Key points
The fetus responds to decreases in arterial partial pressure of oxygen by redirecting the blood flow mainly to the brain and the heart, at a cost to other peripheral organs like the kidneys.
Renal hypoxia and ischaemia stimulate inflammatory and apoptotic responses.
Ketamine, an NMDA receptor antagonist, is able to reduce renal immune and inflammatory gene expressions stimulated by hypoxia.
Ketamine may have therapeutic potential for protection against ischaemic renal damage in fetuses subjected to acute hypoxia.
Abstract
Acute fetal hypoxia is a form of fetal stress that stimulates renal vasoconstriction and ischaemia as a consequence of the physiological redistribution of combined ventricular output. Because of the potential ischaemia–reperfusion injury to the kidney, we hypothesized that it would respond to hypoxia with an increase in the expression of inflammatory genes, and that ketamine (an N‐methyl‐d‐aspartate receptor antagonist) would reduce or block this response. Hypoxia was induced for 30 min in chronically catheterized fetal sheep (125 ± 3 days), with or without ketamine (3 mg kg−1) administered intravenously to the fetus 10 min prior to hypoxia. Gene expression in fetal kidney cortex collected 24 h after the onset of hypoxia was analysed using ovine Agilent 15.5k array and validated with qPCR and immunohistochemistry in four groups of ewes: normoxic control, normoxia + ketamine, hypoxic control and hypoxia + ketamine (n = 3–4 per group). Significant differences in gene expression between groups were determined with t‐statistics using the limma package for R (P ≤ 0.05). Enriched biological processes for the 427 upregulated genes were immune and inflammatory responses and for the 946 downregulated genes were metabolic processes. Ketamine countered the effects of hypoxia on upregulated immune/inflammatory responses as well as the downregulated metabolic responses. We conclude that our transcriptomics modelling predicts that hypoxia activates inflammatory pathways and reduces metabolism in the fetal kidney cortex, and ketamine blocks or ameliorates this response. The results suggest that ketamine may have therapeutic potential for protection from ischaemic renal damage.
Key points
The fetus responds to decreases in arterial partial pressure of oxygen by redirecting the blood flow mainly to the brain and the heart, at a cost to other peripheral organs like the kidneys.
Renal hypoxia and ischaemia stimulate inflammatory and apoptotic responses.
Ketamine, an NMDA receptor antagonist, is able to reduce renal immune and inflammatory gene expressions stimulated by hypoxia.
Ketamine may have therapeutic potential for protection against ischaemic renal damage in fetuses subjected to acute hypoxia.
Abbreviations
- BCL3
B‐cell lymphoma 3‐encoded protein
- CCL
chemokine (C‐C motif) ligand
- CD14
CD14 molecule
- CSF1
macrophage colony stimulating factor 1
- CXCL10
chemokine (C‐X‐C motif) ligand 10
- HC
hypoxic control
- HK
hypoxia + ketamine
- Iba‐1
ionized calcium binding adaptor molecule 1
- IL
interleukin
- IRF1
interferon regulatory factor 1
- KEGG
Kyoto encyclopedia of genes and genomes
- MAPK
mitogen‐activated protein kinases
- NC
normoxic control
- NFκB
nuclear factor kappa‐light‐chain‐enhancer of activated B cells
- NK
normoxia + ketamine
- NMDA
N‐methyl‐D‐aspartate
- TLR
toll like receptor
- TNFα
tumour necrosis factor‐alpha
Introduction
Throughout pregnancy, the fetus maintains a state of oxygen homeostasis within the uterine environment. Hypoxia, a relatively common fetal stressor, has the potential to damage developing organs. The physiological response to hypoxia in utero features a redistribution of combined ventricular output towards the heart and brain and away from organs – including the kidney – that are not needed for short‐term survival (Cohn et al. 1974; Green et al. 1997; Luciano et al. 1998). While there is much known about the physiology of redistribution of fetal blood flow during the response to acute hypoxia, less is known about the cellular response, and specifically whether the response is potentially damaging. Whether there are strategies preserving kidney integrity after hypoxia in utero is relatively unknown (Giussani et al. 1994; Giussani & Davidge, 2013).
Recently, we have applied transcriptomic and systems biology methodology to elucidate the effect of hypoxia on the developing fetal brain (Wood et al. 2014). We reported an increase in inflammatory pathways in the fetal hypothalamus (Wood et al. 2013). We have reported (in abstract) that pretreatment with ketamine decreases brain inflammatory responses (Chang & Wood, 2013). Ketamine, an N‐methyl‐d‐aspartate (NMDA) receptor antagonist, is commonly used in veterinary medicine, pediatric surgery, and neonatal intensive care for its anaesthetic and analgesic properties (Mellon et al. 2007). Clinically, ketamine has been shown to be an anti‐inflammatory agent acting by decreasing serum levels of interleukin 6 (IL‐6) and tumour necrosis factor‐alpha (TNFα) in patients undergoing cardiac surgeries and liver transplantation (Roytblat et al. 1998; Zeyneloglu et al. 2005; Yang et al. 2006). In addition, in vitro studies have shown that lipopolysaccharide‐activated macrophages (Wu et al. 2008) and glial cells (Tanaka et al. 2013) have decreased IL‐6, TNFα, and IL‐1β gene expression when treated with ketamine. Consequently, ketamine might be more broadly useful as a means to decrease peripheral tissue inflammation and reduce or prevent adverse outcomes as it blunts inflammatory responses globally, and not solely in the brain.
NMDA receptors are not only found throughout the nervous system, but are also present in various extraneuronal tissues such as the renal cortex and medulla, and in the atriums, ventricles, aorta and pulmonary artery of the cardiovascular system (Leung et al. 2002). An upregulation and activation of renal NMDA receptors was found in ischaemia–reperfusion‐ and hypoxia–reoxygenation‐induced acute renal injury (Yang et al. 2008; Pundir et al. 2013). Intense activation of NMDA receptors can evoke significant bursts of reactive oxygen species production, calcium ion overload and damage renal tubules, resulting in induction of apoptosis and necrosis of tubular cells, which may contribute to renal dysfunction (Yang et al. 2008). In these acute kidney injury models, various NMDA receptor antagonists were found to attenuate ischaemia–reperfusion‐induced injuries and significantly reduce oxidative stress (Pundir et al. 2013). Ketamine, by blocking NMDA receptors and their activation, may potentially be beneficial by decreasing renal damage in the developing fetus. For these reasons, we hypothesized that ketamine would reduce the expression of renal inflammatory genes in the late gestation ovine fetus exposed to acute hypoxia. The experiments reported herein were designed to test the hypotheses that acute hypoxia induces inflammatory pathways in the fetal kidney and that ketamine ameliorates or eliminates this response.
Methods
Ethical approval
These experiments were approved by the University of Florida Animal Care and Use Committee and were performed in accordance with the Guiding Principles for the Care and Use of Vertebrate Animals in Research and Training of the American Physiological Society. Fifteen chronically catheterized singleton (n = 1 in normoxic ketamine group) or twin (n = 14) ovine fetuses were studied at the gestational age of 122 ± 5 days (full term = 145–147 days), which is approximately equivalent to 28–30 weeks in humans.
Fetal ovine surgical procedures
Ewes were fasted for 24 h before surgery, and the fetal ovine surgical procedures were performed on 116 ± 3 days of gestation. The surgical procedures for chronic catherization of the fetal and maternal femoral arteries and veins and aminiotic fluid were described in detail previously (Wood et al. 1990; Powers & Wood, 2007). Briefly, the ewes were given 750 mg ampicillin (Polyflex, Boehringer Ingelheim VetMedica, Inc., St Joseph, MO, USA), anaesthetized and intubated with 0.5–2% isoflurane with oxygen and vascular catheters were placed. Before the uterus was sutured closed, 500 mg ampicillin was injected into the amniotic fluid. A small tracheostomy catheter was placed in the ewe to produce maternal induced ventilatory hypoxia, which in turn resulted in fetal hypoxic hypoxia (Matsumoto et al. 2001). The maternal and fetal surgical procedures were the same for both normoxic and hypoxic groups. A minimum of 5 days of recovery preceded experimentation. Postoperatively ewes were monitored twice daily for food intake, body temperature and signs of infection or distress. During the postoperative recovery period, the ewes were treated with prophylactic antibiotic (ampicillin 15–20 mg kg−1, i.m. twice daily for 5 days) and analgesic (flunixine meglumin, 0.5 mg kg−1, i.m. once daily for 2–3 days).
In vivo experimental procedures
During the experiments, the ewes were conscious and freestanding in their pens with access to food and water. The fifteen fetuses (7 males, 8 females) were randomly assigned to one of the four groups (n = 3–4 per group): normoxic control (NC), normoxia + ketamine (NK), hypoxic control (HC), and hypoxia + ketamine (HK). In the ketamine‐treated groups, ketamine (3 mg kg−1) was administered via the fetal femoral venous catheter, 10 min prior to the hypoxic stimulus. The dose of ketamine was based upon the clinically used dose in the neonatal intensive care units (Anand, 2007). Hypoxia was induced by infusing nitrogen gas directly into the maternal tracheostomy tube at flow rates sufficient to decrease maternal pressure of oxygen () by 50%. As reported previously, maternal ventilatory hypoxia decreases both maternal and fetal (Chang & Wood, 2015). To closely monitor the changes in the composition of blood gases (ABL80 Radiometer, Copenhagen, Denmark), both maternal and fetal arterial blood samples were drawn anaerobically (1 ml) every 10 min (Tables 1 and 2). At the end of the 30 min period of normoxia or hypoxia, the ewe returned to breathing room air. The ewes and fetuses were killed with an overdose of sodium pentobarbital 24 h after the initial stimulation of hypoxic stress, and various fetal tissues were snap frozen and stored at −80°C until analysis. The kidney cortex analysed were from a subset of experiments from which blood gas data had been previously reported (Chang & Wood, 2015).
Table 1.
Fetal blood gases and pH values
| (mmHg) | (mmHg) | pH | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (min) | NC | NK | HC | HK | NC | NK | HC | HK | NC | NK | HC | HK |
| −10 | 18 ± 1 | 19 ± 2 | 17 ± 1 | 16 ± 1 | 53 ± 1 | 51 ± 2 | 55 ± 3 | 55 ± 1 | 7.38 ± 0.01 | 7.39 ± 0.02 | 7.35 ± 0.01 | 7.37 ± 0.01 |
| 0 | 18 ± 1 | 17 ± 1 | 17 ± 2 | 16 ± 1 | 53 ± 3 | 53 ± 2 | 57 ± 2 | 58 ± 1 | 7.38 ± 0.01 | 7.38 ± 0.02 | 7.35 ± 0.01 | 7.35 ± 0.01 |
| 5 | 17 ± 1a | 16 ± 1a | 11 ± 1b | 11 ± 2a,b | 54 ± 2 | 55 ± 1 | 55 ± 1 | 56 ± 2 | 7.38 ± 0.01 | 7.36 ± 0.01 | 7.36 ± 0.01 | 7.37 ± 0.01 |
| 10 | 17 ± 2a | 17 ± 1a | 10 ± 1b | 8 ± 1a,b | 56 ± 3 | 55 ± 2 | 53 ± 1 | 13 ± 2 | 7.37 ± 0.01 | 7.37 ± 0.02 | 7.37 ± 0.01 | 7.38 ± 0.01 |
| 20 | 16 ± 2a | 17 ± 1a | 10 ± 1b | 7 ± 1a,b | 54 ± 2 | 55 ± 2 | 52 ± 1 | 52 ± 3 | 7.38 ± 0.01 | 7.36 ± 0.02 | 7.36 ± 0.01 | 7.36 ± 0.01 |
| 30 | 17 ± 2a | 17 ± 1a | 9 ± 1b | 9 ± 1a,b | 55 ± 1 | 55 ± 2 | 54 ± 2 | 53 ± 2 | 7.37 ± 0.01a | 7.37 ± 0.02a,b | 7.33 ± 0.03a,b | 7.32 ± 0.02b |
These values are a subset of previously reported values (Chang & Wood, 2015). Statistical significance between groups was declared at P ≤ 0.05 (two‐way ANOVA), and Duncan's test was performed to determine differences between groups (different letters are statistically significantly different from each other, P < 0.05). NC (n = 3), NK (n = 4), HC (n = 4), HK (n = 4); all values are expressed as means ± SEM.
Table 2.
Maternal blood gases and pH values
| (mmHg) | (mmHg) | pH | ||||
|---|---|---|---|---|---|---|
| Time (min) | Normoxia | Hypoxia | Normoxia | Hypoxia | Normoxia | Hypoxia |
| −10 | 103 ± 3 | 97 ± 4 | 38 ± 2 | 39 ± 1 | 7.47 ± 0.01 | 7.47 ± 0.01 |
| 0 | 105 ± 4 | 99 ± 3 | 37 ± 2 | 39 ± 2 | 7.48 ± 0.01 | 7.47 ± 0.01 |
| 5 | 105 ± 4a | 59 ± 5b | 37 ± 2a | 32 ± 1b | 7.47 ± 0.01b | 7.55 ± 0.02a |
| 10 | 98 ± 7a | 55 ± 6b | 39 ± 2a | 33 ± 2b | 7.46 ± 0.01b | 7.54 ± 0.02a |
| 20 | 103 ± 2a | 45 ± 8b | 38 ± 2a | 31 ± 1b | 7.47 ± 0.01b | 7.54 ± 0.01a |
| 30 | 104 ± 2a | 51 ± 7b | 38 ± 2a | 32 ± 1b | 7.48 ± 0.01b | 7.54 ± 0.01a |
These values are a subset of previously reported values (Chang & Wood, 2015). Statistical significance between groups was declared at P ≤ 0.05 (two‐way ANOVA), and Duncan's test was performed to determine differences between groups (different letters are statistically significantly different from each other, P < 0.05). Normoxia (n = 4), hypoxia (n = 5); all values are expressed as means ± SEM.
Microarray procedures
The RNA was extracted from fetal kidney cortex with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and purified with RNeasy Plus Mini Kit (Qiagen, Valencia, CA, USA), resulting in RNA with RNA integrity number values between 7.7 and 9.1, and were labelled with cyanine 3 CTP with the Quick Amp Labelling Kit (Cat. No. 5190‐0442, Agilent Technologies, Santa Clara, CA, USA), according to the manufacturer's protocol. The specific activity of the labelled cRNAs ranged from 10.4 to 12.7 pmol Cy3 (μg RNA)−1 and yields were from 5.8 to 7.9 μg. The cRNA samples were hybridized and processed for one‐channel Ovine Gene Expression Microarray (8 × 15 K slide) – 8 arrays with 15208 oligomers each (Cat. No. G4813A‐019921, Agilent Technologies, Santa Clara, CA, USA) as described earlier (Rabaglino et al. 2012; Wood et al. 2013). The slides were scanned with Microarray Scanner System (G2505‐90021, Agilent) and the measured fluorescence was detected and converted using Agilent Feature Extraction 9.1 software at the Genomics Division of the University of Florida's Interdisciplinary Centre for Biotech Research. Microarray data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE66920.
Microarray data analysis
Raw data was processed with the R software (RCoreTeam, 2013), employing the limma package (Ritchie et al. 2015) to perform background correction and data normalization using the quantile normalization method (Smyth et al. 2005). Probes that were at least 10% brighter than the negative controls on at least three arrays were retained for further analysis. The processed microarray data were statistically analysed with the limma package as well, employing a moderated t test that uses an empirical Bayes method for small sample size per group (P ≤ 0.05; Smyth, 2004). The effect of hypoxia was analysed by comparing the hypoxic group to normoxic group as the control group. Then, the hypoxic + ketamine group was compared to the hypoxic group to evaluate the effect of ketamine under hypoxic conditions. Finally, the normoxic + ketamine group was compared to the normoxic group to evaluate the effect of ketamine under physiological conditions.
Cytoscape and various plug‐ins (GeneMANIA, BiNGO) were used to analyse gene network inference and clustering analysis with the significantly differentially expressed genes (Shannon et al. 2003; Maere et al. 2005; Cline et al. 2007; Warde‐Farley et al. 2010; Zhang et al. 2014). The functional annotation of gene ontogeny for significantly up‐ and downregulated genes were analysed via Database for Annotation, Visualization and Integrated Discovery Bioinformatics Resources 6.7 (Huang da et al. 2009 a,b). The KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways were analysed with WEB‐based GEne SeT AnaLysis Toolkit (WebGestalt) (Zhang et al. 2005; Wang et al. 2013).
Real‐time PCR
cDNA made from the same RNA used for the microarray analyses were used for qPCR validation of the microarray findings. The primers were designed based on the known Ovis aries and Bos taurus genomes (Table 3) for SYBR green detection in qPCR. As the housekeeping control, ovine β‐actin primers and probe were used (Wood et al. 2013). The RNA expression was normalized by the difference in threshold cycle (ΔCt) between the triplicate mean Ct for each gene and the triplicate mean Ct for β‐actin mRNA from the same sample, and these ΔCt values compared between treatment groups.
Table 3.
Primers for real‐time PCR validation
| Official | ||||
|---|---|---|---|---|
| symbol | Gene name | Forward primer | Reverse primer | Species |
| TLR2 | Toll‐like receptor 2 | GATTCTGCTGGAGCCCATTG | TCATGATCTTCCGCAGCTTACA | Ovis aries |
| NFκB | Nuclear factor kappa‐light‐chain‐enhancer of activated B cells | TCCCACAGATGTTCACAAACAGT | GACGCTCAATCTTCATCTTGTGAT | Ovis aries |
| IL‐6 | Interleukin 6 | ATGCTTCCAATCTGGGTTCAA | TCCAGAAGACCAGCAGTGGTT | Ovis aries |
| IL‐1β | Interleukin 1β | CGTGGCCATGGAGAAGCT | GGTCATCATCACGGAAGACATGT | Ovis aries |
| CSF1 | Macrophage colony‐stimulating factor | GACTGGAACATTTTCAGCAAGAACT | TCAGGCTTGGTCACCACATC | Bos taurus |
| CD14 | Monocyte/macrophage surface antigens | CCTAAAGGACTGCCGACCAA | GCGGCTCCCTGCTTAGCT | Ovis aries |
| TNFα | Tumour necrosis factor | CCCTTCCACCCCCTTGTT | ATGTTGACCTTGGTCTGGTAGGA | Ovis aries |
| BCL3 | B‐cell CLL/lymphoma 3 | CATGGAACACCCCCTGTCA | GGCGTATCTCCATCCTCATCA | Ovis aries |
| CCL4 | Chemokine ligand 4 | TCTTACACCCTGCGGCAGAT | GGCTGCTGGTCTCGTAGTAGTCA | Ovis aries |
| CCL5 | Chemokine ligand 5 | CCAGCAGCAAGTGCTCCAT | CGCACACCTGACGGTTCTT | Ovis aries |
| CXCL10 | Chemokine ligand 10 | TTGAACTGATTCCTGCAAGTCA | TTCCTTTTCATTGTGGCAATAATCT | Ovis aries |
| IRF1 | Interferon regulatory factor 1 | CCCAGGGCTGATCTGGATTA | GCGTGCTTCCATGGGATCT | Ovis aries |
Immunohistochemistry
Samples of fetal kidney were immersion fixed in 4% buffered paraformaldehyde overnight, then transferred to 70% reagent alcohol until paraffin embedded. These tissue samples were taken simultaneous with the tissue used for the mRNA isolation and analysis. Histological sections were cut at 5 μm, and immunostained for Iba‐1 using primary antibody from Waco Pure Chemical Industries (Richmond, VA; USA 1:200 dilution, Cat. No. 019–19741), and peroxidase–PAP sandwich technique (Vectastain, Vector Labs, Burlingame, CA, USA). All sections were counterstained with methyl green. Resident macrophages, identified as Iba‐1‐positive cells, were quantified as number of cells in a ×40 visual field by an investigator who was blinded to identity or experimental group of the histological section. Five fields were analysed for each section of kidney cortex, and the mean was calculated for statistical analysis.
Statistics
The experimental design involved a double factorial randomized complete design. Statistical analyses were conducted using different statistical software depending on the required statistical tests. For the microarray data analysis, the R software was used with the limma package (see detailed analysis above). For the qPCR and immunohistochemistry experiments, a normal probability plot test was performed to confirm normality in data distribution. qPCR data were analysed by two‐way analysis of variance with stimulus (hypoxia/normoxia) and treatment (ketamine/control) as factors and to allow for analysis of replicate fields; immunohistochemistry was analysed using two‐way nested design ANOVA to account for variability between visual fields within animal using the Mixed Procedure of SAS/STAT 9.3 (SAS Institute Inc., Cary, NC, USA). The criterion for statistical significance was P ≤ 0.05. When significance was found, groups were further analysed by Duncan's post hoc test to detect differences among them. The qPCR and immunohistochemistry data are presented as mean values ± standard error of the mean (SEM).
Results
Acute maternal ventilatory hypoxia induced significant changes in gene expression. From a total of 1373 significantly differentially regulated genes, 427 were upregulated and 946 were downregulated by hypoxia alone (Fig. 1 A). Ketamine modified the transcriptomic response to hypoxia: 260 genes were upregulated (relative to hypoxia without ketamine), and 193 genes were downregulated (relative to hypoxia without ketamine; Fig. 1 B). Ketamine alone in normoxic conditions differentially regulated 190 genes (97 upregulated and 93 downregulated; Fig. 1 C). Analysis of the 97 upregulated genes did not reveal any statistically significant overrepresented biological pathways, and the 93 downregulated genes were related to cellular transcription processes (data not shown).
Figure 1. Volcano plot illustrating the relationship of gene expression in fetal kidney cortex measured by log2 of fold‐change .
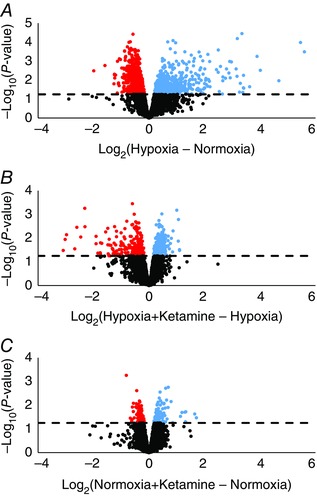
Significantly upregulated (blue) and downregulated (red) genes under hypoxia (A), hypoxia + ketamine (B), and normoxia + ketamine (C). The dashed line indicates whether the gene is statistically significant (above), P ≤ 0.05, or not significant (below). Non‐significant genes are indicated by black circles, and the scale for the y‐axis varies between plots.
Gene ontology analysis of the entire set of 1373 genes differentially expressed in response to hypoxia, revealed upregulation of immune (108 genes, adjusted (adj.) P value = 1.13E‐31) and inflammatory (62 genes, adj. P value = 1.82E‐22) responses, and downregulated cellular metabolic pathways (461 genes, adj. P value = 2.13E‐09) (Table 4). After administration of ketamine, hypoxia upregulated metabolic processes (129 genes, adj. P value = 2.44E‐02), cellular repair, and DNA replication; and downregulated vascular development (34 genes, adj. P value = 2.59E‐12), angiogenesis (27 genes, adj. P value = 3.28E‐11), and response to bacterial and virulent factors.
Table 4.
Top 10 gene ontology biological processes that were significantly overrepresented in the kidney cortex during acute hypoxic stress, with or without ketamine
| No. of | Adjusted | ||
|---|---|---|---|
| Group | Biological process | genes involved | P values |
| Hypoxia upregulated | Immune response | 108 | 1.13E‐31 |
| Immune system process | 133 | 1.11E‐26 | |
| Defense response | 101 | 4.53E‐26 | |
| Inflammatory response | 62 | 1.82E‐22 | |
| Regulation of immune system process | 82 | 1.10E‐21 | |
| Response to other organism | 63 | 5.05E‐19 | |
| Innate immune response | 60 | 1.01E‐18 | |
| Response to biotic stimulus | 63 | 5.32E‐18 | |
| Regulation of defense response | 52 | 7.66E‐18 | |
| Regulation of response to stimulus | 136 | 8.40E‐18 | |
| Hypoxia downregulated | Cellular macromolecule metabolic process | 461 | 2.13 E‐09 |
| Macromolecule metabolic process | 483 | 1.92 E‐07 | |
| Nucleic acid metabolic process | 322 | 1.92 E‐07 | |
| Chromosome organization | 78 | 1.92 E‐07 | |
| Regulation of macromolecule metabolic process | 305 | 1.83 E‐07 | |
| Cellular component organization | 302 | 1.83E‐07 | |
| Cellular component organization at cellular level | 256 | 1.68 E‐07 | |
| Regulation of nitrogen compound metabolic process | 266 | 1.68 E‐07 | |
| Peptidyl‐lysine acetylation | 26 | 1.68 E‐07 | |
| Regulation of nucleobase‐containing compound metabolic process | 263 | 1.12 E‐07 | |
| Hypoxia + ketamine upregulated | Cellular macromolecule metabolic process | 129 | 2.44 E‐02 |
| Response to DNA damage stimulus | 23 | 2.44 E‐02 | |
| Organelle organization | 51 | 4.44 E‐02 | |
| Chromosome organization | 23 | 4.44 E‐02 | |
| Double‐strand break repair | 8 | 4.44 E‐02 | |
| DNA‐dependent DNA replication | 8 | 4.26 E‐02 | |
| Cell cycle checkpoint | 12 | 5.71 E‐02 | |
| Cellular protein metabolic process | 69 | 5.83 E‐02 | |
| S phase | 9 | 5.83 E‐02 | |
| Proteasome regulatory particle assembly | 2 | 5.83 E‐02 | |
| Hypoxia + ketamine downregulated | Negative regulation of biological process | 92 | 1.07 E‐13 |
| Negative regulation of cellular process | 87 | 1.07 E‐13 | |
| Vasculature development | 34 | 2.59 E‐12 | |
| Response to lipopolysaccharide | 23 | 2.59 E‐12 | |
| Blood vessel development | 33 | 2.71 E‐12 | |
| Blood vessel morphogenesis | 31 | 2.80 E‐12 | |
| Response to molecule of bacterial origin | 23 | 5.43 E‐12 | |
| Positive regulation of biological process | 92 | 1.24 E‐11 | |
| Angiogenesis | 27 | 3.28 E‐11 | |
| Response to chemical stimulus | 79 | 5.06 E‐11 |
KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis indicated that hypoxia upregulated pathways involving cytokine–cytokine receptor interaction (30 genes, adj. P value = 3.39E‐20), toll‐like receptor signalling pathway (21 genes, adj. P value = 7.00E‐20), and chemokines signalling pathways (19 genes, adj. P value = 1.69E‐12) (Table 5). Hypoxia downregulated pathways involving cell proliferation and embryonic development, such as the Wnt signalling cascade (19 genes, adj. P value = 3.93E‐8). The presence of ketamine during hypoxic stimulation resulted in the upregulation of metabolic pathways (23 genes, adj. P value = 4.89E‐5), and downregulation of toll‐like receptor (11 genes, adj. P value = 4.52E‐11), mitogen‐activated protein kinase (MAPK) signalling pathways (15 genes, adj. P value = 5.80E‐11), and cytokine–cytokine receptor interaction (12 genes, adj. P value = 6.88E‐8).
Table 5.
Top 10 enriched KEGG pathways that were significantly overrepresented in the kidney cortex during acute hypoxic stress, with or without ketamine
| No. of | Adjusted | ||
|---|---|---|---|
| Group | Pathway name | genes involved | P values |
| Hypoxia upregulated | Cytokine–cytokine receptor interaction | 30 | 3.39E‐20 |
| Osteoclast differentiation | 23 | 3.39E‐20 | |
| Toll‐like receptor signalling pathway | 21 | 7.00E‐20 | |
| Chagas disease (American trypanosomiasis) | 18 | 1.08E‐15 | |
| NOD‐like receptor signalling pathway | 14 | 2.06E‐14 | |
| Malaria | 13 | 8.56E‐14 | |
| MAPK signalling pathway | 23 | 1.96E‐13 | |
| Cytosolic DNA‐sensing pathway | 13 | 2.43E‐13 | |
| Rheumatoid arthritis | 15 | 4.55E‐13 | |
| Chemokine signalling pathway | 19 | 1.69E‐12 | |
| Hypoxia downregulated | Pathways in cancer | 45 | 1.85E‐20 |
| Chronic myeloid leukemia | 14 | 3.89E‐08 | |
| Melanogenesis | 16 | 3.89E‐08 | |
| Wnt signalling pathway | 19 | 3.93E‐08 | |
| Pancreatic cancer | 13 | 1.12E‐07 | |
| TGF‐beta signalling pathway | 14 | 1.12E‐07 | |
| Colorectal cancer | 12 | 1.84E‐07 | |
| Ubiquitin mediated proteolysis | 17 | 1.84E‐07 | |
| Cell cycle | 16 | 2.74E‐07 | |
| Mismatch repair | 8 | 3.03E‐07 | |
| Hypoxia + ketamine upregulated | Metabolic pathways | 23 | 4.89E‐05 |
| Nucleotide excision repair | 4 | 6.20E‐03 | |
| Mismatch repair | 3 | 8.30E‐03 | |
| DNA replication | 3 | 1.86E‐02 | |
| SNARE interactions in vesicular transport | 3 | 1.86E‐02 | |
| Drug metabolism; other enzymes | 3 | 2.89E‐02 | |
| mTOR signalling pathway | 3 | 2.89E‐02 | |
| Pyrimidine metabolism | 4 | 2.89E‐02 | |
| Melanogenesis | 4 | 2.89E‐02 | |
| Calcium signalling pathway | 5 | 3.16E‐02 | |
| Hypoxia + ketamine downregulated | Osteoclast differentiation | 15 | 8.21E‐15 |
| Pathways in cancer | 19 | 1.54E‐13 | |
| Leishmaniasis | 11 | 1.74E‐12 | |
| Rheumatoid arthritis | 11 | 1.89E‐11 | |
| Toll‐like receptor signalling pathway | 11 | 4.52E‐11 | |
| Malaria | 9 | 4.52E‐11 | |
| Chagas disease (American trypanosomiasis) | 11 | 4.81E‐11 | |
| MAPK signalling pathway | 15 | 5.80E‐11 | |
| NOD‐like receptor signalling pathway | 8 | 3.77E‐09 | |
| Cytokine–cytokine receptor interaction | 12 | 6.88E‐08 |
Comparison of the hypoxia upregulated genes (hypoxic control vs. normoxic control: HC‐NC) with the hypoxia + ketamine downregulated genes (hypoxia + ketamine vs. hypoxic control: HK‐HC) revealed the genes that were sensitive to both hypoxia and ketamine; there were 88 genes shared between these groups (Fig. 2). Network inference of these gene sets (hypoxia: 437, hypoxia + ketamine: 260, common genes: 88), generated a merged network in which genes responding to hypoxia (HC‐NC) clustered together (blue and yellow nodes: Fig. 2) and a second network of hypoxia + ketamine downregulated genes (HK‐HC) that was distinct (red and yellow nodes, Fig. 2), but overlapping. The overlapping genes – those that were both increased by hypoxia and decreased during hypoxia by ketamine – tended to cluster with the network of hypoxia‐sensitive genes (yellow nodes, Fig. 2). The common genes were further analysed to determine gene ontology and KEGG pathways (Table 6). The most highly statistically significant overrepresented KEGG pathways were found to be involved in cytokine–cytokine receptor interaction (30 genes, adj. P value = 3.39E‐20), toll‐like receptor (21 genes, adj. P value = 7.00E‐20), MAPK (23 genes, adj. P value = 1.96E‐13), and chemokine‐signalling pathways (19 genes, adj. P value = 1.69E‐12). The overrepresented biological processes with higher significance were negatively involved in regulation of cellular processes (54 genes, adj. P value = 5.52E‐13). The top ranked molecular function terms were cytokine receptor binding and activity (11 genes, adj. P value = 5.41E‐06), nucleic acid binding transcription factor activity (19 genes, adj. P value = 3.00E‐04), and DNA regulations. Of the 88 common genes, most were located in the cellular components nucleus or membrane‐bounded organelle.
Figure 2. Venn diagram and network analysis of significant gene expression upregulated by hypoxia, but downregulated by ketamine during hypoxia .
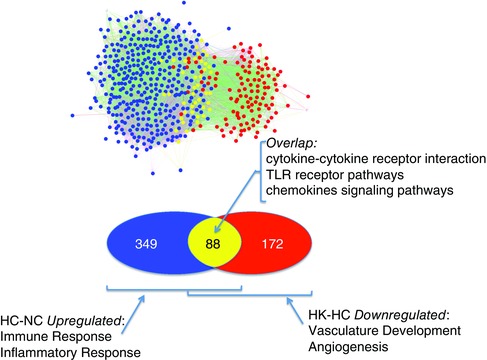
Venn and network inference analyses showing the number of significant genes that was upregulated by hypoxia (blue), downregulated with hypoxia and ketamine (red), and the common genes involved in both groups (yellow). Hypoxia (HC‐NC) and hypoxia + ketamine (HK‐HC).
Table 6.
Top 10 gene ontology biological processes and enriched KEGG pathways that were significantly upregulated in the kidney cortex during acute hypoxic stress, but were significantly downregulated with hypoxia + ketamine
| Analysis | Pathways, processes, functions, components | No. of genes involved | Adjusted P values |
|---|---|---|---|
| KEGG pathways | Osteoclast differentiation | 23 | 3.39E‐20 |
| Cytokine–cytokine receptor interaction | 30 | 3.39E‐20 | |
| Toll‐like receptor signalling pathway | 21 | 7.00E‐20 | |
| Chagas disease (American trypanosomiasis) | 18 | 1.08E‐15 | |
| NOD‐like receptor signalling pathway | 14 | 2.06E‐14 | |
| Malaria | 13 | 8.56E‐14 | |
| MAPK signalling pathway | 23 | 1.96E‐13 | |
| Cytosolic DNA‐sensing pathway | 13 | 2.43E‐13 | |
| Rheumatoid arthritis | 15 | 4.55E‐13 | |
| Chemokine signalling pathway | 19 | 1.69E‐12 | |
| Biological process | Negative regulation of biological process | 56 | 5.55E‐13 |
| Negative regulation of cell communication | 25 | 1.31E‐10 | |
| Negative regulation of cellular process | 54 | 5.52E‐13 | |
| Negative regulation of response to stimulus | 28 | 2.03E‐11 | |
| Negative regulation of signal transduction | 24 | 2.25E‐10 | |
| Negative regulation of signalling | 25 | 1.31E‐10 | |
| Regulation of cell communication | 42 | 1.09E‐10 | |
| Regulation of response to stimulus | 46 | 3.32E‐11 | |
| Regulation of signal transduction | 39 | 1.31E‐10 | |
| Regulation of signalling | 42 | 1.09E‐10 | |
| Molecular function | Protein binding | 72 | 4.08E‐07 |
| Cytokine receptor binding | 11 | 5.41E‐06 | |
| Cytokine activity | 9 | 2.00E‐04 | |
| Nucleic acid binding transcription factor activity | 19 | 3.00E‐04 | |
| Sequence‐specific DNA binding transcription factor activity | 19 | 3.00E‐04 | |
| Regulatory region DNA binding | 10 | 8.00E‐04 | |
| Regulatory region nucleic acid binding | 10 | 8.00E‐04 | |
| Interleukin‐1 receptor binding | 3 | 1.00E‐03 | |
| Sequence‐specific DNA binding | 14 | 1.00E‐03 | |
| RNA polymerase II core promoter proximal region sequence‐specific DNA binding transcription factor activity | 5 | 2.70E‐03 | |
| Cellular component | Cytosol | 31 | 2.00E‐04 |
| IκB–NFκB complex | 3 | 2.00E‐04 | |
| Extracellular space | 15 | 3.00E‐03 | |
| Cell surface | 10 | 9.80E‐03 | |
| Extracellular region part | 16 | 9.80E‐03 | |
| External side of plasma membrane | 6 | 2.03E‐02 | |
| Nucleus | 48 | 2.09E‐02 | |
| Intracellular part | 81 | 2.29E‐02 | |
| Intracellular | 82 | 2.98E‐02 | |
| Membrane‐bounded organelle | 67 | 3.05E‐02 |
Performing the converse comparison – genes that are downregulated by hypoxia (HC‐NC) versus genes that were upregulated during hypoxia by ketamine (HK‐HC) – revealed reciprocal effects on cellular metabolism. Hypoxia decreased expression of 946 genes (blue and yellow nodes, Fig. 3). Ketamine during hypoxia increased expression of 260 genes (red and yellow nodes, Fig. 3). Again, some of these genes were shared by these two groups (n = 130 yellow nodes, Fig. 3) that tended to cluster with the HC‐NC network. Ketamine tended to reverse the effect of hypoxia on metabolism, although analysis of the overlapping genes alone did not yield any significant gene ontology terms (Fig. 3).
Figure 3. Venn diagram and network analysis of significant gene expression downregulated by hypoxia, but upregulated by ketamine during hypoxia .
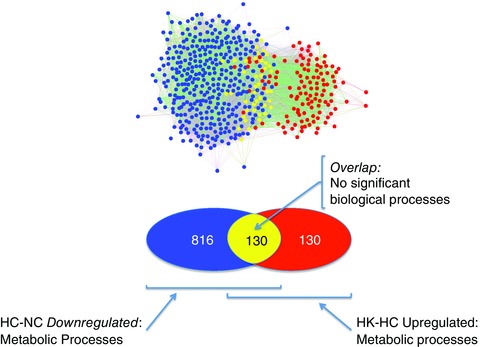
Venn and network inference analyses showing the number of significant genes that was downregulated by hypoxia (blue), upregulated with hypoxia and ketamine (red), and the common genes involved in both groups (yellow). Hypoxia (HC‐NC) and hypoxia + ketamine (HK‐HC).
To validate the microarray results, qPCR was performed on statistically significantly expressed genes in the 88 common genes shared by upregulated hypoxia and downregulated hypoxia + ketamine genes (Fig. 4). The genes selected were involved in inflammatory and immune response pathways (TLR2, NFκB, IL‐6, IL‐1β, CSF1, CD14, TNFα, BCL3, CCL4, CCL5, CXCL10, and IRF1). Expression of all of these genes were significantly increased by hypoxia (comparison of NC vs. HC was significant), and the increase in expression of these genes was inhibited by ketamine (comparison of NC vs. HK was not significant) when the microarray results were analysed, with the exception of CCL4. As shown in Fig. 4, the qPCR gene expression pattern for these inflammatory/immune genes resembles the microarray gene expression: upregulation by hypoxia and prevention of upregulation by ketamine. Again, as measured by real‐time qPCR, the expression of all measured genes was increased by hypoxia (comparison of NC vs. HC was significant), and with the exception of CCL4, the increase in expression of all genes was inhibited by ketamine (comparison of NC vs. HK was not significant) when the qPCR results were analysed. The inhibition of CCL5, CXCL10, IL‐6, and NFκB by ketamine was not complete (comparison of NC vs. HK was not significant, and comparison of HC vs. HK was also not significant): this incomplete inhibition was reflected in insignificant P values (0.06 < P < 0.13) for the hypoxia × ketamine interaction term in the ANOVA (as reported in the legend to Fig. 4).
Figure 4. Ketamine ameliorates the increases in gene expression of inflammatory and immune related genes caused by hypoxia .
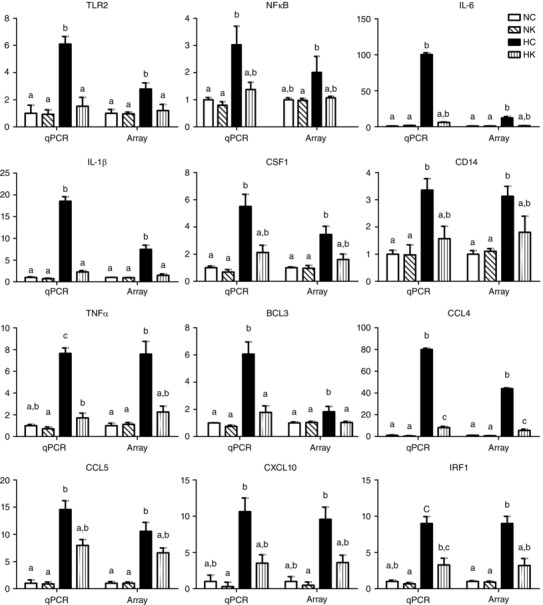
RNA gene expression was measured by microarray and validated by real‐time qPCR for inflammatory and immune related genes. All values are represented as a fold‐change from normoxic control group (NC, normoxia control; NK, normoxia + ketamine; HC, hypoxia control; HK, hypoxia + ketamine). Statistical significance between groups was accepted at P ≤ 0.05 (interaction term in two‐way ANOVA), and Duncan's test was performed to determine differences between groups (for results of Duncan's test, bars with different letters are statistically significantly different from each other, P < 0.05). For gene expression with P > 0.05 between groups (interaction term in two‐way ANOVA), the P values listed here are for real‐time qPCR validation: CXCL10 (P = 0.06); and for the arrays: CXCL10 (P = 0.07), CCL5 (P = 0.08), IL‐6 (P = 0.099), NFκB (P = 0.13). Data are presented as means ± SEM, and the y‐axis scale varies between plots.
We also validated the results of the transcriptomics modelling by immunostaining for macrophages in the same kidneys that were used for microarray analysis. As shown in Fig. 5, hypoxia significantly increased the number of macrophages in the kidney cortex, and ketamine significantly reduced the number of macrophages (P < 0.0001 for main effect of hypoxia; P = 0.0005 for main effect of ketamine; and P = 0.0070 for hypoxia × ketamine interaction).
Figure 5. Local macrophages expressing Iba‐1 immunoreactivity in the kidney cortex of NC, NK, HC and HK fetuses .
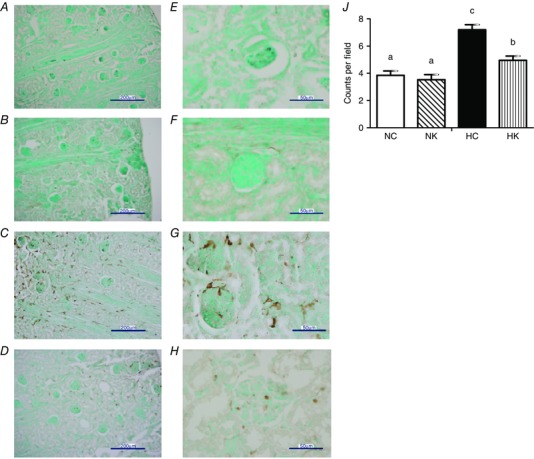
A and E, NC fetuses; B and F, NK fetuses; C and G, HC fetuses; D and H, HK fetuses. Sections were counterstained with methyl green. Macrophage counts per field at ×40 in the kidney cortex (J). Data are expressed as means ± SEM. Different letters indicate statistically significant difference (HC vs. HK P < 0.0001; HC vs. NC P < 0.0001; HC vs. NK P < 0.0001; HK vs. NC P = 0.018; HK vs. NK P = 0.005). Scale bars 200 μm (A–D), 50 μm (E–H). Abbreviations: NC, normoxia control; NK, normoxia + ketamine; HC, hypoxia control; HK, hypoxia + ketamine.
Discussion
Utilizing systems modelling techniques, we have found that 24 h after hypoxia, the genomic expression of immune and inflammatory pathways in the ovine fetal kidney cortex were upregulated, and that pretreatment with ketamine prevented or downregulated these responses. The results of this modelling suggest that hypoxia increases and ketamine decreases inflammation in the kidneys. The renal inflammation response to hypoxia is known and has been reported by others (Ikeda et al. 2000; O'Connell et al. 2003; Xia et al. 2015). The mechanism of the anti‐inflammatory action of ketamine is unclear but might be inferred from known actions of ketamine.
Hypoxia is a relatively common fetal stressor. Severe or prolonged hypoxia can cause growth retardation of the fetus and ischaemia–reperfusion injury to the fetal organs (Zhang, 2005; Giussani & Davidge, 2013). Acute hypoxaemia, especially when combined with acidaemia, stimulates a redistribution of combined fetal cardiac output with resultant decreased fetal renal blood flow by as much as 20% (Cohn et al. 1974). Combinations of decreased arterial oxygen content and decreased renal blood flow cause hypoxic and ischaemic damage to the kidneys, particularly to the renal tubules (Faber & Anderson, 1997; Gibson & Lumbers, 1999). Partial umbilical cord occlusion for 60 min resulting in a plasma pH of < 6.9, which is a severe asphyxic insult, caused renal tubular necrosis in near‐term ovine fetuses (Ikeda et al. 2000), and complete umbilical cord occlusion for 30 min caused renal distal tubule apoptosis in mid‐gestation fetuses 72 h after the occlusion (O'Connell et al. 2003). Hypoxia has also been shown to have an adverse effect on renal development by stimulating inflammatory, immune, and apoptotic processes in the fetal kidneys (Xia et al. 2015). It is conceivable that acute renal damage to the fetus could have long‐term effects on the proper development of renal tubules and vasculature.
In addition to localization within the brain, NMDA receptors are located throughout the kidney in various parts of the nephron: glomerulus (Zhang et al. 2010), collecting ducts (Sproul et al. 2011), epithelial proximal tubular cells (Bozic et al. 2011), and podocytes (Kottgen & Benzing, 2011), which suggest an important role for NMDA receptors in the regulation of renal function. Renal NMDA receptors increase renal vasodilatation via the nitric oxide pathway (Deng et al. 2002), suggesting that NMDA receptors are involved in maintaining renal haemodynamics. In models of renal ischaemia–reperfusion and hypoxia–reoxygenation, NMDA receptors were upregulated in the kidney cortex and the medulla (Yang et al. 2008; Pundir et al. 2013; Liu et al. 2014); activation of NMDA receptors is associated with increased oxidative stress and renal damage (Pundir et al. 2013). Excessive stimulation of NMDA receptors in the kidney can be toxic and cause apoptosis in proximal tubule‐like opossum kidney and distal‐tubule‐like Madin‐Darby canine kidney cells (Leung et al. 2008). In podocytes, over stimulation of the glutamatergic signalling pathway may weaken the integrity of the glomerular filtration barrier therefore causing glomerular damage (Kottgen & Benzing, 2011). Moreover, in both in vitro and in vivo models, the adverse effects of excessive activation of NMDA receptors were attenuated with NMDA receptor antagonists (Leung et al. 2008; Yang et al. 2008; Pundir et al. 2013).
It is known that ischaemia–reperfusion causes apoptosis and inflammation in the kidneys. Kruger et al. showed that, during a renal ischaemia–reperfusion event, activated caspases trigger an inflammatory response in the kidneys, by increasing IL‐1β and IL‐18, which lead to renal ischaemic injury (Melnikov et al. 2001, 2002). Our microarray results show that hypoxia alone induces significant upregulation of inflammatory mediators, such as, IL‐1β, IL‐18, IL‐1, IL‐6, IL‐8, TNFα, TLR2, and TLR4. In the proximal tubule epithelial cells, hypoxic events stimulate toll‐like receptor 4 (TLR4)‐mediated pathways to increase inflammatory (TNFα, IL‐8) and apoptotic (caspases 3, 8 and 9) markers (Liu et al. 2014). In another study, the magnitude of the increase in TLR4 gene and protein expression has been shown to correlate with the degree of ischaemia injury in the kidneys (Kruger et al. 2009). In a renal specific TLR4 knock‐out mouse model, the endothelial adhesion molecules, selectin E and ICAM1 were not expressed, suggesting that TLR4 is one of the main players in recruiting inflammatory cytokines (Chen et al. 2011). Our array analysis shows that hypoxia upregulates selectin P expression (log2 fold‐change = 3.89, adj. P value = 5.69E‐04), which is a related cell adhesion molecule that has an essential role in the recruitment of inflammatory cells and toll‐like receptors (TLR2, log2 fold‐change = 1.48, adj. P value = 9.41E‐03; TLR4, log2 fold‐change = 0.97, adj. P value = 1.78E‐02). The increase in selectin P expression might be a consequence of the upregulation of toll‐like receptors following hypoxia (Semple & Freedman, 2010; Rondina & Garraud, 2014).
The results of our experiment indicate that ketamine, a non‐competitive NMDA receptor antagonist, was able to cause changes in renal gene expression that are consistent with decreased fetal renal inflammatory and immunological responses to acute hypoxia. By finding the common set of genes that is both upregulated by hypoxia and downregulated by ketamine treatment before hypoxia, we were able to model the action of ketamine on the fetal renal cortex after hypoxia. With the list of 88 common genes (cytokines, chemokines, interleukins, glucose transporters), we analysed common pathways and biological processes and found that the most significant pathways involved inflammatory, immune, and metabolic responses. In normoxic conditions, the ovine fetal kidney reabsorbs glucose and releases it into the bloodstream; however, under acute hypoxic conditions, carbohydrate metabolism becomes anaerobic releasing lactate into the bloodstream (Iwamoto & Rudolph, 1985). Interestingly, our array results suggest that during hypoxia, local glucose transporters (SLC2A1, SLC2A3) are upregulated (SLC2A1, log2 fold‐change = 0.89, adj. P value = 2.47E‐03; SLC2A3, log2 fold‐change = 1.94, adj. P value = 9.66E‐04) and that ketamine suppresses this response (SLC2A3, log2 fold‐change = −1.49, adj. P value = 3.87E‐03; SLC2A1, log2 fold‐change = −0.61, adj. P value = 1.59E‐02). These results might be consistent with local vasodilatory effects of ketamine in the kidneys, sustaining the glucose supply and attenuating the changes in gene expression of renal glucose transporters. Furthermore, hypoxia upregulates SLC9A3, the gene encoding NHE3 (sodium–hydrogen exchanger; log2 fold‐change = 0.31, adj. P value = 4.07E‐02); ketamine downregulates the response (log2 fold‐change = −0.28, adj. P value = 4.55E‐02). It is possible that the upregulation of SLC9A3 might be secondary to lactic acidosis during hypoxia. Taken together, the pattern of gene expression suggests that ketamine ameliorates the fetal renal response to hypoxia.
A potential mechanism of both the effect of hypoxia to increase and the effect of ketamine to decrease markers of inflammation is an effect of hypoxia and of ketamine on the number and function of macrophages in the kidney cortex. As shown in Fig. 5, hypoxia increases the number of macrophages in the renal cortex, and ketamine reduces this response. While macrophages have been detected in the developing fetal human and mouse kidneys (Yasui et al. 1997), the function of local renal macrophages in the late gestation fetus still needs elucidating. Our discovery of the effect of hypoxia to increase renal macrophage count corroborates the results of the transcriptomics modelling and reinforces the notion that even transient hypoxia is damaging to the fetal kidney.
Our data suggest that ketamine reduces at least some of the negative consequences of hypoxic stress in fetal kidneys. In both sheep and humans, kidney maturation is complete by the time of parturition (Gimonet et al. 1998); therefore, an individual's nephron endowment cannot increase throughout life. Damage to the kidneys in utero therefore would create a functional deficiency that could not be recovered in postnatal life. For example, hypoxic damage might alter the course of nephron development or vascular regulatory mechanisms, having lifelong consequences for renal blood volume regulatory mechanisms (Gonzalez‐Rodriguez et al. 2013). Our results suggest that blockade of NMDA receptors might be clinically useful in sparing hypoxic damage to the developing kidneys. Nevertheless, it is also possible that blockade of NMDA receptor might have detrimental effects, including possible alterations in the pattern of kidney development in immature fetuses. Further experimentation will be needed to more fully understand the effect of ketamine on the fetal kidney.
Additional information
Competing interests
All authors declare that there are no competing financial, personal or professional interests.
Author contributions
E.I.C., C.E.W.: conception and design of the experiments, collection, analysis and interpretation of data, drafting the article, revising article critically for important intellectual content. M.B.R., M.A.Z: collection, analysis and interpretation of data, revising article critically for important intellectual content. E.M.R., M.K.: interpretation of data, revising article critically for important intellectual content. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed. All authors have approved the final version of the manuscript and agree to be accountable for all aspects of the work.
Funding
This research was supported by the National Institutes of Health (NIH) Grant HD33053 (C.E.W.), and by the NIH Training in Endocrine, Metabolic, and Prenatal Basis of Chronic Kidney Disease Predoctoral Fellowship T32DK076541 (E.I.C. and C.E.W.).
Acknowledgements
The authors thank Ms Xiaoying (Lisa) Fang, Ms Kristina Steinfeldt, Ms Heidi Straub, and Mr Thomas J. Arndt for their expert technical assistance. Also, we thank the Genomics Division of the University of Florida's Interdisciplinary Centre for Biotech Research for the use of the Agilent Bioanalyzer and Agilent scanner.
References
- Anand KJ (2007). Pharmacological approaches to the management of pain in the neonatal intensive care unit. J Perinatol 27 (Suppl. 1), S4–S11. [DOI] [PubMed] [Google Scholar]
- Bozic M, de Rooij J, Parisi E, Ortega MR, Fernandez E & Valdivielso JM (2011). Glutamatergic signalling maintains the epithelial phenotype of proximal tubular cells. J Am Soc Nephrol 22, 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EI & Wood CE (2013). Ketamine decreases apoptotic and inflammatory gene expression in fetal hippocampus exposed to global acute hypoxic hypoxia In Reproductive Sciences, Vol. 20, pp. F089. [Google Scholar]
- Chang EI & Wood CE (2015). Ketamine attenuates the ACTH response to hypoxia in late‐gestation ovine fetus. Neonatology 107, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, John R, Richardson JA, Shelton JM, Zhou XJ, Wang Y, Wu QQ, Hartono JR, Winterberg PD & Lu CY (2011). Toll‐like receptor 4 regulates early endothelial activation during ischemic acute kidney injury. Kidney Int 79, 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila‐Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T & Bader GD (2007). Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2, 2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn HE, Sacks EJ, Heymann MA & Rudolph AM (1974). Cardiovascular responses to hypoxemia and acidemia in fetal lambs. Am J Obstet Gynecol 120, 817–824. [DOI] [PubMed] [Google Scholar]
- Deng A, Valdivielso JM, Munger KA, Blantz RC & Thomson SC (2002). Vasodilatory N‐methyl‐d‐aspartate receptors are constitutively expressed in rat kidney. J Am Soc Nephrol 13, 1381–1384. [DOI] [PubMed] [Google Scholar]
- Faber JJ & Anderson DF (1997). Angiotensin mediated interaction of fetal kidney and placenta in the control of fetal arterial pressure and its role in hydrops fetalis. Placenta 18, 313–326. [DOI] [PubMed] [Google Scholar]
- Gibson KJ & Lumbers ER (1999). Effects of bilateral nephrectomy and angiotensin II replacement on body fluids in foetal sheep. Clin Exp Pharmacol Physiol 26, 765–773. [DOI] [PubMed] [Google Scholar]
- Gimonet V, Bussieres L, Medjebeur AA, Gasser B, Lelongt B & Laborde K (1998). Nephrogenesis and angiotensin II receptor subtypes gene expression in the fetal lamb. Am J Physiol Renal Physiol 274, F1062–F1069. [DOI] [PubMed] [Google Scholar]
- Giussani DA & Davidge ST (2013). Developmental programming of cardiovascular disease by prenatal hypoxia. J Dev Orig Health Dis 4, 328–337. [DOI] [PubMed] [Google Scholar]
- Giussani DA, Spencer JA & Hanson MA (1994). Fetal cardiovascular reflex responses to hypoxaemia. Fetal Matern Med Rev 6, 17–37. [Google Scholar]
- Gonzalez‐Rodriguez P, Jr. , Tong W, Xue Q, Li Y, Hu S & Zhang L (2013). Fetal hypoxia results in programming of aberrant angiotensin ii receptor expression patterns and kidney development. Int J Med Sci 10, 532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LR, Bennet L, Robson S & Hanson MA (1997). The role of carotid chemoreceptors in the effects of hypoxia on renal blood flow in the late gestation sheep fetus. Exp Physiol 82, 183–192. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT & Lempicki RA (2009. a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT & Lempicki RA (2009. b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Murata Y, Quilligan EJ, Parer JT, Murayama T & Koono M (2000). Histologic and biochemical study of the brain, heart, kidney, and liver in asphyxia caused by occlusion of the umbilical cord in near‐term fetal lambs. Am J Obstet Gynecol 182, 449–457. [DOI] [PubMed] [Google Scholar]
- Iwamoto HS & Rudolph AM (1985). Metabolic responses of the kidney in fetal sheep: effect of acute and spontaneous hypoxemia. Am J Physiol Renal Physiol 249, F836–F841. [DOI] [PubMed] [Google Scholar]
- Kottgen M & Benzing T (2011). Strangers on a train: atypical glutamate receptors in the kidney glomerulus. Focus on “Functional NMDA receptors with atypical properties are expressed in podocytes”. Am J Physiol Cell Physiol 300, C9–C10. [DOI] [PubMed] [Google Scholar]
- Kruger B, Krick S, Dhillon N, Lerner SM, Ames S, Bromberg JS, Lin M, Walsh L, Vella J, Fischereder M, Kramer BK, Colvin RB, Heeger PS, Murphy BT & Schroppel B (2009). Donor Toll‐like receptor 4 contributes to ischemia and reperfusion injury following human kidney transplantation. Proc Natl Acad Sci USA 106, 3390–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JC, Ragland N, Marphis T & Silverstein DM (2008). NMDA agonists and antagonists induce renal culture cell toxicity. Med Chem 4, 565–571. [DOI] [PubMed] [Google Scholar]
- Leung JC, Travis BR, Verlander JW, Sandhu SK, Yang SG, Zea AH, Weiner ID & Silverstein DM (2002). Expression and developmental regulation of the NMDA receptor subunits in the kidney and cardiovascular system. Am J Physiol Regul Integr Comp Physiol 283, R964–R971. [DOI] [PubMed] [Google Scholar]
- Liu XJ, Tan Y, Geng YQ, Wang Z, Ye JH, Yin XY & Fu B (2014). Proximal tubule toll‐like receptor 4 expression linked to inflammation and apoptosis following hypoxia/reoxygenation injury. Am J Nephrol 39, 337–347. [DOI] [PubMed] [Google Scholar]
- Luciano R, Gallini F, Romagnoli C, Papacci P & Tortorolo G (1998). Doppler evaluation of renal blood flow velocity as a predictive index of acute renal failure in perinatal asphyxia. Eur J Pediatr 157, 656–660. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K & Kuiper M (2005). BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. [DOI] [PubMed] [Google Scholar]
- Matsumoto LC, Cheung CY & Brace RA (2001). Increased urinary flow without development of polyhydramnios in response to prolonged hypoxia in the ovine fetus. Am J Obstet Gynecol 184, 1008–1014. [DOI] [PubMed] [Google Scholar]
- Mellon RD, Simone AF & Rappaport BA (2007). Use of anaesthetic agents in neonates and young children. Anesth Analg 104, 509–520. [DOI] [PubMed] [Google Scholar]
- Melnikov VY, Ecder T, Fantuzzi G, Siegmund B, Lucia MS, Dinarello CA, Schrier RW & Edelstein CL (2001). Impaired IL‐18 processing protects caspase‐1‐deficient mice from ischemic acute renal failure. J Clin Invest 107, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov VY, Faubel S, Siegmund B, Lucia MS, Ljubanovic D & Edelstein CL (2002). Neutrophil‐independent mechanisms of caspase‐1‐ and IL‐18‐mediated ischemic acute tubular necrosis in mice. J Clin Invest 110, 1083–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell AE, Boyce AC, Lumbers ER & Gibson KJ (2003). The effects of asphyxia on renal function in fetal sheep at midgestation. J Physiol 552, 933–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MJ & Wood CE (2007). Ketamine inhibits fetal ACTH responses to cerebral hypoperfusion. Am J Physiol Regul Integr Comp Physiol 292, R1542–R1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pundir M, Arora S, Kaur T, Singh R & Singh AP (2013). Effect of modulating the allosteric sites of N‐methyl‐d‐aspartate receptors in ischemia–reperfusion induced acute kidney injury. J Surg Res 183, 668–677. [DOI] [PubMed] [Google Scholar]
- Rabaglino MB, Richards E, Denslow N, Keller‐Wood M & Wood CE (2012). Genomics of estradiol‐3‐sulfate action in the ovine fetal hypothalamus. Physiol Genom 44, 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RCoreTeam (2013). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W & Smyth GK (2015). limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondina MT & Garraud O (2014). Emerging evidence for platelets as immune and inflammatory effector cells. Front Immunol 5, 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roytblat L, Talmor D, Rachinsky M, Greemberg L, Pekar A, Appelbaum A, Gurman GM, Shapira Y & Duvdenani A (1998). Ketamine attenuates the interleukin‐6 response after cardiopulmonary bypass. Anesth Analg 87, 266–271. [DOI] [PubMed] [Google Scholar]
- Semple JW & Freedman J (2010). Platelets and innate immunity. Cell Mol Life Sci 67, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B & Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, Article3. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Michaud J & Scott HS (2005). Use of within‐array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21, 2067–2075. [DOI] [PubMed] [Google Scholar]
- Sproul A, Steele SL, Thai TL, Yu S, Klein JD, Sands JM & Bell PD (2011). N‐Methyl‐d‐aspartate receptor subunit NR3a expression and function in principal cells of the collecting duct. Am J Physiol Renal Physiol 301, F44–F54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Kai S, Matsuyama T, Adachi T, Fukuda K & Hirota K (2013). General anaesthetics inhibit LPS‐Induced IL‐1β expression in glial cells. PLoS One 8, e82930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z & Zhang B (2013). WEB‐based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 41, W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde‐Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD & Morris Q (2010). The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38, W214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CE, Kane C & Raff H (1990). Peripheral chemoreceptor control of fetal renin responses to hypoxia and hypercapnia. Circ Res 67, 722–732. [DOI] [PubMed] [Google Scholar]
- Wood CE, Rabaglino MB, Chang EI, Denslow N, Keller‐Wood M & Richards E (2013). Genomics of the fetal hypothalamic cellular response to transient hypoxia: endocrine, immune, and metabolic responses. Physiol Genom 45, 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CE, Rabaglino MB, Richards E, Denslow N, Zarate MA, Chang EI & Keller‐Wood M (2014). Transcriptomics of the fetal hypothalamic response to brachiocephalic occlusion and estradiol treatment. Physiol Genom 46, 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GJ, Chen TL, Ueng YF & Chen RM (2008). Ketamine inhibits tumor necrosis factor‐alpha and interleukin‐6 gene expressions in lipopolysaccharide‐stimulated macrophages through suppression of toll‐like receptor 4‐mediated c‐Jun N‐terminal kinase phosphorylation and activator protein‐1 activation. Toxicol Appl Pharmacol 228, 105–113. [DOI] [PubMed] [Google Scholar]
- Xia S, Lv J, Gao Q, Li L, Chen N, Wei X, Xiao J, Chen J, Tao J, Sun M, Mao C, Zhang L & Xu Z (2015). Prenatal exposure to hypoxia induced beclin 1 signalling‐mediated renal autophagy and altered renal development in rat fetuses. Reprod Sci 22, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CC, Chien CT, Wu MH, Ma MC & Chen CF (2008). NMDA receptor blocker ameliorates ischemia–reperfusion‐induced renal dysfunction in rat kidneys. Am J Physiol Renal Physiol 294, F1433–F1440. [DOI] [PubMed] [Google Scholar]
- Yang Z, Chen ZQ & Jiang XQ (2006). [Effects of subanesthetic dose of ketamine on perioperative serum cytokines in orthotopic liver transplantation]. Nan Fang Yi Ke Da Xue Xue Bao 26, 802–804, 817. [PubMed] [Google Scholar]
- Yasui M, Tanaka H & Seino Y (1997). The role of tissue‐fixed macrophages in apoptosis in the developing kidney. Nephron 77, 325–332. [DOI] [PubMed] [Google Scholar]
- Zeyneloglu P, Donmez A, Bilezikci B & Mercan S (2005). Effects of ketamine on serum and tracheobronchial aspirate interleukin‐6 levels in infants undergoing cardiac surgery. J Cardiothorac Vasc Anaesth 19, 329–333. [DOI] [PubMed] [Google Scholar]
- Zhang B, Kirov S & Snoddy J (2005). WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33, W741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yi F, Xia M, Boini KM, Zhu Q, Laperle LA, Abais JM, Brimson CA & Li PL (2010). NMDA receptor‐mediated activation of NADPH oxidase and glomerulosclerosis in hyperhomocysteinemic rats. Antioxid Redox Signal 13, 975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L (2005). Prenatal hypoxia and cardiac programming. Reprod Sci 12, 2–13. [DOI] [PubMed] [Google Scholar]
- Zhang XF, Dai DQ, Ou‐Yang L & Yan H (2014). Detecting overlapping protein complexes based on a generative model with functional and topological properties. BMC Bioinformatics 15, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]


