Abstract
The complex and dynamic vaginal microbial ecosystem is critical to both health and disease of the host. Studies focusing on how vaginal microbiota influences HIV-1 infection may face limitations in selecting proper animal models. Given that northern pig-tailed macaques (Macaca leonina) are susceptible to HIV-1 infection, they may be an optimal animal model for elucidating the mechanisms by which vaginal microbiota contributes to resistance and susceptibility to HIV-1 infection. However, little is known about the composition and temporal variability of vaginal microbiota of the northern pig-tailed macaque. Here, we present a comprehensive catalog of the composition and temporal dynamics of vaginal microbiota of two healthy northern pig-tailed macaques over 19 weeks using 454-pyrosequencing of 16S rRNA genes. We found remarkably high proportions of a diverse array of anaerobic bacteria associated with bacterial vaginosis. Atopobium and Sneathia were dominant genera, and interestingly, we demonstrated the presence of Lactobacillus-dominated vaginal microbiota. Moreover, longitudinal analysis demonstrated that the temporal dynamics of the vaginal microbiota were considerably individualized. Finally, network analysis revealed that vaginal pH may influence the temporal dynamics of the vaginal microbiota, suggesting that inter-subject variability of vaginal bacterial communities could be mirrored in inter-subject variation in correlation profiles of species with each other and with vaginal pH over time. Our results suggest that the northern pig-tailed macaque could be an ideal animal model for prospective investigation of the mechanisms by which vaginal microbiota influence susceptibility and resistance to HIV-1 infection in the context of highly polymicrobial and Lactobacillus-dominated states.
Keywords: Macaque, Vaginal microbiome, Bacterial vaginosis, Temporal dynamics, Networks
Abbreviations: BV: bacterial vaginosis; CST: community state type; OTUs: operational taxonomic units; NHP: nonhuman primate; NPTM: northern pig-tailed macaques (Macaca leonina)
INTRODUCTION
The complex and dynamic vaginal microbial ecosystem is thought to have been shaped by long-term co-evolutionary processes between vaginal microbiota and the host, and is critical to both host health and disease (Brotman, 2011; Farage et al, 2010; Ma et al, 2012; Macklaim et al, 2012; White et al, 2011). The dynamic equilibrium of the vaginal microbial ecosystem can be altered by external factors, such as sexual activity (Schwebke et al, 1999), vaginal douching (Brotman et al, 2008), catamenial products (Hickey et al, 2013), smoking (Brotman et al, 2014), contraceptive use (Van De Wijgert et al, 2013), antibiotics (Cruciani et al, 2012) and vaginal microbicides (Ravel et al, 2012), and by internal factors, such as menses and hormonal changes during normal menstrual cycles (Gajer et al, 2012), resulting in microbial imbalances or dysbiosis in the vagina (Huang et al, 2014).
Bacterial vaginosis (BV), an imbalance or dysbiosis of vaginal microbiota, is a highly prevalent clinical condition among reproductive-age women (Fredricks et al, 2005; Hickey et al, 2012; Ma et al, 2012; Ravel et al, 2013). It is characterized by a quantitative and qualitative shift in composition of vaginal microbiota from a predominance of Lactobacillus species to a predominance of a diverse array of anaerobic bacteria (Hickey et al, 2012; Ma et al, 2012; Ravel et al, 2013). Although the mechanisms have not been established, current epidemiological evidence suggests that BV increases women’s susceptibility to HIV-1 infection (Atashili et al, 2008; Cohen et al, 2012; Myer et al, 2005). In light of the link between BV and susceptibility to HIV-1 infection, understanding the dynamic shift in vaginal microbiota over time and the factors that influence it is an important component of developing vaginal microbiota-targeted strategies to help prevent vaginal HIV transmission and is necessary for characterizing healthy (resistant to infection) and dysbiotic (susceptible to infection) states (Reid, 2014; Schellenberg & Plummer, 2012).
Due to the many uncontrollable factors described above, it is very difficult to resolve these issues using human vaginal microbiota only. Conversely, due to their phylogenetic proximity to human beings, nonhuman primates can be important for hypothesis testing for the dynamics of vaginal microbiota thought to be associated with resistance and susceptibility to HIV-1 infection under highly controlled experimental conditions not achievable in human studies. Pig-tailed macaques share many similarities to humans in anatomy and physiology of the vagina, and are widely used as animal models for HIV sexual transmission and vaginal microbicide research (Baroncelli et al, 2008; Hatziioannou & Evans, 2012; Veazey, 2013). It should be noted that pig-tailed macaques are the only Old World monkey known to be susceptible to HIV-1 infection with AIDS-like symptoms (Agy et al, 1992; Bosch et al, 2000; Hu, 2005; Kent et al, 1995). According to current primate taxonomy based on morphological characteristics and phylogeographic studies (Campbell et al, 2007; Gippoliti, 2001; Groves, 2001; Kuang et al, 2009; Malaivijitnond et al, 2012; Rosenblum et al, 1997), pig-tailed macaques are split into three species: Northern pig-tailed macaque (Macaca leonina, NPTM), located from about 8°N in peninsular Thailand, through Burma and Indochina into Bangladesh, India extending as far north as Brahmaputra, and the southernmost Yunnan province, China; Sunda pig-tailed macaque (Macaca nemestrina), distributed in the Malay Peninsula from about N7°30', Sumatra, Bangka and Borneo; and the Mentawai macaque (Macaca pagensis), living in the Mentawai Islands (Campbell et al, 2007; Groves, 2001; Kuang et al, 2009; Lei et al, 2014). In a previous report, we found that the TRIM5-Cyclophilin A (TRIM5-CypA) fusion protein in NPTM is dysfunctional in blocking HIV-1 infection, which may explain why pig-tailed macaques are susceptible to HIV-1 infection (Kuang et al, 2009). As mentioned above, the NPTM appears to be an optimal animal model for elucidating the mechanisms by which vaginal microbiota influences resistance and susceptibility to HIV-1 infection. However, very little is known about the composition and temporal variability of vaginal microbiota in NPTM.
The primary objective of this study was to characterize the changes in the composition of the vaginal microbiota of two healthy NPTM followed longitudinally over 19 weeks using a cultivation-independent method based on 454 pyrosequencing of 16S rRNA genes. Secondly, we aimed to quantitatively analyze temporal variability of the vaginal microbiota within individual macaques using ecology and network analysis.
MATERIALS AND METHODS
Experimental animals
Healthy, sexually mature, non-pregnant and non-hormone-treated female NPTM (n=2, 5 years old) were used in the study. Macaques were individually housed in standard stainless steel primate cages in compliance with the Guide for the Care and Use of Laboratory Animals at Kunming Primate Research Center, accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care. Housing rooms were environmentally controlled to maintain a temperature range of 18 °C to 29 °C, relative humidity of 30% to 70%, and a 12:12 light-dark cycle. Macaques were fed twice daily with monkey chow and a variety of fresh fruits and vegetables. All protocols and procedures (SYDW-2011016) used in this study were approved by the Kunming Institute of Zoology Institutional Animal Care and Use Committee. The macaques were trained for all procedures by giving fruit as a reward, and were negative for SIV, HIV-2, SRV, STLV, and CHV-1.
Study design and vaginal swabs collection
The protocol for obtaining vaginal swabs was as follows. Four vaginal swabs were collected from each macaque using individual sterile vaginal swabs. The temporal order of vaginal swab collection was as follows. Three vaginal swabs were utilized for genomic DNA extraction. The last vaginal swab was utilized for measuring vaginal pH. For investigating long interval temporal dynamic changes in the vaginal microbiome, the two female NPTM were studied from 17 November 2011 through to 28 March 2012. Vaginal swabs were collected every other day in the first week, then once weekly over 7 weeks and once bi-weekly over 9 weeks. Overall, 108 vaginal swabs were collected over 19 weeks. Prior to obtaining vaginal swabs, each macaque was gently restrained and placed in ventral recumbency on a surgical table, with the pelvis elevated approximately 60 degrees from horizontal to help visualize the vulva. The area around the vulva was carefully wiped with 0.9% sterile sodium chloride solution in a single upward motion going from vagina to anus, followed by a sterile gauze wiped in the same direction so as not to contaminate with perineal material and stool. A sterile nose speculum was used on the macaques with narrow vaginal openings to facilitate atraumatic insertion of a sterile vaginal swab into the vagina and exclude the possibility of contamination of the sample by perineal material and stool during entry and withdrawal of the vaginal swab. For sampling, vaginal swabs were twisted to collect the vaginal fluid on all sides on the tip of the swab and wiped in five full circles around the vaginal wall for 1 min before being carefully removed. The vaginal swab used for genomic DNA extraction was immediately separated from the stick with sterile scissors and placed in a sterile 5 mL cryovial (Greiner, Germany) on ice, then stored at -80 °C until analysis. The vaginal swab used for measuring vaginal pH was carefully rolled on a pH-indicator strip until the surface was covered completely with vaginal fluid.
Metadata acquisition
Vaginal pH of each macaque was determined by rolling the vaginal swab on a pH-indicator strip (Merck, Germany) with a resolution of 0.5 pH units, and was measured by comparing the color change on the pH-indicator strip with the color chart provided by the manufacturer.
DNA extraction and purification
Genomic DNA extractions from frozen vaginal swabs were performed according to Ravel et al (2011) with minor modification, using a two-step cell lysis procedure based on enzymatic and mechanical lysis. Briefly, the swabs were immersed in 600 µL of 1× phosphate-buffered saline (PBS) (Gibco, Invitrogen, USA) on ice and vortexed vigorously for 5 min to resuspend the cells. A 500 µL aliquot was transferred to a sterile 2.0 mL tube containing 0.1 mm zirconia/silica beads (BioSpec Products, USA) and stored on ice before genomic DNA extraction. An enzymatic solution composed of 50 µL of lysozyme (10 mg/mL; Sigma-Aldrich, USA), 7.5 µL of mutanolysin (20 000 U/mL; Sigma-Aldrich), 4 µL of lysostaphin (3 000 U/mL; Sigma-Aldrich), and 38.5 µL of TE buffer (10 mmol/L Tris-HCl and 1 mmol/L EDTA pH 8.0; Sigma-Aldrich) was added to the tube and mixed. The mixture was incubated for 1 h at 37 °C. Microbial cells were mechanically disrupted using TissueLyser II (Qiagen, Germany) at 28 Hz for 2.5 min. Then, 20 µL of proteinase K (20 mg/mL; Qiagen), 4 µL of RNase A (100 mg/mL; Qiagen), and 600 µL of buffer AL (Qiagen) were added, vortexed thoroughly, and incubated for 30 min at 56 °C. The lysate was processed using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s protocols, but omitting the lysis steps. The DNA was eluted into 150 µL of buffer AE (Qiagen). The concentration of DNA was determined using a NanoDrop ND-2000 spectrophotometer (Thermo Electron Corporation, USA).
PCR amplification and pyrosequencing of barcoded 16S rRNA genes
The PCR amplification and pyrosequencing of barcoded 16S rRNA genes were performed according to our previously described method (Zhang et al, 2013, 2014). PCR amplification of the V1-V2 hypervariable regions of the 16S rRNA genes was performed with barcoded universal bacterial primers 27F and 338R containing Primer A or Primer B. The primers were as follows: 27F (Primer B) 5′-CTATGCGCCTTGCCAGCCCGC TCAGTCAGAGTTTGATCCTGGCTCAG-3′ and 338R (Primer A) 5′-CGTATCGCCTCCCTCGCGCCATCAGNNNNNNNNNNCAT GCTGCCTCCCGTAGGAGT-3′, where the underlined letters denote Primer A and Primer B, universal bacterial primers 27F and 338R are in italic, and 10Ns represents a barcode unique for each sample. The PCR reactions were performed in 96-well PCR microplates (Axygen, USA) using 25 µL (total reaction volume) mixtures containing 2.5 µL of 10× PCR Gold buffer (Applied Biosystems, USA), 1.5 µL of MgCl2 (25 mmol/L, Applied Biosystems), 2.0 µL of dNTP blend (2.5 mmol/L each; Applied Biosystems), 0.25 µL of primers 27F and 338R (20 µmol/L each), 0.25 µL of AmpliTaq Gold DNA polymerase (5 U/µL; Applied Biosystems), and 50 ng of template DNA or RNAse/DNAse free water in case of negative controls. The negative controls were included in each 96-well PCR microplate. The microplates were then placed on a PCR-Cooler (Eppendorf, Germany) during PCR reaction mixture preparation. Reactions were run in a GeneAmp PCR System 9700 (Applied Biosystems) with the following cycling parameters: 10 min initial denaturing at 95 °C followed by 30 cycles of denaturing at 95 °C for 30 s, annealing at 55 °C for 30 s, and elongation at 72 °C for 90 s, with a final extension at 72 °C for 10 min. Triplicate PCR reactions were performed independently for each sample. The barcoded amplicons from the triplicate reactions were then pooled together and subsequently detected by 1.5% agarose gel electrophoresis. Barcoded amplicons were stained with ethidium bromide and quantitated digitally on a ChemiDoc XRS system (BioRad, USA) following electrophoresis. According to the manufacturer’s protocols, barcoded amplicons were purified using a MinElute Gel Extraction Kit (Qiagen). The purified barcoded amplicons were then quantified with a Quant-iT PicoGreen dsDNA assay kit (Invitrogen, USA). Amplicon pyrosequencing was performed using Primer A and Primer B in the 454 Genome Sequencer FLX Titanium platforms (Roche, USA), in accordance with the manufacturer’s protocols, at the Kunming Biological Diversity Regional Center of Large Apparatus and Equipment, Chinese Academy of Sciences.
Sequence analysis and taxonomic assignments
Sequence analysis and taxonomic assignments were performed according to our previously described method (Zhang et al, 2013, 2014). Raw sequencing reads were quality trimmed using the QIIME pipeline v1.4.0 (Caporaso et al, 2010b) according to the following criteria: (a) exact matches to the barcode sequence and primer, (b) minimum and maximum length of 200 bp and 400 bp, (c) no ambiguous bases allowed, and (d) minimum quality score allowed in read=25. The 16S rRNA gene sequences were binned on the basis of the unique sequence barcodes associated with the unique primer used for each sample and trimmed by removal of the barcode and primer sequences. Sequencing errors were removed from filtered sequences using Denoiser 0.91 (Reeder & Knight, 2010). The denoised sequence dataset was clustered into operational taxonomic units (OTUs) using CD-hit (Li & Godzik, 2006) with the criterion of a minimum identity of 97%. A representative sequence was chosen from each OTU by selecting the longest sequence with the largest number of hits to other sequences in the OTU. Representative sequences were aligned to the Greengenes database (Desantis et al, 2006; McDonald et al, 2012) using PyNAST (Caporaso et al, 2010a) with a minimum alignment length of 150 and a minimum identity of 75%. Using ChimeraSlayer (Haas et al, 2011), chimera sequences arising from the PCR amplification of multiple templates or parent sequences were detected and excluded from the aligned representative sequences. Sequences that met quality control criteria were classified to genus using Greengenes (Desantis et al, 2006; McDonald et al, 2012). We used the representative sequences to BLAST against assembled refseq genomes of microbes in GenBank to search the best hits for species. Searches against the database were conducted with MegaBLAST. A sequence read was annotated as the best hit to the database when the total score was highest, the E-value was lowest, and we obtained at least 97% identical between query and subject. The Good’s coverage of each sample was calculated as C=[1-(n/N)]×100%, where n is the number of singleton phylotypes per sample and N is the total number of sequence reads in that sample (Good, 1953). Samples that did not have at least 300 sequences after quality filtering and OTU assignment were excluded from cluster analysis of samples and the following analyses.
Clustering analysis of vaginal bacterial communities
According to bacterial composition and abundance, a cluster of vaginal bacterial communities with similar observed phylotype composition and relative abundances are assigned as a vaginal bacterial community state type (Gajer et al, 2012). Ward linkage hierarchical clustering of the Bray-Curtis distances between vaginal bacterial communities was performed. A Ward linkage hierarchical cluster dendrogram was generated by first generating a Bray-Curtis distance matrix with the dist function and passing this matrix to the hclust function in the R package vegan (Dixon, 2003; R Development Core Team, 2012). A heatmap was generated with the heatmap.2 function of the R package gplots (R Development Core Team, 2012). The Shannon diversity index (Shannon, 1948), which accounts for both the number and relative abundances of phylotypes in a community, was calculated with QIIME pipeline v1.4.0 (Caporaso et al, 2010b) to quantitatively measure vaginal bacterial community diversity. Spearman’s rank correlation coefficients were used to assess correlation between the number of sequences/vaginal pH and the Shannon indices across all samples of the two subjects. Spearman’s rank correlation coefficients were calculated using GraphPad Prism 5. Statistical significance was defined as P<0.05.
Temporal dynamics analysis of vaginal bacterial communities
The median Shannon diversity index and vaginal pH of the dynamic profile of the vaginal bacterial community of each macaque were calculated using GraphPad Prism 5. The profile of the vaginal bacterial community state type is defined as a sequence of vaginal bacterial CST assigned to each community in time series data. The proportion of time that each vaginal bacterial community state type was observed within the profile was calculated using GraphPad Prism 5. Bar plots of phyla and associated heatmaps of phylotypes were constructed for comparing the relative abundances of phyla and phylotypes across samples at all time points from each macaque.
Taylor’s power law analysis for assessing the stability of community diversity and species aggregation over time of vaginal bacterial communities
The stability of community diversity over time was characterized with Taylor’s power law as per Ma (2012). Taylor’s power law has been extensively used to describe the spatial distribution patterns of many species (Taylor, 1961, 1984; Taylor & Taylor, 1977), and can be expressed by: V=aMb, where M and V are population mean (density or abundance) and variance, respectively. Parameter a is considered a sampling factor, and parameter b is an index of aggregation characteristic of the species. Parameter b>1 indicates aggregation, b=1 is indicative of randomness, and b<1 indicates regularity (Ma, 2012; Taylor, 1961, 1984; Taylor & Taylor, 1977). Ma (2012) extended Taylor’s power law for characterizing the microbial communities (Zhang et al, 2014) into a two-step process: the first step computes the pairs of mean and variance from the abundance data, and the second step fits the model. By regressing the natural logarithm of the variance on the natural logarithm of the mean, the resulting regression equation is as follows: ln(V)=ln(a)+bln(M) (Ma, 2012). To examine the community diversity stability over time for a subject, M and V at each time point were computed by averaging the abundances of all species, and total n pairs of M and V per subject were computed for the n time point. To examine the species aggregation stability over time for a subject, M and V of each species were computed by averaging the abundances of all time points, and total S pairs of M and V per subject were computed for the S species. The above calculations and model fittings were performed using R package stats (R Development Core Team, 2012).
Network analyses of correlation profiles of taxa with each other and with vaginal pH
Subject-level network analysis, which finds connections among species and between species and vaginal pH in a time series of vaginal bacterial community samples within a subject (subject A or subject B), was performed by constructing a network in which nodes corresponded to a species or vaginal pH and edges corresponded to pairs of species and vaginal pH that were significantly correlated. Correlation was assessed by Spearman’s rank correlation coefficients between the relative abundance of pairs of species whose mean relative abundances across all samples of each subject were above 0.1% and/or vaginal pH within the subject across the time series of the vaginal bacterial community samples. Spearman’s rank correlation coefficients were calculated using the rcorr function in the R package Hmisc (R Development Core Team, 2012). Custom Perl scripts were used for text manipulation of the resulting coefficients and associated P-values. The P-values on each edge were adjusted to FDR (false discovery rate) for multiple statistical testing when evaluating these correlations using the Benjamini-Hochberg method (Benjamini & Hochberg, 1995) and a significant q value was set to 0.05. Based on Spearman’s rank correlation coefficients above 0.7 or below -0.7, a network of correlation profiles between species and vaginal pH was generated with Cytoscape v2.8.3 (Shannon et al, 2003; Smoot et al, 2011). Network statistics used to describe global and local network properties were calculated with a NetworkAnalyzer plug-in (Assenov et al, 2008) for Cytoscape. We treated edges as undirected when calculating network statistics, because an edge represents a mutual interaction. The global network properties calculated were the number of nodes and edges and centralization. The local network properties calculated were betweenness centrality and degree of node. We also performed network intersection between the correlation networks of subject A and subject B with NetworkAnalyzer (Assenov et al, 2008).
RESULTS
Samples and datasets
A total of 27 NPTM vaginal samples were collected. The vaginal pH of the samples was measured. The composition and abundance of the sampled vaginal bacterial communities were determined by pyrosequencing the V1-V2 hypervariable regions of the 16S rRNA genes. The sequences dataset consisting of 27 362 high-quality classifiable 16S rRNA gene reads was yielded from the 27 samples, with an average of 1 013±117 (SE) reads per sample. We identified 336 OTUs according to 97% sequence similarity at the species level. Singletons—a sequence that only occurred once in the 27 samples—were next removed, representing a total of 109 OTUs. A total of 227 OTUs remained, with an average of 39±3 (SE) OTUs per sample, representing 76 genera from 10 bacterial phyla. Six bacterial phyla (Fusobacteria, Actinobacteria, Firmicutes, Bacteroidetes, Proteobacteria, and Tenericutes) made up 99.94% of sequences. All samples the in dataset had an average of Good’s coverage (estimated percentage of the total species represented in a sampling site) of 98.3%±0.16% (SE), indicating that the sequencing depth was sufficient for reliable analysis of these vaginal microbiota samples.
Hierarchical clustering analysis of vaginal bacterial communities
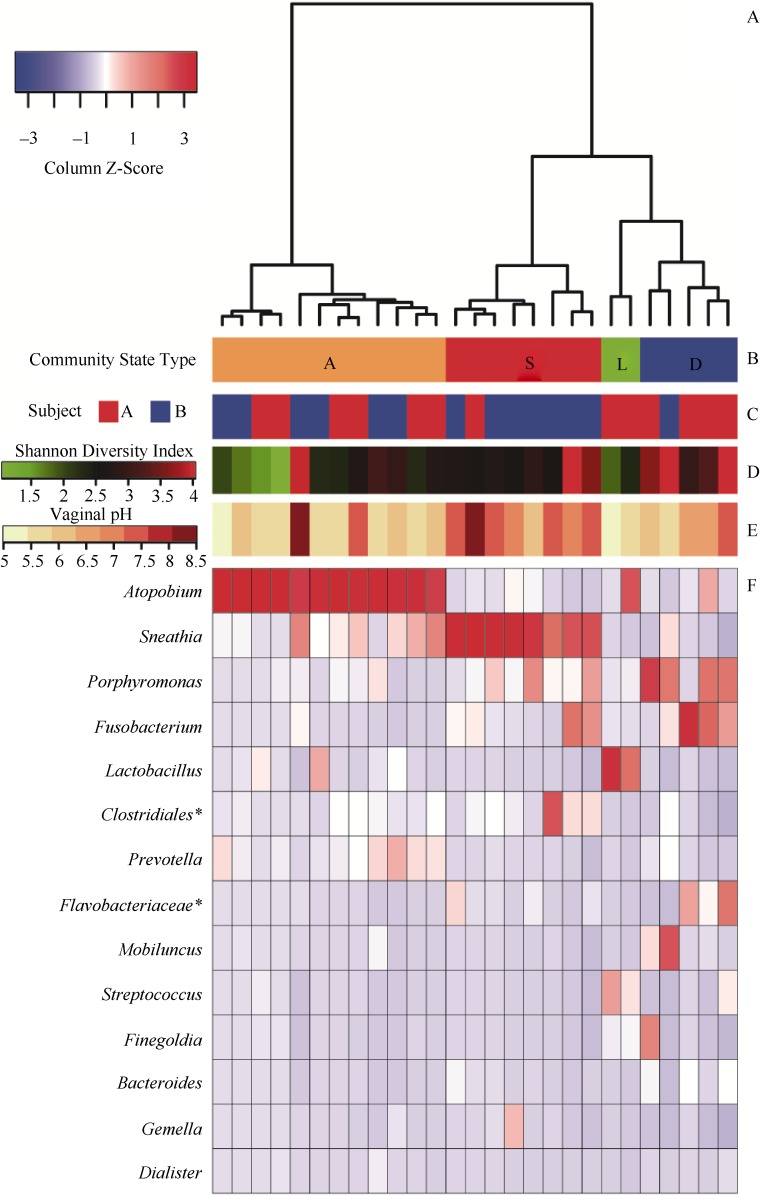
To characterize the composition and abundance of vaginal bacterial communities of NPTM using Ward linkage hierarchical clustering of the Bray-Curtis distances between communities, the vaginal bacterial communities were clustered into four groups based on the relative abundances of the 14 most abundant phylotypes (mean relative abundance of each phylotype across all samples of two subjects was above 1%) (Figure 1A,B,C,F). These were referred to as community state types (CSTs) according to the nomenclature established by Gajer et al (2012). The four CSTs, designated as CST A, CST S, CST L, and CST D, contained 12 (44.44%), 8 (29.63%), 2 (7.41%), and 5 (18.52%) of the 27 samples, respectively. It is worth nothing that the communities of these CSTs, except for CST L, were from both subjects.
Figure 1.
Clustering analysis of vaginal bacterial communities
A: Ward linkage hierarchical cluster dendrogram of the Bray-Curtis distances between bacterial communities in 27 samples from two northern pig-tailed macaques (Macaca leonina). B: Color bar indicating vaginal bacterial CST wherein each sample was assigned to one of four CSTs (CST A, CST S, CST L, and CST D). C: Lower colored blocks indicate the subject from which the sample was obtained. D: Shannon diversity indices calculated for the 27 samples (color key is indicated in the middle left side). E: Vaginal pH measurements for the 27 samples (color key is indicated in the middle left side). F: Heatmap of the relative abundances of the 14 most abundant phylotypes (mean relative abundance of each phylotype across all samples of the two subjects was above 1%). Column Z-score indicates differences between samples in terms of relative abundances of phylotypes associated with the samples. Individual cells are color-coded according to Z-scores to show the normalized abundance of a phylotype in one sample relative to the mean abundance across all phylotypes of the column. Relative intensity of the colors indicates how many standard deviations the observed phylotype abundance is above or below the mean. White color indicates relative abundance of phylotypes with column average. Blue color indicates relative abundances less than average abundance. Red color indicates relative abundance above column average. Phylotypes are listed in order of dominance, with the most dominant on the top. *represents unclassified genera of order/family.
CST A, CST S, and CST L were dominated by Atopobium, Sneathia, and Lactobacillus, respectively (Figure 1B), while communities clustered in CST D were not dominated by one phylotype. CST A had a modest median Shannon diversity index (2.33±0.74) and the second lowest median vaginal pH (5.5±0.37) (Figure 1D,E). CST S had the second highest median Shannon diversity index (2.73±0.25) but had the highest median vaginal pH (7.5±0.37). In contrast to CST A and CST S, CST L had the lowest median Shannon diversity index (2.11±0.37) and the lowest median vaginal pH (5.25±0.37). CST D had modest proportions of Porphyromonas and Fusobacterium, along with low proportions of Mobiluncus, Prevotella, Bacteroides and some unclassified genera of the family Flavobacteriaceae. CST D had the highest median Shannon diversity index (3.64±0.55) and the second highest median vaginal pH (6.5±0.74) (Figure 1D,E).
We assessed the association between the Shannon indices of vaginal bacterial communities and vaginal pH. No correlation was found between the number of bacterial sequences of the 16S rRNA gene of each sample and the Shannon diversity indices of the vaginal bacterial communities (Spearman correlation coefficient=0.022, P=0.913>0.05), which confirmed that microbial diversity was not influenced by the number of sequences itself. Furthermore, the Shannon indices of vaginal bacterial communities were positively correlated with vaginal pH (Spearman correlation coefficient=0.503, P=0.007), indicating that vaginal pH may be a physiological factor influencing the vaginal bacterial community.
Temporal dynamics of vaginal bacterial communities
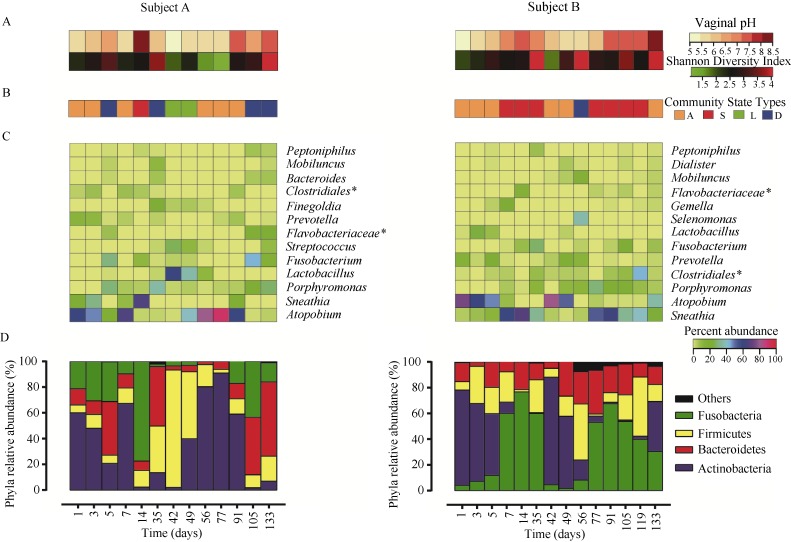
To characterize the temporal dynamics of the vaginal bacterial communities of the individual monkeys, our analyses were respectively conducted at the levels of alpha diversity, CST, genus, and phylum. We noticed remarkable inter- and intra-subject variations in the Shannon diversity index, vaginal pH, CST, and the relative abundances of abundant genera and phyla in subject A and subject B (Figure 2A,B,C,D).
Figure 2.
Temporal dynamics of vaginal bacterial communities
A: Vaginal pH measurements and Shannon diversity indices calculated for each of the 13 samples of subject A and 14 samples of subject B (color key is indicated in the top right corner). B: Profiles of CST for the two northern pig-tailed macaques over time. Each cell represents one sample in the time series. The CSTs in the time series for each macaque are color-coded (lower colored blocks indicate CST wherein each sample was assigned to one of the four CSTs). C: For each macaque, a heatmap was constructed from the relative abundances of phylotypes that comprised at least 1% of the sequences found in that macaque (color key is indicated in the middle right side). Phylotypes are listed in order of dominance, with the most dominant on the bottom. *represents unclassified genera of order/family. D: For each macaque, the bar plot represents the relative abundances of phyla that comprised at least 1% of the total sequences found in that macaque. Phyla that comprised less than 1% of the total sequences found in that macaque were grouped into the “Others” category.
At the alpha diversity level (Figure 2A), both subject A and subject B had a Shannon diversity index<2.0 for at least one time-point tested and a Shannon diversity index>2.0 for the majority of the study. The median Shannon diversity index of the dynamic profile of the vaginal bacterial community of subject A (2.62±0.50) was lower than that of subject B (2.80± 0.60). Similarly, there was no consistent trend in vaginal pH changes between subject A and subject B (Figure 2A). Subject A had pH values<6.5 at most time-points tested (84.62%), while subject B had a more alkaline pH of 7.0 at half the time-points tested. The median vaginal pH of the dynamic profile of the vaginal bacterial community of subject A (6.0±0.74) was lower than that of subject B (6.5±1.48).
At the CST level (Figure 2B), the distribution of CST in the dynamic profiles of the vaginal bacterial communities of subject A and subject B was different. Importantly, only CST A was overrepresented in the dynamic profiles of the vaginal bacterial communities of both subject A (46.15%) and subject B (42.86%).
CST S was overrepresented in subject B (50%), but not in subject A (7.7%), and CST L was not observed in subject B at all (Figure 2C). In contrast to CST S, CST D was overrepresented in subject A (30.77%), but not in that of subject B (7.14%). Interestingly, it is only transitions of vaginal bacterial communities from the CST A to CST A that was more observed than other kinds of transitions of vaginal bacterial communities in subject A and subject B.
At the levels of genus and phylum (Figure 2C,D), we found that the composition and relative abundances of the vaginal bacterial communities across subjects and time points changed markedly over short and long periods. In subject A, the dominant phyla were Actinobacteria (38%), Firmicutes (22.26%), Bacteroidetes (20%), and Fusobacteria (19.5%). The dominant phyla of subject B were similar, but the relative abundances of shared dominant phyla were different. The dominant phyla of subject B were Fusobacteria (34.23%), Actinobacteria (28.21%), Firmicutes (18.68%), and Bacteroidetes (17.15%). It is worth noting that Atopobium (genus of Actinobacteria) and Sneathia (genus of Fusobacteria) were the dominant genera in subject A (36.44% and 11.14%, respectively) and subject B (26.31% and 28.76%, respectively).
Together, these results indicate that the temporal changes in composition of the vaginal bacterial community of the individual monkeys were highly individualized.
Taylor’s power law analysis of temporal variability of vaginal bacterial communities
The Shannon diversity index is a quantitative measure of species diversity, though it can only measure static diversity at a specific time point. Thus, it is difficult to study temporal changes in community diversity. To gain a clearer picture of the temporal variability in the vaginal bacterial communities, we examined the degree of stability of community diversity over time and stability of species aggregation over time (Ma, 2012). Power law parameter b is a measure of instability: the larger the b-value, the higher the instability of community diversity or instability of species aggregation over time of a subject.
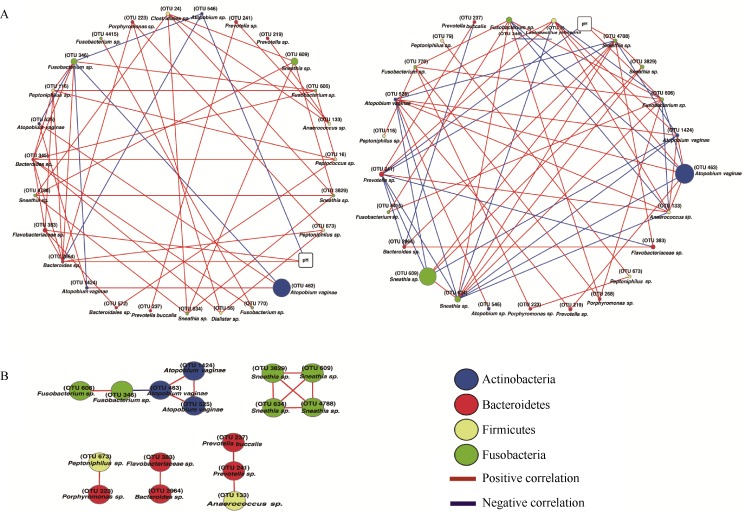
Figure 3.
Integration of vaginal pH with microbial co-occurrence networks using time-series data of each northern pig-tailed macaque
Note, the nodes in the correlation network correspond to OTUs (ellipse nodes) and vaginal pH (round rectangle nodes), whereas the edges represent strong and significant evidence for correlation between nodes. Size of the nodes indicates relative abundances of OTUs (mean relative abundance of each OTU across all samples of each subject was above 0.1%) observed within a subject over time.
Differences in the stability of community diversity and species aggregation over time between subject A and subject B were observed. As seen in Table 1, the stability of community diversity over time in subject A was higher than that in subject B. Similarly, the stability of species aggregation over time in subject A was also higher than that in subject B (Table 2). Notably, the degree of variability in stability of species aggregation over time between subject A and subject B was considerably smaller than that of stability of community diversity over time.
Table 1.
Community diversity stability over time for each northern pig-tailed macaque (Macaca leonina)
| Subject | b | SE (b) | ln (a) | SE ln (a) | R2 | P | n |
|---|---|---|---|---|---|---|---|
| A | 1.5753 | 0.3243 | 4.5357 | 0.5186 | 0.6531 | 0.0005049 | 13 |
| B | 2.0384 | 0.1975 | 3.7854 | 0.3472 | 0.8904 | 2.537e-07 | 14 |
Table 2.
Species aggregation stability over time for each northern pig-tailed macaque (Macaca leonina)
| Subject | b | SE (b) | Ln (a) | SE ln (a) | R2 | P | Species |
|---|---|---|---|---|---|---|---|
| A | 1.74695 | 0.02313 | 1.80973 | 0.04177 | 0.9729 | < 2.2e-16 | 160 |
| B | 1.76344 | 0.02455 | 1.87320 | 0.04399 | 0.9647 | < 2.2e-16 | 190 |
Together, these results suggest that the temporal variability in the vaginal bacterial community of the individual monkeys was considerably individualized.
Network analysis of temporal variability of vaginal bacterial communities
To ascertain the effect of vaginal pH on the temporal variability of vaginal bacterial communities and disentangle correlation profiles of species with each other and with vaginal pH over time for each subject, network analysis was conducted as per Duran-Pinedo et al (2011).
Firstly, by comparing the relative abundances of OTUs across all samples of the two subjects, we assessed inter-subject variation at the species level. We found that 123 OTUs were shared between subject A and subject B. Subject A and subject B had 37 and 67 unique OTUs, respectively. The summed relative abundances of unique OTUs across all samples of subject A (1.95%) were lower than that of subject B (8.49%). Thus, total abundance of shared OTUs accounted for the majority of total abundance of all OTUs in subject A and subject B. To reduce the complexity of the network and avoid the potential impact of inter-subject variation on network analysis, only OTUs with mean relative abundances above 0.1% across all samples of each subject were considered for network analysis. The resulting correlation network (species and vaginal pH) of subject A consisted of 27 nodes and 50 edges, including 44 positive correlations and 6 negative correlations. For subject B, the resulting correlation network (species and vaginal pH) consisted of 25 nodes and 63 edges, including 44 positive correlations and 6 negative correlations.
Next, we compared the differences in centralization between the correlation networks of subject A and subject B. Network centralization, as an important parameter of the global properties of network, is used for describing whether the network is dominated by several hub nodes or not: the lower the centralization, the higher the resilience of a network to environmental change. We found that the correlation network of subject A (0.178) had lower centralization compared with the correlation network of subject B (0.270), indicating that the network of subject A was more resilient than subject B to changes in relative abundances of taxa or vaginal pH over time.
In order to quantify the importance of a node, such as vaginal pH or species, the degree (number of connections a node has to other nodes in the network) and betweenness centrality (amount of control a node exerts over the interactions of other nodes in the network) of a given node were calculated. Both degree and betweenness centrality are two measures of the importance of a node: the larger the values of degree and betweenness centrality, the higher the importance of the node. The betweenness centrality of vaginal pH in the correlation network of subject A (0.007) was lower than that of subject B (0.032), while the degree of vaginal pH in the correlation network of each subject was equal to 3. These results indicate that the influence of vaginal pH on the correlation network of subject A was weaker than that on the correlation network of subject B.
Four species, including (OTU 345) Bacteroides sp, (OTU 346) Fusobacterium sp, (OTU 2964) Bacteroides sp, and (OTU 24) Clostridiales sp in the correlation network of subject A (Table 3) and six species, including (OTU 634) Sneathia sp, (OTU 606) Fusobacterium sp, (OTU 241) Prevotella sp, (OTU 346) Fusobacterium sp, (OTU 463) Atopobium vaginae, and (OTU 4788) Sneathia sp in the correlation network of subject B (Table 4) were defined as hub species whose degree and betweenness centrality were both higher than the mean degree and mean betweenness centrality of all nodes in the correlation network of each subject. Interestingly, there were no consistent connections between vaginal pH and hub species between subject A and subject B. In the correlation network of subject A, vaginal pH was positively correlated with (OTU 2964) Bacteroides sp, while vaginal pH was positively correlated with (OTU 606) Fusobacterium sp and (OTU 4788) Sneathia sp and negatively with (OTU 241) Prevotella sp in the correlation network of subject B.
Table 3.
Betweenness centrality and degree of hub species in correlation network of subject A
| OTUs | Betweenness centrality | Degree |
|---|---|---|
| (OTU 345) Bacteroides sp | 0.36373 | 8 |
| (OTU 346) Fusobacterium sp | 0.34571 | 8 |
| (OTU 2964) Bacteroides sp | 0.26201 | 8 |
| (OTU 24) Clostridiales sp | 0.57143 | 5 |
| Mean of network | 0.1036±0.03413 (SE) | 3.704±0.3767(SE) |
Table 4.
Betweenness centrality and degree of hub species in correlation network of subject B
| OTUs | Betweenness centrality | Degree |
|---|---|---|
| (OTU 634) Sneathia sp | 0.220773 | 12 |
| (OTU 606) Fusobacterium sp | 0.2565217 | 9 |
| (OTU 241) Prevotella sp | 0.1682367 | 9 |
| (OTU 346) Fusobacterium sp | 0.1881642 | 8 |
| (OTU 463) Atopobiumvaginae | 0.09275362 | 9 |
| (OTU 4788) Sneathia sp | 0.08828502 | 9 |
| Mean of network | 0.07116±0.01828 (SE) | 5.280±0.6417 (SE) |
We found that the intersection connections between the correlation networks of subject A and subject B were comprised of 16 nodes and 15 edges. Interestingly, all edges, except the negative correlations between (OTU 346) Fusobacterium sp and (OTU 463) Atopobium vaginate, were positively correlated. In the correlation network of both subjects, (OTU 346) Fusobacterium sp was a hub species and (OTU 463) Atopobium vaginae was the most abundant species.
Overall, these data suggest that inter-subject variability in vaginal bacterial communities over time could be mirrored in inter-subject variation in correlation profiles of species with each other and with vaginal pH over time. These results may be associated with the influence of vaginal pH and hub species on the correlation network of each subject.
DISCUSSION
To the best of our knowledge, this is the first longitudinal study on the vaginal microbiota in normal NPTM where samples have been collected over 19 weeks and the composition and temporal variability of vaginal microbiota have been characterized using a combination of 454 pyrosequencing of the 16S rRNA genes and well-established methods of ecology and network analysis. The major findings of this study were: (1) vaginal microbiota in NPTM had high proportions of a diverse array of anaerobic bacteria associated with BV, decreased proportions of Lactobacillus, but higher microbial diversity, and Atopobium and Sneathia were the dominant genera; (2) temporal dynamics of the NPTM vaginal microbiota were considerably individualized; and (3) vaginal pH may influence the temporal dynamics of the vaginal microbiota. This work emphasizes the importance of prospective investigation in capturing temporal dynamics of vaginal microbiota in its full complexity and ascertaining the influencing factors.
Efforts to reveal the mechanisms by which vaginal microbiota impacts resistance to and risk of HIV infection are hampered by the dynamic shifts in vaginal microbiota over time, and the factors that influence this are poorly understood (Brotman, 2011; Nardis et al, 2013; Petrova et al, 2013; Saxena et al, 2012; Schellenberg & Plummer, 2012). To date, comprehensive understanding of the dynamic landscape of vaginal microbiota remains poorly elucidated due to the inherent limitations of culture-dependent methods (Farage et al, 2010), low-resolution molecular techniques (Farage et al, 2010), and cross-sectional analyses (Ravel et al, 2011). While most attention has focused on cross-sectional profiles of vaginal microbiota, there is a distinct lack of information regarding the dynamic landscape of vaginal microbiota (Hickey et al, 2012; Ma et al, 2012). As high-throughput sequencing-based techniques for microbiota analysis have been developed and used, vaginal microbiota has been revealed as considerably more complex and dynamic (Witkin & Ledger, 2012).
Recently, the vaginal microbiota of healthy, asymptomatic, non-pregnant women of reproductive age has been examined in cross-sectional and longitudinal cohorts using cultivation-independent methods based on 454 pyrosequencing of 16S rRNA genes (Gajer et al, 2012; Ravel et al, 2011). These studies demonstrated that vaginal microbiota could be clustered into six community state types (CSTs), four of which were dominated by one of four Lactobacillus spp., including L. crispatus (CST I), L. gasseri (CST II), L. iners (CST III) and L. jensenii (CST V), with the remaining two (CST IV-A and IV-B) characterized by a diverse array of anaerobic bacteria associated with BV (Gajer et al, 2012; Ma et al, 2012; Ravel et al, 2011). CST IV-A and IV-B differed in both composition and relative abundance of phylotypes (Gajer et al, 2012). For example, CST IV-A had modest proportions of Lactobacillus spp. and low proportions of Prevotella, Peptoniphilus, Anaerococcus, Corynebacterium, Streptococcus, or Finegoldia, whereas CST IV-B was characterized by high proportions of Atopobium and the presence of Sneathia, Prevotella, Parvimonas, Gardnerella, Mobiluncus or Peptoniphilus (Gajer et al, 2012).
Importantly, we found that Atopobium and Sneathia were dominant genera in NPTM, and CST A and CST IV-B were very similar in composition and relative abundance of phylotypes. Moreover, in our study, OTU 463 (97% similarity to Atopobium vaginae) was the dominant Atopobium species. Atopobium vaginae is a gram-positive facultative anaerobic bacterial species and has been highly predictive of both asymptomatic BV (healthy asymptomatic women) and symptomatic BV (women with BV) (Menard et al, 2008; Rodriguez Jovita et al, 1999; Shipitsyna et al, 2013). Several identified bacteria in our study, including Sneathia, Prevotella, and Mobiluncus, have also been reported to be highly predictive for BV (Fettweis et al, 2014; Fredricks et al, 2005). Thus, with respect to specific composition at the genus level, the vaginal microbiota of NPTM contained many bacteria associated with BV in women. It is important to note that the vaginal microbiota of NPTM had lower proportions of Lactobacillus but higher microbial diversity in comparison with that of the majority of women, which is most commonly dominated by Lactobacillus spp and has low microbial diversity (Ravel et al, 2011). These findings are in agreement with a few exploratory studies that have described differences between human and NHP species (Spear et al, 2010, 2012; Yildirim et al, 2014). Although these studies have found trends toward decreased proportions of Lactobacillus and increased microbial diversity, we are the first to demonstrate the presence of Lactobacillus-dominated vaginal microbiota in NHP species. Taken together, the vaginal microbiota of NPTM can closely resemble asymptomatic and symptomatic BV, characterized by the decrease in proportions of Lactobacillus and increase in microbial diversity, suggesting that NPTM could be an optimal animal model for prospective investigations on the mechanisms by which vaginal microbiota influence susceptibility and resistance to HIV-1 infection in the context of highly polymicrobial and Lactobacillus-dominated states.
To date, the vaginal microbiota of ten NHP species has been characterized using cultivation-independent methods based on 454 pyrosequencing of the 16S rRNA genes (Spear et al, 2010, 2012; Yildirim et al, 2014). Of high relevance to our study are the findings observed in Sunda pig-tailed macaques (Macaca nemestrina) (Spear et al, 2012). We observed that these two host species were similar in composition of most of the abundant genera and phyla, but differed in the relative abundances of those genera and phyla, suggesting differences in host species analyzed. For example, with respect to dominant genera, the major difference between the two species was that NPTM had high proportions of Atopobium (31.19%) within the phylum Actinobacteria and modest proportions of Fusobacterium (6.85%) within the phylum Fusobacteria, while Sunda pig-tailed macaques had high proportions of Fusobacterium (13.3%) and modest proportions of Atopobium (3.6%). This may be explained by the negative correlation between the dominant (OTU 346) Fusobacterium sp and the dominant Atopobium species, (OTU 463) Atopobium vaginae, in our study. In addition, Sneathia within the phylum Fusobacteria was the dominant genus in both NPTM (20.28%) and Sunda pig-tailed macaques (18.9%). Interestingly, Sneathia has been consistently found to be the abundant genus in ten nonhuman primate species, in healthy, asymptomatic women, and in women with BV (Fettweis et al, 2014; Yildirim et al, 2014). The presence of non-Lactobacillus-dominant CST in healthy, asymptomatic, non-pregnant women of reproductive age and in nonhuman primates has challenged the notion that healthy vaginal microbiota is necessarily associated with vaginal microbiota characterized by Lactobacillus-dominance (Gajer et al, 2012; Hickey et al, 2012; Ma et al, 2012; Ravel et al, 2011; Spear et al, 2010, 2012; Yildirim et al, 2014). Given that humans are different from NHP species in terms of vaginal microbial diversity, comparative analysis of composition and dynamics of vaginal microbiota may provide a better understanding of how microbial community formation and function are conserved across human and NHP species and how high diversity vaginal microbiota promotes host health, which will have important implications for host health and for selecting NHP species models with similar vaginal microbial community structure to humans for HIV-1 vaginal transmission.
Although many cross-sectional studies have captured static snapshots of vaginal microbiota, what is considered representative at any given time remains unclear. Thus, cross-sectional studies may be insufficient to capture the dynamic shifts in vaginal microbiota over time. To our knowledge, only four prior longitudinal studies have revealed temporal dynamics of vaginal microbiota in humans (Gajer et al, 2012; Ravel et al, 2013; Chaban et al, 2014) and PTM (Spear et al, 2012) using cultivation-independent methods based on 454 pyrosequencing of 16S rRNA genes and cpn60 genes. Generally, however, temporal dynamics of vaginal microbiota in primates remain poorly understood. In this work, we found that, at the level of CST, alpha diversity, genus, phylum, and species, both temporal changes in composition and temporal variability in the vaginal microbiota of each monkey were considerably personalized. Our results corroborated earlier findings that the temporal dynamics of vaginal microbiota in women with asymptomatic and symptomatic BV and PTM are highly personalized (Gajer et al, 2012; Ravel et al, 2013; Spear et al, 2012). Although these studies found trends toward personalized temporal dynamics in the vaginal microbiota at the levels of CST, alpha diversity, genus, and phylum, we are the first to apply theories of macro-ecology and networks to interpret the differences in personalized temporal dynamics of vaginal microbiota at the species level. Clearly, further longitudinal investigations into comparisons of personalized temporal dynamics of vaginal microbiota measured before and after vaginal HIV-1 infection, coupled with metadata and the adoption of ecological perspectives, will be critical for understanding the temporal relationship between vaginal microbiota and susceptibility to virus infection, and will contribute to the development of more personalized medicine (Ma, 2012; Ravel et al, 2013).
How the normal variability in microbiota through time may arise in response to dynamic changes in physiological factors is a central question in understanding crosstalk between vaginal microbiota and physiological factors. Vaginal pH is determined by the interplay between host and vaginal microbiota (Witkin & Ledger, 2012). Importantly, we found inter-subject variation in correlation profiles of species with each other and with vaginal pH over time. These results suggest that the biologically plausible mechanisms by which vaginal pH could influence normal variability in vaginal microbiota through time affected the specific species colonizing the vagina, which in turn affected the species that had close relationships with them. Furthermore, consistent with previous studies (Spear et al, 2010), the vaginal pH in 92.6% of the longitudinally collected samples from the present study (pH≥5.5) was higher than that of reproductive-age women (Ravel et al, 2011). Interestingly, vaginal pH was found to differ among reproductive-age women with different ethnicities (Ravel et al, 2011). It is believed that estrogen, glycogen, and Lactobacillus are the driving forces for interspecies differences in vaginal pH (Farage et al, 2010; Mirmonsef et al, 2012), but other host factors such as sex hormones or breeding and ovulatory seasonality may play an important role in affecting this variation (Campbell et al, 2007; Gajer et al, 2012).
Despite the limited sample size, we provided a comprehensive catalog of composition and baseline characterization of the normal temporal variability in vaginal microbiota in NPTM. Further studies are required to replicate these results in larger studies. Ultimately, despite the significant advances in the past few years, future progress in studies on vaginal microbiota is expected in many directions (Ma et al, 2012; Macklaim et al, 2012), ranging from data collection of vaginal microbiota to insights into how host genetics influence composition and temporal patterns of vaginal microbiota dynamics, as well as functions of vaginal CST. Further functional studies to explore the impact of composition and temporal dynamics of vaginal microbiota on maladies such as HIV-1/AIDS (Petrova et al, 2013; Saxena et al, 2012) and sexually transmitted diseases (Brotman, 2011; Nardis et al, 2013) could foster therapies aimed at manipulating the vaginal microbiota of the host to help form efficient first lines of defense against pathogens and regulate the vaginal mucosal immune system to prevent infection-related morbidity in women.
ACKNOWLEDGMENTS
We thank Yu-Qi HE, Xiao-Lu LI, and Zhao-Li DING (Kunming Biological Diversity Regional Center of Large Apparatus and Equipment, Chinese Academy of Sciences) for sequencing support; Hong DUAN and Yi SUN (First People’s Hospital of Yunnan Province) for support with measuring levels of serum sex hormones; and Gui LI (Experimental Animal Core Facility, Kunming Institute of Zoology, Chinese Academy of Sciences) for sample collection support. We are indebted to our colleagues: Zheng-Xi DAI, Jian-Bao HAN, Rong-Hua LUO, Ping-Xian XU, and Liu-Meng YANG for all their help. We also thank Dr. Christine WATTS for assistance with editing the manuscript.
Funding Statement
This work was supported in part by the Key Scientific and Technological Program of China (2012ZX10001-007,2013ZX10001-002), the National Basic Research Program of China (2012CBA01305), the National Natural Science Foundation of China (81172876,81273251,U1202228), and the Knowledge Innovation Program of CAS (KSCX2-EW-R-13,KJZD-EW-L10-02)
REFERENCES
- [1].Agy MB, Frumkin LR, Corey L, Coombs RW, Wolinsky SM, Koehler J, Morton WR, Katze MG. 1992. Infection of Macaca nemestrina by human immunodeficiency virus type-1. Science, 257(5066): 103-106. [DOI] [PubMed] [Google Scholar]
- [2].Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M. 2008. Computing topological parameters of biological networks. Bioinformatics, 24(2): 282-284. [DOI] [PubMed] [Google Scholar]
- [3].Atashili J, Poole C, Ndumbe PM, Adimora AA, Smith JS. 2008. Bacterial vaginosis and HIV acquisition: a meta-analysis of published studies. AIDS, 22(12): 1493-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Baroncelli S, Negri DRM, Michelini Z, Cara A. 2008. Macaca mulatta, fascicularis and nemestrina in AIDS vaccine development. Expert Review of Vaccines, 7(9): 1419-1434. [DOI] [PubMed] [Google Scholar]
- [5].Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 57(1): 289-300. [Google Scholar]
- [6].Bosch ML, Schmidt A, Chen J, Florey MJ, Agy M, Morton WR. 2000. Enhanced replication of HIV-1 in vivo in pigtailed macaques (Macaca nemestrina). Journal of Medical Primatology, 29(3-4): 107-113. [DOI] [PubMed] [Google Scholar]
- [7].Brotman RM. 2011. Vaginal microbiome and sexually transmitted infections: an epidemiologic perspective. The Journal of Clinical Investigation, 121(12): 4610-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brotman RM, Ghanem KG, Klebanoff MA, Taha TE, Scharfstein DO, Zenilman JM. 2008. The effect of vaginal douching cessation on bacterial vaginosis: a pilot study. American Journal of Obstetrics and Gynecology, 198(6): 628.e1- 628e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Brotman RM, He X, Gajer P, Fadrosh D, Sharma E, Mongodin EF, Ravel J, Glover ED, Rath JM. 2014. Association between cigarette smoking and the vaginal microbiota: a pilot study. BMC Infectious Diseases, 14: 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Campbell CJ, Fuentes A, Mackinnon KC, Panger M, Bearder SK. 2007. Primates in Perspective. New York: Oxford University Press. [Google Scholar]
- [11].Caporaso JG, Bittinger K, Bushman FD, Desantis TZ, Andersen GL, Knight R. 2010a. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics, 26(2): 266-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, Mcdonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010b. QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7(5): 335-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chaban B, Links MG, Jayaprakash TP, Wagner EC, Bourque DK, Lohn Z, Albert AY, Van Schalkwyk J, Reid G, Hemmingsen SM, Hill JE, Money DM. 2014. Characterization of the vaginal microbiota of healthy Canadian women through the menstrual cycle. Microbiome, 2: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cohen CR, Lingappa JR, Baeten JM, Ngayo MO, Spiegel CA, Hong T, Donnell D, Celum C, Kapiga S, Delany S, Bukusi EA. 2012. Bacterial vaginosis associated with increased risk of female-to-male HIV-1 transmission: a prospective cohort analysis among African couples. PLoS Medicine, 9(6): e1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cruciani F, Brigidi P, Calanni F, Lauro V, Tacchi R, Donders G, Peters K, Guaschino S, Vitali B. 2012. Efficacy of rifaximin vaginal tablets in treatment of bacterial vaginosis: a molecular characterization of the vaginal microbiota. Antimicrobial Agents and Chemotherapy, 56(8): 4062-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Desantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology, 72(7): 5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dixon P. 2003. VEGAN, a package of R functions for community ecology. Journal of Vegetation Science, 14(6): 927-930. [Google Scholar]
- [18].Duran-Pinedo AE, Paster B, Teles R, Frias-Lopez J. 2011. Correlation network analysis applied to complex biofilm communities. PLoS One, 6(12): e28438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Farage MA, Miller KW, Sobel JD. 2010. Dynamics of the Vaginal Ecosystem-Hormonal Influences. Infectious Diseases: Research and Treatment, 3: 1-15. [Google Scholar]
- [20].Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, Strauss JF, III, Jefferson KK, Buck GA. 2014. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology, 160(Pt 10): 2272-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fredricks DN, Fiedler TL, Marrazzo JM. 2005. Molecular identification of bacteria associated with bacterial vaginosis. The New England Journal of Medicine, 353(18): 1899-1911. [DOI] [PubMed] [Google Scholar]
- [22].Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, Koenig SS, Fu L, Ma ZS, Zhou X, Abdo Z, Forney LJ, Ravel J. 2012. Temporal dynamics of the human vaginal microbiota. Science Translational Medicine, 4(132): 132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gippoliti S. 2001. Notes on the taxonomy of Macaca nemestrina leonina Blyth, 1863 (Primates: Cercopithecidae). Hystrix-Italian Journal of Mammalogy, 12(1): 51-54. [Google Scholar]
- [24].Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika, 40(3-4): 237-264. [Google Scholar]
- [25].Groves CP. 2001. Primate Taxonomy. Washington,DC: Smithsonian Institution Press. [Google Scholar]
- [26].Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, Desantis TZ, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Research, 21(3): 494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hatziioannou T, Evans DT. 2012. Animal models for HIV/AIDS research. Nature Reviews Microbiology, 10(12): 852-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hickey RJ, Abdo Z, Zhou X, Nemeth K, Hansmann M, Osborn TW, III, Wang F, Forney LJ. 2013. Effects of tampons and menses on the composition and diversity of vaginal microbial communities over time. BJOG: An International Journal of Obstetrics & Gynaecology, 120(6): 695-706. [DOI] [PubMed] [Google Scholar]
- [29].Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ. 2012. Understanding vaginal microbiome complexity from an ecological perspective. Translational Research, 160(4): 267-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hu SL. 2005. Non-human primate models for AIDS vaccine research. Current Drug Target-Infectious Disorders, 5(2): 193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Huang B, Fettweis JM, Brooks JP, Jefferson KK, Buck GA. 2014. The changing landscape of the vaginal microbiome. Clinics in Laboratory Medicine, 34(4): 747-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kent SJ, Corey L, Agy MB, Morton WR, Mcelrath MJ, Greenberg PD. 1995. Cytotoxic and proliferative T cell responses in HIV-1-infected Macaca nemestrina. The Journal of Clinical Investigation, 95(1): 248-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kuang YQ, Tang X, Liu FL, Jiang XL, Zhang YP, Gao G, Zheng YT. 2009. Genotyping of TRIM5 locus in northern pig-tailed macaques (Macaca leonina), a primate species susceptible to Human Immunodeficiency Virus type 1 infection. Retrovirology, 6: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lei AH, Zhang GH, Tian RR, Zhu JW, Zheng HY, Pang W, Zheng YT. 2014. Replication potentials of HIV-1/HSIV in PBMCs from northern pig-tailed macaque (Macaca leonina). Zoological Research, 35(3): 186-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li WZ, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics, 22(13): 1658-1659. [DOI] [PubMed] [Google Scholar]
- [36].Ma B, Forney LJ, Ravel J. 2012. Vaginal microbiome: rethinking health and disease. Annual Review of Microbiology, 66: 371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ma ZS. http://adsabs.harvard.edu/abs/2012arXiv1205.3504M. [Google Scholar]
- [38].Macklaim JM, Cohen CR, Donders G, Gloor GB, Hill JE, Parham GP, Ravel J, Spear G, Van De Wijgert J, Reid G. 2012. Exploring a road map to counter misconceptions about the cervicovaginal microbiome and disease. Reproductive Sciences, 19(11): 1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Malaivijitnond S, Arsaithamkul V, Tanaka H, Pomchote P, Jaroenporn S, Suryobroto B, Hamada Y. 2012. Boundary zone between northern and southern pig-tailed macaques and their morphological differences. Primates, 53(4): 377-389. [DOI] [PubMed] [Google Scholar]
- [40].McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME Journal, 6(3): 610-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Menard JP, Fenollar F, Henry M, Bretelle F, Raoult D. 2008. Molecular quantification of Gardnerella vaginalis and Atopobium vaginae loads to predict bacterial vaginosis. Clinical Infectious Diseases, 47(1): 33-43. [DOI] [PubMed] [Google Scholar]
- [42].Mirmonsef P, Gilbert D, Veazey RS, Wang J, Kendrick SR, Spear GT. 2012. A comparison of lower genital tract glycogen and lactic acid levels in women and macaques: implications for HIV and SIV susceptibility. AIDS Research and Human Retroviruses, 28(1): 76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Myer L, Denny L, Telerant R, Souza M, Wright TC, Jr., Kuhn L. 2005. Bacterial vaginosis and susceptibility to HIV infection in South African women: a nested case-control study. The Journal of Infectious Diseases, 192(8): 1372-1380. [DOI] [PubMed] [Google Scholar]
- [44].Nardis C, Mosca L, Mastromarino P. 2013. Vaginal microbiota and viral sexually transmitted diseases. Annali di Igiene: Medicina Preventiva e di Comunità, 25(5): 443-456. [DOI] [PubMed] [Google Scholar]
- [45].Petrova MI, Van Den Broek M, Balzarini J, Vanderleyden J, Lebeer S. 2013. Vaginal microbiota and its role in HIV transmission and infection. FEMS Microbiology Reviews, 37(5): 762-792. [DOI] [PubMed] [Google Scholar]
- [46].Ravel J, Brotman RM, Gajer P, Ma B, Nandy M, Fadrosh DW, Sakamoto J, Koenig SS, Fu L, Zhou X, Hickey RJ, Schwebke JR, Forney LJ. 2013. Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome, 1(1): 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, Mcculle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proceedings of the National Academy of Sciences of the United States of America, 108(Suppl 1): 4680-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ravel J, Gajer P, Fu L, Mauck CK, Koenig SS, Sakamoto J, Motsinger-Reif AA, Doncel GF, Zeichner SL. 2012. Twice-daily application of HIV microbicides alter the vaginal microbiota. mBio, 3(6): pii: e00370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nature Methods, 7(9): 668-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Reid G. 2014. Modulating the vaginal microbiome: the need for a bridge between science and practice. Seminars in Reproductive Medicine, 32(1): 28-34. [DOI] [PubMed] [Google Scholar]
- [51].Rodriguez Jovita M, Collins MD, Sjoden B, Falsen E. 1999. Characterization of a novel Atopobium isolate from the human vagina: description of Atopobium vaginae sp. nov. International Journal of Systematic Bacteriology, 49(Pt 4): 1573-1576. [DOI] [PubMed] [Google Scholar]
- [52].Rosenblum LL, Supriatna J, Melnick DJ. 1997. Phylogeographic analysis of pigtail macaque populations (Macaca nemestrina) inferred from mitochondrial DNA. American Journal of Physical Anthropology, 104(1): 35-45. [DOI] [PubMed] [Google Scholar]
- [53].R Development Core Team. 2012. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- [54].Saxena D, Li YH, Yang LY, Pei ZH, Poles M, Abrams WR, Malamud D. 2012. Human microbiome and HIV/AIDS. Current HIV/AIDS Reports, 9(1): 44-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schellenberg JJ, Plummer FA. 2012. The Microbiological Context of HIV Resistance: Vaginal Microbiota and Mucosal Inflammation at the Viral Point of Entry. International Journal of Inflammation, 2012: Article ID 131243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Schwebke JR, Richey CM, Weiss HL. 1999. Correlation of behaviors with microbiological changes in vaginal flora. The Journal of Infectious Diseases, 180(5): 1632-1636. [DOI] [PubMed] [Google Scholar]
- [57].Shannon CE. 1948. A mathematical theory of communication. Bell System Technical Journal, 27(3): 379-423. [Google Scholar]
- [58].Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11): 2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shipitsyna E, Roos A, Datcu R, Hallén A, Fredlund H, Jensen JS, Engstrand L, Unemo M. 2013. Composition of the vaginal microbiota in women of reproductive age--sensitive and specific molecular diagnosis of bacterial vaginosis is possible?. PLoS One, 8(4): e60670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics, 27(3): 431-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Spear GT, Gilbert D, Sikaroodi M, Doyle L, Green L, Gillevet PM, Landay AL, Veazey RS. 2010. Identification of rhesus macaque genital microbiota by 16S pyrosequencing shows similarities to human bacterial vaginosis: implications for use as an animal model for HIV vaginal infection. AIDS Research and Human Retroviruses, 26(2): 193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Spear GT, Kersh E, Guenthner P, Vishwanathan SA, Gilbert D, Zariffard MR, Mirmonsef P, Landay A, Zheng L, Gillevet P. 2012. Longitudinal assessment of pigtailed macaque lower genital tract microbiota by pyrosequencing reveals dissimilarity to the genital microbiota of healthy humans. AIDS Research and Human Retroviruses, 28(10): 1244-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Taylor LR. 1961. Aggregation, variance and the mean. Nature, 189(4766): 732-735. [Google Scholar]
- [64].Taylor LR. 1984. Assessing and interpreting the spatial distributions of insect populations. Annual Review of Entomology, 29(1): 321-357. [Google Scholar]
- [65].Taylor LR, Taylor RAJ. 1977. Aggregation, migration and population mechanics. Nature, 265(5593): 415-421. [DOI] [PubMed] [Google Scholar]
- [66].Van De Wijgert JH, Verwijs MC, Turner AN, Morrison CS. 2013. Hormonal contraception decreases bacterial vaginosis but oral contraception may increase candidiasis: implications for HIV transmission. AIDS, 27(13): 2141-2153. [DOI] [PubMed] [Google Scholar]
- [67].Veazey RS. 2013. Animal models for microbicide safety and efficacy testing. Current Opinion in HIV and AIDS, 8(4): 295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].White BA, Creedon DJ, Nelson KE, Wilson BA. 2011. The vaginal microbiome in health and disease. Trends in Endocrinology and Metabolism, 22(10): 389-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Witkin SS, Ledger WJ. 2012. Complexities of the uniquely human vagina. Science Translational Medicine, 4(132): 132fs11. [DOI] [PubMed] [Google Scholar]
- [70].Yildirim S, Yeoman CJ, Janga SC, Thomas SM, Ho M, Leigh SR, Consortium PM, White BA, Wilson BA, Stumpf RM. 2014. Primate vaginal microbiomes exhibit species specificity without universal Lactobacillus dominance. The ISME Journal, 8(12): 2431-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zhang ZG, Geng JW, Tang XD, Fan H, Xu JC, Wen XJ, Ma ZS, Shi P. 2014. Spatial heterogeneity and co-occurrence patterns of human mucosal-associated intestinal microbiota. The ISME Journal, 8(4): 881-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhang ZG, Zhai HQ, Geng JW, Yu R, Ren HQ, Fan H, Shi P. 2013. Large-scale survey of gut microbiota associated with MHE Via 16S rRNA-based pyrosequencing. The American Journal of Gastroenterology, 108(10): 1601-1611. [DOI] [PubMed] [Google Scholar]