in PBS for 20 min. Discs were washed three times with PBT (0.15% Triton X, 0.2% NP40, 0.2% Tween 20 in PBS) for 5 min. After blocking in a PBT solution containing 3% bovine serum albumin (Sigma-Aldrich) and 5% goat normal serum (Vector Laboratories) for 1 h, discs were incubated overnight at 4°C with primary antibodies. Following six washes with PBT for 5 min, discs were incubated in the dark with fluorescently-labelled secondary antibodies for 1 h at room temperature and washed three times for 5 min with PBT. Samples were then stained with Hoechst 33258 (0.5 μg/mL) (Molecular Probe) for 15 min, followed by six washes for 5 min with PBT. Before mounting in Vectashield (Vector Laboratories), samples were soaked in 50% glycerol in PBS for 30 min. As for primary antibodies, culture supernatants were used. Anti-Ubx (FP3.38; 1:2) and anti-Scr (6H4B; 1:2) supernatants were prepared in our lab [64], whereas anti-Antp (4C13; 1:4) was obtained from Developmental Studies Hybridoma Bank. The secondary antibodies were conjugated with Rhodamine Red-X and Cy5 for the anti-mouse and anti-rabbit antibodies, respectively (Jackson ImmunoResearch). All confocal images were taken using an LSM510Meta microscope (Zeiss) at 20X magnification. Images were aligned and processed using Adobe Photoshop CS3.
Chromatin immunoprecipitation assays (ChIP)
ChIP of imaginal discs was performed as described previously [65] with some modifications. Details are available in the supplementary information (S1 Text). Specific antibodies for Ser5P (H14, Covance), Ser2P (H5, Covance), rabbit H3K4me3 (ab8580, Abcam), rabbit H3K27me3 (39156, Active motif), and anti-Flag (F-3165, M2, Sigma) were used for ChIP experiments. Quantitative PCR was performed in triplicate using SYBR Green at standard settings (ABI 7500, Applied Bioscience). Data are represented as percentage DNA present in input material. Bar graphs in Figs 1, 2 and 7 represent the averages ± SD values obtained from two independent experiments repeated twice.
RT-qPCR
Total RNAs from 20–30 wing, second leg or eye-antenna imaginal discs were prepared from L3 of different genotypes by RNeasy protocol (QIAGEN). After reverse-transcription, cDNAs were measured by quantitative PCR according to the vendor’s protocol, using SYBR Green at standard settings (ABI 7500, Applied Bioscience). Two separate experiments with each in triplicate were performed. Primers are listed in S4 Table.
Supporting Information
(A) Restricted Ubx induction in wing discs. Four Gal4 lines (actin, dpp, A9, engrailed) were used to drive Trx and GFP simultaneously. Their effects on GFP and Ubx expression in discs and adult wings are shown. GFP (green) and Ubx signals (red) are shown in separate panels or together with DNA staining (blue). Adult wings resulting from dpp-, A9-, and engrailed-driven Trx are shown. actin-driven Trx causes pupal lethality. A schematic representation of anterior/posterior (A/P) and dorsal/ventral (D/V) compartments and their boundaries is shown. White dots outline the pouch region. Scale bars in this and subsequent confocal images represents 50 μm. (B) Restricted Scr induction in leg discs. The patterns of Scr induction in L1, L2 and L3 leg discs from actin>GFP (left panel) or actin>Trx, GFP (right panel) larvae are compared. Ectopic Scr signals are seen in regions forming adult tarsal segments. Individual or merged images of Scr (red), GFP (green) and DNA (blue) are shown. Anterior/Posterior (A/P) compartments are indicated.
(TIF)
Both Scr and Antp contain two separate promoters and start sites. The direction of transcription is indicated by rightward arrows. The non-coding and coding exons are indicated by grey and orange boxes, respectively. Introns are indicated by thin lines. Approximate locations of primers used in this study are indicated by red (ChIP) or blue bars (RT-qPCR) below the gene map. Magnified views of the TSS regions of Ubx and Antp are shown. P1 and P2 correspond to the proximal regions of promoters 1 and 2, respectively. +0.1K and +1K correspond to regions about 100 bp and 1 kb downstream of the TSS, respectively. 3’ corresponds to the gene terminus. bxd-1, bxd-2 and bx correspond to Ubx distal regulatory elements. P1, P2 and 3’ end primers were used to measure RNA-Pol II occupancy, whereas other primers were used to measure tri-methylated H3 or H3.3. Primer sequences are listed in S4 Table. The upstream-most start sites are used as the reference point for the scales shown above genes.
(PDF)
(A) Analyses of Ser5P enrichment in Scr or Ubx in different leg discs. ChIP analyses of promoter and 3’ regions of Scr or Ubx were performed with a Ser5P antibody for samples prepared from WT L2 (left) or L3 discs (right). (B) Analyses of Ser5P and Ser2P enrichment of Scr or Ubx in wing or eye-antenna discs. Chromatin samples prepared from wing (left) or eye-antenna discs (right) of the following genetic backgrounds were subjected to ChIP analyses with antibodies against Ser5P or Ser2P: WT, dpp>Trx, Hira-/Y, Hira-/Y;dpp>Trx. The bar graphs represent the averages ± SD of two independent ChIP experiments, each with two separate qPCR reactions. The scale on the y-axis corresponds to 1% of input chromatin. Analyzed regions are described in S2 Fig.
(PDF)
Based on the wing phenotype induced by dpp>Trx, a modifier screening was conducted with co-expression of the RNAi lines listed in S1 Table. In no case was the small wing induced by dpp>Trx further reduced. The effects of RNAi lines were evaluated by the degree of recovery to full wing size as indicated by the number of “+” signs. The “-” sign indicates no effect. Scale bar, 0.3 mm.
(PDF)
Transformation of adult L2/3 to L1 was scored in a heterozygous Pc background with other mutants. Each bar represents the percentage of ESCT transformation with respect to the number of ESCT (x-axes). Note that the percentage transformation increased with increased ESCT when Pc and other mutants were combined. p- and W-value were calculated based on one-sided Wilcoxon rank sum test.
(TIF)
(A) Adult leg phenotype. WT, Hira and dom adults mutants exhibit sex combs only on the first leg (L1, arrow). (B) Extra sex combs on adult legs. The sex combs induced in adult L2 (upper) and L3 (lower) by dpp>Trx are enhanced in Hira and dom mutants. The average number of sex comb teeth is shown and the numbers of legs scored are in parentheses. (C) Ectopic Scr expression in leg discs. Upper panel, Scr expression induced in L2 discs by dpp>Trx in WT, Hira or dom mutant backgrounds. Lower panel, merged images for Scr (red), GFP (green) and DNA (blue) as described previously. (B, C) Experiments were carried out at 21°C. (D) Bar graph depicts phenotypic variation as described previously.
(TIF)
(A) Schematic diagram of components employed in the G-TRACE strategy [49]. UAS-RFP and UAS-FLP are directly under the control of dpp-Gal4, which is active primarily in larval discs. Nuclear EGFP (nEGFP) is linked to the Ubi-p63E promoter with a cassette containing a transcriptional termination signal flanked by FRT. Through the removal of FRT by FLP recombinase, rearranged Ubi-p63E-nEGFP is constitutively activated, thus marking all descendent cells. (B) Lineage tracing of dpp-Gal4 in imaginal discs. Experiments were performed in the presence of tub-Gal80ts as described in Fig 3C. Gal80ts is inactivated for different lengths at 29°C and then re-activated by shifting to 21°C for indicated hours. dpp-Gal4 becomes activated reciprocally. Due to its rapid degradation, strong RFP signals can be seen only in cells that remain active for dpp-Gal4. In contrast, all cells that have ever been active for dpp-Gal4 are marked by GFP signals. Hours refer to the duration of chase described in Fig 3C. Individual RFP (red), GFP (green) or a merge panel with DNA (blue) are shown for leg (left), wing (middle) and eye-antenna discs (right). Arrowheads indicate regions with deviant Hox expression in Fig 3A. Note that most GFP signals appear during the 48–72 h interval.
(TIF)
Bar graphs are as described previously. Hours indicate the duration of Trx pulse (see Fig 3C). At any given time points, the sex combs induced in adult L2 and L3 by dpp>Trx are enhanced in Hira mutants.
(TIF)
Bar graph is as described previously. Hours indicate the duration of Trx pulses (see Fig 5A). For both 24 and 48 h, the sex combs induced in adult L2 and L3 by dpp>Trx are enhanced in Hira mutants.
(TIF)
(A) Stability of ectopic Scr in L2 discs. Pulse-chase experiments were performed for animals carrying tub-Gal80ts; dpp-Gal4, UAS-Scr in WT (left) or Hira backgrounds (right) with different lengths of chase as indicated in Fig 3C. Scr staining in L2 discs is shown alone or together with DNA. Note that similar results were observed in WT or Hira mutants. (B) Stability of ectopic Antp in eye-antenna discs. Similar pulse-chase experiments were performed for animals carrying tub-Gal80ts; dpp-Gal4, UAS-Antp. Antp staining in eye-antenna discs is shown alone or together with DNA (blue). In all cases, ectopic Scr and Antp signals disappeared within 12 h of chase.
(TIF)
Experiment was carried out as in Fig 3C. Pulse-chase experiments were performed for animals carrying tub-Gal80ts; dpp-Gal4; UAS-Trx, GFP. Duration of the chase is indicated. Trx (red) and GFP (green) signals in wing, eye-antenna and L2 discs are shown alone or together with DNA (blue). Note that endogenous Trx expression is uniform in all discs and that induced Trx signals disappear faster than GFP.
(TIF)
(A) Ectopic Hox expression in Pc4 heterozygote mutants. Antp, Ubx and Scr expression patterns in Pc-/+ (left) or Pc-/+;dpp>Trx (right) backgrounds are shown for eye-antenna, wing and L2 disc, respectively. Arrowheads indicate regions with deviant Hox expression in Fig 3A. For comparison, Hox expression induced by dpp>Trx alone is shown (Figs 2C, 2D and S6C). (B) Enhanced adult phenotypes in Pc4 heterozygote mutants. Arista-to-leg transformation is seen in all Pc-/+;dpp>Trx adults (left panel). The percentages of transformed dpp>Trx adults are indicated in parentheses. Strong transformation of L2/L3 is seen in Pc-/+;dpp>Trx adults (right panel). The average numbers of sex comb teeth in Pc-/+;dpp>Trx are indicated for L2 and L3. The corresponding numbers in Pc-/+ mutants are indicated in parentheses. Scale bar, 0.1 mm.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We are very grateful to the following people for flies: K. Ahmad, S. Hirose, B. Loppin and H. Sun. We thank Bloomington Drosophila Stock Center, Drosophila Genetic Resource Center, Vienna Stock Center and Fly Core in Taiwan. We thank W. Chiang and P. Ho for the pilot experiments and data presented in Table 1. We also thank the imaging and genomic core facilities, IMB, Academia Sinica for technical support. We thank Y. Chang, T. Hsieh, J. Kung, J. Leu and J. Wu for helpful discussion and editing assistance from J. O’Brien.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants to D.-H. Huang from Academia Sinica, Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–31. 10.1038/Nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–45. 10.1016/j.cell.2010.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell. 2013;153:976–87. 10.1016/j.cell.2013.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaertner B, Johnston J, Chen K, Wallaschek N, Paulson A, Garruss AS, et al. Poised RNA polymerase II changes over developmental time and prepares genes for future expression. Cell Rep. 2012;2:1670–83. 10.1016/j.celrep.2012.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet. 2012;13:233–45. 10.1038/Nrg3163 [DOI] [PubMed] [Google Scholar]
- 6.McClure KD, Schubiger G. Transdetermination: Drosophila imaginal disc cells exhibit stem cell-like potency. Int J Biochem Cell Biol. 2007;39:1105–18. 10.1016/j.biocel.2007.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. 10.1038/Nrg1726 [DOI] [PubMed] [Google Scholar]
- 8.Mann RS, Morata G. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu Rev Cell Dev Biol. 2000;16:243–71. 10.1146/annurev.cellbio.16.1.243 [DOI] [PubMed] [Google Scholar]
- 9.Worley MI, Setiawan L, Hariharan IK. Regeneration and transdetermination in Drosophila imaginal discs. Annu Rev Genet. 2012;46:289–310. 10.1146/annurev-genet-110711-155637 [DOI] [PubMed] [Google Scholar]
- 10.Hadorn E. Imaginal discs: Transdetermination In: Ashburner M and Wright TRF. editors. Genetics and biology of Drosophila. New York: Academic Press; 1978. pp. 556–617 [Google Scholar]
- 11.Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15:340–56. 10.1038/Nrm3789 [DOI] [PubMed] [Google Scholar]
- 12.Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–54. 10.1101/gad.388706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V. Alternative epigenetic chromatin states of Polycomb target genes. PLoS Genet. 2010;6: e1000805 10.1371/journal.pgen.1000805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, et al. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012;150:922–33. 10.1016/j.cell.2012.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–45. 10.1016/j.cell.2007.02.009 [DOI] [PubMed] [Google Scholar]
- 16.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–11. 10.1038/Ng.2007.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–6. 10.1038/Ng.2007.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chopra VS, Hong JW, Levine M. Regulation of Hox gene activity by transcriptional elongation in Drosophila. Curr Biol. 2009;19:688–93. 10.1016/j.cub.2009.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–11. 10.1016/j.cell.2011.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chopra VS, Hendrix DA, Core LJ, Tsui C, Lis JT, Levine M. The polycomb group mutant esc leads to augmented levels of paused Pol II in the Drosophila embryo. Mol Cell. 2011;42:837–44. 10.1016/j.molcel.2011.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–54. 10.1101/gad.2005511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, et al. H2AZ is enriched at Polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–61. 10.1016/j.cell.2008.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–62. 10.1038/nature06929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–14. 10.1101/gad.1259805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–11. 10.1126/science.1134004 [DOI] [PubMed] [Google Scholar]
- 26.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–91. 10.1016/j.cell.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–20. 10.1016/j.cell.2013.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–9. 10.1038/Ncb1674 [DOI] [PubMed] [Google Scholar]
- 29.Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–90. 10.1038/Nature04059 [DOI] [PubMed] [Google Scholar]
- 30.Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011;44:928–41. 10.1016/j.molcel.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 31.Chen P, Zhao J, Wang Y, Wang M, Long H, Liang D, et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013;27:2109–24. 10.1101/gad.222174.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pchelintsev NA, McBryan T, Rai TS, van Tuyn J, Ray-Gallet D, Almouzni G, et al. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep. 2013;3:1012–9. 10.1016/j.celrep.2013.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonnefoy E, Orsi GA, Couble P, Loppin B. The essential role of Drosophila HIRA for de novo assembly of paternal chromatin at fertilization. PLoS Genet. 2007;3:1991–2006. 10.1371/journal.pgen.0030182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakai A, Schwartz BE, Goldstein S, Ahmad K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr Biol. 2009;19:1816–20. 10.1016/j.cub.2009.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA. 2010;107:14075–80. 10.1073/pnas.1008850107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–60. 10.1101/gr.101477.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21:175–86. 10.1016/j.gde.2011.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–5. 10.1038/nature09725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brand AH, Perrimon N. Targeted gene-expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–15. [DOI] [PubMed] [Google Scholar]
- 40.Staehling-Hampton K, Jackson PD, Clark MJ, Brand AH, Hoffmann FM. Specificity of bone morphogenetic protein-related factors: cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ. 1994;5(6):585–93. [PubMed] [Google Scholar]
- 41.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. 10.1016/0092-8674(92)90471-N [DOI] [PubMed] [Google Scholar]
- 42.Zeng WL, Andrew DJ, Mathies LD, Horner MA, Scott MP. Ectopic expression and function of the Antp and Scr homeotic genes—the n-terminus of the homeodomain is critical to functional specificity. Development. 1993;118(2):339–52. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol. 2002;22:2918–27. 10.1128/mcb.22.9.2918-2927.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, et al. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–14. 10.1101/gad.1091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22(14):1921–33. 10.1101/gad.1643208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–200. 10.1016/S1097-2765(02)00542-7 [DOI] [PubMed] [Google Scholar]
- 47.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–7. 10.1038/Ng1637 [DOI] [PubMed] [Google Scholar]
- 48.Ruhf ML, Braun A, Papoulas O, Tamkun JW, Randsholt N, Meister M. The domino gene of Drosophila encodes novel members of the SWI2/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development. 2001;128:1429–41. . [DOI] [PubMed] [Google Scholar]
- 49.Evans CJ, Olson JM, Ngo KT, Kim E, Lee NE, Kuoy E, et al. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods. 2009;6:603–5. 10.1038/nmeth.1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiang A, Oconnor MB, Paro R, Simon J, Bender W. Discrete Polycomb-binding sites in each parasegmental domain of the bithorax complex. Development. 1995;121(6):1681–9. [DOI] [PubMed] [Google Scholar]
- 51.Orlando V, Jane EP, Chinwalla V, Harte PJ, Paro R. Binding of Trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 1998;17:5141–50. 10.1093/emboj/17.17.5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13(11):2553–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fritsch C, Brown JL, Kassis JA, Muller J. The DNA-binding Polycomb group protein Pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999;126(17):3905–13. [DOI] [PubMed] [Google Scholar]
- 54.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–4. 10.1126/science.1186777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kraushaar DC, Jin WF, Maunakea A, Abraham B, Ha M, Zhao KJ. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 2013;14 10.1186/gb-2013-14-10-r121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simon JA, Kingston RE. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. 10.1038/nrm2763 [DOI] [PubMed] [Google Scholar]
- 57.Cavalli G, Paro R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999;286:955–8. 10.1126/science.286.5441.955 [DOI] [PubMed] [Google Scholar]
- 58.Schneiderman JI, Orsi GA, Hughes KT, Loppin B, Ahmad K. Nucleosome-depleted chromatin gaps recruit assembly factors for the H3.3 histone variant. Proc Natl Acad Sci USA. 2012;109:19721–6. 10.1073/pnas.1206629109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin CY, Zang CZ, Wei G, Cui KR, Peng WQ, Zhao KJ, et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–5. 10.1038/Ng.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011;30(14):2817–28. 10.1038/emboj.2011.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, et al. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol. 2011;31(21):4310–8. 10.1128/MCB.06092-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tie F, Banerjee R, Saiakhova AR, Howard B, Monteith KE, Scacheri PC, et al. Trithorax monomethylates histone H3K4 and interacts directly with CBP to promote H3K27 acetylation and antagonize Polycomb silencing. Development. 2014;141(5):1129–39. 10.1242/dev.102392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakayama T, Nishioka K, Dong YX, Shimojima T, Hirose S. Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev. 2007;21:552–61. 10.1101/Gad.1503407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang YL, Peng YH, Pan IC, Sun DS, King B, Huang DH. Essential role of Drosophila Hdac1 in homeotic gene silencing. Proc Natl Acad Sci USA. 2001;98:9730–5. 10.1073/pnas.171325498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–14. 10.1006/meth.1996.0407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Restricted Ubx induction in wing discs. Four Gal4 lines (actin, dpp, A9, engrailed) were used to drive Trx and GFP simultaneously. Their effects on GFP and Ubx expression in discs and adult wings are shown. GFP (green) and Ubx signals (red) are shown in separate panels or together with DNA staining (blue). Adult wings resulting from dpp-, A9-, and engrailed-driven Trx are shown. actin-driven Trx causes pupal lethality. A schematic representation of anterior/posterior (A/P) and dorsal/ventral (D/V) compartments and their boundaries is shown. White dots outline the pouch region. Scale bars in this and subsequent confocal images represents 50 μm. (B) Restricted Scr induction in leg discs. The patterns of Scr induction in L1, L2 and L3 leg discs from actin>GFP (left panel) or actin>Trx, GFP (right panel) larvae are compared. Ectopic Scr signals are seen in regions forming adult tarsal segments. Individual or merged images of Scr (red), GFP (green) and DNA (blue) are shown. Anterior/Posterior (A/P) compartments are indicated.
(TIF)
Both Scr and Antp contain two separate promoters and start sites. The direction of transcription is indicated by rightward arrows. The non-coding and coding exons are indicated by grey and orange boxes, respectively. Introns are indicated by thin lines. Approximate locations of primers used in this study are indicated by red (ChIP) or blue bars (RT-qPCR) below the gene map. Magnified views of the TSS regions of Ubx and Antp are shown. P1 and P2 correspond to the proximal regions of promoters 1 and 2, respectively. +0.1K and +1K correspond to regions about 100 bp and 1 kb downstream of the TSS, respectively. 3’ corresponds to the gene terminus. bxd-1, bxd-2 and bx correspond to Ubx distal regulatory elements. P1, P2 and 3’ end primers were used to measure RNA-Pol II occupancy, whereas other primers were used to measure tri-methylated H3 or H3.3. Primer sequences are listed in S4 Table. The upstream-most start sites are used as the reference point for the scales shown above genes.
(PDF)
(A) Analyses of Ser5P enrichment in Scr or Ubx in different leg discs. ChIP analyses of promoter and 3’ regions of Scr or Ubx were performed with a Ser5P antibody for samples prepared from WT L2 (left) or L3 discs (right). (B) Analyses of Ser5P and Ser2P enrichment of Scr or Ubx in wing or eye-antenna discs. Chromatin samples prepared from wing (left) or eye-antenna discs (right) of the following genetic backgrounds were subjected to ChIP analyses with antibodies against Ser5P or Ser2P: WT, dpp>Trx, Hira-/Y, Hira-/Y;dpp>Trx. The bar graphs represent the averages ± SD of two independent ChIP experiments, each with two separate qPCR reactions. The scale on the y-axis corresponds to 1% of input chromatin. Analyzed regions are described in S2 Fig.
(PDF)
Based on the wing phenotype induced by dpp>Trx, a modifier screening was conducted with co-expression of the RNAi lines listed in S1 Table. In no case was the small wing induced by dpp>Trx further reduced. The effects of RNAi lines were evaluated by the degree of recovery to full wing size as indicated by the number of “+” signs. The “-” sign indicates no effect. Scale bar, 0.3 mm.
(PDF)
Transformation of adult L2/3 to L1 was scored in a heterozygous Pc background with other mutants. Each bar represents the percentage of ESCT transformation with respect to the number of ESCT (x-axes). Note that the percentage transformation increased with increased ESCT when Pc and other mutants were combined. p- and W-value were calculated based on one-sided Wilcoxon rank sum test.
(TIF)
(A) Adult leg phenotype. WT, Hira and dom adults mutants exhibit sex combs only on the first leg (L1, arrow). (B) Extra sex combs on adult legs. The sex combs induced in adult L2 (upper) and L3 (lower) by dpp>Trx are enhanced in Hira and dom mutants. The average number of sex comb teeth is shown and the numbers of legs scored are in parentheses. (C) Ectopic Scr expression in leg discs. Upper panel, Scr expression induced in L2 discs by dpp>Trx in WT, Hira or dom mutant backgrounds. Lower panel, merged images for Scr (red), GFP (green) and DNA (blue) as described previously. (B, C) Experiments were carried out at 21°C. (D) Bar graph depicts phenotypic variation as described previously.
(TIF)
(A) Schematic diagram of components employed in the G-TRACE strategy [49]. UAS-RFP and UAS-FLP are directly under the control of dpp-Gal4, which is active primarily in larval discs. Nuclear EGFP (nEGFP) is linked to the Ubi-p63E promoter with a cassette containing a transcriptional termination signal flanked by FRT. Through the removal of FRT by FLP recombinase, rearranged Ubi-p63E-nEGFP is constitutively activated, thus marking all descendent cells. (B) Lineage tracing of dpp-Gal4 in imaginal discs. Experiments were performed in the presence of tub-Gal80ts as described in Fig 3C. Gal80ts is inactivated for different lengths at 29°C and then re-activated by shifting to 21°C for indicated hours. dpp-Gal4 becomes activated reciprocally. Due to its rapid degradation, strong RFP signals can be seen only in cells that remain active for dpp-Gal4. In contrast, all cells that have ever been active for dpp-Gal4 are marked by GFP signals. Hours refer to the duration of chase described in Fig 3C. Individual RFP (red), GFP (green) or a merge panel with DNA (blue) are shown for leg (left), wing (middle) and eye-antenna discs (right). Arrowheads indicate regions with deviant Hox expression in Fig 3A. Note that most GFP signals appear during the 48–72 h interval.
(TIF)
Bar graphs are as described previously. Hours indicate the duration of Trx pulse (see Fig 3C). At any given time points, the sex combs induced in adult L2 and L3 by dpp>Trx are enhanced in Hira mutants.
(TIF)
Bar graph is as described previously. Hours indicate the duration of Trx pulses (see Fig 5A). For both 24 and 48 h, the sex combs induced in adult L2 and L3 by dpp>Trx are enhanced in Hira mutants.
(TIF)
(A) Stability of ectopic Scr in L2 discs. Pulse-chase experiments were performed for animals carrying tub-Gal80ts; dpp-Gal4, UAS-Scr in WT (left) or Hira backgrounds (right) with different lengths of chase as indicated in Fig 3C. Scr staining in L2 discs is shown alone or together with DNA. Note that similar results were observed in WT or Hira mutants. (B) Stability of ectopic Antp in eye-antenna discs. Similar pulse-chase experiments were performed for animals carrying tub-Gal80ts; dpp-Gal4, UAS-Antp. Antp staining in eye-antenna discs is shown alone or together with DNA (blue). In all cases, ectopic Scr and Antp signals disappeared within 12 h of chase.
(TIF)
Experiment was carried out as in Fig 3C. Pulse-chase experiments were performed for animals carrying tub-Gal80ts; dpp-Gal4; UAS-Trx, GFP. Duration of the chase is indicated. Trx (red) and GFP (green) signals in wing, eye-antenna and L2 discs are shown alone or together with DNA (blue). Note that endogenous Trx expression is uniform in all discs and that induced Trx signals disappear faster than GFP.
(TIF)
(A) Ectopic Hox expression in Pc4 heterozygote mutants. Antp, Ubx and Scr expression patterns in Pc-/+ (left) or Pc-/+;dpp>Trx (right) backgrounds are shown for eye-antenna, wing and L2 disc, respectively. Arrowheads indicate regions with deviant Hox expression in Fig 3A. For comparison, Hox expression induced by dpp>Trx alone is shown (Figs 2C, 2D and S6C). (B) Enhanced adult phenotypes in Pc4 heterozygote mutants. Arista-to-leg transformation is seen in all Pc-/+;dpp>Trx adults (left panel). The percentages of transformed dpp>Trx adults are indicated in parentheses. Strong transformation of L2/L3 is seen in Pc-/+;dpp>Trx adults (right panel). The average numbers of sex comb teeth in Pc-/+;dpp>Trx are indicated for L2 and L3. The corresponding numbers in Pc-/+ mutants are indicated in parentheses. Scale bar, 0.1 mm.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.