Abstract
Human chromosome 14q32.2 carries paternally expressed genes including DLK1 and RTL1, and maternally expressed genes including MEG3 and RTL1as, along with the germline-derived DLK1-MEG3 intergenic differentially methylated region (IG-DMR) and the postfertilization-derived MEG3-DMR. Consistent with this, paternal uniparental disomy 14 (upd(14)pat), and epimutations (hypermethylations) and microdeletions affecting the IG-DMR and/or the MEG3-DMR of maternal origin, result in a unique phenotype associated with characteristic face, a small bell-shaped thorax with coat-hanger appearance of the ribs, abdominal wall defects, placentomegaly and polyhydramnios. Recently, the name ‘Kagami–Ogata syndrome' (KOS) has been approved for this clinically recognizable disorder. Here, we review the current knowledge about KOS. Important findings include the following: (1) the facial ‘gestalt' and the increased coat-hanger angle constitute pathognomonic features from infancy through childhood/puberty; (2) the unmethylated IG-DMR and MEG3-DMR of maternal origin function as the imprinting control centers in the placenta and body respectively, with a hierarchical interaction regulated by the IG-DMR for the methylation pattern of the MEG3-DMR in the body; (3) RTL1 expression level becomes ~2.5 times increased in the absence of functional RTL1as-encoded microRNAs that act as a trans-acting repressor for RTL1; (4) excessive RTL1 expression and absent MEG expression constitute the primary underlying factor for the phenotypic development; and (5) upd(14)pat accounts for approximately two-thirds of KOS patients, and epimutations and microdeletions are identified with a similar frequency. Furthermore, we refer to diagnostic and therapeutic implications.
Introduction
Human chromosome 14q32.2 harbors an imprinted region.1, 2 Consistent with this, paternal uniparental disomy 14 (upd(14)pat) results in a unique phenotypic constellation, and maternal uniparental disomy 14 (upd(14)mat) leads to less characteristic but clinically discernible features.2, 3 Upd(14)pat- and upd(14)mat-compatible phenotypes are also caused by epimutations and microdeletions affecting the maternally and paternally derived chromosome 14q32.2 imprinted region, respectively.2, 4, 5, 6, 7, 8, 9 However, until recently, there were no pertinent names for the clinically recognizable disorders. Although the terms ‘upd(14)pat syndrome' and ‘upd(14)mat syndrome' were utilized previously,5, 7 both are apparently inappropriate as ‘upd(14)pat/mat syndrome' can be brought about by (epi)genetic mechanisms other than upd(14)pat/mat.2, 4, 5, 6, 7, 8, 9
Recently, the names ‘Kagami–Ogata syndrome' (KOS) (OMIM 608149) and ‘Temple syndrome' (TS) (OMIM 616222) have been proposed for upd(14)pat/upd(14)mat and related conditions, respectively.10, 11 The syndrome names have been approved by the European Network for Human Congenital Imprinting Disorders (EUCID.net) (www.imprinting-disorders.eu) on the basis of their significant contribution to the elucidation of clinical and molecular characteristics of the two disorders. Here, we review the current knowledge about KOS.
Clinical findings
Patients
After identification of upd(14)pat by Wang et al.12 in 1991, and that of epimutations and microdeletions affecting the 14q32.2 imprinted region of maternal origin by Kagami et al.2 in 2008, a total of 35 Japanese and 18 non-Japanese patients have been identified with KOS, excluding one Japanese patient with a ring 14 chromosome missing multiple nonimprinted genes2 and including two unpublished Japanese patients with epimutations (Supplementary Table S1). The patients have upd(14)pat, epimutations or microdeletions, and there has been no KOS patient with a single gene mutation or a duplication of the paternally derived 14q32.2 imprinted region. Upd(14)pat and epimutations reported to date are invariably sporadic, whereas microdeletions of variable sizes are identified as a sporadic form or as a familial form transmitted from the mother.
Phenotypic summary
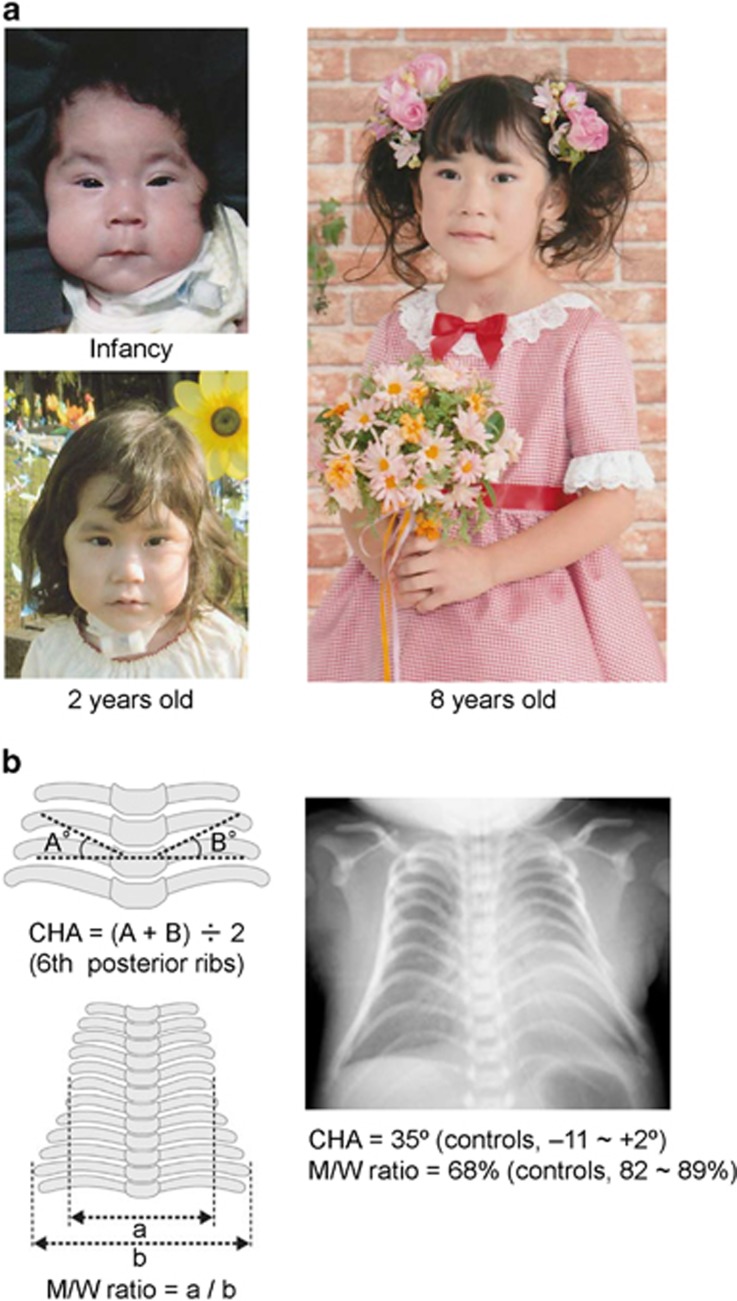
Comprehensive clinical studies have recently been performed for Japanese patients.10 The results, in conjunction with clinical findings of non-Japanese patients (Supplementary Table S1), are summarized as follows: (1) phenotypes are similar irrespective of the underlying cause and ethnicity; (2) the facial ‘gestalt' with full cheeks and protruding philtrum and the increased coat-hanger angle to the ribs constitute unique pathognomonic features from infancy through childhood/puberty (and probably in adulthood as well) in KOS, and the decreased ratio of the mid to widest thorax diameter (M/W) is also specific to KOS in infancy, although it becomes within the normal range after infancy (Figure 1, Supplementary Figure S1) (facial photographs have also been published in many papers);10, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 (3) abdominal wall defects including omphalocele, placentomegaly and polyhydramnios represent characteristic but not specific features in KOS, as they are also observed in several disorders such as Beckwith–Wiedemann syndrome and androgenetic mosaicism;22, 23 (4) KOS is also associated with rather common but important features such as prenatal overgrowth/overweight, developmental delay and feeding difficulties; (5) polyhydramnios is ascribed to placentomegaly and feeding difficulty (impaired swallowing), and the relevance of both placental and body factors explains why polyhydramnios can be present in patients lacking placentomegaly or feeding difficulties; (6) hepatoblastoma has been identified in three infantile patients; and (7) mortality is ~30%, and has invariably occurred before 4 years of age.
Figure 1.
Unique pathognomonic features in Kagami–Ogata syndrome (KOS). (a) Photographs of a patient with a maternally inherited 411 354 bp microdeletion involving DLK1, the IG-DMR, the MEG3-DMR, MEG3, RTL1/RTL1as, MEG8 and a centromeric part of snoRNAs (Deletion-4 in Figure 3).2 IG-DMR, intergenic differentially methylated region. The facial ‘gestalt' with full cheeks and protruding philtrum is observed from infancy through childhood. (b) Chest roentgenogram of a hitherto unreported Japanese neonatal patient with an epimutation. The CHA (coat-hanger angle) to the ribs is increased, and the M/W ratio (the ratio of the mid to widest thorax diameter) is decreased. Normal values are based on our previous report.10
In addition, two negative findings would also be worth pointing out. First, hypothyroidism has not been described in KOS patients.2, 10 This finding, together with lack of hyperthyroidism in TS patients,11 would argue against DIO3 being an imprinted gene, because DIO3 functions as an inactivator of thyroid hormones.24 Second, there is no KOS patient with diabetes mellitus, although positive associations have been identified between paternally inherited rs941576 in this imprinted region and type I diabetes mellitus25 and between downregulation of MEGs in this imprinted region and type II diabetes mellitus.26 Thus, the relevance of this imprinted region to diabetes mellitus would be minor, if any.
Human chromosome 14q32.2 imprinted region
Imprinted genes
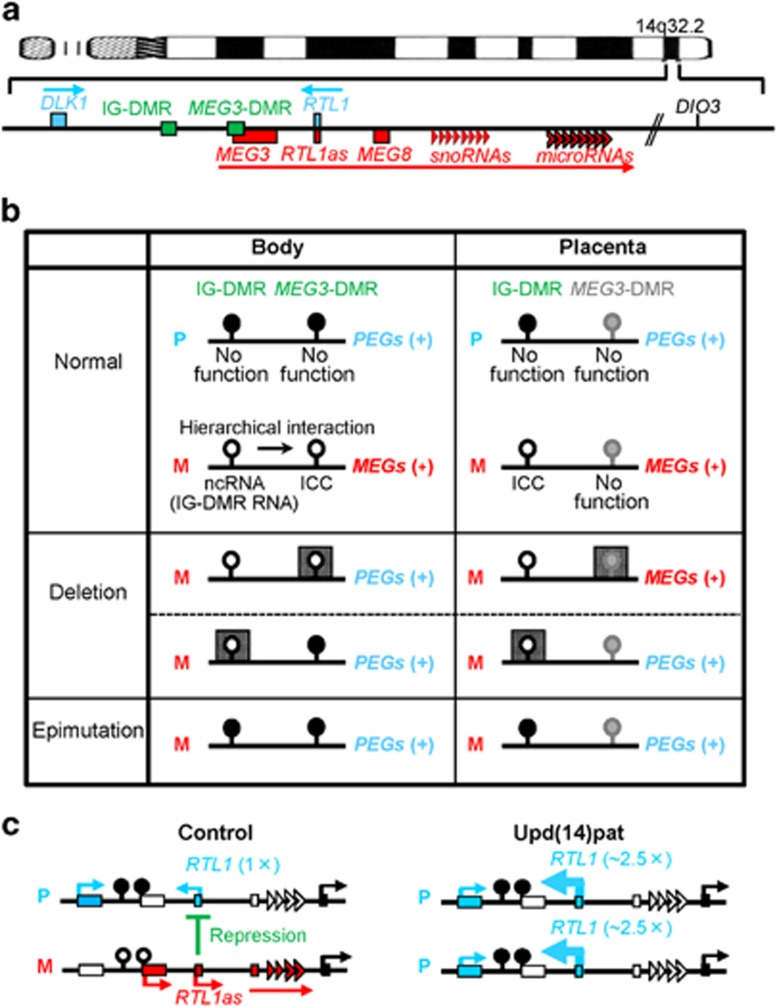
This region harbors protein-coding paternally expressed genes (PEGs) such as DLK1 and RTL1, and noncoding maternally expressed genes (MEGs) such as MEG3 (alias, GTL2), RTL1as (RTL1 antisense), MEG8, snoRNAs and microRNAs (Figure 2a).1, 2 Parent-of-origin-specific expression patterns have been confirmed in somatic and placental cells of control subjects and KOS patients.2, 4, 27 For DIO3, biparental expression has been indicated at least in the placenta,27 consistent with lack of thyroid diseases in KOS and TS.10, 11
Figure 2.
The human chromosome 14q32.2 imprinted region. (a) Schematic representation of the physical map of this region. PEGs are shown in blue, MEGs in red, a probably non-imprinted gene (DIO3) in black and the DMRs in green. (b) Methylation patterns of the DMRs. Black, white and gray painted circles represent methylated DMRs, unmethylated DMRs and non-DMRs, respectively. The arrow indicates a hierarchical interaction between the IG-DMR and the MEG3-DMR that appears to be medicated by cis-acting ncRNAs (the IG-DMR RNA) that exerts an enhancer-like function for the MEG3 promoter and prevents the MEG3-DMR from methylation. The deleted regions are shown with stippled squares. (c) Interaction between RTL1 and RTL1as. In control subjects, RTL1as-encoded microRNAs function as a trans-acting repressor for RTL1. In upd(14)pat patients, RTL1 expression level becomes ~5 times increased because of two copies of functional RTL1 and no functional RTL1as (shown with thick arrows). DMR, differentially methylated region; ICC, imprinting control center; IG-DMR, intergenic differentially methylated region; M, maternally derived chromosome; MEGs, maternally expressed genes; ncRNA, noncoding RNA; P, paternally derived chromosome; PEGs, paternally expressed genes; upd(14)pat, paternal uniparental disomy 14.
Differentially methylated regions (DMRs)
This region also carries the intergenic DMR (IG-DMR) between DLK1 and MEG3 and the MEG3-DMR at the MEG3 promoter region (Figure 2a).2, 28 Both DMRs are methylated after paternal transmission and unmethylated after maternal transmission in the body; in the placenta, the IG-DMR alone remains as a DMR, and the MEG3-DMR is grossly hypomethylated regardless of parental origin (Figure 2b).2, 4, 27 These findings suggest that the IG-DMR is a germline-derived primary DMR, whereas the MEG3-DMR is a postfertilization-derived secondary DMR.1, 29
Imprinting control centers (ICCs)
The MEG3-DMR and the IG-DMR of maternal origin function as ICCs in the body and the placenta, respectively, with a hierarchical interaction between the two DMRs in the body (Figure 2b).4 Indeed, loss of a 4303-bp segment encompassing the unmethylated MEG3-DMR of maternal origin has resulted in maternal to paternal epigenotypic alteration (absent MEG expression and biparental PEG expression) in the body, thereby leading to a typical KOS body phenotype in the presence of the differentially methylated IG-DMR; the placenta was apparently normal, consistent with the non-DMR pattern of the MEG3-DMR in the placenta.4 Similarly, loss of an 8558-bp segment involving the unmethylated IG-DMR of maternal origin has caused maternal to paternal epigenotypic alteration in the placenta and an epimutation (hypermethylation) of the MEG3-DMR in the body, thereby leading to typical KOS placental and body phenotypes.4 It is likely that the unmethylated IG-DMR controls the imprinting pattern directly in the placenta, and indirectly in the body by hierarchically regulating the methylation pattern of the MEG3-DMR. In this regard, it is postulated that epimutations (hypermethylations) of the IG-DMR of maternal origin also lead to maternal to paternal epigenotypic alterations, as reported in other imprinting disorders (Figure 2b).3, 30 Furthermore, as the epimutations identified in KOS and TS patients have invariably affected both the IG-DMR and the MEG3-DMR, with no isolated epimutation of the IG-DMR or the MEG3-DMR,2, 6, 7, 8, 9, 10, 11 this would be compatible with the notion that the methylation pattern of the MEG3-DMR is determined by that of the IG-DMR.
Although it remains to be elucidated how the two unmethylated DMRs of maternal origin function as ICCs with an interaction in the body, significant progress has been made for this issue. It is likely that nearly all MEGs, which are transcribed in the same orientation from the forward strand with a strikingly similar tissue expression pattern, comprise a long multicistronic noncoding RNA transcript (Figure 2a),31, 32 and that this transcript is expressed by the MEG3 promoter at the MEG3-DMR in somatic cells and by a different promoter in placental cells. Indeed, it is difficult to postulate a parent-of-origin-specific function for the placental MEG3 promoter that resides in a rather hypomethylated region irrespective of the parental origin.2, 4, 27 Furthermore, Kota et al.33 have shown that the IG-DMR of maternal origin harbors bidirectionally expressed cis-acting relatively short (mostly <500 bp and up to 750 bp) noncoding RNAs named the ‘IG-DMR RNA' that exerts an enhancer-like function for the MEG3 promoter as recently identified enhancer RNAs34, 35 and protects the Gtl2/Meg3-DMR from de novo methylation (Figure 2b). These findings would explain why the MEG3-DMR can control nearly all the MEGs expression in the body under the hierarchical regulation of the IG-DMR, although it remains unknown how the IG-DMR regulates the MEG expression in the placenta. For PEGs, the biallelic expression of DLK1 and absence of noncoding RNA in birds and fish that are free from genomic imprinting imply that the paternal-type imprinting pattern with positive DLK1 expression is the default situation of this domain.36, 37 Thus, loss or epimutation of the IG-DMR and the MEG3-DMR of maternal origin would result in the positive PEG expression as the default condition in the placenta and the body, respectively.
For the MEG3-DMR, it contains two putative CTCF-binding sites with a DMR-compatible methylation pattern.1, 4, 38 Thus, it may be possible that preferential binding of CTCF protein with versatile functions to the unmethylated CTCF-binding sites activates all MEGs as a large transcription unit. In addition, as mouse Gtl2/Meg3-DMR is associated with parental origin-specific histone acetylation,39 CTCF protein binding may inhibit all PEGs by affecting histone modification.1, 4, 38, 39, 40 However, although the CTCF protein can bind to the two putative binding sites,38 previous studies have failed to show a preferential binding of CTCF protein to the unmethylated binding sites.41 Thus, the relevance of the CTCF-binding sites to the ICC function awaits further investigations.
It should be pointed out that the expression patterns of the 14q32.2 imprinted genes are also influenced by a trans-acting factor(s) other than the ICCs. It has recently been shown that IPW, a long noncoding RNA in the Prader–Will syndrome (PWS) critical region, functions as a regulator for the expression of the imprinted genes at chromosome 14q32.2.42 Thus, the imprinting regulation mechanisms would be much more complex.
RTL1/RTL1as interaction and expression dosage of the imprinted genes
RTL1as-encoded microRNAs function as a trans-acting repressor for RTL1 expression (Figure 2c). Actually, quantitative PCR analyses have indicated ~5 times, rather than 2 times, increased RTL1 expression level in fresh placentas with upd(14)pat, as well as doubled DLK1 expression level and absent MEGs expression.27 This suggests that the RTL1 expression level is ~2.5 times increased in the absence of functional RTL1as. Immunohistochemical examinations have also identified markedly increased RTL1 protein expression and moderately increased DLK1 protein expression in the vascular endothelial cells and pericytes of chorionic villi with upd(14)pat.27 The repressive effect of Rtl1as on Rtl1 expression has also been demonstrated in mice.43, 44 No other gene–gene interaction has been demonstrated in this imprinted region.
Major phenotypic determinant(s)
Expression dosage of the imprinted genes
Expression patterns in upd(14)pat, epimutations and various microdeletions are shown in Figure 3. Upd(14)pat and epimutations are accompanied by essentially identical expression patterns (that is, 2 times DLK1 and ~5 times RTL1 expression dosages, and absent MEGs expression), although the possible presence of trisomic cells in trisomy rescue (TR)-type upd(14)pat, that of normal cells in postfertilization mitotic error (PE)-type upd(14) and that of normal cells escaping hypermethylation in epimutations remain possible. In contrast, microdeletions are accompanied by variable expression patterns of the imprinted genes, depending on the size of the deleted regions. Thus, (epi)genotype–phenotype correlations in patients with microdeletions are informative in considering the major phenotypic determinant(s).
Figure 3.
Expression patterns of the imprinted genes and methylation patterns (modified from our previous report).10 For explanations, see the legend for Figure 2. MEG3 is not expressed in Deletions-6–7, because MEG3 exons 1–3 are deleted.
(Epi)genotype–phenotype correlations in the body
Body phenotypes are comparable among patients with different causes, and excessive RTL1 expression (~2.5 times or ~5 times) and absent MEGs expression are shared in common (Figure 3 and Supplementary Table S1). In contrast, as patients with Deletions-2 and -4 are associated with typical KOS phenotype in the presence of a normal (1 time) DLK1 expression dosage, this argues against a major effect of doubled DLK1 expression dosage on phenotypic development. Thus, there are two possibilities with regard to the major phenotypic determinant(s) in KOS.
First, excessive RTL1 expression may constitute the major phenotypic determinant(s).10 In support of this, mouse Rtl1 is expressed in the fetal ribs, brain and skeletal muscles,10, 45 although assessment of body phenotype remains poor in Rtl1as knockout mice, which died within a day, with 2.5–3.0 times Rtl1 expression.44 In this case, it is assumed that clinical features are more severe in patients with ~5 times RTL1 expression than in those with ~2.5 times RTL1 expression. Although our initial studies supported such an RTL1 dosage effect on the phenotypic severity,2 the difference in the RTL1 expression dosage turned out to have no discernible clinical effects after analyzing long-term clinical courses.10 This may be explained by assuming the presence of a threshold for the RTL1 expression level or the buffering effects of multiple genetic and environmental factors for the difference in the RTL1 expression dosage.
Second, absent MEGs expression may play a crucial role in the phenotypic development, independently of the excessive RTL1 expression resulting from loss of functional RTL1as. Indeed, as mouse Gtl2/Meg3 is expressed in multiple fetal tissues including the primordial cartilage,46 absent MEG3 expression may be involved in the phenotypic development. Null expression of snoRNAs and/or miRNAs may also have clinical effects, as observed in exceptional PWS patients lacking only snoRNAs including SNORD116.47 Furthermore, absence of functional IG-DMR RNA may also lead to abnormal phenotypes by affecting the expression patterns of the imprinted genes,33 although the IG-DMR RNA has not been studied in KOS patients.
The two possibilities are not mutually exclusive, and excessive RTL1 expression coexists with absent expression of MEGs including RTL1as. Thus, further studies are required to elucidate which is the more critical determinant. For example, clinical studies in patients with tiny deletions involving maternally inherited MEG3, RTL1as, snoRNAs and/or miRNAs, but not the DMRs, will serve to clarify this matter.
(Epi)genotype–phenotype correlations in the placenta
The above notion is also applicable to Deletions-1–4, because Deletions-1–4 are associated with placentomegaly and polyhydramnios, in the presence of excessive RTL1 expression (~2.5 times or ~5 times), normal or doubled DLK1 expression and absent MEGs expression in the placenta (Figure 3 and Supplementary Table S1). In particular, excessive RTL1 expression would play a crucial role in the development of placental features, because Rtl1as knockout mice have placentomegaly (~150%) in the presence of 2.5–3.0 times Rtl1 expression and apparently normal expression of other imprinted genes in this domain.44
For the placental phenotype, two findings should be pointed out. First, Deletion-5 was free from placentomegaly,5 although ~2.5 times RTL1 expression dosage and absent expression of nearly all MEGs are predicted in the placenta as well as in the body (Figure 3). However, this would not pose a major problem, because placentomegaly is not an invariable feature even in patients with upd(14)pat with ~5 times RTL1 expression dosage and null MEG expression. In this context, polyhydramnios in the patient with Deletion-5 would primarily be because of defective swallowing of amniotic fluid, as indicated by the presence of feeding difficulties.5
Second, the two siblings with Deletion-6 had polyhydramnios, whereas the patient with Deletion-7 apparently had neither placentomegaly nor polyhydramnios, despite the similar deletion sizes (the deletion size according to hg19: Deletion-6, Chr14: 101 291 322–101 297 145 bp; and Deletion-7, Chr14:101 291 225–101 295 527 bp) (Figure 3).4, 5 As MEGs in the placenta would be transcribed by a cis-acting promoter other than the MEG3 promoter in the presence of the IG-DMR, the placentas with Deletions-6 and -7 are predicted to be accompanied by normal expression levels of most MEGs including RTL1as, except for MEG3 that is disrupted by the microdeletions, and normal expression levels of PEGs including RTL1. This notion explains lack of placentomegaly and polyhydramnios in Deletion-7, and excludes a positive role of MEG3 deficiency in the development of aberrant placental phenotype. For Deletion-6, polyhydramnios would be explained by body factors, because one of two siblings with Deletion-6 manifests feeding difficulty and both siblings have muscular hypotonia that can cause polyhydramnios.5, 48 In addition, placentomegaly is not described in the two siblings with Deletion-6, although no description of a particular phenotype does not necessarily indicate the lack of a corresponding phenotype.
Underlying factors for KOS and upd(14)pat
Underlying causes for KOS
Relative frequency of underlying causes is shown in Table 1. Upd(14)pat accounts for approximately two-thirds of KOS patients, and epimutations and microdeletions are identified with a similar frequency. Notably, upd(14)pat has predominantly been found in Japanese patients with normal karyotype and in non-Japanese patients with abnormal karyotype. This would primarily be because of the difference in molecular methods employed before and after the identification of the DMRs.2 Indeed, Japanese patients have been found by methylation analysis of the DMRs that is carried out irrespective of the karyotype, whereas non-Japanese patients have primarily been detected by genotyping analysis that is preferentially performed for chromosomal abnormalities as a risk factor for upd(14)pat.49 This notion would also explain why epimutations have been found in Japanese patients only.
Table 1. Underlying factors for Kagami–Ogata syndrome and upd(14)pat.
| Japanese (n=35) | Non-Japanese (n=18) | Total (n=53) | |
|---|---|---|---|
| Underlying causes for KOS | |||
| Upd(14)pat | 23 | 14 | 37 |
| Normal karyotype | 20 | 5 | 25 |
| Abnormal karyotype | 3 | 9 | 12 |
| Epimutation | 7a | 0 | 7 |
| Microdeletion | 5b | 4c | 9 |
| Underlying mechanisms for upd(14)pat | |||
| Normal karyotype | |||
| TR/GC | 7 | 0 | 7 |
| MR/PE | 12 | 2 | 14 |
| PE | 1 | 2 | 3 |
| No detailed information | 0 | 1 | 1 |
| Abnormal karyotype | |||
| Robertsonian translocation | 2d | 3e | 5 |
| Isochromosome for 14q | 0 | 5f | 5 |
| Unknown/other karyotype | 1 | 1g | 2 |
Abbreviations: GC, gamete complementation; KOS, Kagami–Ogata syndrome; MR, monosomy rescue; PE, postfertilization mitotic error; TR, trisomy rescue; upd(14)pat, paternal uniparental disomy 14.
Including two hitherto unreported patients.
Including sibling cases; thus, four microdeletions have been found in five patients.
Including sibling cases; thus, three microdeletions have been detected in four patients.
45,XX,rob(13;14)(q10;q10) (n=1) and 45,XX,rob(14;21)(q10;q10q) (n=1); parental karyotype has not been examined.
45,XX,rob(13;14)(q10;q10) (n=3); the same Robertsonian translocations have been found in the fathers of the three patients.
45,XX,i(14q) (n=4) and 45,XY,i(14q) (n=1); parental karyotypes are invariably normal in the five patients.
46,XX[6]/47,XX,+mar[44]. Although the marker chromosome is derived from chromosome 14, it does not involve the 14q32.2 imprinted region, and full isodisomy for chromosome 14 has been shown by microsatellite analysis.
The relative frequency is similar to that observed in TS,11 but is different from that in other imprinted disorders. For example, microdeletions at chromosome 15q11.2–q13 imprinted region are most frequent in PWS and Angelman syndrome, and epimutations of the H19-DMR and the KvDMR1 at chromosome 11p15 are most prevalent in Silver–Russell syndrome and Beckwith–Wiedemann syndrome, respectively.3, 30 Although the preponderance of microdeletions in PWS/Angelman syndrome is explained by the presence of low-copy repeats flanking the imprinted region,50 it is unknown why upd and epimutations are predominant in KOS/TS and Silver–Russell syndrome/Beckwith–Wiedemann syndrome, respectively.
Underlying mechanisms for upd(14)pat
Upd(14)pat is frequently identified in patients with normal karyotype (Table 1). Upd(14)pat in such patients is primarily caused by TR, gamete complementation, monosomy rescue (MR) and PE.30 TR/gamete complementation is accompanied by at least one heterodisomic locus, MR by full isodisomy and PE by full or segmental isodisomy.30 In this regard, the predominance of MR/PE-type upd(14)pat in Japanese patients would be related to the recent increase in the maternal childbearing age,51 because MR-type upd(14)pat is mediated by a nullisomic oocyte that is produced by maternal age-dependent nondisjunction at meiosis 1 as well as by maternal age-independent nondisjunction at meiosis 2.52 Indeed, maternal childbearing age is significantly increased in Japanese patients with MR/PE-type upd(14)pat.30 Consistent with this, maternal childbearing age is also significantly advanced in PWS patients with M1 nondisjunction-mediated TR/gamete complementation-type upd(15)mat.51
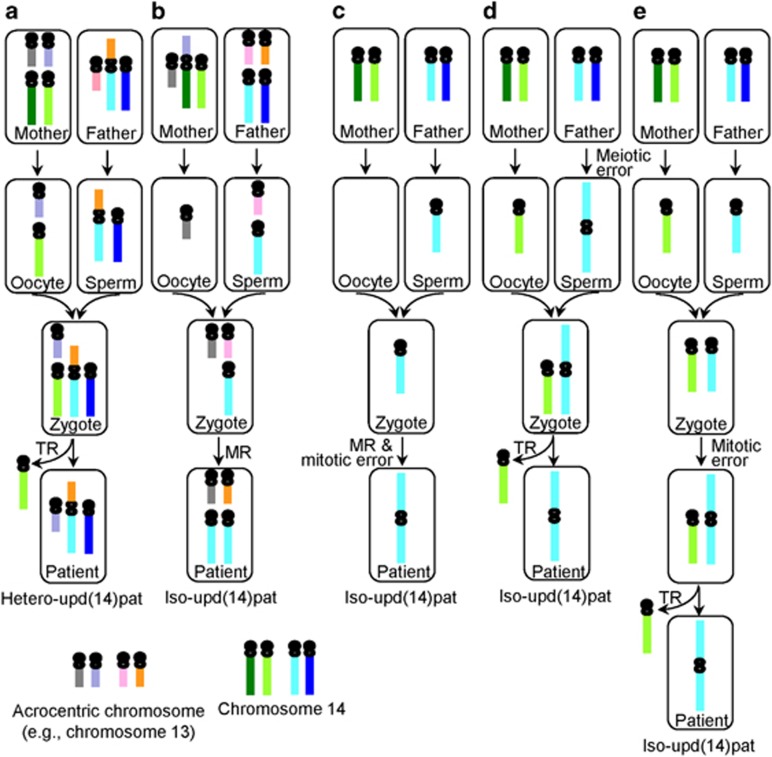
Upd(14)pat is also found in patients with abnormal karyotype (Table 1). The Robertsonian 13q;14q translocations in three non-Japanese patients with hetero-upd(14)pat were transmitted from their fathers to the patients,12, 13, 53 although Robertsonian translocations could occur as a de novo event. This indicates the production of TR-mediated upd(14)pat in these patients (Figure 4a). Here, it appears worth pointing out that a maternal Robertsonian translocation could be a risk factor for iso-upd(14)pat, because it constitutes a predisposing factor for the production of a nullisomic oocyte and resultant monosomic zygote that could be subject to MR for a paternally derived chromosome 14 (Figure 4b). However, a maternal Robertsonian translocation has not been found to date, probably because parental karyotyping has not been performed in patients with normal karyotype. In contrast, the i(14q) chromosomes with full iso-upd(14)pat in five non-Japanese patients were formed as a de novo event.14, 15, 16, 54, 55 It is likely that the i(14q) chromosome is generated by MR in association with isochromosome formation (centromeric misdivision or U-type sister chromatid exchange) during mitosis, or by TR after isochromosome formation during meiosis or mitosis (Figures 4c–e).49, 56, 57 In addition, i(14q) chromosome with hetero-upd(14)pat could be produced by U-type exchange between non-sister chromatids during meiosis.56
Figure 4.
Schematic representation of the generation of upd(14)pat in patients or parents with Robertsonian translocation or i(14q). MR, monosomy rescue; TR, trisomy rescue; upd(14)pat, paternal uniparental disomy 14. (a) Hetero-upd(14)pat mediated by paternal Robertsonian translocation and post-zygotic TR. (b) Iso-upd(14)pat mediated by maternal Robertsonian translocation and post-zygotic MR. (c) Iso-upd(14)pat generated by concomitant occurrence of post-zygotic MR and isochromosome formation. (d) Iso-upd(14)pat generated by sequential occurrence of meiotic isochromosome formation and post-zygotic TR. (e) Iso-upd(14)pat generated by sequential occurrence of mitotic isochromosome formation and TR.
Diagnostic and therapeutic implications
Clinical diagnostic implications
The facial gestalt and the increased coat-hanger angle are mandatory for the clinical diagnosis of KOS, and the decreased M/W ratios also constitute a diagnostic indication in infancy.10 Furthermore, when several characteristic features appear in association with the pathognomonic features, this confirms the diagnosis of KOS. Differential diagnosis includes Beckwith–Wiedemann syndrome and androgenetic mosaicism.22, 23
Molecular diagnostic implications
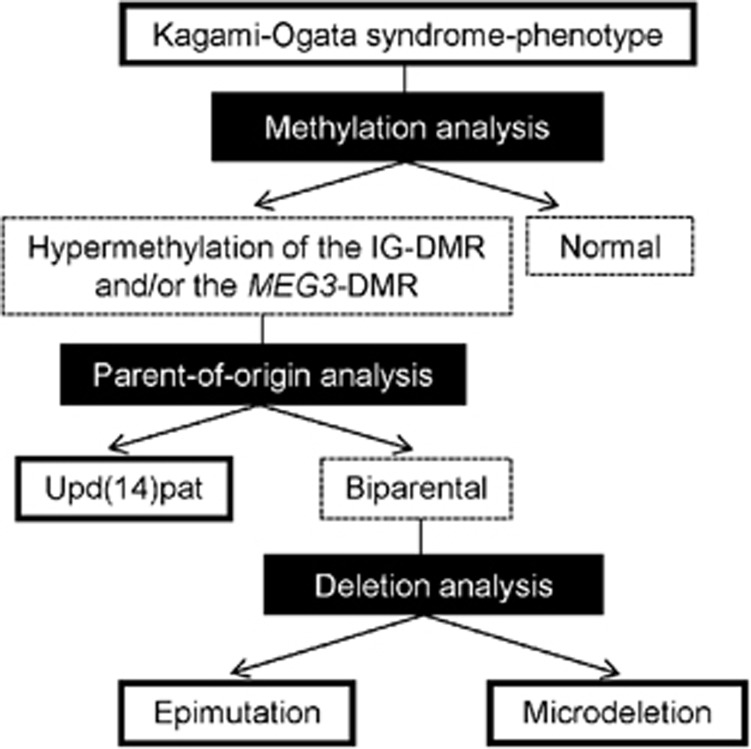
The flowchart of molecular diagnosis is shown in Figure 5. As all the patients with KOS identified to date have hypermethylation of the IG-DMR and/or the MEG3-DMR, methylation patterns of the two DMRs should be analyzed as the first step. When hypermethylation is found for either of the two DMRs of maternal origin, this confirms the diagnosis of KOS. When hypermethylation is absent, clinical diagnosis should be reconsidered. However, it would be suggested to examine a possible deletion involving RTL1as but not the DMRs, or a possible mutation of RTL1/RTL1as, because loss of RTL1as, gain-of-function mutation of RTL1 and loss-of-function mutation of RTL1as could lead to excessive RTL1 expression. In addition, there might be microdeletions affecting MEG3, MEG8, snoRNAs and/or miRNAs but not the DMRs.
Figure 5.
Molecular diagnostic flowchart. Methylation analysis is performed by combined bisulfite restriction analysis, bisulfite sequencing, multiple ligation probe amplification (MLPA) using SALSA MLPA kit ME032 UPD7/UPD14 (MRC-Holland, Amsterdam, The Netherlands) or pyrosequencing. Parent-of-origin analysis is carried out by microsatellite analysis or single-nucleotide polymorphism (SNP) array. Deletion analysis is performed by MLPA, fluorescence in situ hybridization or array comparative genomic hybridization. Methylation and deletion analyses need DNA samples of patients alone, whereas parent-of-origin analysis requires DNA samples of patients and their parents. MLPA is utilized for deletion analysis and methylation analysis before and after digestion of the genomic DNA samples with a methylation-sensitive restriction enzyme HhaI, respectively.
If the diagnosis of KOS is confirmed, upd(14)pat should be examined as the second step. When upd(14)pat is identified, karyotyping is recommended to examine the possibility of Robertsonian translocation or i(14q) chromosome. If the patient has such an abnormal chromosome, a recurrent risk of KOS should be considered.
When upd(14)pat is excluded, possible deletion of the IG-DMR and/or the MEG3-DMR should be investigated. This permits the molecular diagnosis of an epimutation when no deletion is detected and that of a microdeletion when a deletion is delineated. If a deletion is identified, it is recommended to examine whether the deletion is formed as a de novo event or derived from the mother. In the latter case, there is a 50% recurrence risk of KOS.
Management
The management for KOS remains symptomatic, including mechanical ventilation, tracheostomy, tube feeding, surgical operation for omphalocele and supportive therapy for developmental delay. In this regard, our thorough survey data in 34 Japanese patients are summarized as follows: (1) mechanical ventilation was required by 32 patients, and was discontinued during infancy in 22 patients, with a median duration of 1 month (range, 0.1–17 months); (2) tracheostomy was performed in approximately one-third of patients; (3) tube feeding was necessary in all but a single patient, and was discontinued in 16 patients, with a median period of ~7.5 months (range, 0.1–89 months); (4) developmental delay was invariably present in 26 patients examined for developmental status, with the median developmental/intellectual quotient of 55 (range, 29–70); and (5) gross motor development was also almost invariably delayed, with head control being achieved at ~7 months (range, 3–14 months), sitting without support at ~12 months (range, 8–27 months) and walking without support at 25.5 months (range, 20–90 months) (except for a single 3-year-old patient who showed head control at 33 months of age because of severe hypotonia) (for the detailed data of individual patient, see Kagami et al.10). In addition, periodical screenings for hepatoblastoma are recommended, including serum α-fetoprotein measurement and abdominal ultrasonography.
It is worth emphasizing that patients with KOS become free from mechanical ventilation, tracheostomy and tube feeding, and there is no report of death at ≥4 years of age.10 According to our survey, all patients go to school by themselves and get on their daily lives from childhood, although they have developmental delay. Explaining such a prognosis to parents when their affected child is in infancy and under intensive management is expected to reduce parents' anxiety and to facilitate the attachment formation between parents and the patient.
Conclusion
We reviewed current knowledge about KOS. Although several issues remain to be clarified, significant progress has been made for the clarification of clinical findings and underlying (epi)genetic factors. We conclude that KOS is a clinically recognizable upd(14)pat and related disorder affecting the maternally derived chromosome 14q32.2 imprinted region.
Acknowledgments
We thank Ms Emma Barber for her editorial assistance. This work was supported by Grants-in-Aid for Scientific Research (A) (25253023) and Research (B) (23390083) from the Japan Society for the Promotion of Science (JSPS), by Grants for Health Research on Children, Youth, and Families (H25-001) and for Research on Intractable Diseases (H22-161) from the Ministry of Health, Labor and Welfare (MHLW), by Grants from the National Center for Child Health and Development (23A-1, 25-10), by Grant from Takeda Science Foundation and by the Grant from the Japan Agency for Medical Research and Development (AMED) (H27-141).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Journal of Human Genetics website (http://www.nature.com/jhg)
Supplementary Material
References
- da Rocha, S. T., Edwards, C. A., Ito, M., Ogata, T. & Ferguson-Smith, A. C. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 24, 306–316 (2008). [DOI] [PubMed] [Google Scholar]
- Kagami, M., Sekita, Y., Nishimura, G., Irie, M., Kato, F., Okada, M. et al. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat. Genet. 40, 237–242 (2008). [DOI] [PubMed] [Google Scholar]
- Hoffmann, K. & Heller, R. Uniparental disomies 7 and 14. Best Pract. Res. Clin. Endocrinol. Metab. 25, 77–100 (2011). [DOI] [PubMed] [Google Scholar]
- Kagami, M., O'Sullivan, M. J., Green, A. J., Watabe, Y., Arisaka, O., Masawa, N. et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet. 6, e1000992 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beygo, J., Elbracht, M., de Groot, K., Begemann, M., Kanber, D., Platzer, K. et al. Novel deletions affecting the MEG3-DMR provide further evidence for a hierarchical regulation of imprinting in 14q32. Eur. J. Hum. Genet. 23, 180–188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple, I. K., Shrubb, V., Lever, M., Bullman, H. & Mackay, D. J. Isolated imprinting mutation of the DLK1/GTL2 locus associated with a clinical presentation of maternal uniparental disomy of chromosome 14. J. Med. Genet. 44, 637–640 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoki, K., Ogata, T., Kagami, M., Tanaka, T. & Saitoh, S. Epimutation (hypomethylation) affecting the chromosome 14q32.2 imprinted region in a girl with upd(14)mat-like phenotype. Eur. J. Hum. Genet. 16, 1019–1023 (2008). [DOI] [PubMed] [Google Scholar]
- Buiting, K., Kanber, D., Martín-Subero, J. I., Lieb, W., Terhal, P., Albrecht, B. et al. Clinical features of maternal uniparental disomy 14 in patients with an epimutation and a deletion of the imprinted DLK1/GTL2 gene cluster. Hum. Mutat. 29, 1141–1146 (2008). [DOI] [PubMed] [Google Scholar]
- Zechner, U., Kohlschmidt, N., Rittner, G., Damatova, N., Beyer, V., Haaf, T. et al. Epimutation at human chromosome 14q32.2 in a boy with a upd(14)mat-like clinical phenotype. Clin. Genet. 75, 251–258 (2009). [DOI] [PubMed] [Google Scholar]
- Kagami, M., Kurosawa, K., Miyazaki, O., Ishino, F., Matsuoka, K. & Ogata, T. Comprehensive clinical studies in 34 patients with molecularly defined UPD(14)pat and related conditions (Kagami-Ogata syndrome). Eur. J. Hum. Genet 23, 1488–1498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannides, Y., Lokulo-Sodipe, K., Mackay, D. J., Davies, J. H. & Temple, I. K. Temple syndrome: improving the recognition of an underdiagnosed chromosome 14 imprinting disorder: an analysis of 51 published cases. J. Med. Genet. 51, 495–501 (2014). [DOI] [PubMed] [Google Scholar]
- Wang, J. C., Passage, M. B., Yen, P. H., Shapiro, L. J. & Mohandas, T. K. Uniparental heterodisomy for chromosome 14 in a phenotypically abnormal familial balanced 13/14 Robertsonian translocation carrier. Am. J. Hum. Genet. 48, 1069–1074 (1991). [PMC free article] [PubMed] [Google Scholar]
- Cotter, P. D., Kaffe, S., McCurdy, L. D., Jhaveri, M., Willner, J. P. & Hirschhorn, K. Paternal uniparental disomy for chromosome 14: a case report and review. Am. J. Med. Genet. 70, 74–79 (1997). [DOI] [PubMed] [Google Scholar]
- Papenhausen, P. R., Mueller, O. T., Sutcliffe, M., Diamond, T. M., Kousseff, B. G. & Johnson, V. P. Uniparental isodisomy of chromosome 14 in two cases: an abnormal child and a normal adult. Am. J. Med. Genet. 59, 271–275 (1995). [DOI] [PubMed] [Google Scholar]
- Walter, C. A., Shaffer, L. G., Kaye, C. I., Huff, R. W., Ghidoni, P. D., McCaskill, C. et al. Short-limb dwarfism and hypertrophic cardiomyopathy in a patient with paternal isodisomy 14: 45,XY,idic(14)(p11). Am. J. Med. Genet. 65, 259–265 (1996). [DOI] [PubMed] [Google Scholar]
- Stevenson, D. A., Brothman, A. R., Chen, Z., Bayrak-Toydemir, P. & Longo, N. Paternal uniparental disomy of chromosome 14: confirmation of a clinically-recognizable phenotype. Am. J. Med. Genet. A 130A, 88–91 (2004). [DOI] [PubMed] [Google Scholar]
- Kurosawa, K., Sasaki, H., Sato, Y., Yamanaka, M., Shimizu, M., Ito, Y. et al. Paternal UPD14 is responsible for a distinctive malformation complex. Am. J. Med. Genet. 110, 268–272 (2002). [DOI] [PubMed] [Google Scholar]
- Coveler, K. J., Yang, S. P., Sutton, R., Milstein, J. M., Wu, Y. Q., Bois, K. D. et al. A case of segmental paternal isodisomy of chromosome 14. Hum. Genet. 110, 251–256 (2002). [DOI] [PubMed] [Google Scholar]
- Curtis, L., Antonelli, E., Vial, Y., Rimensberger, P., Le Merrer, M., Hinard, C. et al. Prenatal diagnostic indicators of paternal uniparental disomy 14. Prenat. Diagn. 26, 662–666 (2006). [DOI] [PubMed] [Google Scholar]
- Mattes, J., Whitehead, B., Liehr, T., Wilkinson, I., Bear, J., Fagan, K. et al. Paternal uniparental isodisomy for chromosome 14 with mosaicism for a supernumerary marker chromosome 14. Am. J. Med. Genet. A 143A, 2165–2171 (2007). [DOI] [PubMed] [Google Scholar]
- Irving, M. D., Buiting, K., Kanber, D., Donaghue, C., Schulz, R., Offiah, A. et al. Segmental paternal uniparental disomy (patUPD) of 14q32 with abnormal methylation elicits the characteristic features of complete patUPD14. Am. J. Med. Genet. A 152A, 1942–1950 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weksberg, R., Shuman, C. & Beckwith, J. B. Beckwith-Wiedemann syndrome. Eur. J. Hum. Genet. 18, 8–14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazawa, K., Nakabayashi, K., Matsuoka, K., Masubara, K., Hata, K., Horikawa, R. et al. Androgenetic/biparental mosaicism in a girl with Beckwith-Wiedemann syndrome-like and upd(14)pat-like phenotypes. J. Hum. Genet. 56, 91–93 (2011). [DOI] [PubMed] [Google Scholar]
- Kohrle, J. Thyroid hormone transporters in health and disease: advances in thyroid hormone deiodination. Best Pract. Res. Clin. Endocrinol. Metab 21, 173–191 (2007). [DOI] [PubMed] [Google Scholar]
- Wallace, C., Smyth, D. J., Maisuria-Armer, M., Walker, N. M., Todd, J. A. & Clayton, D. G. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat. Genet. 42, 68–71 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameswaran, V., Bramswig, N. C., McKenna, L. B., Penn, M., Schug, J., Hand, N. J. et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 19, 135–145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami, M., Matsuoka, K., Nagai, T., Yamanaka, M., Kurosawa, K., Suzumori, N. et al. Paternal uniparental disomy 14 and related disorders: placental gene expression analyses and histological examinations. Epigenetics 7, 1142–1150 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, S. K., Wylie, A. A., Coveler, K. J., Cotter, P. D., Papenhausen, P. R., Sutton, V. R. et al. Epigenetic detection of human chromosome 14 uniparental disomy. Hum. Genet. 22, 92–97 (2003). [DOI] [PubMed] [Google Scholar]
- Nowak, K., Stein, G., Powell, E., He, L. M., Naik, S., Morris, J. et al. Establishment of paternal allele-specific DNA methylation at the imprinted mouse Gtl2 locus. Epigenetics 6, 1012–1020 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami, M., Kato, F., Matsubara, K., Sato, T., Nishimura, G. & Ogata, T. Relative frequency of underlying genetic causes for the development of UPD(14)pat-like phenotype. Eur. J. Hum. Genet. 20, 928–932 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierling, S., Dalbert, S., Schoppenhorst, S., Tsai, C. E., Oliger, S., Ferguson-Smith, A. C. et al. High-resolution map and imprinting analysis of the Gtl2-Dnchc1 domain on mouse chromosome 12. Genomics 87, 225–235 (2007). [DOI] [PubMed] [Google Scholar]
- Benetatos, L., Hatzimichael, E., Londin, E., Vartholomatos, G., Loher, P., Rigoutsos, I. et al. The microRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol Life Sci. 70, 795–814 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota, S. K., Llères, D., Bouschet, T., Hirasawa, R., Marchand, A., Begon-Pescia, C. et al. ICR noncoding RNA expression controls imprinting and DNA replication at the Dlk1-Dio3 domain. Dev. Cell 31, 19–33 (2014). [DOI] [PubMed] [Google Scholar]
- Kim, T. K., Hemberg, M., Gray, J. M., Costa, A. M., Bear, D. M., Wu, J. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Garcia-Bassets, I., Benner, C., Li, W., Su, X., Zhou, Y. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, S., Han, J.Y. & Lee, K. Cloning of avian Delta-like 1 homolog gene: the biallelic expression of Delta-like 1 homolog in avian species. Poult Sci. 89, 948–955 (2010). [DOI] [PubMed] [Google Scholar]
- Glazov, E.A., McWilliam, S., Barris, W.C. & Dalrymple, B.P. Origin, evolution, and biological role of miRNA cluster in DLK-DIO3 genomic region in placental mammals. Mol. Biol. Evol. 25, 939–948 (2008). [DOI] [PubMed] [Google Scholar]
- Rosa, A. L., Wu, Y. Q., Kwabi-Addo, B., Coveler, K. J., Reid. Sutton, V. & Shaffer, L. G. Allele-specific methylation of a functional CTCF binding site upstream of MEG3 in the human imprinted domain of 14q32. Chromosome Res. 13, 809–818 (2005). [DOI] [PubMed] [Google Scholar]
- Carr, M. S., Yevtodiyenko, A., Schmidt, C. L. & Schmidt, J. V. Allele-specific histone modifications regulate expression of the Dlk1-Gtl2 imprinted domain. Genomics 89, 280–290 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, L. B., Pan, H., Hannenhalli, S., Cheng, Y., Ma, J., Fedoriw, A. et al. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development 135, 2729–2738 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray, E. N. & Schmidt, J. V. Identification of imprinting regulators at the Meg3 differentially methylated region. Genomics 100, 184–194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer, Y., Sagi, I., Yanuka, O., Eiges, R. & Benvenisty, N. The noncoding RNA IPW regulates the imprinted DLK1-DIO3 locus in an induced pluripotent stem cell model of Prader-Willi syndrome. Nat Genet. 46, 551–557 (2014). [DOI] [PubMed] [Google Scholar]
- Seitz, H., Youngson, N., Lin, S. P., Dalbert, S., Paulsen, M., Bachellerie, J. P. et al. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 34, 261–262 (2003). [DOI] [PubMed] [Google Scholar]
- Sekita, Y., Wagatsuma, H., Nakamura, K., Ono, R., Kagami, M., Wakisaka, N. et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 40, 243–248 (2008). [DOI] [PubMed] [Google Scholar]
- Brandt, J., Schrauth, S., Veith, A. M., Froschauer, A., Haneke, T., Schultheis, C. et al. Transposable elements as a source of genetic innovation: expression and evolution of a family of retrotransposon-derived neogenes in mammals. Gene 345, 101–111 (2005). [DOI] [PubMed] [Google Scholar]
- da Rocha, S.T., Tevendale, M., Knowles, E., Takada, S., Watkins, M. & Ferguson-Smith, A. C. Restricted co-expression of Dlk1 and the reciprocally imprinted non-coding RNA, Gtl2: implications for cis-acting control. Dev. Biol. 306, 810–823 (2007). [DOI] [PubMed] [Google Scholar]
- Sahoo, T., del Gaudio, D., German, J. R., Shinawi, M., Peters, S. U., Person, R. E. et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 40, 719–721 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, F. G., Leveno, K. J., Bloom, S. L., Spong, C. Y., Dashe, J. S., Hoffman, B. L. et al. Williams Obstetrics 24th edn 231–239 (McGraw-Hill Education, New York, NY, USA, 2014). [Google Scholar]
- Berend, S. A., Horwitz, J., McCaskill, C. & Shaffer, L. G. Identification of uniparental disomy following prenatal detection of Robertsonian translocations and isochromosomes. Am. J. Hum. Genet. 66, 1787–1793 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Y., Walkowicz, M. J., Buiting, K., Johnson, D. K., Tarvin, R. E., Rinchik, EM. et al. The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum. Mol. Genet. 8, 533–542 (1999). [DOI] [PubMed] [Google Scholar]
- Matsubara, K., Murakami, N., Nagai, T. & Ogata, T. Maternal age effect on the development of Prader-Willi syndrome resulting from upd(15)mat through meiosis 1 errors. J. Hum. Genet. 56, 566–571 (2011). [DOI] [PubMed] [Google Scholar]
- Pellestor, F., Andreo, B., Anahory, T. & Hamamah, S. The occurrence of aneuploidy in human: lessons from the cytogenetic studies of human oocytes. Eur. J. Med. Genet. 49, 103–116 (2006). [DOI] [PubMed] [Google Scholar]
- Yano, S., Li, L., Owen, S., Wu, S. & Tran, T. A further delineation of the uniparental disomy (UPD 14): the fifth reported liveborn case. Am. J. Hum. Genet. 69, A739 (2001). [Google Scholar]
- Klein, J., Shaffer, L., McCaskill, C., Sheerer, L., Otto, C. & Main, D. Delineation of the paternal disomy 14 syndrome: identification of a case by prenatal diagnosis. Am. J. Hum. Genet. 65, A179 (1999). [Google Scholar]
- McGowan, K. D., Weiser, J. J., Horwitz, J., Berend, S. A., McCaskill, C., Sutton, V. R. et al. The importance of investigating for uniparental disomy in prenatally identified balanced acrocentric rearrangements. Prenat. Diagn. 22, 141–143 (2002). [PubMed] [Google Scholar]
- Baumer, A., Basaran, S., Taralczak, M., Cefle, K., Ozturk, S., Palanduz, S. et al. Initial maternal meiotic I error leading to the formation of a maternal i(2q) and a paternal i(2p) in a healthy male. Cytogenet. Genome. Res. 118, 38–41 (2007). [DOI] [PubMed] [Google Scholar]
- Wang, J. C., Vaccarello-Cruz, M., Ross, L., Owen, R., Pratt, V. M., Lightman, K. et al. Mosaic isochromosome 15q and maternal uniparental isodisomy for chromosome 15 in a patient with morbid obesity and variant PWS-like phenotype. Am. J. Med. Genet. A 161A, 1695–1701 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.