Abstract
Copper can be found in the environment at concentrations ranging from a shortage up to the threshold of toxicity for plants, with optimal growth conditions situated in between. The plant stem plays a central role in transferring and distributing minerals, water and other solutes throughout the plant. In this study, alfalfa is exposed to different levels of copper availability, from deficiency to slight excess, and the impact on the metabolism of the stem is assessed by a non-targeted proteomics study and by the expression analysis of key genes controlling plant stem development. Under copper deficiency, the plant stem accumulates specific copper chaperones, the expression of genes involved in stem development is decreased and the concentrations of zinc and molybdenum are increased in comparison with the optimum copper level. At the optimal copper level, the expression of cell wall-related genes increases and proteins playing a role in cell wall deposition and in methionine metabolism accumulate, whereas copper excess imposes a reduction in the concentration of iron in the stem and a reduced abundance of ferritins. Secondary ion mass spectrometry (SIMS) analysis suggests a role for the apoplasm as a copper storage site in the case of copper toxicity.
Keywords: ATX1, Cellulose synthase, Copper deficiency, Copper excess, Ionomics, Stem
Introduction
Copper (Cu) homeostasis in plant cells is a tightly controlled process that ensures a sufficient delivery of Cu to Cu-binding proteins but also avoids the harmful effect that Cu excess may have through the generation of reactive oxygen species (ROS) (Drazkiewicz et al. 2004). Under physiological conditions, the transition metal Cu is found in two common oxidation states Cu(I) and Cu(II). In higher plants, the main function of Cu is the transport of electrons in mitochondria and chloroplasts. Cu acts as a cofactor for numerous enzymes involved in the control of the cellular redox state and the modeling of the cell wall (Cohu and Pilon 2010). As for all other microelements, plant Cu requirements are low, and symptoms of Cu toxicity, such as root growth inhibition, can appear at concentrations as low as 1 µM (Chen et al. 2002).
Under Cu deficiency, a sufficient delivery of Cu to developing organs and maturing seeds is maintained through the activation of genes involved in Cu acquisition and redistribution, whose expression is mainly controlled by the SPL7 (SQUAMOSA promoter-binding protein-like7) transcription factor, which is a central regulator of Cu homeostasis (Yamasaki et al. 2009, Garcia-Molina et al. 2013). In eukaryotes, Cu homeostasis is mainly maintained by two Cu transporter families, P1B-type ATPases and high affinity Cu transport proteins from the Ctr/COPT family whose members have diverse functions, from Cu acquisition to Cu delivery and redistribution between green tissues, senescing tissues and reproductive organs (Abdel-Ghany and Pilon 2008, Garcia-Molina et al. 2013). Intracellular Cu transport through the Golgi apparatus, vesicles or other compartments is generally mediated by a system involving one Cu chaperone (ATX1) and one P1B-ATPase. In a recent model proposed by Flores and Unger (2013), Cu transporters deliver Cu to ATX1 proteins, possibly through the interaction with the C-terminal domain of the transporter, allowing the further delivery to the Cu pumps (Shin et al. 2012, Flores and Unger 2013). In Arabidopsis, Cu deficiency causes chlorotic symptoms that appear first at the tip of the young leaves and lead to the formation of necrotic lesions (Abdel-Ghany and Pilon 2008). Plants grown under Cu deficiency also show impairment in the photosynthetic electron transport chain and a reduction in non-photochemical quenching, which is consistent with an impaired plastocyanin function (Abdel-Ghany and Pilon 2008).
In contrast, Cu excess triggers phenotypical changes such as a reduction of root and shoot growth, a decrease in cell viability in root tips and the premature induction of root lignification. Supra-optimal Cu concentrations furthermore cause changes in the mineral profile of plants, for instance a decreased soil to shoot translocation of potassium (K) and changes in the root to shoot calcium (Ca) ratio (Lequeux et al. 2010). Young bean plants exposed to Cu excess were reported to accumulate this element in the cell wall of leaves and to induce the formation of thicker tracheids. This latter development of xylem vessels correlates with an increased activity of key enzymes involved in the biosynthesis of lignin, namely guaiacol peroxidase, coniferyl alcohol peroxidase, NADH oxidase and phenylalanine-ammonia lyase (PAL) (Bouazizi et al. 2011). In radish, increasing but environmentally relevant concentrations of Cu (1–8 µM) induced an increased activity of anionic and cationic peroxidases (Pxds) and increased lignification (Chen et al. 2002). An excess of Cu2+ ions can induce the generation of ROS, causing cellular damage. Fry et al. (2002) suggested that Cu excess can generate the non-enzymatic production of ·OH radicals in the cell wall, thereby causing cell wall loosening (Fry et al. 2002). It was thus postulated that ·OH-mediated loosening and Pxd-mediated tightening reactions would compete for available H2O2, and that the net effect would depend on the activity of Pxds (Fry 1998). The tight spatio-temporal regulation of Cu homeostasis may also participate in the integral cellular circadian system, by influencing the expression of key oscillator components through the activity of the SPL7 transcription factor (Andrés-Colás et al. 2010).
In alfalfa, the stem represents more than half of the total biomass of mature plants. During shoot development, the stem undergoes specific maturation processes leading to accumulation of cell wall polysaccharides and lignin (Dien et al. 2006). Cu availability may influence the cell wall properties of the stem and consequently the global quality of alfalfa. Nevertheless, the precise impact of Cu availability on cell wall differentiation still remains poorly understood. In this study, alfalfa was grown in hydroponics in six different conditions of Cu availability which mimic the global range of Cu nutrition tolerable for alfalfa growth, from deficiency (3 nM Cu) to slight toxicity (10 µM Cu). The metabolic responses of the stem were compared to understand how Cu nutrition impacts the development of this organ. Proteomics was used as the core approach and was complemented by the expression analysis of key genes involved in the deposition of the cell wall. The metabolic modifications are discussed according to the evolution of the ionome of the different plant organs and are illustrated by secondary ion mass spectrometry (SIMS) imaging.
An important limitation in the common proteomics studies is the almost systematic use of parametrical univariate statistical tests to determine the differentially abundant proteins although most restrictive assumptions of applicability are not tested (Dupae et al. 2014). To prevent such limitations, Dupae et al. recommend the combined use of non-parametrical tests and multivariate analysis such as PCA (principal component analysis) to extract relevant information. Different tools allowing a precise description of the principal components have been developed, such as the R package FactoMineR (Lê et al. 2008), and were used in the present study to differentiate between experimental groups in terms of correlated abundance rather than absolute abundance and to create clusters of similar behavior (Dupae et al. 2014). By applying the same approach to ionomics and gene expression analysis, this study highlights the evolution of key metabolic processes occurring in alfalfa stem which are modulated by Cu nutrition, with a particular emphasis on cell wall-related pathways.
Results
Copper concentrations of 300 nM to 3 µM Cu are optimal for alfalfa growth
When grown at the two lowest Cu concentrations (3 and 30 nM), alfalfa plants showed symptoms of mineral deficiency such as the presence of leaf chlorosis, necrosis of the leaf fringes, early leaf fall and a lower leaf density (Supplementary Fig. S1). From 300 nM to 3 µM Cu the plants were more homogeneous in their development, developed a higher leaf surface area and the leaves tended to remain greener. At the highest Cu concentration (10 µM Cu), the most striking changes in plant phenotype were interveinal chlorosis (Fig. 1).
Fig. 1.
Leaf phenotype of plants cultivated at 10 µM Cu, 7 months after germination. Note the presence of interveinal chlorosis.
Root copper availability influences the accumulation of a panel of micro- and macroelements throughout the plant
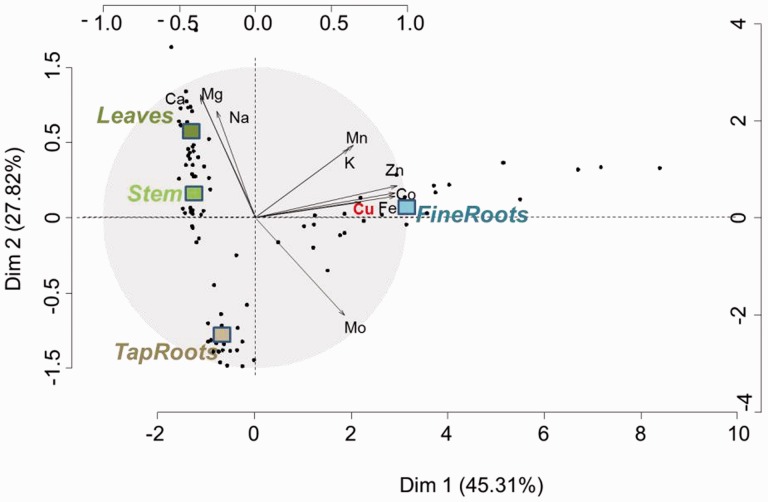
The distribution of 10 elements [Ca, cobalt (Co), Cu, iron (Fe), K, magnesium (Mg), manganese (Mn), molybdenum (Mo), sodium (Na) and zinc (Zn)] was assessed on four plant parts, the fine roots, the taproot, the stem and the leaves, and the results were analyzed using PCA (Fig. 2; Table 1; Supplementary Table S1A–D). The fine roots which were in direct contact with the nutrient solution were characterized by the highest variations in mineral concentration in response to the Cu treatment (Fig. 2; Supplementary Table S1A). In particular, a 325-fold increase in Cu concentration was reported in the fine roots of the plants cultivated at 10 µM Cu (Cu6) in comparison with those grown at 3 nM Cu (Cu 1), whereas this ratio reached 82.31, 50.31 and 21.70 for the taproot, the stem and the leaves, respectively (Table 1). These variations in mineral concentration measured in the fine roots according to the treatment were confirmed by the wide dispersion of these samples on the first axis of the PCA. In fact, this first axis (Dim 1, Fig. 2) which accounted for 45.31% of the total variability observed among all samples, discriminated the fine roots from the other three organs (leaves, stems and taproot). This axis was mainly explained by Zn, Fe and Co which positively correlated by >90% with this axis, indicating higher concentrations for these metals in the fine roots, and this independent of the Cu treatment. The second dimension of the PCA (Dim 2) was mainly explained by Mg (R2 = 0.81, P < 0.05), Ca (R2 = 0.80, P < 0.05) and Na (R2 = 0.71, P < 0.05), which accumulated to a higher extent in the leaves than in the taproot. Consistent with the importance of Mo for symbiotic nitrogen fixation, this element accumulated in the below-ground parts of the plants (Fig. 2). The clustering of the different parts of alfalfa following PCA revealed, however, an organ-specific global pattern for mineral distribution in this species. The elements Fe, Co, Cu, Zn and Mo were at a higher concentration in the root environment (Table 2), whereas the concentrations of Na, Mg and Ca were higher in the shoot organs (Table 2). Intermediate behavior was observed for Mn and K. However, the distribution of the elements among the plants can vary with the treatment. In particular, the highest variations in mineral distribution in response to the treatment are observed for Cu, Fe, Mn, Zn, Co and Mo in the fine roots (SD Cu 15.8%; SD Fe 12.4%; SD Mn 8.2%; SD Zn 7.2%, SD Co 6.4%; SD Mo 5.4%, Table 2) for Cu, Mn and Fe in the leaves (SD Cu 8.1%; SD Mn 7.0%; SD Fe 5.4%, Table 2) and for Fe, Cu and Zn in the stem (SD Fe 5.5%; SD Cu 5.2%; SD Zn 5.1%, Table 2). In contrast, the distribution of the elements in the taproot appeared more homogeneous.
Fig. 2.
PCA of alfalfa mineral content. The mineral content of the 10 elements Mn, Fe, Co, Cu, Zn, Mo, Na, Mg, K and Ca is analyzed in four replicates and in four organs (fine roots, taproot, stem and leaves). Six conditions of copper availability are tested (3 nM, 30 nM, 300 nM, 1 µM, 3 µM and 10 µM). Black dots represent each sample (96 in total). Squares represent the barycenter of the 24 samples per organ.
Table 1.
Copper concentrations in the different plant organ (µg g–1 DW) expressed per organ and per condition [(Cu1) 3 nM, (Cu2) 30 nM, (Cu3) 300 nM, (Cu4) 1 µM, (Cu5) 3 µM, (Cu6) 10 µM]
| Fine roots |
Taproot |
Stem |
Leaves |
|||||
|---|---|---|---|---|---|---|---|---|
| Average | SD | Average | SD | Average | SD | Average | SD | |
| Cu1 | 4.7 | 0.4 | 2.4 | 0.3 | 1.9 | 0.2 | 2.5 | 0.2 |
| Cu2 | 8.0 | 0.6 | 3.5 | 0.6 | 3.8 | 0.7 | 5.1 | 0.8 |
| Cu3 | 27.7 | 4.5 | 11.2 | 2.8 | 11.4 | 3.4 | 13.8 | 1.5 |
| Cu4 | 59.1 | 13.2 | 21.0 | 2.1 | 23.1 | 3.8 | 20.9 | 2.4 |
| Cu5 | 95.2 | 22.9 | 30.5 | 5.6 | 30.7 | 7.2 | 21.4 | 3.1 |
| Cu6 | 1536.3 | 348.3 | 196.5 | 17.1 | 96.5 | 24.0 | 53.2 | 6.5 |
Table 2.
Global distribution of Ca, Co, Cu, Fe, K, Mg, Mn, Mo, Na and Zn (in %) among the different organs of alfalfa
| Ca | Co | Cu | Fe | K | Mg | Mn | Mo | Na | Zn | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fine roots | Average | 14.5 | 75.9 | 51.1 | 55.3 | 37.9 | 20.8 | 51.3 | 39.0 | 21.8 | 54.8 |
| SD | 1.7 | 6.4 | 15.8 | 12.4 | 4.0 | 1.3 | 8.2 | 5.4 | 1.7 | 7.9 | |
| Tap root | Average | 6.2 | 7.5 | 16.6 | 13.4 | 14.9 | 18.5 | 6.9 | 40.0 | 12.5 | 11.2 |
| SD | 0.9 | 2.2 | 3.4 | 1.9 | 1.3 | 1.4 | 1.7 | 2.7 | 2.0 | 1.2 | |
| Stem | Average | 22.3 | 7.1 | 15.7 | 14.7 | 23.4 | 28.9 | 7.2 | 13.2 | 36.5 | 20.2 |
| SD | 1.3 | 2.8 | 5.2 | 5.5 | 1.9 | 1.7 | 1.2 | 2.4 | 3.8 | 5.1 | |
| Leaves | Average | 57.0 | 9.6 | 16.6 | 16.5 | 23.8 | 31.8 | 34.6 | 7.8 | 29.2 | 13.8 |
| SD | 1.7 | 1.9 | 8.1 | 5.4 | 1.8 | 1.7 | 7.0 | 1.4 | 2.7 | 3.5 |
Note that for each mineral, the sum of all averages is equal to 100%.
SDs indicate the variability observed among the six conditions of copper nutrition.
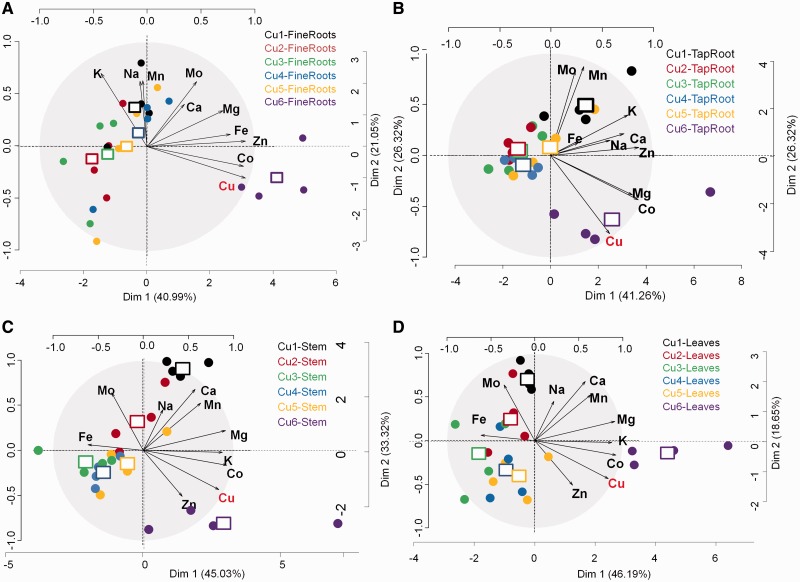
PCAs carried out by organ revealed that samples collected from plants cultivated in conditions 2–5 (30 nM Cu to 3 µM Cu) cluster, thus indicating a relatively similar distribution of the 10 elements in the organs of the plants cultivated in these four treatments (Fig. 3). In contrast, the mineral composition of all organs of plants cultivated in condition 6 (10 µM Cu) was shifted from the others, which highlights that the mineral homeostasis was disturbed at this Cu concentration and suggests that the threshold value for Cu toxicity was reached. Similarly, and most specifically in the above-ground organs, the samples originating from the plants cultivated at the lowest concentration (3 nM) cluster away from the samples grown in the four intermediate conditions. This indicates a second switch in mineral homeostasis when Cu concentrations were <30 nM.
Fig. 3.
PCA of alfalfa mineral content carried out by plant organ (A, fine roots; B, taproot; C, stem; D, leaves). The mineral content of the 10 elements Mn, Fe, Co, Cu, Zn, Mo, Na, Mg, K and Ca is analyzed in four replicates. Six conditions of copper availability are tested [(Cu1) 3 nM, (Cu2) 30 nM, (Cu3) 300 nM, (Cu4) 1 µM, (Cu5) 3 µM and (Cu6) 10 µM]. Black dots represent each sample (24 in total). Squares represent the barycenter of the four replicates per copper condition.
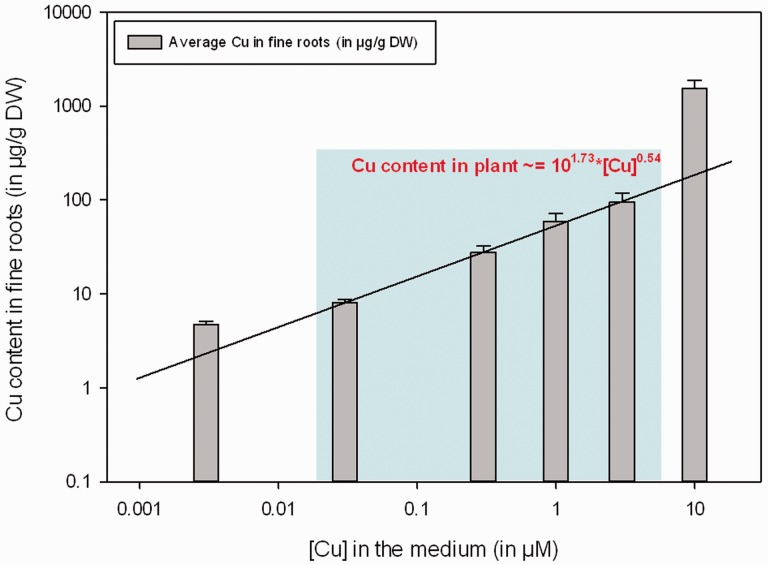
From condition 2 (30 nM Cu) to condition 5 (3 µM Cu), each increase in Cu concentration in the medium is followed by an increase in Cu concentration in all organs, but more specifically in the fine roots where it can be roughly modeled by the following function (Fig. 4).
Independently of the considered organ, Cu and Co concentrations strongly correlated (Pearson correlation coefficient >0.80; P < 0.05, Supplementary Table S1C). In the fine roots, Cu also correlated with Zn (Pearson correlation coefficient = 0.86; P < 0.05), but this was not the case in other organs. Although Cu concentrations positively correlated with Fe in the fine roots of the hydroponically grown alfalfa (Pearson correlation coefficient = 0.6; P < 0.05), an inverse trend was recorded in the green tissues (Pearson correlation coefficient less than –0.4; P < 0.05) (Supplementary Table S1C). In particular, the mean Fe content of the stem and the leaves was reduced by 54% and 37%, respectively, when the Cu concentration increased from 3 µM (Cu5) to 10 µM (Cu6) (Supplementary Table S1A, B).
Fig. 4.
Copper content measured in the fine roots according to the concentration of copper in the medium. The blue frame refers to the range of copper concentration in which the content of copper can be modeled by the function: Copper in plant (in µg g–1 DW) = 101.73 × [Copper in the medium (in µM)]0.54.
In the stem, the samples grown in the two extreme conditions were grouped in the right panel of the score plot (Dim 1 vs. Dim 2). This clustering of conditions Cu1 and Cu6 was attributed to the similarities in ionome shared by these two conditions, and in particular a higher accumulation of Mg than in the intermediate conditions. However, the separation of these samples along Dim2 revealed that plants grown in Cu excess accumulated more Co and Cu but less Fe than most of the other treatments, whereas Cu deprivation induced an accumulation of Mo (and to a lower extent Na) in the stem (Supplementary Table S1A, B; Fig. 3C).
Copper deficiency causes a decreased expression of cell wall-associated genes in the stem
The expression of different genes involved in cell wall biosynthesis in alfalfa stem, namely cellulose synthase (CesA) genes, two genes involved in the phenylpropanoid pathway [PAL and cinnamyl alcohol dehydrogenase (CAD)], sucrose synthase (SuSy) as well as class III Pxds known to be involved in lignification in Arabidopsis thaliana (Pxd42 and Pxd53) (Nielsen et al. 2001, Yokoyama and Nishitani 2006, Marjamaa et al. 2009, Saathoff et al. 2013) was quantified by quantitative real-time PCR (RT-qPCR) and analyzed using PCA (Fig. 5; Supplementary Table S2A).
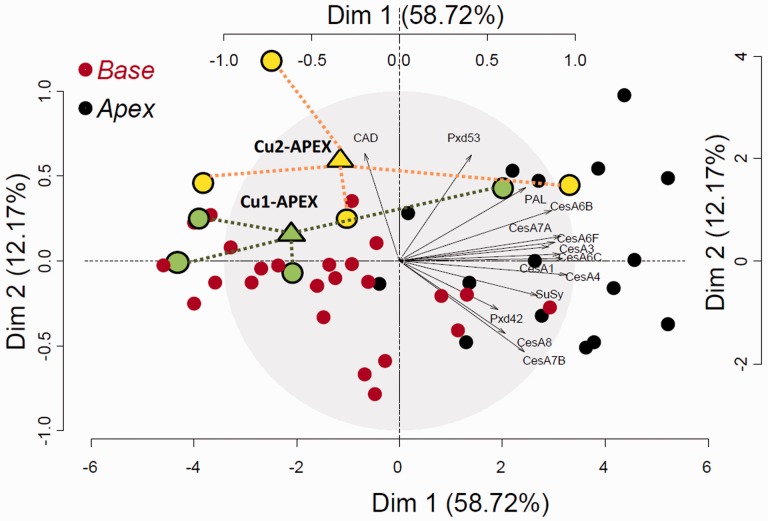
Fig. 5.
PCA of the gene expression data. Red dots refer to basal samples, and black dots refer to apical samples. Green and yellow circles represent the apical stem individuals from plants grown at Cu1 and Cu2, respectively. Triangles refer to the barycenter of the latter. The expression profile of 14 cell wall-related genes analyzed on 48 samples is represented.
Most genes involved in cellulose biogenesis exhibited an increased expression in the apical region of the stem in comparison with the base of the stem, which may reflect a global decrease in metabolic activities resulting from both the cell differentiation and the lignification processes (Printz et al. 2015). The first principal component of the PCA represented the majority of the variance observed on the measurements (58.72%). This axis was mainly explained by the difference in gene expression between the basal and the apical part of the stem. However, 75% of the apical samples collected from the two lowest conditions of Cu availability (3 nM Cu and 30 nM) clustered in the left panel of the PCA (Fig. 5, yellow and green filled circles), where almost all samples from the basal stem regions were located. This co-segregation suggests that low Cu availability influences the expression of these cell wall-related genes more specifically in the apical part of the stem.
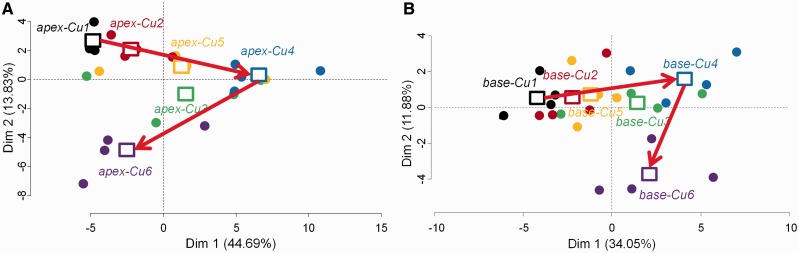
Additional PCAs were performed separately on the basal and on the apical samples (Fig. 6A, B). In both cases, the first dimension of the PCA represented > 55% of the total variability observed among the samples. In the apical third, most of the samples originating from the plants cultivated at the two lowest conditions of Cu availability (3 nM, Cu1; and 30 nM, Cu2) were grouped in the left panel, which confirms that Cu deficiency causes important changes in the expression of cell wall-related genes in the youngest region of the stem (Fig. 6A). In particular, samples that cluster in the left panel of the PCA tend to have a lower expression of the cell wall-associated genes CesA4, CesA1, CesA7A, CesA6C, CesA6F, SuSy, CesA3, CesA6B and CesA7B [‘dimdesc’ function of the FactoMineR R package, P-value < 0.05; correlation with the axis higher than |0.7| (Supplementary Table S2B)]. This lower expression of some cell wall-related genes of the samples cultivated in Cu1 and Cu2 is confirmed at the basal extremity of the stem (Fig. 6B).
Fig. 6.
Principal component analysis (PCA) of the gene expression data carried out on (A) the apical samples and (B) the basal samples. Squares represent the barycenter of the four replicates per copper condition. Represented is the expression profile of 14 cell wall-related genes analyzed on 24 samples each. (Cu1) 3 nM Cu, (Cu2) 30 nM Cu, (Cu3) 300 nM Cu, (Cu4) 1 µM Cu, (Cu5) 3 µM Cu and (Cu6) 10 µM Cu.
The stem proteome is influenced by copper availability
Proteins were extracted from two regions of the stem of alfalfa, the apical and the basal extremities, according to a trichloroacetic acid (TCA)/phenol–SDS protein extraction method (Printz et al. 2015). Among the 236 spots of interest picked in this study (P-value for multiple comparison of means inferior to 0.05), 109 contained one unique protein that was reliably identified (see the Materials and Methods). This allowed discrimination of 51 proteins which were differentially expressed in response to the treatment in the apical part, 35 in the basal part and 91 proteins which followed a differential accumulation according to the ‘treatment’ effect of the global analysis of variance (ANOVA).
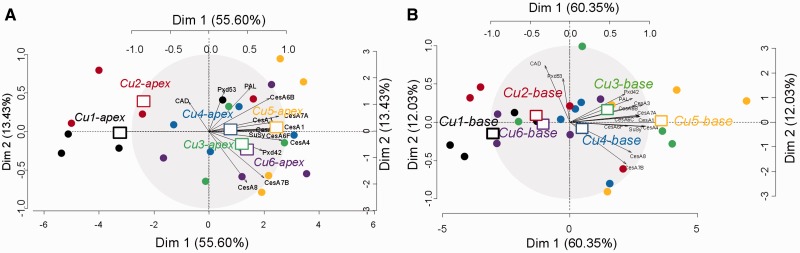
To investigate the effect of Cu on the stem metabolism in the two different stem segments, two PCAs were separately carried out on the two subsets of significant proteins identified in the apical part (51) and in the basal part (35) (Fig. 7A, B). In both cases, the two first dimensions of the PCA discriminated the plants grown in the condition of Cu deficiency Cu1 (3 nM Cu) from the samples grown in Cu4 (1 µM Cu). Along this axis, all samples grown in conditions Cu2 (30 nM Cu), Cu3 (300 nM Cu), Cu5 (3 µM Cu) and Cu6 (10 µM Cu) clustered intermediately, which suggests that the abundance of the proteins that highly contributed to this dimension fluctuated between the two extreme values reached in Cu1 and in Cu4 (Fig. 7A, B). However, the proximity of the apical samples grown in Cu6 (10 µM Cu) to those grown in Cu1 (3 nM Cu) along the first dimension indicated that the proteomes of the apical stem from plants grown in the two extreme conditions shared similarities. The shift of the samples grown in Cu6 (10 µM Cu) on the second dimension of each PCA nonetheless indicated that the behavior of some proteins was specific to the condition of high Cu availability in the apical and in the basal region (Fig. 7A, B). To select the proteins that highly contributed to the two first dimensions of the PCA, we applied the ‘dimdesc’ function of the FactoMineR R package at the threshold of 0.05. A total of 39 and 19 proteins, respectively, were detected as high contributors to Dim 1 (Cu1/Cu6 vs. Cu4; Fig. 7A) and Dim 2 (Cu6-specific) in the apical part of the stem, whereas 22 and eight proteins, respectively, significantly contributed to these two principal components in the most lignified, basal region (in that case Dim 1 represents Cu1 vs. Cu4, and Dim 2 represents the Cu6-specific protein profile, Fig. 7B).
Fig. 7.
PCA of the differentially regulated proteins identified in (A) the apical samples and (B) the basal samples. Squares represent the barycenter of the four replicates per copper condition. Represented is the expression profile of 51 and 35 proteins (apex and base) analyzed on 24 samples each. (Cu1) 3 nM Cu, (Cu2) 30 nM Cu, (Cu3) 300 nM Cu, (Cu4) 1 µM Cu, (Cu5) 3 µM Cu and (Cu6) 10 µM Cu. Red arrows link the barycenter of the samples grown under copper deprivation (Cu1), optimal copper (Cu4) and copper excess (Cu6).
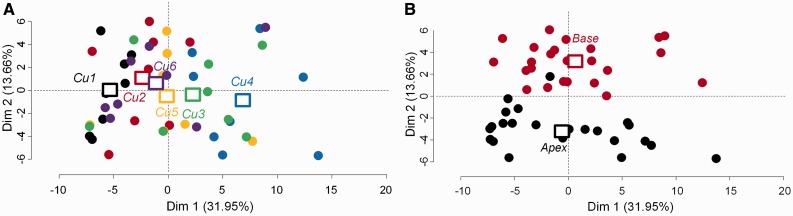
Similarly, we performed a third PCA on the 91 proteins which were initially selected according to the ‘treatment’ effect of the global ANOVAs (P-value < 0.05) (Fig. 8). Again, the first dimension of the PCA (31.95%) allowed separation of the plants cultivated in Cu1 from those cultivated in Cu4. The second dimension of the PCA (13.66%) clearly differentiated the apical from the basal samples (Fig. 8). High contributors to this axis, a set of 56 proteins, correspond to proteins having an opposite behavior in the base in comparison with the apex. The third dimension of the PCA (8.79%) showed the clustering of the Cu6 samples away from all other samples (not shown). Consequently, the three main factors influencing our data set were, in decreasing order, the difference between the plants grown under low and intermediate Cu availability (31.95%), the stem region (13.66%) and the threshold level of Cu tolerance above which the plant switched its metabolism to cope with an excessive availability of Cu (8.79%).
Fig. 8.
PCA showing the distribution of the individuals based on the set of 91 proteins differentially regulated by the treatment. Squares represent the barycenter of the eight replicates per copper condition and (B) the barycenter of the 24 replicates per stem region. Represented is the expression profile of 91 proteins analyzed on 48 samples. In (A) (Cu1) 3 nM Cu, (Cu2) 30 nM Cu, (Cu3) 300 nM Cu, (Cu4) 1 µM Cu, (Cu5) 3 µM Cu and (Cu6) 10 µM Cu; in (B) red dots refer to apical samples and black dots to basal samples.
From this third PCA, 64 and 33 proteins, respectively, were selected for their high contribution to Dim1 (Cu1 vs. Cu4; 31.95%) and Dim3 (Cu6; 8.79%) at the threshold of 0.05 and were integrated in the previous set of 62 proteins, leading to a global set of 92 non-redundant proteins which followed significant variations in abundance in response to variations in Cu availability in the medium.
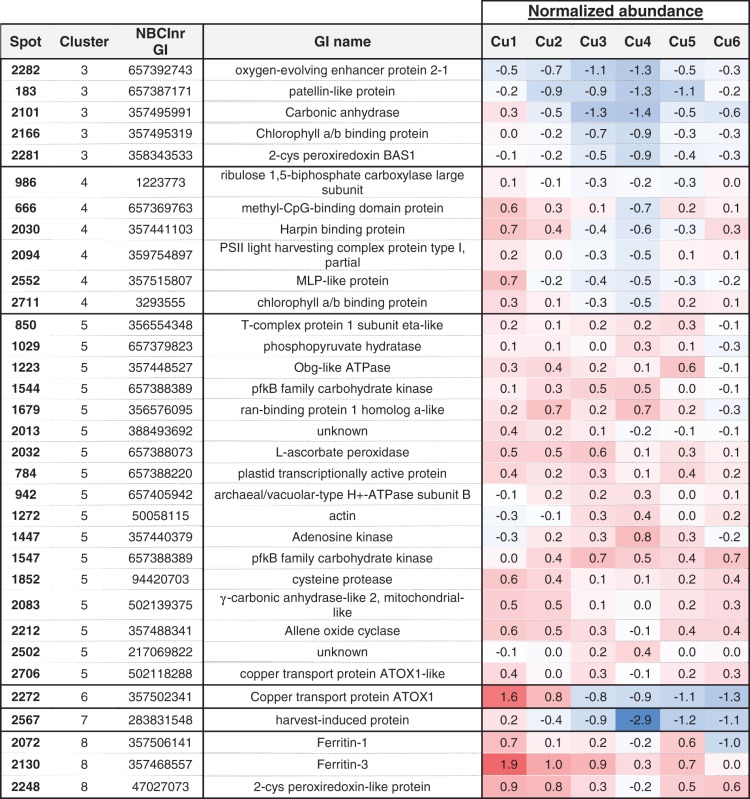
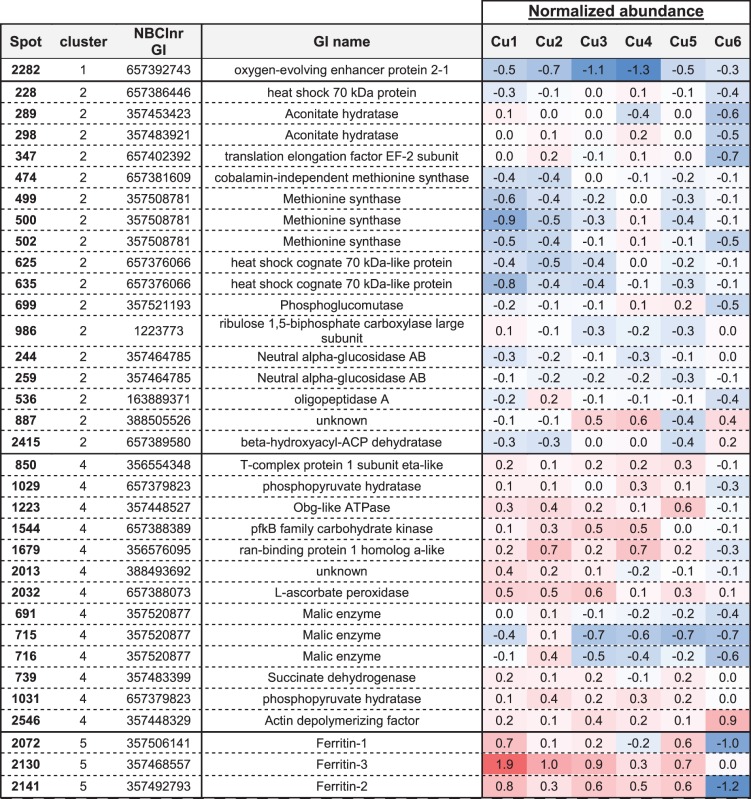
Among these, 78 proteins differentiated the plants cultivated in Cu deficiency (Cu1) from those grown in optimal conditions (Cu4) (Table 3). By applying the Hierarchical Clustering on Principle Components (‘HCPC’) R function, the 78 proteins were assigned to nine clusters. In the first two protein clusters are proteins involved in the cytoskeleton, lignin and methionine metabolism, tubulin alpha NCBInr gi:557159508, caffeoyl-CoA 3-O-methyltransferase (CCOMT) NCBInr gi:363541987 and various methionine synthase isoforms NCBInr gi:657381609 and 357508781, whose abundance is reduced in the apical and in the basal stem regions under limited Cu availability in comparison with the optimal Cu level. Also, a tendency of a lower expression of genes coding for these methionine synthase isoforms (gi:657381609 and 357508781) and S-adenosylmethionine synthase (NCBInr gi:223635319) was reported under Cu deficiency (Supplementary Fig. S2). In the apical regions, a lower abundance of SuSy (NCBInr gi:657398021 and gi:3915046), rhamnose biosynthetic-like enzyme (NCBInr gi:657399031) and S-adenosylmethionine synthase (NCBInr gi:223635319) was additionally reported at low Cu availability in comparison with the optimal level of Cu nutrition. Interestingly, proteins from these clusters (1 and 2) are also those which display a similar lower abundance in the case of Cu excess in comparison with the optimal Cu level in the apical stem region. These two protein clusters thus allowed a separation of the stem samples depending on whether the plants were cultivated under non-optimal or optimal conditions of growth, but did not allow differentiation of the plants grown under Cu deficiency from those grown under Cu excess. Interestingly, the stem proteome profile which is specific to Cu deficiency included a high abundance of the Cu transport protein ATX1 (NCBInr gi:357502341), which was further confirmed by RT-qPCR, and a high abundance of ferritins 1 and/or 3 (NCBInr gi:357506141 and gi:357468557) in the whole plant stem in comparison with all other conditions. In the apical region, Cu deficiency also led to a higher accumulation of hypothetical protein MTR_1g082420 (NCBInr gi:357441569), which is a putative chloroplast-located protein with phosphatase activity, and a higher accumulation of harvest-induced protein (NCBInr gi:283831548) which has a bet-v1 ligand-binding domain. When excessive Cu is applied (Cu6), the proteome profile that emerges combines the low abundance of methionine synthase isoforms (most specifically in the apical region as already depicted in the case of Cu deprivation) in comparison with the optimal Cu range, and also a lower abundance of translation elongation factor EF-2 subunit (NCBInr gi:657402392), phosphoglucomutase (NCBInr gi:357521193), heat shock protein (HSP; NCBInr gi:357495169), ran-binding protein 1 homolog a-like (NCBInr gi:356576095), a lower presence of the Cu transport protein ATX1 (NCBInr gi:357502341) and a sudden reduction in ferritins 1, 2 and/or 3 (NCBInr gi: 357506141, gi:357492793 and gi:357468557) (Supplementary Table S3E, F). A reduced abundance of ferritin transcripts was observed for ferritin 2 in the apical part and for ferritin 3 in the basal region (Supplementary Fig. S2). Specific for the basal part, Cu excess triggers in addition a lower accumulation of a protein disulfide-isomerase (NCBInr gi:266743) and a higher presence of actin depolymerizing factor (NCBInr gi:357448329) (Table 4). Although the P1B-type ATPase HMA5 is involved in the transport of Cu, no significant change in expression was observed.
Table 3.
Clustering of the proteins that highly contribute to differentiate the plants grown under copper deficiency (Cu1) from those grown at optimal conditions (Cu4)
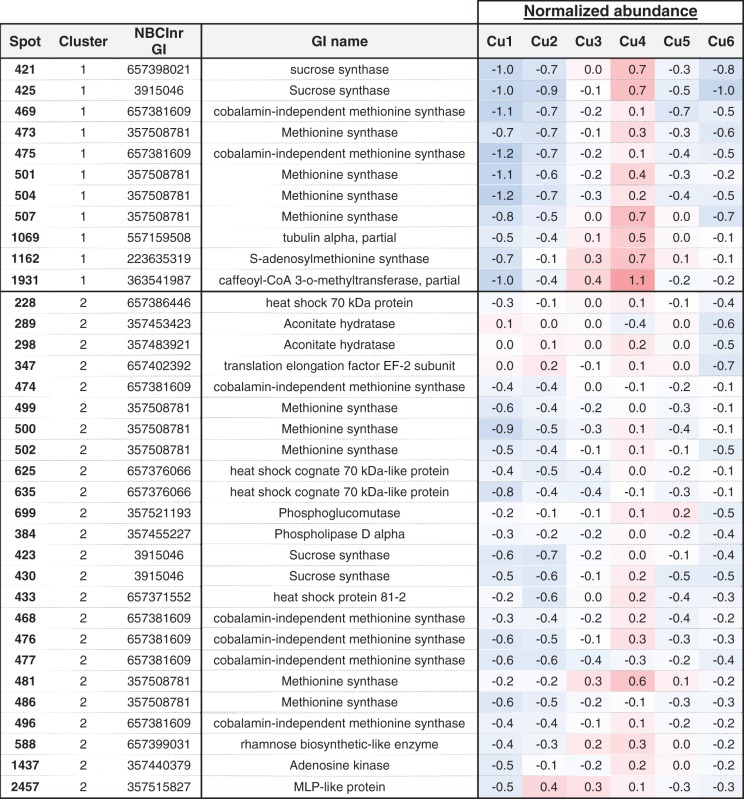 |
 |
Represented are the proteins whose abundance varied significantly with the treatment (P-value ANOVA < 0.05, global data set) and that highly contribute to differentiate Cu1 from Cu4 after applying the ‘dimdesc’ function of the R package FactoMineR at the threshold of 0.05.
Normalized abundances are represented by a color gradient from blue (negative values) to red (positive values). Additional data are available in Supplementary Table S3.
Table 4.
Clustering of the proteins that highly contribute to differentiate the plants grown at light copper toxicity (Cu6)
 |
Represented are the proteins whose abundance varied significantly with the treatment (P-value ANOVA < 0.05, global data set) and that highly contribute to differentiate Cu1 from Cu4 after applying the ‘dimdesc’ function of the R package FactoMineR at the threshold of 0.05.
Normalized abundances are represented by a color gradient from blue (negative values) to red (positive values). Additional data are available in Supplementary Table S3.
The extracellular matrix of the stem accumulates copper under high copper availability in the medium
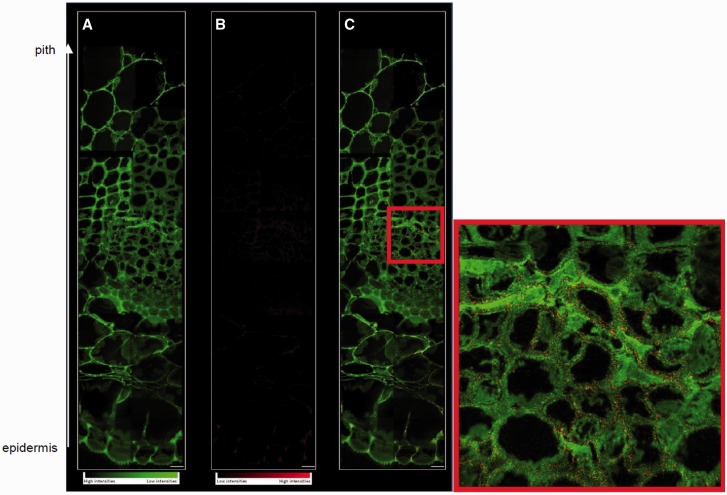
Microelemental imaging of non-hyperaccumulating plant species is challenging especially when plants are grown in conditions of low mineral availability (Moore et al. 2014, Zhao et al. 2014). SIMS imaging was performed on the sections obtained from three basal stem regions sampled from plants cultivated in low (Cu2; 30 nM Cu), optimal (Cu4; 1 µM Cu) and high (Cu6; 10 µM Cu) Cu availability. However, the signal recorded for 63Cu did not reach the limit of detection for the plants cultivated in Cu2 and in Cu4. In Cu6, the 63Cu signal is clearly visible in the extracellular matrix at the periphery of the cells (Fig. 9).
Fig. 9.
NanoSIMS analysis of the peripheral region to the central region of a basal stem section prepared by freezing followed by freeze substitution. (A) 12C14N– image in green scale color from low to high intensity, (B) copper distribution 63Cu– in red scale color, (C) color overlay for CN and Cu. Scale bar = 5 µm. In the red square is a focus of the cambial region of the stem.
Discussion
The aim of the current study is to understand how balancing the availability of Cu in the root environment can affect the metabolic dynamism of the stem. More specifically it aims at understanding which processes involved in cell wall development are modulated by Cu nutrition. A global proteome approach was performed and was completed by targeted genomics and ionomics studies. Analyses were performed on alfalfa plants grown immediately from sowing in six conditions of Cu availability ranging from 3 nM to 10 µM. This range was selected from a first screening during which plants were germinated for 1 month at different Cu concentrations ranging from 3 nM to 30 µM.
From the physiological observations, three distinct responses can be distinguished, whether the plants were cultivated at a low Cu availability (3 and 30 nM Cu) and show symptoms of leaf chlorosis, leaf tip necrosis, early leaf fall and a low leaf density, whether they were cultivated at an optimal range of Cu (300 nM to 3 µM Cu) or whether they were cultivated above the threshold of tolerance for this mineral (threshold set between 3 and 10 µM Cu) which is characterized by the development of interveinal chlorosis (Fig. 1; Supplementary Fig. S1). From 30 nM to 3 µM Cu, the evolution of Cu in the fine roots follows a logarithmic increase as a function of the concentration of Cu in the medium, which indicates that the same increase of Cu in the medium induces a greater variation of Cu in the roots at low Cu availability than at high Cu availability and suggests a higher efficiency of the Cu transport system when Cu availability is low. Outside this range (at 3 nM Cu and at 10 µM Cu, respectively), the variation of Cu in the roots as a function of the concentration of Cu in the medium does not follow this logarithmic trend, which suggests that other mechanisms of Cu uptake/transport are involved (Fig. 4).
Ionomics was performed on four plant organs, namely the fine roots, the taproot, the stem and the leaves. Modulating Cu availability induces broader variations in mineral homeostasis in the fine roots than in the other plant organs. This is confirmed by the important shift of the fine root samples on the first axis of the PCA (Fig. 2) which explains 45.31% of the total variability observed among the samples. The uptake/translocation of Cu is broadly regulated in the fine roots, as suggested by the clustering of these samples on the right panel of the global PCA. This segregation of the fine root samples highlighted the filtering role of the endoderm where highly specific transporters such as members of the P1B-type ATPase protein family are located (Deng et al. 2013). Similar filtering occurs for Fe, Co and Zn. In our study, the good correlation coefficient observed between Co and Cu led us to hypothesize the existence of a link between Cu and Co homeostasis.
The ionome analysis performed on each plant organ (Fig. 3) suggests that the plants developed a tissue-specific pattern of mineral homeostasis which is modulated according to the availability of Cu. In particular, the clear segregation of the stem samples cultivated at the lowest and at the highest conditions of Cu availability on the two first axes of the stem PCA (78.35% of cumulated weight) showed that the stem metal homeostasis is greatly impacted by variations in Cu nutrition. The grouping of the stem samples cultivated in the two most extreme conditions on the first dimension of the PCA (Fig. 3C) is attributed to the similarities in ionome shared by the plants grown at these two concentrations, such as, for example, a tendency to accumulate a higher content of Mg. However, their distinct clustering along the second axis of the PCA indicated that the two most extreme conditions also exhibited differences. In the case of Cu excess, the stems tend to accumulate Cu and Co and to lower the content of Fe, while in the case of Cu deficiency the two elements Mo and Na tend to accumulate more. As suggested by the similarities between the PCAs, the mineral profile appears conserved in green tissues. In particular, accumulation of Mo in the case of Cu deprivation has already been observed in Brassica napus and Dioscorea esculenta (O’Sullivan and Ernest 2007, Billard et al. 2014).
To assess the effect that Cu nutrition may exert on the dynamism of the cell wall, we focused our analysis on the apical and the basal thirds of the stem which were previously shown to have a distinct structure and metabolism (Printz et al. 2015). A panel of 14 genes involved in cell wall synthesis (specifically cellulose and lignin) were selected and their expression level was followed by RT-qPCR. In accordance with Printz et al. (2015), multivariate analysis indicated that the genes involved in cellulose synthesis tend to be more expressed in the apical extremities of the stem. However, at the two lowest concentrations the apical fringes shared higher similarities in the gene expression profile with the basal mature segment than with the actively growing apical stem regions (Fig. 5). Consequently, deficiency in Cu caused a reduction in plant growth and led to a lower expression of key genes involved in cell wall biogenesis, and this most specifically in the most sensitive apical region of the stem. By performing the PCA by stem region, we noted, however, that the treatment also induced variations in the expression of cell wall-related genes in the basal extremity of the stems, as indicated by the tendency of all the genes except CAD and Pxd53 to be more highly expressed in the plants grown at 3 µM Cu (Cu5) in comparison with those grown at 3 nM Cu (Cu1) (Fig. 6B).
Non-targeted proteomics was performed on the apical and the basal stem regions in order to understand which cellular metabolism altered by Cu nutrition may explain the described phenotype. Our first focus is on the proteins reaching an extreme (highest or lowest) abundance at the optimal range of Cu availability (300 nM to 3 µM Cu) (Supplementary Table S3A–F). Proteins reaching the highest abundance at this optimal Cu availability were identified as cytosolic proteins such as SuSy, rhamnose biosynthetic-like enzyme, methionine synthase, S-adenosylmethionine synthase, CCOMT, HSPs, adenosine kinase and partial tubulin-α (Supplementary Table S3E, F). Two putative cell wall-related proteins, one stem 28 kDa glycoprotein and one heat shock cognate 70 kDa-like protein as well as one putative chloroplast heat shock protein 81-2 displayed similar variations.
All these proteins respond similarly to both Cu deficiency and excess. A possible mediation may occur through microRNA signaling (Abdel-Ghany and Pilon 2008). Although similar changes were observed in the basal region, some variations were restricted to the apical third (SuSys, stem 28 kDa glycoprotein, rhamnose biosynthetic-like enzyme and S-adenosylmethionine synthase), confirming the higher sensitivity of the youngest regions. At xylem sap pH, Cu transport is mainly mediated by Cu complexes such as Cu-nicotianamine (Cu-NA) which are believed to be the preferred orchestrator (together with histidine) of stem Cu transport (Liao et al. 2000, Irtelli et al. 2009). Since NA results from the condensation of three molecules of S-adenosylmethionine by NA synthase, the production of NA is closely linked to the activities of both methionine synthase and S-adenosylmethionine synthase. The low abundance of these latter under low Cu availability may be linked to lower needs for Cu chelators since the amount of Cu transferred to the stem is significantly reduced compared with the optimal condition (Table 1; Fig. 3). Under high Cu availability, reducing the amount of NA may, in contrast, be seen as a strategy to limit the translocation of the element by restricting it to the roots (Fig. 2) (Burkhead et al. 2009).
In plants, the development of the stem is closely linked to the differentiation of the cell wall which undergoes specific stages of maturation. During the first phase of growth, the youngest stem region can elongate due to the flexibility of the primary wall which surrounds the cells. This primary cell wall further undergoes a broad secondary development conferring rigidity to the stalk and making the upright growth of the plant possible. With increasing stem maturity, the proportions of cellulose and lignin in the stem increase progressively, whereas the proportion of pectin decreases (Printz et al. 2015). Lignin in dicotyledonous plants is mostly composed of guaiacyl (G) and syringyl (S) units, and their synthesis is regulated by the activity of various enzymes among which CCOMT may play an important role, notably in the synthesis of the G subunits (Guo et al. 2001). Interestingly, CCOMT has S-adenosylmethionine as substrate, and non-optimal Cu availability induces a lower accumulation of methionine synthase and S-adenosylmethionine synthase in both the apical and the basal extremities of the stem, suggesting that Cu limitation or excess impact the production of the G subunits by limiting the accumulation of at least three enzymes acting upstream of their synthesis. The relationship between methionine synthase, S-adenosylmethionine synthase, CCOMT and lignin metabolism was interestingly already observed during compression wood formation in pine (Villalobos et al. 2012).
Specifically in the apical region, a rhamnose biosynthetic-like enzyme and isoforms of SuSy followed similar variations (spot 598). Since the monomer rhamnose forms part of pectins and hemicelluloses, changes in the abundance of enzymes involved in the synthesis of this hexose such as the rhamnose biosynthetic-like enzyme may influence the structure of the polysaccharidic network of the cell wall. In addition, the synthesis of cellulose requires the activity of multiple CesA proteins grouped in rosettes and involved in catalyzing the polymerization of glucan chains (Li et al. 2014). It has been shown that the membrane-associated rosettes consist of both a structural unit and a catalytic unit which includes SuSy as a major component (Fujii et al. 2010). In 2011, Brill et al. further identified a cell wall-localized SuSy putatively involved in providing UDP-glucose for cellulose and callose synthesis from extracellular sugars (Brill et al. 2011). Interestingly, most of the genes related to cell wall biogenesis, including SuSy, tend to be more highly expressed at concentrations of Cu approaching the ‘optimal’ range (300 nM to 3 µM Cu). Altogether, this suggests that Cu nutrition can influence the dynamism of the cell wall, in particular in the apical part of the stem, by modulating the expression and the production of various enzymes involved in this process at the transcriptional and translational levels.
Cu availability also influences the stem photosynthetic metabolism. A panel of chloroplastic proteins (oxygen-evolving enhancer protein 2-1, carbonic anhydrase, Chl a/b-binding protein, RuBisCo subunits and PSII light-harvesting complex proteins) were of lower abundance at 1 µM Cu than in the two extreme culture conditions (Tables 3, 4; Supplementary Table S3E, clusters 3 and 4). Also characteristic of the Cu4 condition was a significantly lower abundance in the apical region of one harvest-induced protein (NCBInr gi:283831548) that contains a ligand-binding bet_v_1 domain susceptible to bind hydrophobic ligands, and the low accumulation of a hypothetical protein MTR_1g082420 (NCBInr gi:357441569), with a putative phosphatase activity and predicted to be located in the chloroplast. Although their role in Cu metabolism/tolerance in not elucidated, their important fluctuations as a response to variations in copper nutrition make them good candidates for functional characterization (Tables 3, 4; Supplementary Table S3E, F).
In plants, Cu transport, translocation and redistribution are mediated by high-affinity transporters and chaperones regulated by a central Cu homeostatic machinery which involves the Cu-responsive transcription factor SPL7 (Tapken et al. 2012). In the case of copper deprivation, such high-affinity Cu transporters include members of the COPT protein family which receive Cu from specific Cu chaperones (ATX1) which are functional homologs of the yeast Anti-oxidant 1 ATOX1 protein (Himelblau et al. 1998, Abdel-Ghany and Pilon 2008). Here, the abundance of one Cu transport protein ATX1 (NCBInr gi:357502341) reached a maximum under the Cu-deficient conditions, decreased with increasing Cu availability and attained a minimum in the case of Cu excess. This protein shares 75% identity with its homolog in A. thaliana ATX1 which is involved in Cu delivery to specific protein targets such as intracellular cuproproteins and transporters (Puig and Peñarrubia 2009). Arabidopsis mutants lacking ATX1 proteins are hypersensitive to Cu excess and more sensitive to Cu deficiency. It was thus postulated that ATX1 is required for tolerance to Cu deficiency in Arabidopsis, by increasing Cu use efficiency. In addition, ATX1 overexpression enhances Cu extraction from soil (Shin et al. 2012). In our study on alfalfa stem, the overabundance of ATX1 in the case of Cu deprivation and its low level in the case of Cu excess confirmed the crucial role of this protein in increasing the Cu use efficiency in the stem, probably helping to drive a higher flux of Cu to the developing leaves in the case of deficiency and to lower it in the case of toxicity. However, no significant variation in the expression of the Cu transporter HMA5 was reported.
Similarly, the abundance of ferritins is reduced with increasing Cu supply (NCBInr gi:57506141, NCBInr gi:357468557 and NCBInr gi:357492793; Supplementary Table S3E, F) in both the apical and the basal segments. In particular, an important drop is observed when reaching Cu toxicity. This variation corroborates with the about 2-fold decrease in Fe content in the stems of plants from this condition in comparison with the other treatments. The role of ferritins in Cu homeostasis is unclear. They are ubiquitous proteins, mainly localized in plastids and mitochondria, able to control the level of Fe in cells and involved in the control of the redox status of the plants (Briat et al. 2010, Strozycki et al. 2010). In particular, A. thaliana mutants lacking ferritins display higher accumulation of ROS probably due to free Fe-induced Fenton reactions and show an impaired plant growth and fertility (Ravet et al. 2009). Tarantino et al. (2003) observed a high promoter activity of AtFer1 (for A. thaliana ferritin-1) in the vicinity of the vessels in the leaves of A. thaliana. Recently, immunolocalization of ferritins in A. thaliana leaves showed the presence of ferritins in xylem-associated cells and suggested a role for ferritins (at least some isoforms) in buffering the excess of Fe during xylem unloading (Roschzttardtz et al. 2013). Our results indicate that Cu excess in the medium triggered a reduction of Fe content in stems, together with a reduction of the amount of ferritins. It is interesting to note that the symptoms of interveinal chlorosis observed at 10 µM Cu were similar to the classical symptoms of Fe deficiency in plants. It also confirms the existence of cross-talk between Cu and Fe uptake/translocation and in particular the influence that Cu availability has on the Fe homeostasis already observed in Arabidopsis (Bernal et al. 2012). It is not clear, however, if the decrease of Fe in the stem resulted only from this reduced amount of ferritins or from disruptions of its transport across the root endodermis. Since Fe and Cu play an essential role in chloroplast metabolism (Ravet and Pilon 2013), it is ultimately not surprising that the modulation of Cu (from deficiency to excess) influenced the accumulation of many chloroplast-localized proteins.
In this study, we aimed to gain insights into the effect of various conditions of Cu availability on alfalfa stem metabolism. By modulating the availability of Cu, three distinct responses were identified depending on whether the plants were grown under Cu deficiency, under a sufficient amount of Cu or under Cu excess. When the availability of Cu dropped below the needs of the plant, the expression of key genes involved in cell wall development decreased, most specifically in the apical extremity of the stem, and this coincides with a lower accumulation of proteins playing a role in this process. In contrast, the abundance of specific Cu chaperones rose to increase the Cu use efficiency. Interestingly, many proteins involved in chloroplast metabolism tend to be of higher abundance in the case of Cu deprivation, although the phenotype of the plants shows signs of chloroplast deregulation. Increasing the Cu availability induced the expression of cell wall-related genes and stimulated the accumulation of proteins playing a role in cell wall deposition and in methionine metabolism. However, when the availability of Cu exceeded the threshold of tolerance for alfalfa, the abundance of ferritins and the concentration of Cu chaperones in the stem suddenly decreased, in contrast to the content of Cu that rapidly increased. The subcellular localization of Cu, as obtained by SIMS imaging (Fig. 9), suggests that apoplastic export mechanisms are activated when toxic levels of Cu are reached, thereby generating a new cellular homeostasis. Metal accumulation in the apoplast has been widely studied in metal-tolerant species such as in Silene paradoxa (Colzi et al. 2011). Intriguingly at high Cu availability, the most Cu-tolerant S. paradoxa ecotypes accumulate less Cu than the sensitive ecotypes, this being apparently linked to a decrease in total pectin content and to an increased degree of pectin methylation in tolerant vs. sensitive plants (Colzi et al. 2011). Globally, the high accumulation of Cu in the alfalfa stem apoplast at high Cu availability may reveal the Cu sensitivity of alfalfa and may be envisaged as a last cellular alarm prior to broad metabolic damage.
Materials and Methods
Plant growth and sampling
Surface-sterilized alfalfa seeds (Medicago sativa L., cv Giulia) were sown on agar plates [10 g l–1 agar, nitrogen-free Broughton and Dilworth (BD) 1 × culture medium] where Cu was added at various total concentrations of CuSO4 [(Cu1) 3 nM, (Cu2) 30 nM, (Cu3) 300 nM, (Cu4) 1 µM, (Cu5) 3 µM, (Cu6) 10 µM]. Each seed was inoculated with 4 µl of inoculation mix [4 g l–1 peat-based inoculant (HiStick®, Becker Underwood) in yeast extract–mannitol (YEM) solution (Broughton and Dilworth 1971)]. After 4 weeks of growth in an incubator (Forma Scientific Diurnal Growth Chamber, Model 3740) [23°C/20°C (day/night), 13 h/11 h (day/night), light intensity of 150 µmol m–2.s–1 (Sylvania Grolux Fluorescent lamps)], inoculated plants were replanted into open-bottom polypropylene pots (L × W × H: 2.8 × 2.8 × 6.2 cm) filled with LD-PE granulates (2–3 mm diameter) prior to being transferred to a hydroponic system (Broughton and Dilworth 1971, Printz et al. 2013a).
The hydroponic system consisted of a permanently aerated BD 1 × nutrient solution, depleted in Cu, complemented with CuSO4 to attain the six Cu concentrations previously described (Cu1–Cu6).
For each treatment, 120 plants were cultivated in 500 ml low-density polyethylene dark tanks (six plants per tank). The volume of the culture medium was adjusted weekly to 400 ml with ultrapure water, and solutions were renewed monthly. Plants were cultivated in growth chambers (Forma Scientific Diurnal Growth Chamber, Model 3740) at a 13 h/11 h (day/night) cycle at a light intensity of 150 µmol m–2 s–1 (Sylvania Grolux Fluorescent lamps) and temperature was set at 23°C/20°C (day/night). No difference in seedling mortality was observed between the six conditions. Homogeneous plants per condition were sampled after 7 months of growth in hydroponics.
For gene expression and proteomics analyses, plant stem portions, namely the basal third and the apical third, were cut with a razor blade and frozen in liquid nitrogen. Either one (proteomics) or two (genomics) plants were used to constitute one biological replicate. Statistical analyses were performed on four replicates for RT-qPCR and proteomics. For mineral content analysis, 5–10 plants were divided into fine roots, taproot, stems and leaves and were used to constitute one replicate per organ. Fine roots and taproot were carefully rinsed with ultrapure water prior to drying in an oven. Four replicates per organ were used for statistical analysis.
Mineral content analysis by ICP-MS
About 250 mg of plant organs—fine roots, taproots, stems and leaves—were oven-dried for 5 d at 70°C, ground and mineralized in 7 ml of nitric acid (67–69%) and 3 ml of hydrogen peroxide (30%) using a Multiwave PRO microwave reaction system (Anton Paar GmbH) according to the manufacturer’s instructions. Sample volumes were adjusted to 25 ml with MilliQ Water (Millipore) and the metal concentration of each organ was determined by inductively coupled plasma-mass spectrometry (ICP-MS), using an Elan DRC-e (PerkinElmer). In total, the concentration of 10 elements (Mn, Fe, Co, Cu, Zn, Mo, Na, Mg, K, Ca) was analyzed (Supplementary Table S1A–D).
Genomics
RNA extraction and cDNA synthesis. Stem regions were ground to a fine powder in liquid nitrogen, using a mortar and pestle, and RNA was extracted as described (Guerriero et al. 2014). A 100 mg aliquot of finely ground sample was weighed and total RNA was extracted using the RNeasy Plant Mini Kit with on-column DNase I treatment (Qiagen). The integrity of the extracted RNA was checked with an Agilent Bioanalyzer [all the RNA integrity numbers (RINs) were > 7.5] and the purity/concentration was measured using a NanoDrop ND-1000 spectrophotometer (A260/280 and A260/230 ratios between 1.9 and 2.2). Subsequently, 1 µg of extracted RNA was reverse transcribed using the Superscript II cDNA Synthesis kit (Invitrogen), according to the manufacturer’s guidelines.
Quantitative real-time PCR and statistical analysis. For RT-qPCR analysis, 10 ng of cDNA were used as template. Reactions were set up on 384-well plates using a liquid handling robot (epMotion, Eppendorf) and the Takyon Low ROX SYBR MasterMix dTTP Blue Kit (Eurogentec) on a ViiA 7 Real-Time PCR System (Applied Biosystems) in a final volume of 10 µl.
The reactions were performed in technical triplicates and repeated on four independent biological replicates. The PCR conditions consisted of an initial denaturation at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 s, and annealing/extension at 60°C for 60 s. A melting curve analysis was performed at the end of the experiment to check the specificity of the amplified products. All of the qPCR amplicons were verified with sequencing on an Applied Biosystems 3500 Genetic Analyser using the BigDye Terminator v3.1 Cycle Sequencing and the BigDye XTerminator Purification kits, according to the manufacturer’s instructions.
The results were analyzed using the qBasePLUS version 2.5 software (Biogazelle; Hellemans et al. 2007) and normalized using translation initiation factor IIA (TFIIA) and elongation initiation factor 4A (eif4A) (Guerriero et al. 2014). The primers used to amplify the reference genes, the CesA genes, PAL, CAD and SuSy, have already been described (Guerriero et al. 2014). Two class III Pxd orthologs shown to be involved in lignification in A. thaliana (i.e. Pxd42 and Pxd53; E-values 0 and 6e-153, identity 84% and 59%, respectively) (Nielsen et al. 2001, Yokoyama and Nishitani 2006, Marjamaa et al. 2009, Saathoff et al. 2013) were targeted. The primers to amplify the Pxds were the following: Pxd42Fwd TGCTGTTCAGAGTTGTGATGC, Pxd42Rev CCAAAGCTTCTGTCGTGTTC (designed on the M. truncatula Refseq accession No. XM_003610383.1) and Pxd53Fwd TGGTCCTGATTGGAAAGTGC, Pxd53Rev TTGAAAGCGGGACTAGGAAG (designed on the M. sativa GenBank accession No. AAB41811.1).
The expression levels are indicated as ‘Normalized relative expression’ (Supplementary Table S2A, B) and have been analyzed by PCA.
To validate our proteomics data, seven genes whose encoded proteins changed significantly in abundance with the treatment, as well as the Cu transporter HMA5 were selected and their expression followed by RT-qPCR. These genes were the following: Medtr7g013660.1, copper chaperone ATX-1; Medtr7g069980.1, ferritin-1; Medtr5g083170.1, ferritin-2; Medtr4g014540.1, ferritin-3; Medtr5g010420.1, heavy metal P-type ATPase; Medtr2g046710.1, S-adenosylmethionine synthase-like protein; Medtr6g027920.2, cobalamin-independent methionine synthase; and Medtr7g086300.1, cobalamin-independent methionine synthase. The primers used to amplify these genes are presented in Supplementary Table S4. Differences in ‘normalized relative expression’ were analyzed by ANOVA followed by Tukey’s HSD test (Supplementary Fig. S2).
Proteomics
Proteomics extraction, labeling, migration and spot detection were performed as described previously (Printz et al. 2013b).
Spot selection for multivariate analysis. Following spot detection using the DeCyder software (v7.0, GE-Healthcare), normalized spot volumes were extracted and standardized according to the spot volumes measured on the internal standard. All volumes were log(2) transformed. Only spots for which a value was obtained on at least three out of every four gels were conserved for statistical analysis. In cases where one missing value was reported, the fourth value was mathematically imputed by implementing the mean log(2)-transformed volume of the 10 spots having the nearest profile of expression on all other gels. Data were finally median normalized (Supplementary Table S3A).
Further processing was performed using R (v3.1.2). Homoscedasticity was tested and, depending on this test, an ANOVA or a test of equal means in a one-way layout in the case of unequal variances were applied. This procedure was performed on (i) the global data set and separately on (ii) the apical samples and (iii) the basal samples (Supplementary Table S3B).
In cases where a treatment effect was reported for a spot (P < 0.05) in at least one of the three statistical processings (treatment effect on the global data set, treatment effect on the apical samples, treatment effect on the basal samples), the spot was checked using the Decyder (v7.0, GE-Healthcare) spot view and selected for picking.
Spot picking, digestion and MS/MS analysis. Selected spots were picked with an Ettan Spotted Picker (GE Healthcare). Digestion was carried out using a Freedom EVO II workstation (Tecan). Briefly, gel plugs were washed for 20 min in a 50 mM ammonium bicarbonate solution in 50% (v/v) MeOH/MQ water and dehydrated for 20 min with 75% acetonitrile (ACN). After dehydration, proteins were digested with trypsin Gold (Promega), 8 µl of a solution containing 5 ng µl–1 trypsin in 20 mM ammonium bicarbonate (overnight, 37°C). After digestion, peptides were extracted from the gel plugs with 50% (v/v) ACN containing 0.1% (v/v) trifluoroacetic acid (TFA) and dried.
Peptides were then solubilized in 0.7 µl of 50% (v/v) ACN containing 0.1% (v/v) TFA and spotted manually on MALDI-TOF (matrix-assisted laser desorption ionization-time of flight) targets. A volume of 0.7 µl of 7 mg ml–1 α-cyano-4-hydroxycinnamic acid in 50% (v/v) ACN containing 0.1% (v/v) TFA was added. A MALDI peptide mass spectrum was acquired using the AB Sciex 5800 TOF/TOF (AB Sciex), and the 10 most abundant peaks, excluding known contaminants, were automatically selected and fragmented. MS analyses were carried out as described previously (Printz et al. 2013b). MS and MS/MS spectra were submitted for NCBInr database-dependent identification using the taxonomy Viridiplantae (2,471,722 sequences) (http://www.ncbi.nlm.nih.gov) downloaded on October 30, 2014 and containing 51,656,746 sequences on an in-house MASCOT server (Matrix Science, www.matrixscience.com). A second search was carried out against an EST (expressed sequence tag) Fabaceae database downloaded on December 17, 2013 and containing 19,932,450 sequences. The parameters used for these searches were mass tolerance MS 100 p.p.m., mass tolerance MS/MS 0.5 Da, fixed modifications cysteine carbamidomethylation, and variable modifications methionine oxidation, and double oxidation of tryptophan and of tryptophan to kynurenine. Proteins were considered as identified when at least two peptides passed the MASCOT-calculated 0.05 threshold scores (respectively a score of 53 for all NCBI Viridiplantae queries and 58 for the EST Fabaceae queries) (Supplementary Table S3C, D). Determination of the subcellular location of proteins was performed using TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP) with the standard search parameters for plants.
Statistical analyses
Results from ionomics, RT-qPCR and proteomics were analyzed by multivariate analyses using the freeware R (v3.1.2) with the additional packages FactoMineR v1.0 and the functions ‘PCA’, ‘HCPC’ and ‘dimdesc’. PCA axes were characterized, and high contributors to the PCA axes were identified with the ‘dimdesc’ function (P < 0.05) as presented in the Results. For proteomics, the multivariate analysis was performed on the spots for which the calculated P-value for multiple comparison of means was inferior to 0.05 and in which one unique significant protein was identified. In particular, this procedure allowed extraction of 78 and 40 proteins that differentiated low, optimal and high Cu availability, and that were further clustered according to their standardized abundance in nine and six groups, respectively, using the ‘HCPC’ R function.
Analyses were performed on four replicates per condition and per stem region for RT-qPCR and proteomics, and on four replicates per organ for mineral content analysis.
SIMS imaging
Sample preparation. The basal third of three fresh stems cultivated in Cu2 (30 nM Cu), Cu4 (1 µM Cu) and Cu6 (10 µM Cu) were cut transversally into sections of about 1 mm thick in phosphate-buffered saline (PBS) buffer. Cryofixation was performed by impact freezing using a Leica EM MM80 (2 min). Following the vitrification of the sample, freeze substitution was performed in a Leica AFS system (1°C h–1 over 8 d until –20°C at 1 × 10–4 Pa) and the samples were further embedded in a Unicryl resin (Electron Microscopy Sciences) at –20°C. After 24 h, the temperature was increased to 20°C and the final polymerization was carried out under UV for 48 h. Embedded samples were sectioned transversally by ultramicrotomy (Leica Ultracut) to 300–400 nm sections and placed on a silicon wafer (Siltronix) for SIMS analysis.
Sample analysis with NanoSIMS 50. The SIMS image acquisitions were performed on a NanoSIMS 50 instrument (CAMECA). For the detection of secondary negative ions (M–), the primary bombardment selected was cesium. The primary ion (Cs+) was accelerated at + 8 kV and the current was measured at 1.5 pA. These conditions allow estimation of the probe size in the range of 100–150 nm (lateral resolution). The samples were polarized at –8 kV and the surface scan was recorded as a matrix of 256 × 256 pixels and a counting time of 30 ms per pixel (∼33 min per image). The emitted secondary negative ions from the nano volume were filtered in mass with an instrument tuned for a mass resolution power M/ΔM of 4,500. The masses recorded and counted simultaneously were the 12C14N– cluster (m = 26,00307 uma), the 32S– ion (m = 31,97207 uma), the 31P16O– cluster (m = 46,96868 uma) and both isotopes 63Cu– ion (62,92959 uma) and 65Cu– ion (64,92779 uma), respectively 69.17% and 30.83% in natural abundance. Thus, the isotopic ratio of Cu was controlled to ensure that no mass interference was induced (Hallégot et al. 2006, Moore et al. 2011, Mueller et al. 2013). The sections of the stem were analyzed from the periphery to the central part in eight pieces of 40 × 40 µm2, allowing an image of 80 × 320 µm2 (512 × 2,056 pixels) to be obtained.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the National Research Fund [FNR Project CANCAN (C13/SR/5774202)].
Supplementary Material
Acknowledgements
We are grateful to Dr. Lucien Hoffmann for the careful reading of the article. We acknowledge in particular Céline Leclercq, Sébastien Planchon and Laurent Solinhac for their day-to-day technical support, Simone Zorzan for valuable discussions concerning statistics, Johanna Ziebel for performing all ICP-MS analyses, and Patrick Grysan, Esther Lentzen and Brahime El Adib for their teachings on SIMS analysis.
Glossary
Abbreviations
- ACN
acetonitrile
- ANOVA
analysis of variance
- BD
Broughton and Dilworth
- CAD
cinnamyl alcohol dehydrogenase
- CesA
cellulose synthase
- CCOMT
caffeoyl CoA 3-O-methyltransferase
- MALDI
matrix-assisted laser desorption ionization
- NA
nicotianamine
- PAL
phenylalanine ammonia-lyase
- PCA
principal component analysis
- Pxd
peroxidase
- ROS
reactive oxygen species
- RT-qPCR
quantitative real-time PCR
- SIMS
secondary ion mass spectrometry
- SuSy
sucrose synthase
- SPL7
SQUAMOSA promoter-binding protein-like7
- TFA
trifluoroacetic acid
- TOF
time of flight
Disclosures
The authors have no conflicts of interest to declare.
References
- Abdel-Ghany S.E., Pilon M. (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283: 15932–15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-Colás N., Perea-García A., Puig S., Peñarrubia L. (2010) Deregulated copper transport affects Arabidopsis development especially in the absence of environmental cycles. Plant Physiol. 153: 170–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal M., Casero D., Singh V., Wilson G.T., Grande A., Yang H., et al. (2012) Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24: 738–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billard V., Ourry A., Maillard A., Garnica M., Coquet L., Jouenne T., et al. (2014) Copper-deficiency in Brassica napus induces copper remobilization, molybdenum accumulation and modification of the expression of chloroplastic proteins. PLoS One 9: e109889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouazizi H., Jouili H., Geitmann A., El Ferjani E. (2011) Cell wall accumulation of Cu ions and modulation of lignifying enzymes in primary leaves of bean seedlings exposed to excess copper. Biol Trace Elem. Res. 139: 97–107. [DOI] [PubMed] [Google Scholar]
- Briat J.F., Ravet K., Arnaud N., Duc C., Boucherez J., Touraine B., et al. (2010) New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Ann. Bot. 105: 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill E., van Thournout M., White R.G., Liewellyn D., Campbell P.M., Engelen S., et al. (2011) A novel isoform of sucrose synthase is targeted to the cell wall during secondary cell wall synthesis in cotton fiber. Plant Physiol. 157: 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton W.J., Dilworth M.J. (1971) Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125: 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhead J.L., Gogolin Reynolds K.A., Abdel-Ghany S.E., Cohu C.M., Pilon M. (2009) Copper homeostasis. New Phytol. 182: 799–816. [DOI] [PubMed] [Google Scholar]
- Chen E.L., Chen Y.A., Chen L.M., Liu Z.H. (2002) Effect of copper on peroxidase activity and lignin content in Raphanus sativus. Plant Physiol. Biochem. 40: 439–444. [Google Scholar]
- Cohu C.M., Pilon M.(2010) Cell biology of copper. In Cell Biology of Metals and Nutrients. Edited by Rüdiger H., Mendel R.R. pp. 55–74. Springer, Berlin. [Google Scholar]
- Colzi I., Doumett S., Del Bubba M., Fornaini J., Arnetoli M., Gabbrielli R., et al. (2011) On the role of the cell wall in the phenomenon of copper tolerance in Silene paradoxa L. Environ. Exp. Bot. 72: 77–83. [Google Scholar]
- Deng F., Yamaji N., Xia J., Ma J.F. (2013) A member of the heavy metal P-type ATPase OsHMA5 is involved in xylem loading of copper in rice. Plant Physiol. 163: 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dien B.S., Jung H.J.G., Vogel K.P., Casler M.D., Lamb J.F.S., Iten L., et al. (2006) Chemical composition and response to dilute-acid pretreatment and enzymatic saccharification of alfalfa, reed canarygrass and switchgrass. Biomass Bioenergy 30: 880–891. [Google Scholar]
- Drazkiewicz M., Skórzynska-Polit E., Krupa Z. (2004) Copper-induced oxidative stress and antioxidant defence in Arabidopsis thaliana. Biometals 17: 379–387. [DOI] [PubMed] [Google Scholar]
- Dupae J., Bohler S., Noben J.P., Carpentier S., Vangronsveld J., Cuypers A. (2014) Problems inherent to a meta-analysis of proteomics data: a case study on the plants’ response to Cd in different cultivation conditions. J. Proteomics 108: 30–54. [DOI] [PubMed] [Google Scholar]
- Flores A.G., Unger V.M. (2013) Atox1 contains positive residues that mediate membrane association and aid subsequent copper loading. J. Membr. Biol. 246: 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry S.C. (1998) Oxidative scission of plant cell wall polysaccharides by ascorbate-induced hydroxyl radicals. Biochem. J. 332: 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry S., Miller J., Dumville J. (2002) A proposed role for copper ions in cell wall loosening. Plant Soil 247: 57–67. [Google Scholar]
- Fujii S., Hayashi T., Mizuno K. (2010) Sucrose synthase is an integral component of the cellulose synthesis machinery. Plant Cell Physiol. 51: 294–301. [DOI] [PubMed] [Google Scholar]
- Garcia-Molina A., Andrés-Colás N., Perea-García A., Neumann U., Dodani S.C., Huijser P., et al. (2013) The Arabidopsis COPT6 transport protein functions in copper distribution under copper-deficient conditions. Plant Cell Physiol. 54: 1378–1390. [DOI] [PubMed] [Google Scholar]
- Guerriero G., Legay S., Hausman J.F. (2014) Alfalfa cellulose synthase gene expression under abiotic stress: a Hitchhiker’s guide to RT-qPCR normalization. PLoS One 9: e103808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D., Chen F., Inoue K., Blount J.W., Dixon R.A. (2001) Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13: 73–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallégot P., Audinot J.N., Migeon H.N. (2006) Direct NanoSIMS imaging of diffusible elements in surfaced block of cryo-processed biological samples. Appl. Surf. Sci. 252: 6706–6708. [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himelblau E., Mira H., Lin S.J., Culotta V.C., Peñarrubia L., Amasino R.M. (1998) Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiol. 117: 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irtelli B., Petrucci W.A., Navari-Izzo F. (2009) Nicotianamine and histidine/proline are, respectively, the most important copper chelators in xylem sap of Brassica carinata under conditions of copper deficiency and excess. J. Exp. Bot. 60: 269–277. [DOI] [PubMed] [Google Scholar]
- Lequeux H., Hermans C., Lutts S., Verbruggen N. (2010) Response to copper excess in Arabidopsis thaliana: impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiol. Biochem. 48: 673–682. [DOI] [PubMed] [Google Scholar]
- Lê S., Josse J., Husson F. (2008) FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 25: 1–18. [Google Scholar]
- Liao M.T., Hedley M.J., Woolley D.J., Brooks R.R., Nichols M.A. (2000) Copper uptake and translocation in chicory (Cichorium intybus L. cv. Grasslands Puna) and tomato (Lycopersicon esculentum Mill. cv. Rondy) plants grown in NFT system. I. Copper uptake and distribution in plants. Plant Soil 221: 135–142. [Google Scholar]
- Li S., Bashline L., Lei L., Gu Y. (2014) Cellulose synthesis and its regulation. Arabidopsis Book 12: e0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjamaa K., Kukkola E.M., Fagerstedt K.V. (2009) The role of xylem class III peroxidases in lignification. J. Exp. Bot. 60: 367–376. [DOI] [PubMed] [Google Scholar]
- Moore K.L., Chen Y., van de Meene A.M.L., Hughes L., Liu W., Geraki T., et al. (2014) Combined NanoSIMS and synchrotron X-ray fluorescence reveal distinct cellular and subcellular distribution patterns of trace elements in rice tissues. New Phytol. 201: 104–115. [DOI] [PubMed] [Google Scholar]
- Moore K.L., Schröder M., Wu Z., Martin B.G.H., Hawes C.R., McGrath S.P., et al. (2011) High-resolution secondary ion mass spectrometry reveals the contrasting subcellular distribution of arsenic and silicon in rice roots. Plant Physiol. 156: 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C.W., Weber P.K., Kilburn M.R., Hoeschen C., Kleber M., Pett-Ridge J. (2013) Advances in the analysis of biogeochemical interfaces: NanoSIMS to investigate soil microenvironments. Adv. Agron. 121: 1–46. [Google Scholar]
- Nielsen K.L., Indiani C., Henriksen A., Feis A., Becucci M., Gajhede, et al. (2001) Differential activity and structure of highly similar peroxidases. Spectroscopic, crystallographic, and enzymatic analyses of lignifying Arabidopsis thaliana peroxidase A2 and horseradish peroxidase A2. Biochemistry 40: 11013–11021. [DOI] [PubMed] [Google Scholar]
- O’Sullivan J.N., Ernest J. (2007) Nutrient deficiencies in lesser yam (Dioscorea esculenta) characterized using constant-water table sand culture. J. Plant Nutr. Soil Sci. 170: 273–282. [Google Scholar]
- Printz B., Guerriero G., Sergeant K., Renaut J., Lutts S., Hausman J.F. (2015) Ups and downs in alfalfa: proteomic and metabolic changes occurring in the growing stem. Plant Sci. 238: 13–25. [DOI] [PubMed] [Google Scholar]
- Printz B., Sergeant K., Guignard C., Renaut J., Hausman J.F. (2013a) Physiological and proteome study of sunflowers exposed to a polymetallic constraint. Proteomics 13: 1993–2015. [DOI] [PubMed] [Google Scholar]
- Printz B., Sergeant K., Lutts S., Guignard C., Renaut J., Hausman J.F. (2013b) From tolerance to acute metabolic deregulation: contribution of proteomics to dig into the molecular response of alder species under a polymetallic exposure. J. Proteome Res. 12: 5160–5179. [DOI] [PubMed] [Google Scholar]
- Puig S., Peñarrubia L. (2009) Placing metal micronutrients in context: transport and distribution in plants. Curr. Opin. Plant Biol. 12: 299–306. [DOI] [PubMed] [Google Scholar]
- Ravet K., Touraine B., Boucherez J., Briat J.F., Gaymard F., Cellier F. (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J. 57: 400–412. [DOI] [PubMed] [Google Scholar]
- Ravet K., Pilon M. (2013) Copper and iron homeostasis in plants: the challenges of oxidative stress. Antioxid. Redox. Signal. 19: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H., Conéjéro G., Divol F., Alcon C., Verdeil J.L., Curie C., et al. (2013) New insights into Fe localization in plant tissues. Front. Plant Sci. 4: 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saathoff A.J., Donze T., Palmer N.A., Bradshaw J., Heng-Moss T., Twigg P., et al. (2013) Towards uncovering the roles of switchgrass peroxidases in plant processes . Front. Plant Sci. 4: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin L.J., Lo J.C., Yeh K.C. (2012) Copper chaperone antioxidant protein1 is essential for copper homeostasis. Plant Physiol. 159: 1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strozycki P.M., Szymanski M., Szczurek A., Barciszewski J., Figlerowicz M. (2010) A new family of ferritin genes from Lupinus luteus—comparative analysis of plant ferritins, their gene structure, and evolution. Mol. Biol. Evol. 27: 91–101. [DOI] [PubMed] [Google Scholar]
- Tapken W., Ravet K., Pilon M. (2012) Plastocyanin controls the stabilization of the thylakoid Cu-transporting P-type ATPase PAA2/HMA8 in response to low copper in Arabidopsis. J. Biol. Chem. 287: 18544–18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantino D., Petit J.M., Lobreaux S., Briat J.F., Soave C., Murgia I. (2003) Differential involvement of the IDRS cis-element in the developmental and environmental regulation of the AtFer1 ferritin gene from Arabidopsis. Planta 217: 709–716. [DOI] [PubMed] [Google Scholar]
- Villalobos D.P., Díaz-Moreno S.M., Said E.S.S., Cañas R.A., Osuna D., Kerckhoven S.H.E.V., et al. (2012) Reprogramming of gene expression during compression wood formation in pine: coordinated modulation of S-adenosylmethionine, lignin and lignan related genes. BMC Plant Biol. 12: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. (2009) SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R., Nishitani K. (2006) Identification and characterization of Arabidopsis thaliana genes involved in xylem secondary cell walls. J. Plant Res. 119: 189–194. [DOI] [PubMed] [Google Scholar]
- Zhao F.J., Moore K.L., Lombi E., Zhu Y.G. (2014) Imaging element distribution and speciation in plant cells. Trends Plant Sci. 19: 183–192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.