Familial platelet disorder/acute myeloid leukemia (FPD/AML) is an autosomal dominant inherited disorder characterized by thrombocytopenia and high propensity to various hematological malignancies. FPD/AML is caused by monoallelic mutations of RUNX1, which are in many cases point mutations disrupting DNA-binding or transactivating capacities of RUNX1, and these mutations are considered to act in dominant-negative manner to various degrees for residual wild-type allele.1, 2 Interestingly, some FPD/AML traits present monoallelic loss of RUNX1 gene by microdeletion of chromosome 21.1 In such cases, no mutations were found in the residual RUNX1 allele, thus suggesting that haploinsufficiency of RUNX1 is sufficient to cause FPD/AML.
Genetic basis of leukemic transformation in FPD/AML patients still remains elusive. We have recently reported recurrent mutation of CDC25C in FPD/AML patients, and suggested that this mutation is one of the early events, which defines pre-leukemic state.3 However, all pedigrees that we examined carried point mutations of RUNX1, and no cases with monoallelic RUNX1 loss were present. We suspected that the dosage of RUNX1 activity may affect the transforming processes, and FPD/AML with haploinsufficient RUNX1 allele might require unique genetic events for transformation that are distinct from cases with RUNX1 point mutation. In order to identify collaborating mutations with haploinsufficient RUNX1 allele, we performed genome-wide mutational analyses of two transformed cases of FPD/AML with monoallelic RUNX1 loss. This study was conducted with approval from the internal review board of Keio University School of Medicine and conformed to the principles outlined in the Declaration of Helsinki for the use of human tissue or subjects. Samples from the patients were collected with written informed consent.
Patient 1
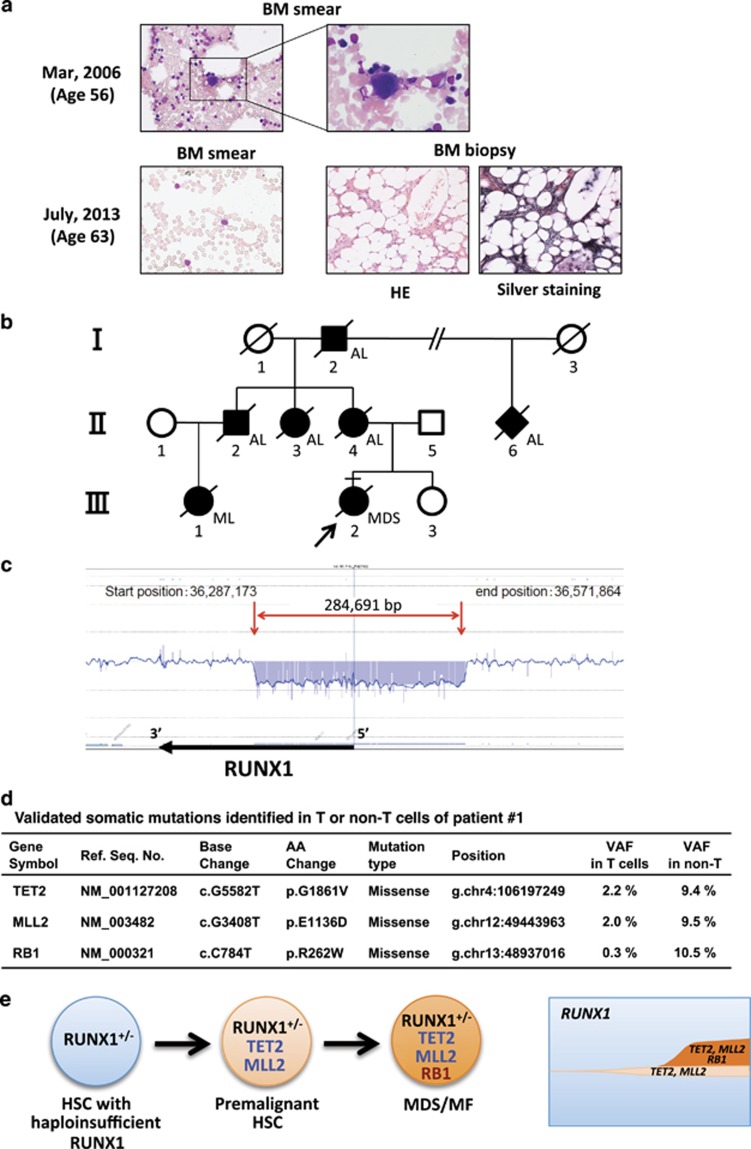
The patient was referred to our hospital for persisting thrombocytopenia from childhood. Bone marrow (BM) examination at the age of 56 revealed normocellular marrow with micromegakaryocytes (Figure 1a), which led to a diagnosis of idiopathic thrombocytopenic purpura. However, she started to develop progressive pancytopenia at the age of 63, when the marrow presented severe hypoplasia with moderate fibrosis and no blast proliferation (Figure 1a). Of note, dysplasia was not evident and the karyotype was normal.
Figure 1.
Clinical, pathological and molecular data of patient 1. (a) Photographs of BM smears and biopsies. BM smears were stained with May-Grunwald Giemsa staining. BM biopsies were stained with Hematoxylin-Eosin (HE) or silver staining. Original magnification; 400 × or 1000 × . (b) Family tree of the pedigree. Filled symbols; affected members, slashed symbols; deceased members, arrow; proband, square; male, circle; female, diamond; sex not determined. AL; acute leukemia, ML; malignant lymphoma. (c) Schematic data of array-comparative genomic hybridization (aCGH) on RUNX1 locus. aCGH was performed using custom-made oligonucleotide microarrays (Agilent Technologies, Santa Clara, CA, USA) covering human chromosome 21, including RUNX1 locus with resolution of several hundred base pairs. A chromosomal region of heterozygous microdeletion is indicated by red arrows. (d) Validated somatic mutations identified in T or non T cells of patient 1. Whole-exome sequencing was performed as described previously.13 Whole-exome capture was accomplished with the cDNA library prepared by SureSelect Human All Exon V5 (Agilent Technologies). Captured targets were subjected to massively parallel sequencing by Illumina HiSeq 2000 (San Diego, CA, USA). Candidate somatic nucleotide variants were validated by deep sequencing. (e) A proposed model of disease progression and clonal architecture of patient 1. HSCs with haploinsufficient RUNX1 first acquire mutations of TET2 and MLL2, which establishes pre-leukemic state. Premalignant HSCs then acquire RB1 mutation to progress into full-blown myelodysplastic syndrome with myelofibrosis (MF).
Family history revealed high penetrance of hematological malignancies such as AML and myelodysplastic syndrome, which suggested an inherited mutation of RUNX1 (Figure 1b). However, no mutations were discovered in the coding region of RUNX1, which suggested the chromosomal microdeletion encompassing RUNX1 locus, as was reported in some cases of FPD/AML.4 Indeed, array-comparative genomic hybridization analysis of the patient's somatic DNA revealed ~285 kb heterozygous deletion including the promoter and the 5′-half of RUNX1 gene (Figure 1c). These data clearly demonstrated that this pedigree was an FPD/AML with haploinsufficient RUNX1 allele.
We tried to dissect the genetic basis underlying BM failure by whole-exome sequencing of genomic DNA extracted from CD3+ T cells and CD3− non T cells of patient's peripheral blood (Figure 1d). This revealed mutations of TET2 and MLL2 genes in peripheral T cells at low variant allele frequency (2.0–2.2%). Interestingly, non T cells harbored mutation of RB1 gene in addition to TET2 and MLL2 at variant allele frequency of ~10%. These data suggest that RUNX1-haploinsufficient hematopoietic stem cells acquired mutations in TET2 and MLL2 genes to establish premalignant hematopoietic stem cells, which then underwent malignant transformation by acquiring RB1 mutation (Figure 1e).
Patient 2
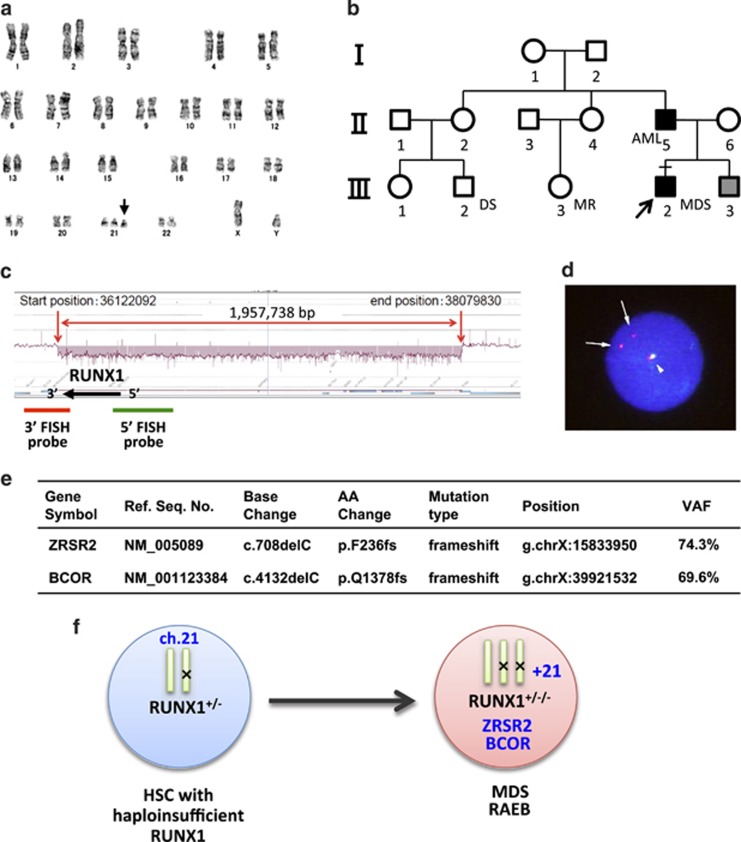
The patient presented thrombocytopenia (46 × 109/l) and was diagnosed with lower-risk myelodysplastic syndrome by BM examination at the age of 29. Two years later, he presented progressive leukocytopenia (2.0 × 109/l) with recurrent bacterial infection. BM aspirates revealed 5.6% of blasts with abnormal karyotype, including trisomy 21 (Figure 2a), which led to a diagnosis of myelodysplastic syndrome: RAEB-1. BM blasts further increased to 13.0% in the following 2 years, and stem cell transplantation was subsequently performed using human leukocyte antigen-matched cord blood as a donor.
Figure 2.
Clinical, cytogenetic and molecular data of patient 2. (a) Karyotype analysis of BM cells. Arrow indicates additional chromosome 21. (b) Family tree of the pedigree. Filled symbols; affected members, gray symbols; members with thrombocytopenia, arrow; proband, square; male, circle; female. DS; Down syndrome-like phenotype, MR; mental retardation. (c) Schematic data of aCGH on RUNX1 locus. aCGH was performed as described in Figure 1. A chromosomal region of heterozygous microdeletion is indicated by red arrows. FISH probes for 5′- or 3′-RUNX1 locus are indicated by green or red lines, respectively. (d) FISH analysis of BM cells for RUNX1 locus. 99.7% (997/1000) of evaluable cells presented trisomy 21 with abnormal signals for RUNX1 locus. Arrows indicate abnormal chromosome 21 hybridized only with 3′-probe (red signals). Arrowhead indicates normal chromosome 21 hybridized to both 5′- and 3′-probes. (e) Validated somatic mutations identified in BMMNCs of patient 2. Whole-exome and deep sequencing was performed as described in Figure 1. (f) A proposed model of disease progression of patient 2. In transformed cells, abnormal chromosome 21 with haploinsufficient RUNX1 allele duplicated, which collaborated with mutations of ZRSR2 and BCOR genes to initiate myelodysplastic syndrome.
Family history showed that his father developed AML and his younger brother presented thrombocytopenia (Figure 2b). Interestingly, two cousins of the proband presented mental retardation and Down syndrome-like phenotype, respectively. High incidence of hematological malignancies and metal retardation suggested a rare type of FPD/AML caused by microdeletion of chromosome 21, as reported previously.5 Indeed, array-comparative genomic hybridization analysis using the patient's somatic DNA confirmed ~2 Mb heterozygous deletion in chromosome 21 encompassing the entire RUNX1 gene and a large genomic region of 5′-RUNX1 (Figure 2c), which indicates that this pedigree is an FPD/AML with haploinsufficient RUNX1 allele.
We next investigated the genetic events critical for myeloid transformation in this patient. As described above, trisomy 21 was noted in the BM cells, which was considered to have contributed to leukemic transformation. Interestingly, fluorescent in situ hybridization analysis of the BM cells revealed duplication of ‘abnormal' chromosome 21 lacking hybridization to the 5′- probe for RUNX1 locus (Figures 2c and d). These data strongly suggested that maintaining haploinsufficient RUNX1 allele with trisomy 21 was critical for transformation. Furthermore, mutations of ZRSR2 and BCOR genes were detected by whole-exome sequence of genomic DNA from BM mononuclear cells (Figure 2e). These data suggest that disruption of ZRSR2 and BCOR genes combined with duplication of abnormal chromosome 21 with RUNX1 deletion is critical for myeloid transformation (Figure 2f).
We have recently reported recurrent CDC25C mutation in ~50% of FPD/AML patients. Hierarchical architecture analysis showed that CDC25C mutation was an early event during transformation, which defines a pre-leukemic clone.3 Mutant CDC25C was shown to confer cells with a proliferative advantage by facilitating their mitotic entry. In the present cases, however, CDC25C mutation was absent, and instead, TET2 mutation or trisomy 21 was identified in each patient. Intriguingly, both of these genetic alterations confer cells with clonal advantage similarly as CDC25C mutation. TET2 mutation has been shown to augment stem cell capacity of hematopoietic stem cells, which then facilitate the expansion of mutated clones.6, 7, 8 Trisomy 21 is a well-known chromosomal abnormality associated with AML,9, 10 and it was shown to confer hematopoietic progenitors with enhanced self-renewal capacity without inducing leukemia.4, 11 Interestingly, trisomy 21 has been reported in some transformed or non-transformed cases of FPD/AML.4, 12 In these cases including ours, duplication occurred invariably with abnormal chromosome 21 carrying RUNX1 mutation, maintaining a single copy of wild-type RUNX1 gene unaffected. This implies that amplification of dose sensitive genes on chromosome 21 combined with RUNX1 haploinsufficiency is a critical step for leukemic transformation in FPD/AML.
Taken together, our study suggests that RUNX1 haploinsufficiency collaborates with genetic alterations conferring clonal advantage such as TET2 mutation or trisomy 21 to establish pre-leukemic state, similarly as RUNX1 point mutation does with CDC25C mutation. This study adds valuable molecular insight into the transforming processes in FPD/AML patients.
Acknowledgments
We thank J Kawakita for excellent technical assistance. This work was supported in part by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author contributions
MS, HK, KY, AY, HK, NW, YS, KC, HT, MK, SM and SO performed research. YH, HH, TK, ST and SO contributed vital new reagents. HN designed and performed research, analyzed the data and wrote the paper.
The authors declare no conflict of interest.
References
- Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet 1999; 23: 166–175. [DOI] [PubMed] [Google Scholar]
- Michaud J, Wu F, Osato M, Cottles GM, Yanagida M, Asou N et al. In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood 2002; 99: 1364–1372. [DOI] [PubMed] [Google Scholar]
- Yoshimi A, Toya T, Kawazu M, Ueno T, Tsukamoto A, Iizuka H et al. Recurrent CDC25C mutations drive malignant transformation in FPD/AML. Nat Commun 2014; 5: 4770. [DOI] [PubMed] [Google Scholar]
- Shinawi M, Erez A, Shardy DL, Lee B, Naeem R, Weissenberger G et al. Syndromic thrombocytopenia and predisposition to acute myelogenous leukemia caused by constitutional microdeletions on chromosome 21q. Blood 2008; 112: 1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzaki E, Morin G, Pollazzon M, Papa FT, Buoni S, Hayek J et al. Syndromic mental retardation with thrombocytopenia due to 21q22.11q22.12 deletion: Report of three patients. Am J Med Genet A 2010; 152A: 1711–1717. [DOI] [PubMed] [Google Scholar]
- Kunimoto H, Fukuchi Y, Sakurai M, Sadahira K, Ikeda Y, Okamoto S et al. Tet2 disruption leads to enhanced self-renewal and altered differentiation of fetal liver hematopoietic stem cells. Sci Rep 2012; 2: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 2011; 20: 25–38. [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011; 20: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010; 116: 354–365. [DOI] [PubMed] [Google Scholar]
- Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood 2000; 96: 4075–4083. [PubMed] [Google Scholar]
- Sikand GS, Taysi K, Strandjord SE, Griffith R, Vietti TJ. Trisomy 21 in bone marrow cells of a patient with a prolonged preleukemic phase. Med Pediatr Oncol 1980; 8: 237–242. [DOI] [PubMed] [Google Scholar]
- Preudhomme C, Renneville A, Bourdon V, Philippe N, Roche-Lestienne C, Boissel N et al. High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. Blood 2009; 113: 5583–5587. [DOI] [PubMed] [Google Scholar]
- Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet 2014; 46: 171–175. [DOI] [PubMed] [Google Scholar]