Abstract
Dubowitz Syndrome is an autosomal recessive disorder characterized by the constellation of mild microcephaly, growth and mental retardation, eczema and peculiar facies, but causes are still unknown. We studied a multiplex consanguineous family with many features of Dubowitz syndrome using whole exome sequencing and identified a splice mutation in NSUN2, encoding a conserved RNA methyltransferase. NSUN2 has been implicated in Myc-induced cell proliferation and mitotic spindle stability, which might help explain the varied clinical presentations that can include chromosomal instability and immunological defects. Patient cells displayed loss of NSUN2-specific methylation at two residues of the aspartate tRNA. Our findings establish NSUN2 as the first causal gene with relationship to the Dubowitz syndrome spectrum phenotype.
Keywords: Dubowitz, NSUN2, MISU, RNA methylation, microcephaly
Dubowitz Syndrome (DS, %223370) is a rare, developmental disorder characterized by small stature, intellectual disability, mild microcephaly, and a distinct facial appearance consisting of blepharophimosis, broad nasal bridge, sloping forehead and micrognathia [1, 2]. Although first reported in 1965 by Dubowitz as a condition characterized by familial low birthweight, dwarfism with unusual facies and skin eruption [3], the phenotypic spectrum has been expanded to include a wide variety of less frequent features such as congenital heart defects, frequent infections and chromosomal instability [4]. DS is distinguished from other blepharophimosis-mental retardation syndromes such as Say-Barber-Biesecker variant of Ohdo syndrome [2], recently linked to mutations in the KAT6B gene encoding a histone acetyltransferase [5]. To date there are approximately 140 DS reported cases with no causative mutations identified. Although there is a presumed recessive mode of inheritance in most cases, there are reports that suggest more complex modes of inheritance can occur [6].
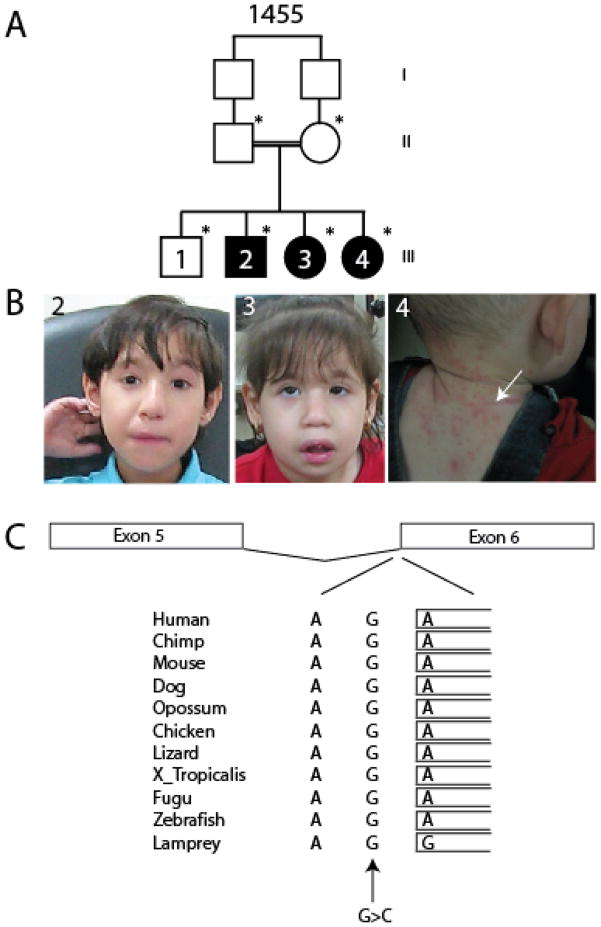
We identified a consanguineous multiplex family 1455 in the United Arab Emirates of Lebanese origin (Fig. 1A). The eldest sibling was healthy with no pathological features. The three affected members displayed features consistent with DS, including low birth weight (5% or less), and oligohydramnios in the two older affected individuals (Table 1), which are compared with typical features of DS [2]. By 1 year of age, growth retardation, and developmental delay were noted. Facial features included blepharophimosis, telecanthus, hypertelorism, high or broad nasal bridge (Fig. 1B). Oral features included smooth philtrum, downturning upper lip and dental anomalies. Eczema was clearly apparent over the chest and back in the youngest affected at the time of evaluation, requiring treatment, although the two older affected individuals did not have a history of notable eczema. The neurological features included microcephaly (−2%ile occipital-frontal head circumference at birth, progressing to −4 SD 2 years of age), axial hypotonia, and autistic features. The eldest affected displayed epileptic seizures, controlled on valproate, a feature not observed in the younger two affected individuals by the time of evaluation. The brain MRI was unremarkable. The remainder of the history and physical was non-contributory, and serum chemistry, immune profiles and routine karyotype were normal. However, notable hoarse voice, triangular face, round nose tip and abnormal ears, occasionally observed in DS, were not reported. We therefore cautiously refer to this condition as a Dubowitz-like condition, although it may in fact represent Dubowitz syndrome, and the pedigree is consistent with a recessive mode of inheritance.
Figure 1.
Pedigree, phenotype, and predicted effect on splicing of the NSUN2 mutation. (A) Family-1455 presents three affected children from a first-cousin marriage. (B) Photograph of affected individuals, demonstrating blepharophimosis, telecanthus, hypertelorism, high/broad nasal bridge, smooth philtrum, and downturning upper lip in 2 and 3. Eczema on the nape and back are visible in 4 (arrow). (C) Invariant conservation across species of the G residue at the predicted exon 6 splice acceptor site (arrow), with mutant G>C base indicated.
Table 1.
| Features | Reported in Dubowitz S. | 1455-3-2 | 1455-3-3 | 1455-3-4 |
|---|---|---|---|---|
| Pregnancy/neonatal | ||||
| IUGR | 69% | 2.200 kg (<5%) | 2.900 kg (5%) | 2.900 kg (5%) |
| Oligohydramnios | 2% | + | + | − |
| Respiratory problems | 15% | − | − | − |
| Feeding difficulties/GER | 22% | − | − | − |
| Frequent infections | 32% | − | − | − |
| Growth retardation (PN) | 86% | + | + | + |
| Developmental/mental delay | 72% | + | + | + |
| Speech delay | 67% | + | + | + |
| Craniofacial | ||||
| Microcephaly | 79% | + | + | + |
| Blepharophimosis | 43% | + | + | + |
| Ptosis | 38% | − | − | − |
| Upward palpebral fissures | 11% | − | − | − |
| Telecanthus | 25% | + | + | + |
| Hypertelorism | 19% | + | + | + |
| Broad/depressed nasal bridge | 22% | high bridge | + | + |
| Anteverted nares | − | − | − | − |
| Low-set/dysplastic ears | 25% | − | − | − |
| Preauricular pits | − | − | − | − |
| Cleft palate | 8% | − | − | − |
| Micrognathia | 57% | − | + | − |
| Tooth problems | 29% | + | + | + |
| Small mouth | Rare | − | − | − |
| Mask like/immobile face | − | − | + | − |
| smooth philtrum | + | + | + | |
| downturning upper lip/absent cupid bow | + | + | + | + |
| Neurological | ||||
| Abnormal brain image | 3% | − | − | − |
| Axial hpotonia | 40% | + | + | + |
| Ocular impairment | 22% | − | − | − |
| Hearing impairment | − | − | − | − |
| Cutaneous | ||||
| Sparse hair/thin eyebrows | 41% | − | + | + |
| Eczema | 42% | − | − | + |
| Thin skin | − | − | − | − |
| Cardiac | 9% | − | − | − |
| Gastrointestinal | ||||
| Gastrectasia | − | − | − | − |
| Constipation | 9% | − | − | + |
| Distension of gall-bladder | − | − | − | − |
| Skeletal/limb | ||||
| Long thumbs | Rare | − | − | − |
| Clinodactyl of 5th fingers | 34% | − | − | − |
| Contractures | − | − | − | − |
| Urogenital abnormalities | 41% | − | − | − |
| Thyroid abnormalities | Rare | + | − | − |
| Other | HA, HD | Self Stimulation, Convulsions | Self Stimulation | Self Stimulation |
| Inheritance | AR | AR | AR | AR |
AR: Autosomal Recessive; HD: Hematological disturbance; HA: Hyperactivity
In order to identify the gene for this condition, we performed whole exome sequencing on two affected siblings. The study was approved by the Ethics Committee and the family provided informed consent for study. Blood DNA was extracted using Qiagen reagents, then subjected to exome capture with the Agilent SureSelect Human All Exome 50 Mb kit, sequenced on an Illumina HiSeq2000 instrument, resulting in ~94% target coverage at > 10X depth. GATK [7] was used for variant identification and intersected with identity-by-descent blocks identified by HomozygosityMapper [8], and then filtered for homozygous variants shared between the two affected individuals using custom Python scripts (available upon request).
We identified a homozygous variant in an intronic region of the gene NSUN2, located one base pair upstream of the start of exon 6 (chr. 5 base position 6,622,214 C>G; hg19) at the canonical splice acceptor site (Fig. 1C). The variant was found to be homozygous in all three affected cases, occurred with a 19.5 cM block of homozygosity in both affected members between chr. 5 base position 2,752,824–22,078,584 (Supplementary Fig. 1), was absent in the single unaffected sibling, and heterozygous in each parent, and thus segregated according to a recessive mode of inheritance.
There were six other shared novel homozygous genetic variants, each leading to amino acid transversions annotated to be potentially damaging, occurring in the CUL4A, RIMS2, TARP, PKHD1L1, CACHD1, and TRHR genes. However, the reference amino acid for each was not fully conserved across evolution, and further upon direct testing was found to fail test of segregation in the family. There were no other likely deleterious variants that segregated according to the presumed recessive mode of inheritance, supporting pathogenicity of the NSUN2 variant. The variant was not encountered in our in-house exome database of ~1000 Middle Eastern individuals, and was not reported in any public databases. We also found the variant absent in a panel of 96 ethnically matched Arab controls. The mutated position was highly conserved across species and annotation with SeattleSeq predicted it to be critical for splicing. Direct Sanger sequence analysis of four additional patients with DS-like phenotypes for all coding and splice sites yielded no obvious mutations, suggesting genetic heterogeneity.
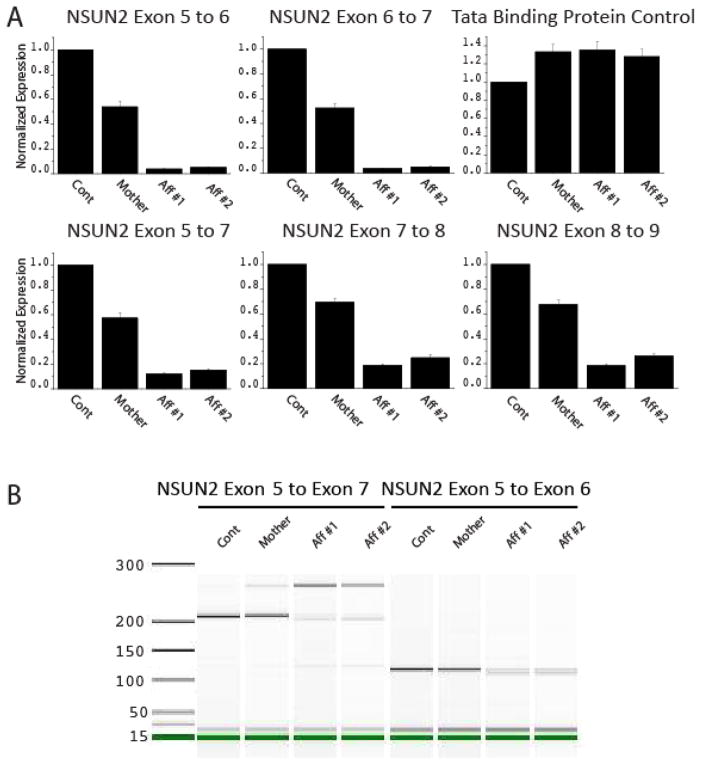
The NSUN2 gene is alternatively spliced with at least four splice variants and three proposed alternative start sites of transcription. To determine whether the exon 6 splice acceptor mutation leads to altered mRNA levels, we performed quantitative RT-PCR. Dermal fibroblasts were obtained from two affected individuals, the mother, and one unrelated control, cultured under similar conditions, and total RNA was reverse transcribed using SuperScript III (Invitrogen). Amplifications from exon 5 to 6 and exon 6 to 7 were compared with a control reaction against TATA binding protein (TBP) using the LightCycler 480 SYBR Green I (Roche) assay. All assays were normalized to GAPDH and then to the expression level of the unrelated control. We found a severe 95% reduction in normalized expression for both products from both affected patients, whereas the obligate carrier mother showed about a 50% reduction in amplification, suggesting a functional consequence of the mutation on the NSUN2 mRNA (Fig. 2A). Because exon 6 is not present in all deposited NSUN2 mRNAs, and its absence maintains the open reading frame, we additionally assessed amplifications from exon 5 to 7, 7 to 8 and 8 to 9. In all reactions, we found comparable 70–80% reduction in NSUN2 mRNA, which excluded the possibility of trivial exon skipping. The results suggest a global reduction in mRNA levels from altered splicing, resulting in either partial or complete loss of mRNA in the two affected individuals.
Figure 2.
Altered NSUN2 mRNA levels and aberrant splicing in affected individuals from Family-1455. (A) Each experiment compares qRT-PCR levels from healthy unrelated control to mother and two affected members. Severely reduced incorporation of exon 6 was detected from exon 5 to 6 and 6 to 7 reactions, whereas mother displayed about 50% of control. TATA Binding Protein qRT-PCR, used as control. Reactions from exons 5 to 7, 7 to 8 and 8 to 9 showed overall reduction in NSUN2 mRNA levels. (B) Agilent Bioanalyzer visualization of aberrant splicing of NSUN2 mRNA in Family-1455. Amplification from exon 5 to 7 produced a 210 bp product in control and mother, severely reduced in the affected individuals, who instead show a 250 bp product, also evident in mother at reduced level, containing an AluY exon.
To determine the effect on splicing, we analyzed RT-PCR products from affected patient and control fibroblasts by Bioanalyzer (Agilent, Inc) electrophoresis. Amplification from exon 5 to 7 demonstrated an aberrant sized product from the two affected individuals of about 250 bp, also evident in the mother’s sample, whereas the predicted size product of 210 bp was notably reduced in the two affected individuals. Products from exon 5 to 6 were reduced without an obvious difference in molecular weight (Fig. 2B). Analysis of the reference genome for possible cryptic splice acceptor and donor sites identified an AluY transposon located between exon 6 and 7. Replacement of exon 6 with a single arm of the transposon is predicted to yield a 250 bp cDNA product from exon 5 to 7. In order to determine if this AluY exon was incorporated in the mRNA, we sequenced the aberrant 250 bp band. As predicted, we identified a sequence corresponding precisely to the 3′ arm of the AluY transposon (Supplementary Fig. 2A). This sequence trace demonstrated splicing from exon 5 to the 3′ AluY arm to exon 7, which is predicted to maintain the reading frame (Supplementary Fig. 2B, Supplementary Fig. 3). Alu repeats have a two-part structure consisting of 5′ and 3′ arms separated by an ‘A’ rich linker region. The 3′ arm is terminated by a PolyA tail [9]. Novel insertions of Alu elements within the genome have been linked to disease, and their incorporation into mRNAs is frequently associated with microRNA-mediated degradation. Indeed dozens of human microRNAs are predicted to target conserved regions of Alu transposons[10].
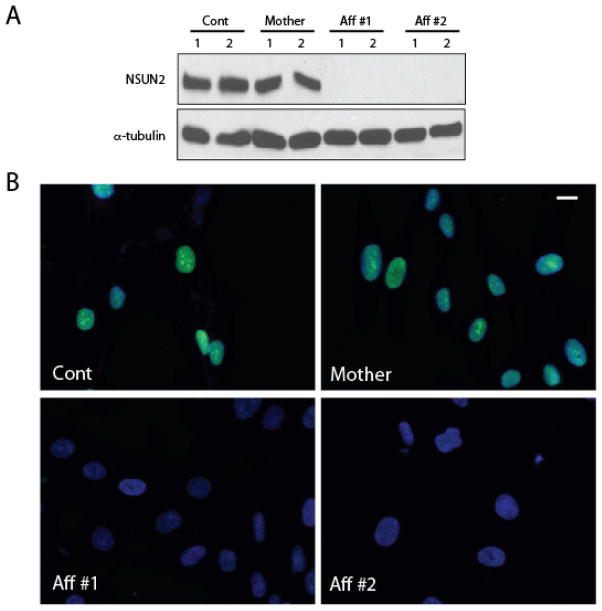
In order to test for an effect on NSUN2 protein expression, we generated whole cell lysates from cultured fibroblasts and performed Western analysis by loading each sample in duplicate. We probed the blot for NSUN2 protein using a rabbit affinity-purified polyclonal antibody (Proteintech Catalog 20854-1), generated against full-length recombinant human protein. In control and mother cell line, we detected robust protein expression at the expected size of about 100 kD, similar to the published molecular weight [11]. Protein loading was controlled by probing the blot with anti-alpha-tubulin. In both patient cell lines, however, there was undetectable protein (Fig. 3A). To confirm these results, we fixed and stained these fibroblasts with NSUN2 antibody. Because NSUN2 is a nuclear and nucleolar protein, we counterstained nuclei with Hoechst. In control and mother lines, NSUN2 staining overlapped precisely with nuclei and nucleoli. In both patient lines, in images acquired with identical settings, there was striking reduction in staining for NSUN2, without detectable signal in either nucleoplasm or nucleoli (Fig. 3B). The data suggests severe reduction in NSUN2 protein levels in patient cells.
Figure 3.
Reduced NSUN2 protein levels in affected cell lines from Family-1455. (A) Western blot from whole cell extracts of control, mother and two affected individuals (Aff #1 and Aff #2), loaded in duplicate, show undetectable protein, compared with alpha-tubulin loading control. (B) Immunofluorescence of same cells showing undetectable NSUN2 reactivity (green) but intact Hoechst (blue) staining in affected cells compared with cells from control and mother. Staining is specific to nuclei and is enriched at nucleoli as previously described. Scale bar 10 uM.
NSUN2 is an RNA methyltransferase known to operate downstream of c-MYC during cell proliferation. It contains a NOL1/NOP2/Sun domain, and was demonstrated to have in vitro activity to methylate cytosine in tRNA and hemimethylated DNA (polydI:dC) [11, 12]. In yeast, the closest orthologue is the gene SUN (Trm4/Ncl1), which is capable of methylating tRNAs at all four known sites (positions 34, 40, 48, 49) and where gene inactivation completely blocks tRNA methylation [13]. A recent study utilizing metazoan cells demonstrated a requirement for NSUN2 in cytosine methylation (m5C) of residue 34 in tRNALeu(CAA) and residue 47 and 48 in tRNAAsp(GTC) [14], whereas residue 38 in tRNAAsp(GTC) is methylated by the alternative methyltransferase DNMT2 [15]. NSUN2 deficient mice lack m5C of tRNALEU(CAA) at position C34. These mice also display a low body weight phenotype and partial alopecia, as well as altered cell cycle timing and self-renewal capacity in epidermal stem cells [16].
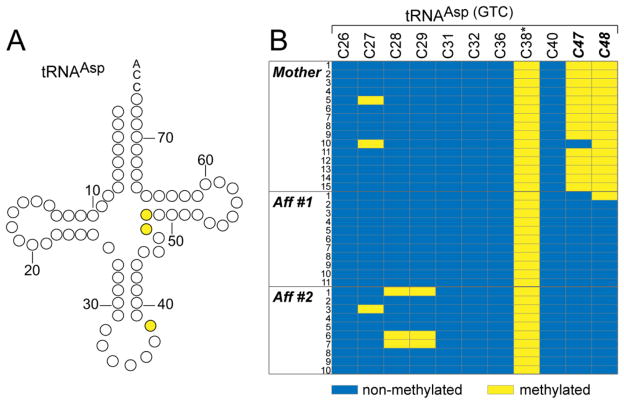
To test for similar defects in human NSUN2-mutant cells, RNA was isolated from dermal fibroblasts, and processed for bisulfite sequencing as previously described [16]. Primers were designed to amplify mature tRNAASP(GTC) (Fig. 4A, Fw: 5′-GTTAGTATAGTGGTGAGTAT-3′, Rev: 5′-CTCCCCATCAAAAAATCAAAC-3′). Visualization of bisulfite sequencing data demonstrated the loss of cytosine-5 methylation at these NSUN2 target sites (C47 and C48) in affected individuals but not the unaffected mother (Figure 4B). Position C38, the target of DNMT2, was methylated in all individuals. These results suggest that loss of the NSUN2 gene results in a loss of site-specific 5-cytosine methylation in patient cells. DNMT2 and NSUN2 are the only known tRNA:m5C methyltransferases known, and although recently demonstrated to protect tRNA from cleavage and degradation [15], the significance of this post-translational modification is not well understood.
Figure 4.
Site-specific loss of cytosine-5 methylation (m5C) in two affected members. (A) Secondary structure of tRNAAsp. Prediction is that C47 and C48 require NSUN2 for m5C, whereas C38, methylated by DNMT2, will be unaffected. (B) RNA bisulfite sequencing of tRNAAsp(GTC) in the mother as a control and two affected members. In affected members m5C is specifically lost at NSUN2-dependent sites C47 and C48 (highlighted in bold) whereas m5C of C38 (highlighted with a star) remains unchanged. Blue indicates cytosine-5 not methylated; yellow indicated cytosine-5 methylated.
NSUN2 has been shown to interact with the mitotic spindle and to play an important role in cell division, and interestingly, its effect on mitotic spindle stability is independent of this methyltransferase activity [17]. Thus it has been hypothesized to play a dual role in mammalian cells. Phosphorylation of NSUN2 by the cell-cycle related kinase Aurora reduces methyltransferase activity [11], further supporting this dual-role hypothesis. Cultured cells depleted of NSUN2 exhibit increased apoptosis, reduced proliferation, and a variety of spindle defects leading to abnormal mitoses [17], which might relate to the frequent reports of potential defects in immunological function and chromosomal instability in DS. Our discovery of a damaging mutation in NSUN2 leading to a DS-like condition further highlights the role of cell proliferation genes in normal brain development and emphasizes the connection between abnormal cell cycle and microcephaly phenotypes.
Supplementary Material
Acknowledgments
We thank J. Meerloo at the UCSD Neurosciences Core for microscopy services (US National Institute of Neurological Disorders and Stroke P30NS047101), Bruno Reversade for sequencing NSUN2 in additional Dubowitz probands (from the www.dubowitzsyndrome.org cohort), Broad Institute (U54HG003067 to Eric Lander) for sequencing support and analysis. This work was supported by the US National Institutes of Health R01NS048453, R01NS052455 and P01HD070494 (J.G.G.), the Daland Fellowship from the American Philosophical Society (J.H.L), the American Heart Association 09POST2250641 (J.E.L.), the Simons Foundation Autism Research Initiative, and the Howard Hughes Medical Institute (J.G.G.).
Footnotes
Web Resources
The URLs for data presented herein are as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
SeattleSeq Annotation, http://gvs.gs.washington.edu/
Genome browser, http://www.genome.ucsc.edu/
References
- 1.Tsukahara M, Opitz JM. Dubowitz syndrome: review of 141 cases including 36 previously unreported patients. Am J Med Genet. 1996;63(1):277–89. doi: 10.1002/(SICI)1096-8628(19960503)63:1<277::AID-AJMG46>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 2.Dentici ML, Mingarelli R, Dallapiccola B. The difficult nosology of blepharophimosis-mental retardation syndromes: report on two siblings. Am J Med Genet A. 2011;155A(3):459–65. doi: 10.1002/ajmg.a.33642. [DOI] [PubMed] [Google Scholar]
- 3.Dubowitz V. Familial Low Birthweight Dwarfism with an Unusual Facies and a Skin Eruption. J Med Genet. 1965;2(1):12–7. doi: 10.1136/jmg.2.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Nemri AR, Kilani RA, Salih MA, Al-Ajlan AA. Embryonal rhabdomyosarcoma and chromosomal breakage in a newborn infant with possible Dubowitz syndrome. Am J Med Genet. 2000;92(2):107–10. doi: 10.1002/(sici)1096-8628(20000515)92:2<107::aid-ajmg5>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 5.Clayton-Smith J, O’Sullivan J, Daly S, Bhaskar S, Day R, Anderson B, Voss AK, Thomas T, Biesecker LG, Smith P, Fryer A, Chandler KE, Kerr B, Tassabehji M, Lynch SA, Krajewska-Walasek M, McKee S, Smith J, Sweeney E, Mansour S, Mohammed S, Donnai D, Black G. Whole-exome-sequencing identifies mutations in histone acetyltransferase gene KAT6B in individuals with the Say-Barber-Biesecker variant of Ohdo syndrome. Am J Hum Genet. 2011;89(5):675–81. doi: 10.1016/j.ajhg.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehes K. Morphogenetic variants in parents of children with recessive malformation syndromes. Kinderarztl Prax. 1990;58(3):125–9. [PubMed] [Google Scholar]
- 7.Depristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seelow D, Schuelke M, Hildebrandt F, Nurnberg P. HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37(Web Server issue):W593–9. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3(5):370–9. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 10.Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22(10):532–6. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Sakita-Suto S, Kanda A, Suzuki F, Sato S, Takata T, Tatsuka M. Aurora-B regulates RNA methyltransferase NSUN2. Mol Biol Cell. 2007;18(3):1107–17. doi: 10.1091/mbc.E06-11-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16(10):971–81. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 13.Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5(8):1105–18. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24(15):1590–5. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011;7(12):e1002403. doi: 10.1371/journal.pgen.1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain S, Benavente SB, Nascimento E, Dragoni I, Kurowski A, Gillich A, Humphreys P, Frye M. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J Cell Biol. 2009;186(1):27–40. doi: 10.1083/jcb.200810180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.