Abstract
The Saccharomyces cerevisiae Forkhead Box (FOX) proteins, Fkh1 and Fkh2, regulate diverse cellular processes including transcription, long-range DNA interactions during homologous recombination, and replication origin timing and long-range origin clustering. We hypothesized that, as stimulators of early origin activation, Fkh1 and Fkh2 abundance limits the rate of origin activation genome-wide. Existing methods, however, are not well-suited to quantitative, genome-wide measurements of origin firing between strains and conditions. To overcome this limitation, we developed qBrdU-seq, a quantitative method for BrdU incorporation analysis of replication dynamics, and applied it to show that overexpression of Fkh1 and Fkh2 advances the initiation timing of many origins throughout the genome resulting in a higher total level of origin initiations in early S phase. The higher initiation rate is accompanied by slower replication fork progression, thereby maintaining a normal length of S phase without causing detectable Rad53 checkpoint kinase activation. The advancement of origin firing time, including that of origins in heterochromatic domains, was established in late G1 phase, indicating that origin timing can be reset subsequently to origin licensing. These results provide novel insights into the mechanisms of origin timing regulation by identifying Fkh1 and Fkh2 as rate-limiting factors for origin firing that determine the ability of replication origins to accrue limiting factors and have the potential to reprogram replication timing late in G1 phase.
Chromosomal DNA replication in eukaryotes follows a spatiotemporal program determined by the selective activation of potential replication origins distributed throughout chromosomes. Origins are “licensed” for one round of replication through the cell cycle-regulated establishment of the pre-RC complex in early G1 phase upon exit from mitosis (for review, see Sclafani and Holzen 2007). Initiation of licensed origins in the ensuing S phase requires stimulation by cell cycle-regulated kinases CDK and DDK (for review, see Labib 2010). The kinetics of initiation at different origins, however, are influenced by local chromatin structure and the subnuclear localization of chromosomal domains, such as subtelomeric and telomeric regions that are heterochromatic, peripherally located, and late-replicating (for review, see Aparicio 2013; Rhind and Gilbert 2013; Smith and Aladjem 2014). Accordingly, histone deacetylases and other chromatin modifiers have been implicated in regulating the timing and efficiency of replication origins in eukaryotic cells (for review, see Smith and Aladjem 2014; Creager et al. 2015).
The exact mechanism through which chromatin structure delays replication initiation remains unclear but is thought to impede the accessibility of trans-activating factors to the licensed origin complex. In this context of generally repressive chromatin, specific mechanisms have recently been identified in yeasts that act to stimulate the early replication of specific sequences. For example, centromeres are early replicating in both budding and fission yeasts, though remarkably, this is achieved through distinct mechanisms that act to efficiently recruit the replication initiation kinase DDK to specific origins (Hayashi et al. 2009; Li et al. 2011; Natsume et al. 2013). In Saccharomyces cerevisiae, the Forkhead Box (FOX) DNA binding proteins Fkh1 and Fkh2 are responsible for stimulating the activation of most noncentromeric, early-firing origins in the genome (Knott et al. 2012). In the absence of FKH1 and FKH2, the majority of these early-firing origins are significantly delayed in activation (thus defined as “Fkh-activated”), while a similar number of normally later-firing origins are advanced in their timing (defined as “Fkh-repressed”).
The exact mechanism of origin regulation by Fkh1 and/or Fkh2 (Fkh1/2) remains unclear, but “activation” appears to involve the preferential recruitment of limiting initiation factors, like Cdc45, to origins with Fkh1/2 bound in cis (Knott et al. 2012). As origin loading of Cdc45 is DDK-dependent, Fkh1/2 may be inferred to enhance DDK recruitment to Fkh-activated origins. In contrast, “repression” by Fkh1/2 was suggested to reflect an indirect consequence of limiting replication factors being sequestered by the activated origins. Furthermore, Fkh1/2 are required for the G1 phase clustering of early (activated) origins, suggesting a function of Fkh1/2 in the spatial organization of origins and the ensuing replication process (Knott et al. 2012). Such a proposed mechanism shares a striking parallel with the mechanism of Fkh1 in regulating donor selection during mating-type switching, which involves homologous DNA recombination between distal chromosomal DNA sequences (for review, see Haber 2012). Fkh1 and Fkh2 have also been extensively characterized as transcriptional regulators, particularly of sets of genes that control cell cycle progression (for review, see Murakami et al. 2010). While it seems likely that the functions of Fkh1/2 in replication and transcription share certain mechanisms, no correlation has been found between transcriptional changes and the alterations in origin usage in fkh1Δ fkh2Δ cells (Knott et al. 2012).
Fkh1/2 binds proximal to most Fkh-activated origins; however, Fkh1/2 binding also occurs near some origins not detectably activated by Fkh1/2. This might reflect the action of mechanisms opposing origin activation at certain loci or limited occupancy of Fkh1/2 at these loci (Ostrow et al. 2014). Indeed, potential Fkh1/2 binding sequences appear to greatly exceed Fkh1/2 abundance. Thus, we hypothesized that Fkh1/2 abundance might limit the level of origin activation genome-wide. However, most replication analysis methods are currently unsuited to quantitatively analyze origin firing levels genome-wide or are lacking in sensitivity (for review, see Gilbert 2010).
Results
Quantitative BrdU immunoprecipitation analyzed by sequencing (qBrdU-seq)
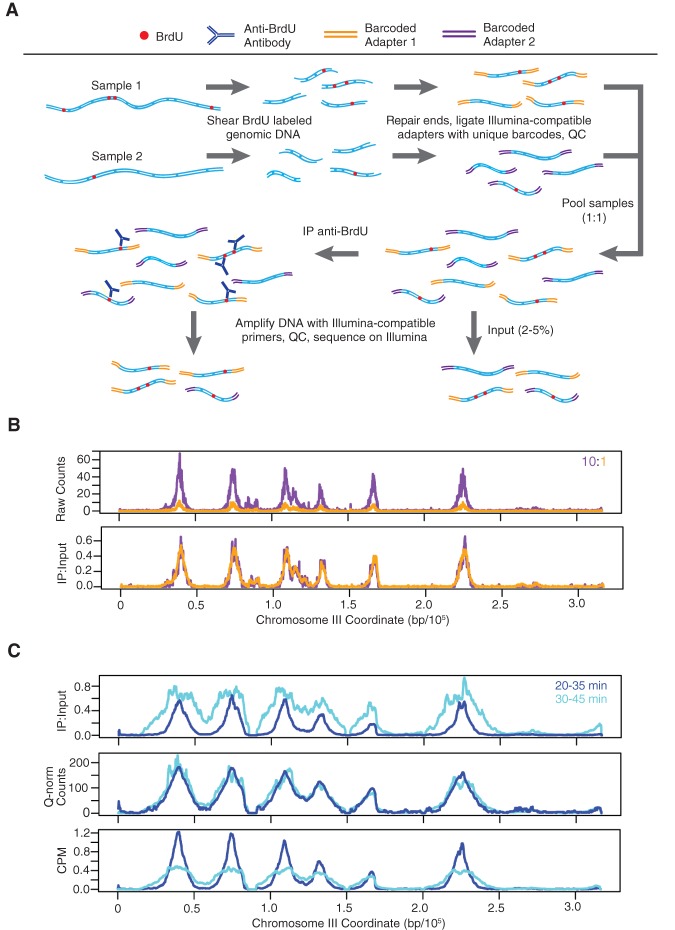
To accurately quantify and directly compare replication levels at origins between different samples, we developed a quantitative method for BrdU immunoprecipitation (IP) analyzed by high-throughput sequencing (Fig. 1A). This method, termed qBrdU-seq, involves pooling of two or more uniquely barcoded samples prior to the BrdU-IP and library amplification steps, along with parallel analysis to quantify the input DNA sample for normalization of the immunoprecipitated material. Thus, the most critical DNA manipulation for library preparation, addition of adapters, which include the barcodes, is carried out prior to depletion of DNA quantity by IP; and numerous operations that may introduce substantial variability between individually prepared samples, including IP, PCR amplification, and quantification of sequencing reads, are conducted under identical and/or more reliable conditions. We confirmed the ability of qBrdU-seq to detect and correct for differences in sample inputs by adding distinct barcodes to equal aliquots of a BrdU-labeled genomic DNA sample and combining the distinctly barcoded samples in different proportions for IP, amplification, and sequence analysis. The results show that normalization of the IP data by the input data (IP:Input) largely corrected for a 10-fold difference in input DNA (Fig. 1B).
Figure 1.
Quantitative BrdU-IP-seq analysis. (A) Scheme of the method. BrdU-labeled genomic DNA from each sample is barcoded by end-ligation of Illumina-compatible linkers. Samples are pooled, a small fraction of this pool is set aside as “Input,” and the remainder is subjected to immunoprecipitation (IP) with anti-BrdU antibody. The IP and Input samples are PCR-amplified with indexed primers and sequenced. IP sample reads are normalized against Input sample reads. (B) Validation of the method. A BrdU-labeled genomic DNA sample was split in two and each aliquot was uniquely barcoded. The distinctly barcoded samples were pooled at a ratio of 1:10 (color-keyed) and processed as above. IP results are shown raw and with normalization against the Input. (C) Comparison to CPM and Quantile normalization (Q-norm). JPy88 (WT) cells were synchronized in G1 phase with α-factor, released into S phase, and aliquots were incubated with BrdU for the indicated time intervals and harvested. The samples were processed as described for qBrdU-seq in A, and the IP sequence reads were analyzed by qBrdU-seq, CPM, or Quantile normalization and plotted as overlays of the time points.
To validate this approach further, we tested qBrdU-seq normalization against two other commonly used approaches: CPM (counts per million bp, a variation of RPKM), which normalizes read depth between samples, and Quantile normalization, which fits the samples to similar distributions (Bolstad et al. 2003; Mortazavi et al. 2008). When applied to the above test sample analyzed at 10-fold different concentrations, both methods successfully corrected the data, as expected because the correctly normalized samples should have identical read depths and distributions (Supplemental Fig. S1A). As a more stringent test, we analyzed a pair of samples (with replicates) with different levels of BrdU incorporation; the samples represent sequential BrdU pulses at the start of S phase with significantly more bulk replication occurring during the second pulse (Supplemental Fig. S1B). Both CPM and Quantile normalization performed poorly on these samples by essentially equalizing the nonequal signals for the two time points (Fig. 1C). The CPM- and Quantile-normalized results show little evidence of replication progression, such as BrdU incorporation at greater distances from the origins in the second time point. In contrast, the IP:Input normalization clearly reflects progression of S phase and greater signal in the latter time point reflective of the bulk DNA analysis (Fig. 1C; Supplemental Fig. S1B). Furthermore, the qBrdU-seq normalization by input correctly scales the IP data to the degree of replication that has occurred in the corresponding input, which is apparent in the flattening and sometimes splitting of the BrdU signal at early origins by the second time point, as the DNA duplication corrects the signal relative to flanking sequences still undergoing replication. These results verify that qBrdU-seq is a superior method for analysis of BrdU-IP-seq data.
Fkh1 and Fkh2 overexpression advances initiation timing globally
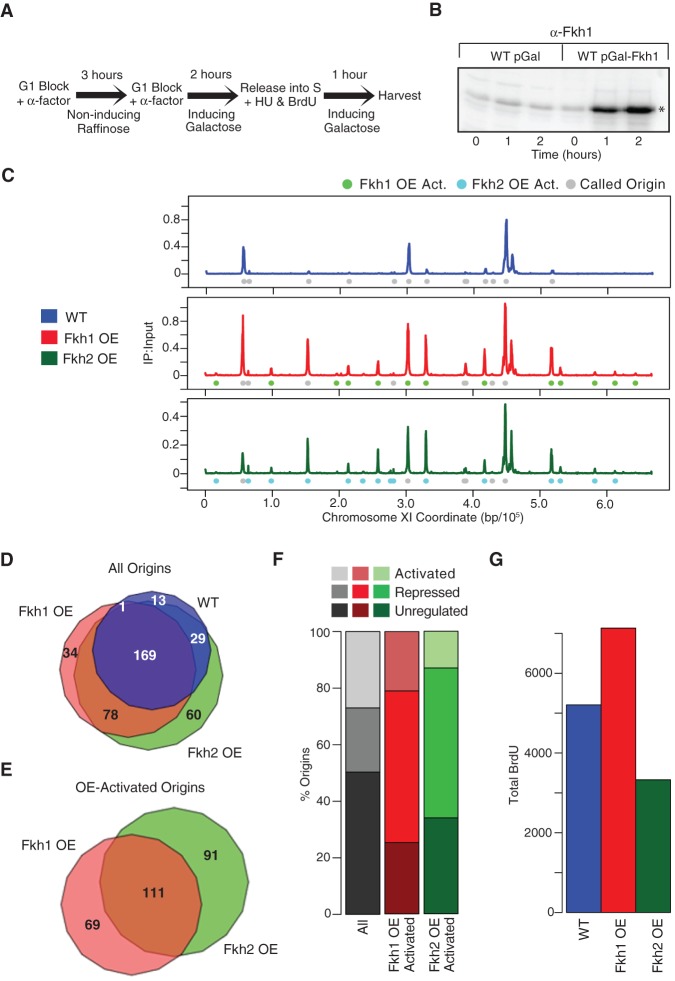
To test the hypothesis that Fkh1/2 is rate-limiting for early origin firing, we overexpressed Fkh1 and Fkh2 and analyzed replication origin dynamics. Wild-type (WT) cells harboring a plasmid vector with FKH1 or FKH2 expression under control of the GAL1/10 promoter or the empty GAL1/10 vector were synchronized in late G1 phase with α-factor under noninducing conditions, followed by incubation under inducing conditions while the late G1 arrest was maintained. After 2 h of induction to allow Fkh1 or Fkh2 accumulation, cells were released from G1 into S phase in the presence of BrdU to label replicating DNA plus hydroxyurea (HU) to arrest replication in early S phase, allowing early but not late origins to initiate replication (Fig. 2A,B).
Figure 2.
Fkh1-OE and Fkh2-OE stimulate origin firing globally. (A) Scheme for overexpression and replication analysis. Cells grown in YEP-raffinose (noninducing) were blocked in G1 phase by incubation with α-factor for 3 h, followed by incubation in YEP-galactose plus α-factor for 2 h to induce expression while maintaining the G1 block. Cells were released from G1 block into S phase by incubation in YEP-galactose, minus α-factor, plus BrdU and HU for 1 h and harvested. (B) Immunoblot analysis of Fkh1 expression in JPy88 (WT) and JPy89 (Fkh1-OE) induced as in part A; Fkh1 is indicated with an asterisk. (C) qBrdU-seq analysis of JPy88, JPy89, and JPy90 (Fkh2-OE) treated as in A was performed in triplicate and averaged data plotted for Chromosome XI. (D,E) Venn diagrams of all origins identified in the indicated sets (D) and of all OE-activated origins (E). (F) Distribution of previously defined origin classes: Fkh-activated, Fkh-repressed, and Fkh-unregulated among Fkh1-OE-activated and Fkh2-OE-activated origins; “All” shows the distribution of all previously defined origins. (G) Total normalized BrdU signal ±5 kb of all called origins for WT, Fkh1-OE, and Fkh2-OE.
Chromosomal plots of the BrdU incorporation data show that both Fkh1 overexpression (Fkh1-OE) and Fkh2 overexpression (Fkh2-OE) caused striking changes in the relative initiation levels of many origins, most notably increasing the activation of many later-firing origins relative to earlier-firing origins (Fig. 2C; Supplemental Fig. S2 for all chromosome plots). For example, plots of BrdU incorporation along Chromosome XI show three main peaks (representing early origins) and several small peaks (representing late or inefficient origins) in WT cells, whereas Fkh1-OE and Fkh2-OE show significantly greater BrdU incorporation at virtually all of the minor peak positions in addition to some origin loci lacking BrdU incorporation in WT cells . To characterize the effects of Fkh1-OE and Fkh2-OE across the genome, we determined whether the level of BrdU incorporation at these origins was significantly altered. The results show 212 origins firing in WT cells, 282 firing in cells with Fkh1-OE, and 336 firing in cells with Fkh2-OE, in total comprising 384 total origins (Fig. 2D). Fkh1-OE and Fkh2-OE significantly increased BrdU incorporation at 180 origins (termed Fkh1-OE-activated) and at 202 origins (Fkh2-OE-activated), respectively, with the majority of these origins belonging to both groups (Fig. 2E). The Fkh1-OE-activated and Fkh2-OE-activated origin sets were predominantly composed of origins previously categorized as Fkh-repressed or Fkh-unregulated (Fig. 2F; Knott et al. 2012). Thus, with increased dosage, Fkh1 and Fkh2 can act broadly across the genome to increase the activation of many origins.
Consistent with the increased BrdU incorporation at many origins, Fkh1-OE increased total BrdU incorporation at origins in HU (Fig. 2G). In contrast, total BrdU incorporation was notably lower in cells with Fkh2-OE compared with WT (Fig. 2G). Analysis of total DNA content by flow cytometry showed that Fkh2-OE caused a delay in S phase entry relative to WT and Fkh1-OE (Supplemental Fig. S1B). Fkh2-OE also delayed the timing of bud emergence compared with WT and Fkh1-OE (Supplemental Fig. S1C), indicating that delayed passage through Start caused the consequent delay in S phase entry. Thus, we conclude that the reduced level of total replication measured by qBrdU-seq in cells with Fkh2-OE results from this cell cycle progression defect rather than a DNA replication defect per se. We also note that this difference in BrdU incorporation due to defective S phase entry was revealed by the qBrdU-seq normalization but masked by CPM normalization (Supplemental Fig. S3). Overall, the effects of Fkh1-OE and Fkh2-OE were qualitatively similar, suggesting that the greater dependence on FKH1 versus FKH2 in deletion analysis reflects their in vivo binding preferences as opposed to inherently different functional capabilities.
We considered the possibility that Fkh1-OE and Fkh2-OE might alter origin timing by affecting the levels of replication proteins, particularly rate-limiting initiation factors identified in recent studies (Patel et al. 2008; Mantiero et al. 2011; Tanaka et al. 2011). However, we observed no change in the levels of several replication initiation proteins that we examined, including Dbf4, the overexpression of which was essential for advancement of late origin firing in all of these previous studies (Supplemental Fig. S4). We also tested whether the expression levels of any DNA replication genes were altered by both Fkh1-OE and Fkh2-OE using RNA-seq analysis and identified only two genes annotated as having a DNA replication initiation function: NOC3, which is up-regulated 1.5-fold, and CTF3, which is down-regulated twofold (Supplemental Table S1). Noc3 has been reported to function in pre-RC assembly (Zhang et al. 2002); however, pre-RC assembly is not thought to be limiting at most origins (Santocanale et al. 1999; Wyrick et al. 2001), and pre-RC levels are unaffected in fkh1Δ fkh2Δ cells (Knott et al. 2012). Ctf3 has been reported to function in plasmid stability; however, this probably reflects its well-established function at kinetochores in chromosome segregation rather than a function in DNA replication (Measday et al. 2002). Furthermore, analysis of the RNA-seq data found no correlation between the alterations of origin activities and local alterations in transcription caused by Fkh1-OE and Fkh2-OE (data not shown), consistent with our previous conclusion that the function of Fkh1 and Fkh2 in regulating replication origin initiation is independent of their effects on transcription (Knott et al. 2012).
We also addressed the possibility that Fkh1-OE and Fkh2-OE might alter the levels of dNTP pools in the cells and allow the higher levels of origin firing observed in early S phase. Total DNA content in HU was little changed due to Fkh1-OE or Fkh2-OE, suggesting little if any change in dNTP pool levels (Supplemental Fig. S5A). In contrast, overexpression of ribonucleotide reductase (RNR3), which has been shown to increase dNTP levels (Chabes and Stillman 2007), substantially increased DNA content in HU-arrested cells (Supplemental Fig. S5B). Analysis by qBrdU-seq showed that Rnr3 overexpression (Rnr3-OE) affected replication differently than Fkh1-OE or Fkh2-OE, specifically by increasing fork progression more than origin firing (Supplemental Fig. S5C). These results support the conclusion that origin stimulation by Fkh1-OE and Fkh2-OE does not involve altered dNTP levels.
Increased rate of origin firing slows replication fork rate
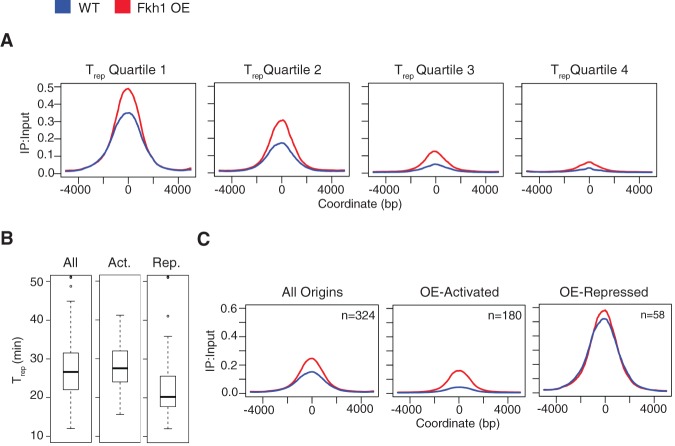
Further analysis was focused on Fkh1, which plays the predominant role at origins (Knott et al. 2012; Ostrow et al. 2014) and which did not exhibit an S phase entry delay like Fkh2 when overexpressed. To determine how the effects of Fkh1-OE on origins relate to the normal replication timing of the affected origins, we plotted the average BrdU incorporation signal for 10-kb regions centered on origins divided into quartiles according to their replication timings (TRep) (Fig. 3A; Raghuraman et al. 2001). This analysis shows increased average BrdU incorporation in all replication timing quartiles, with the greatest relative effects in the later quartiles. Accordingly, Fkh1-OE-activated origins are mainly comprised of later-firing origins (Fig. 3B). Thus, Fkh1-OE most strongly affects those origins that are not already activated at normal Fkh1 dosage (Fig. 2F) and many additional origins are susceptible to advancement by Fkh1; however, its normal dosage limits its functional targets.
Figure 3.
Fkh1-OE stimulates origin firing. (A) Origins were divided into quartiles according to TRep, and BrdU incorporation was averaged across 10-kb regions aligned by the peak summit of each origin. (B) TRep distributions of all origins and those classified as Fkh1-OE-activated and Fkh1-OE-repressed. (C) Average BrdU incorporation for 10-kb regions aligned by the peak summit of all origins and those classified as Fkh1-OE-activated and Fkh1-OE-repressed.
Despite the overall increase in origin firing due to Fkh1-OE (Figs. 2G, 3A), 58 origins were identified as having significantly reduced BrdU incorporation (Fkh1-OE-repressed). These origins were mostly early origins (Fig. 3B). However, closer examination shows that Fkh1-OE increased the average BrdU peak height at these origins, reflecting an increased level of origin firing, while causing a small but significant decrease in the area under the BrdU peaks at these origins, which is due to a slightly narrower average peak (Fig. 3C). This suggested a decrease in the progression of replication forks from these origins, which would be consistent with a titration of limiting factors away from these replication forks to the additional replication forks established at origins advanced by Fkh1-OE.
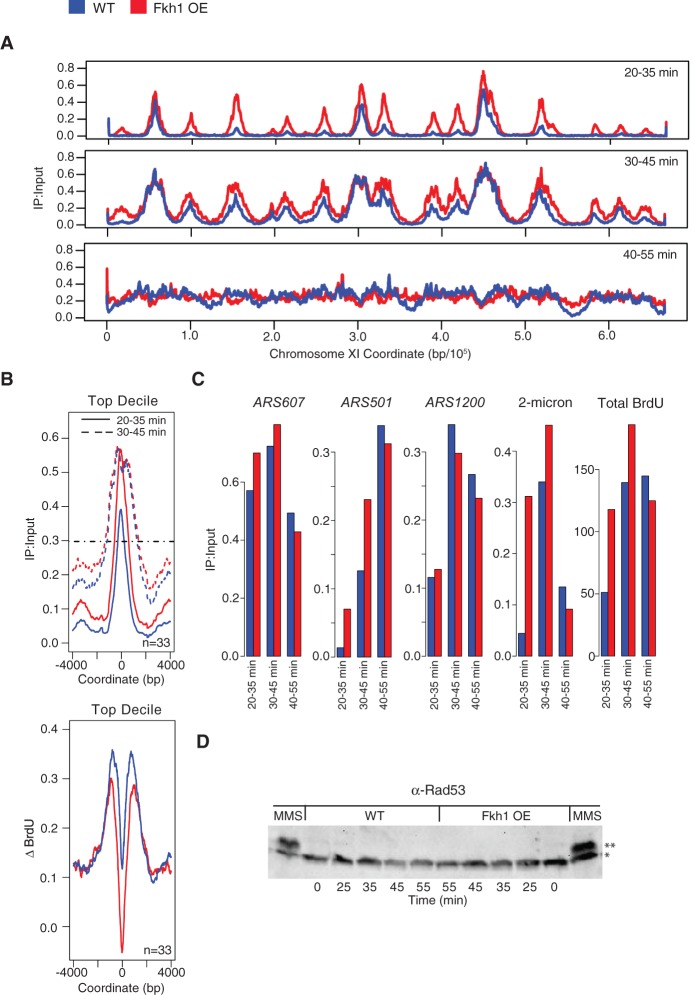
Because the available nucleotide pools primarily determine the extent of replication in HU, effects of Fkh1-OE may have been obscured by the use of HU. Therefore, we analyzed Fkh1-OE effects on replication in cells released into S phase without HU. Cells were subjected to galactose induction as above, released into S phase, and pulsed with BrdU at different times after release. qBrdU-seq analysis was carried out and the BrdU signal was plotted for each interval. The replication profiles reveal the increased firing of many replication origins early in the time-course, including the earliest origins (Fig. 4A; Supplemental Fig. S6 for all chromosome plots). Strikingly, despite the taller and wider BrdU peaks in cells with Fkh1-OE at 35 min, the distribution of BrdU incorporation appeared to equalize by 45 min, suggesting a slowing of DNA synthesis at replication forks in cells with Fkh1-OE.
Figure 4.
Fkh1-OE increases early origin firing and decreases replication fork rate. Strains JPy88 (WT) and JPy89 (Fkh1-OE) arrested and induced as described in Figure 2A were released into S phase without HU, and aliquots were incubated with BrdU for the indicated intervals and harvested. (A) qBrdU-seq analysis was performed in duplicate and plotted for Chromosome XI. (B) Analysis of the earliest decile of origins: Average BrdU incorporation is plotted for the indicated time intervals (upper panel), and the horizontal dashed line indicates the position at which fork rate measurements were calculated; the difference in BrdU incorporation between the first two intervals is plotted (lower panel). (C) Quantification of BrdU maximum peak height for the indicated loci for each time interval. Total BrdU (summed max peak heights) includes all origins including all copies of 2-micron and rDNA (ARS1200) origins. (D) Immunoblot analysis of Rad53; Rad53 is indicated with an asterisk, and phosphorylated Rad53 is indicated with two asterisks. “MMS” indicates sample was released after G1 block and Gal-induction into the presence of 0.033% MMS for 1 h prior to harvest.
To examine the rate of fork progression more precisely, we plotted the average BrdU signal for the first two time points for the earliest-firing decile of origins (Fig. 4B). This plot clearly shows that the leading edge of replication emanating from early origins is advanced in cells with Fkh1-OE at 35 min; however, by the second time point, the distance from the origins has converged. We also plotted the difference in BrdU signal between the first and second pulse intervals as an indication of the area replicated during the second time point relative to the first. Both plots show that these replication forks covered less distance during the second pulse interval in cells with Fkh1-OE (Fig. 4B). We estimated the rate of fork progression based on the differences in peak widths between the two time points as 975 bp/min for WT and 790 bp/min for Fkh1-OE (Fig. 4B). Thus, an increased rate of origin firing is accompanied by a decreased rate of DNA synthesis at replication forks, which can explain the overall similar rates of bulk DNA replication in cells with and without Fkh1-OE as measured by flow cytometry (Supplemental Fig. S1B).
We considered that decreased replication initiation of the multicopy ribosomal DNA (rDNA) origins might also compensate for the increased firing of single-copy late origins, resulting in the similar overall replication rate. We plotted BrdU peak signals for several representative origins from the time-course data, including ARS607 (early, single-copy) (Friedman et al. 1997; Yamashita et al. 1997), 2-micron (early, ∼60 episomal copies) (Zakian et al. 1979; Futcher and Cox 1984), ARS501 (late, single-copy) (Ferguson et al. 1991), and rDNA (timing uncharacterized, ∼150 tandem copies) (Fig. 4C; Kobayashi et al. 1998). The plots show expected differences in the peak firing time signals from early and late origins, further validating the ability of qBrdU-seq to robustly compare the levels of BrdU incorporation between loci, temporally. Fkh1-OE increased early firing of all these origins with the exception of the rDNA, which was decreased. These results are consistent with recent reports that rDNA origins are in competition with other origins for initiation (Kwan et al. 2013; Yoshida et al. 2014). Nevertheless, this decrease of rDNA origin firing is insufficient to compensate for increased firing of other origins as shown by the increase in total origin firing, which includes 2-micron and rDNA scaled to include all copies (Fig. 4C). These results further support the conclusion that slower fork progression balances the overall replication rate in cells with excess origin firing.
Higher rate of origin firing by Fkh1-OE does not cause detectable replication stress
We wondered whether the higher rate of origin firing resulting from Fkh1-OE causes replication stress. Although chronic Fkh1- and Fkh2-OE causes lethality (Postnikoff et al. 2012), cells remain viable under the acute OE used here and do not exhibit rapid or uniform cell cycle arrest, requiring several generations to arrest cell division (Supplemental Fig. S7). We also examined activation of checkpoint kinase Rad53 through its phosphorylation, which is detectable as a mobility shift in gel electrophoresis. In cells released into S phase without HU present, we did not detect Rad53 phosphorylation with or without Fkh1-OE in the time frame of these experiments or in longer time-courses corresponding to the eventual growth arrest caused by chronic Fkh1-OE (Fig. 4D; Supplemental Fig. S7; data not shown). Checkpoint signaling to activate Rad53 remained functional, as treatment with the DNA damaging agent methyl-methane-sulfonate (MMS) robustly induced Rad53 phosphorylation in cells with Fkh1-OE (Fig. 4D). These findings are consistent with the normal S phase length with Fkh1-OE indicated by flow cytometry (Supplemental Fig. S1B) and suggest that normal cells can compensate for the increased level of origin initiation without an overt checkpoint response.
Overexpressed Fkh1 acts through direct binding to affected origins
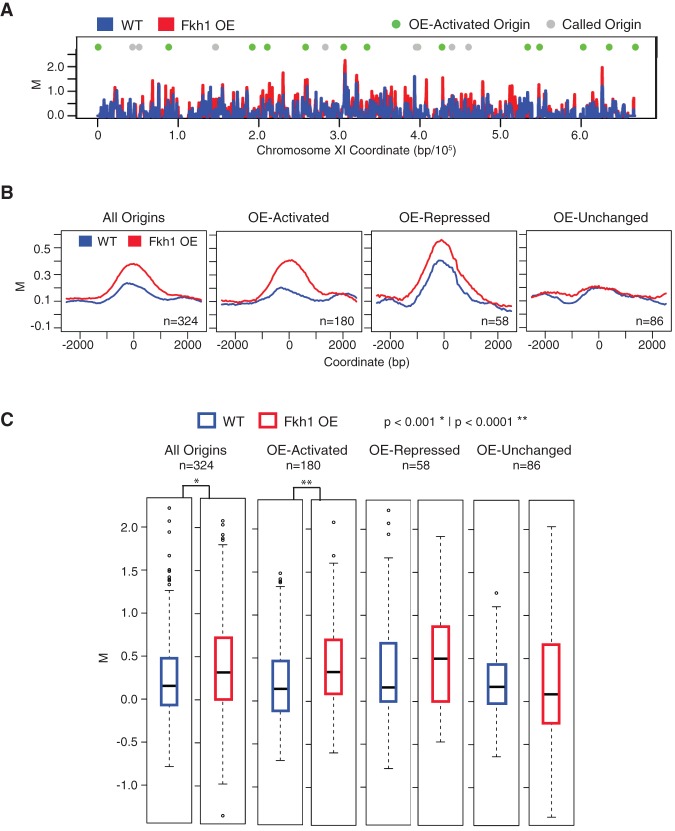
Origin activation by Fkh1 involves direct binding in cis to the origin (Knott et al. 2012). Thus, we expected that origin activation by Fkh1-OE would also reflect its direct binding proximal to the affected origins, likely due to the presence of Fkh1 recognition sequences that are infrequently bound in the absence of OE. To confirm this supposition, we examined the binding of Fkh1 in cells with and without Fkh1-OE using chromatin immunoprecipitation analyzed by whole genome tiling microarray (ChIP-chip). Endogenous and overexpressed FKH1 was Myc epitope-tagged, and cells were analyzed in G1 phase after induction or not of Fkh1-OE. Fkh1 binding along Chromosome XI shows increased binding at several Fkh1-OE-activated origins in Fkh1-OE cells (Fig. 5A). Fkh1 binding near origins was further analyzed by plotting the average Fkh1-Myc ChIP signal for 5-kb regions centered on different origin classes (Fig. 5B). These data were also plotted as box-and-whisker plots to show the data point distributions (Fig. 5C). Fkh1-OE-activated origins show minimal enrichment of Fkh1 in cells without OE; and with Fkh1-OE, the average Fkh1 ChIP signal increases at these origins. Fkh1-OE-repressed origins show a smaller increase of Fkh1 binding with Fkh1-OE; however, these origins also show a higher level of binding than other origin classes without OE (Fig. 5B,C), consistent with the fact that many of these origins are normally Fkh-activated and Fkh1-bound (Knott et al. 2012; Ostrow et al. 2014). Origins that were not significantly activated or repressed by Fkh1-OE (Fkh1-OE-unchanged) show minimal enrichment of Fkh1 binding with or without Fkh1-OE (Fig. 5B,C). Thus, increased Fkh1 binding at origins with low occupancy at normal Fkh1 expression levels correlates with the increased firing of these origins, consistent with overexpressed Fkh1 acting directly on origins.
Figure 5.
Fkh1-OE increases Fkh1 occupancy at stimulated origins. Strains JPy105 (WT, Fkh1-Myc) and JPy106 (Fkh1-Myc-OE) arrested and induced (but not released into S phase) as described in Figure 2A were subjected to ChIP-chip analysis of Fkh1. (A) ChIP signal along Chromosome XI is plotted with origins indicated above. (B) The average ChIP signal is plotted for 5-kb regions aligned on origins of the indicated groups. (C) Box-and-whisker plots of the distributions of ChIP signals for individual origins within the indicated origin groups.
Fkh1-OE reprograms origin timing in heterochromatin in late G1 phase
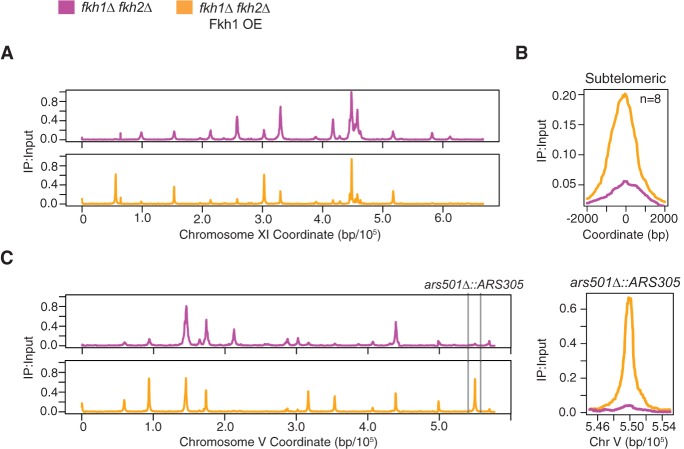
Previous studies have suggested that origin initiation timing is established in the late M to early G1 period of the cell cycle (Raghuraman et al. 1997; Dimitrova and Gilbert 1999). However, the above results demonstrate that the replication timing of many origins throughout the genome can be advanced by de novo Fkh1 binding during arrest in late G1 phase. To examine this “reprogramming” of origin timing more rigorously and rule out that the presence of a normal dosage of Fkh1 and Fkh2 prior to induction of OE in G1 phase contributed to subsequent origin activation by OE, we performed the Fkh1-OE induction in G1 phase in fkh1Δ fkh2Δ cells. Fkh1-OE restores origin firing of Fkh-activated and Fkh1-OE-activated origins, yielding a pattern very similar to that of WT cells with Fkh1-OE (Fig. 6A; cf. Fig. 2B), thus supporting the conclusion that Fkh1 can reprogram origin firing in late G1. This result also demonstrates that Fkh1-OE can stimulate additional origin firing independently of Fkh2.
Figure 6.
Fkh1 stimulates origin firing in subtelomeric heterochromatin. Strains OAy1071 (fkh1Δ fkh2Δ GAL-FKH1) and OAy1073 (fkh1Δ fkh2Δ) were induced and released into HU as described in Figure 2A and subjected to qBrdU-seq analysis. (A) Data are plotted for Chromosome XI. (B) Averaged BrdU signal for subtelomeric origins detected to fire. (C) Data are plotted for Chromosome V, highlighting the subtelomeric ars501Δ::ARS305 locus.
To scrutinize this activity of Fkh1 within heterochromatin, we more closely examined the activity of origins within subtelomeric heterochromatin. With induction of Fkh1-OE in G1-blocked fkh1Δ fkh2Δ cells, subtelomeric origins showed increased early initiation in the ensuing S phase, suggesting that Fkh1 can reprogram origin firing in G1 phase within established subtelomeric heterochromatin (Fig. 6B). The average level of subtelomeric origin activity in HU was still relatively low compared with early origins (Fig. 6A), suggesting only a limited ability of Fkh1 to stimulate origins within this chromatin domain. However, ChIP-chip shows that Fkh1/2 occupancy is low in subtelomeric domains (Ostrow et al. 2014), suggesting that the limited effect of Fkh1-OE reflects a dearth of Fkh1 binding sites associated with subtelomeric origins.
To further test whether Fkh1-OE can efficiently stimulate early activation within a subtelomeric domain, we replaced ARS501 with ARS305, a Fkh-activated origin with well-defined Fkh1 binding sequences. The ARS501 locus was chosen because it was the subject of the previous study concluding that initiation timing is established prior to the late G1 arrest point (Raghuraman et al. 1997) and thus presented a relatively well-characterized context in which to test Fkh1's ability to alter the established timing program. In fkh1Δ fkh2Δ cells, ARS305 at the ARS501 locus (ars501Δ::ARS305) did not initiate replication in HU, consistent with delayed activation (Fig. 6C). Induction of Fkh1-OE in these cells after arresting in late G1 resulted in robust activation of ars501Δ::ARS305 in HU to a level comparable to early origins (Fig. 6C). Thus, Fkh1 can reprogram the initiation timing of an origin within heterochromatin in late G1 phase.
Discussion
Fkh1/2 occupancy at replication origins is limiting for origin firing in early S phase
This study shows that Fkh1 and Fkh2 have a general stimulatory function in the activation of replication origins in S. cerevisiae. Overexpression of either Fkh1 or Fkh2 stimulates the earlier activation of many late (or dormant) origins. This function is consistent with our previous study indicating that FKH1 and FKH2 are responsible for the timing of most early origins in the yeast genome. This finding also supports our previous conclusion that origin repression by Fkh1/2 (i.e., origins that fire earlier in fkh1Δ fkh2Δ) was an indirect effect of its activation of other origins, which depletes limiting initiation factors. Indeed, the data show that many origins previously defined as Fkh-repressed are activated by increased Fkh1/2 dosage; however, normal Fkh1/2 dosages limit occupancy, and hence activation, at these origins. Despite the ability of Fkh2-OE to stimulate origin firing, deletion of FKH2 does not de-regulate origin firing, whereas deletion of FKH1 de-regulates many early origins (Knott et al. 2012). Thus, specificity in Fkh1 and Fkh2 binding preferences and occupancy levels at origins at normal dosages underlies their differential requirements in deletion studies. Indeed, Fkh1 binding is more frequently detected at Fkh-activated origins than Fkh2 (Ostrow et al. 2014). Whereas it might seem counterintuitive that Fkh2-OE should activate more origins than Fkh1-OE, we suggest that this reflects the normally greater abundance of Fkh1 and its preference for binding near origins, such that Fkh2-OE increases the number of Fkh1/2-bound origins more than Fkh1-OE (Ghaemmaghami et al. 2003; Kulak et al. 2014; Ostrow et al. 2014). Together with the ChIP-chip analysis showing increased Fkh1 occupancy at Fkh1-OE-activated origins, the results support the conclusion that the relative binding occupancy of Fkh1/2 is a direct determinant of origin activation timing.
Several recent studies have shown that overexpression of replication initiation proteins advances the firing of late origins in early S phase, indicating that these factors are present in rate-limiting amounts (Mantiero et al. 2011; Tanaka et al. 2011). This suggests a model in which those origins best able to recruit the limiting factors will initiate early with higher probability (Mantiero et al. 2011; Bechhoefer and Rhind 2012). Fkh1/2 appears to function in this recruitment mechanism, as the binding of Cdc45 to early origins in G1 phase is dependent on FKH1 and FKH2 (Knott et al. 2012). However, Fkh1-OE does not increase the dosage of limiting initiation proteins, suggesting instead that Fkh1/2 enables origins to more effectively recruit the available factors to trigger initiation (see below).
qBrdU-seq accurately determines relative amounts of DNA replication
The results of several studies have indicated that increased late origin firing depresses the level of early origin firing, at least in HU (Mantiero et al. 2011; Tanaka et al. 2011; Peace et al. 2014; Yoshida et al. 2014); however, the prior methods either did not allow a direct determination of replication levels genome-wide or involved statistical normalization between high-throughput data sets. Thus, the observation that Fkh1-OE and Fkh2-OE advanced the activation of many late origins prompted us to determine whether there was a diminution of early origin firing as a consequence of advanced late origin firing. We developed qBrdU-seq to correctly quantify the relative levels of origin firing between experiments by enabling direct normalization of the results without reliance on specific inferences or statistical methods for normalization, which depend on potentially flawed assumptions about the quality and distribution of the data. Our method is similar to the recently developed Bar-ChIP method for analysis of ChIP-seq data (Chabbert et al. 2015). Comparison of qBrdU-seq to commonly used normalization methods demonstrated the superiority of this new approach. The method also eliminates multiple technical sources of variation by processing pooled samples, which simplifies processing and reduces reagent costs while facilitating sample replication. The qBrdU-seq analysis showed that Fkh1-OE stimulated more total initiations in early S phase, especially at normally later origins (Figs. 2, 4).
Origin firing and replication fork progression
While Fkh1-OE increased the overall origin initiation rate in early S phase (Figs. 2G, 4C), the overall length of S phase based on bulk DNA measurement by flow cytometry was similar to WT cells (Supplemental Fig. S1B), suggesting that some compensation occurs to slow replication in cells with Fkh1-OE. The qBrdU-seq analysis demonstrated that the higher overall initiation rate in cells with Fkh1-OE was not accompanied by a reduction of early origin firing, measured as the BrdU peak height. Indeed, despite the substantial increase in late origin firing in HU and in the earliest time point in the time-course experiment, the earliest origins still showed higher initiation levels than without OE (Figs. 3A, 4A,C). Instead, the qBrdU-seq revealed that DNA synthesis at replication forks emanating from the earliest origins was reduced (Fig. 4B). As a result, cells without Fkh1-OE were largely able to catch up in the replication program. This effect on replication forks is probably not limited to replication forks from the earliest origins, but those are the most easily measured due to the difficulty of tracking forks from later origins.
The slower fork progression as a result of higher levels of origin firing is in accord with our recent study demonstrating an inverse relationship between origin firing levels and fork rate in cells with different initiation levels (Zhong et al. 2013). In addition, previous studies involving the combined overexpression of multiple replication initiation proteins resulting in advanced late origin firing also showed decreased quantities and lengths of replication intermediates from early origins (Mantiero et al. 2011; Tanaka et al. 2011); however, the presence of HU in those experiments complicated a definitive conclusion about the quantitative effects on overall origin firing as well as fork rate. The qBrdU-seq method allows direct comparison of relative replication levels at all loci across the genome.
Interestingly, the increased origin firing and consequent fork slowing occurs without triggering detectable replication stress signaling via Rad53 phosphorylation (Fig. 4D). The previous studies involving overexpression of replication initiation factors came to different conclusions regarding Rad53 activation in cells exhibiting advanced late origin firing (Mantiero et al. 2011; Tanaka et al. 2011). Mantiero et al. observed transient, low-level activation of Rad53 in cells overexpressing Sld2, Sld3, Dbf4, and Dpb11 in addition to overexpressing Cdc45 and Sld7 or having a deletion of RPD3. In the latter case, the cells appeared to complete bulk DNA replication more rapidly, which may explain the induction of a replication stress response. In both of those overexpression regimens, deletion of SML1, which increases dNTP pools, suppressed Rad53 activation, indicating that the cause of Rad53 activation was related to dNTP depletion. In contrast, Tanaka et al. observed no Rad53 activation in cells overexpressing Cdc45, Sld3, and Sld7, which advanced late origin firing but did not alter the overall rate of S phase according to bulk DNA content analysis. It would appear that the conditions of Mantiero et al. cause a greater increase in the replication capacity than the conditions of Tanaka et al. Fkh1-OE also stimulates additional origin firing without significantly increasing the total rate of S phase or inducing replication stress but does so without increasing the levels of other replication initiation proteins, suggesting that it acts to increase the ability of replication origins to accrue the available initiation factors rather than altering the overall capacity for replication.
Fkh1 programs replication origin timing in G1 phase
We further addressed the idea that Fkh1 acts to enhance origins’ ability to accrue replication factors by examining Fkh1's ability to stimulate an origin in subtelomeric chromatin, which is generally repressive for origin function. Given the established view that replication timing is established in early G1 phase (Raghuraman et al. 1997; Dimitrova and Gilbert 1999), it is remarkable that induction of Fkh1-OE in G1-arrested cells is able to stimulate the earlier activation of many late origins throughout the genome, including in subtelomeric regions (Fig. 6). We examined Fkh1's ability to stimulate origin function within heterochromatin in a well-defined system with ARS305, a Fkh-activated origin, inserted into the subtelomeric ARS501 locus, which is programmed to replicate late. Indeed, ars501Δ::ARS305 failed to initiate early in the absence of Fkh1/2 function but showed robust activation in early S phase upon induction of Fkh1-OE in late G1 (Fig. 6C). Thus, Fkh1 can reprogram origin timing within established subtelomeric heterochromatin. In contrast, overexpression of limiting replication factors only weakly activated heterochromatic origins when the chromatin structure was altered by deletion of the histone deacetylase RPD3 (Mantiero et al. 2011). These findings further support the conclusion that Fkh1 stimulates origin initiation by altering origin properties that regulate the accrual of replication initiation factors. Together with our recent finding that Fkh1/2 binds origins in G1 phase (Ostrow et al. 2014), Fkh1 binding appears to define the timing decision point for many origins in S. cerevisiae. That Fkh1 can reprogram timing in late G1 also argues against the idea that events associated with the timing of ORC binding or efficiency of MCM loading determine subsequent origin timing (Wu and Nurse 2009; Yang et al. 2010).
Fkh1/2 likely alters the chromatin and subnuclear localization of origins. Our previous study showed that Fkh1 and Fkh2 are required for the spatial clustering of early replication origins in G1 phase (Knott et al. 2012). While it remains unclear how this function of Fkh1/2 is executed, the FHA domain of Fkh1 plays a critical role in donor preference in mating-type switching, which involves a long-range interaction between distal chromosomal sequences. The mechanistic implication is that the phosphopeptide binding function of the FHA domain mediates interaction between chromatin-bound Fkh1 at the recombination enhancer and an as yet to be identified phosphoprotein at the distal MAT locus. Origin regulation also requires the Fkh1 FHA domain suggesting that Fkh1 bound at origins functions in a similar manner (SK Villwock and OM Aparicio, in prep.), albeit presumably targeting a different phosphoprotein(s), to recruit origins into clusters. Alternatively, the FHA domain may recruit a rate-limiting initiation factor, which results in the assembly of the origin into a cluster or nascent replication factory. Additionally, Fkh1 and Fkh2 have been reported to recruit chromatin remodelers and histone deacetylases, which are likely to play a role in determining the subnuclear organization of their target loci (Sherriff et al. 2007; Veis et al. 2007; Voth et al. 2007; Linke et al. 2013). Future studies will examine the role of chromatin modifiers and remodelers as effectors of Fkh1 and whether Fkh1 regulates subnuclear localization of DNA loci.
Methods
Plasmid and strain constructions and culture methods
Plasmid pCD43, herein referred to as pGAL, contains the GAL1/10 promoter inserted into pRS316 (Shaywitz et al. 1995). The FKH1 ORF was PCR-amplified (with HindIII and EcoRI overhangs added) from genomic DNA and inserted into HindIII- and EcoRI-digested pCD43, yielding pGAL-FKH1. The FKH2 ORF was PCR-amplified (with SpeI and NotI overhangs added) from genomic DNA and inserted into SpeI- and NotI-digested pCD43, yielding pGAL-FKH2. FKH1-MYC9 was PCR-amplified from genomic DNA of yeast strain ZOy14 with KpnI and EcoRI overhangs and inserted into KpnI- and EcoRI-digested pCD43, yielding pGAL-FKH1-MYC9. The 2.2 kb SalI-SpeI GAL-FKH1 fragment from pGAL-FKH1 was inserted into SalI-SpeI-digested pRS405, yielding p405-GAL-FKH1. pΔ501-ARS305 has a 540-bp PCR fragment containing ARS305 inserted into SpeI-SacI-digested pARS501 (a gift from B. Ferguson) using the In-Fusion HD Cloning Kit (Clontech). pJBN161 contains GAP-RNR3 and LEU2 (Desany et al. 1998).
All yeast strains are congenic with W303 and most are derived from BrdU-incorporating strains CVy61 and CVy63 (Viggiani and Aparicio 2006); genotypes of all strains are given in Supplemental Table S2. PCR-amplified DNA sequences were confirmed by DNA sequencing (Retrogen). Plasmids and integrating constructs were introduced into yeast by lithium acetate transformation and selection on appropriate medium (Gietz and Schiestl 2007). Strains OAy1071 and OAy1073 were derived from crosses of strains OAy1069 and OAy1070; relevant genotypes were confirmed by PCR analysis. In the parent strains, ars305Δ::BrdU-Inc (URA3) was introduced with BglII-digested p306-ars305Δ-BInc, which integrates and replaces 159 bp encompassing ARS305 (Zhong et al. 2013). ARS305 was swapped for ARS501 in two steps: ars501Δ::URA3 (Candida albicans) was constructed by PCR using long-oligonucleotides and pAG61 as the template (Addgene). Next, ARS305 was introduced with HindIII-digested pΔ501-ARS305, which replaces URA3 (C. albicans) and was selected for on 5-FOA. The resulting ars501Δ::ARS305, replaces ∼270 bp encompassing ARS501 with ∼540 bp encompassing ARS305. GAL-FKH1 (LEU2) was integrated at the leu2 locus by loop-in of BstEII-digested p405-GAL-FKH1.
For late G1 block, induction, and release, cells (preselected on SD−URA for plasmid as appropriate) were inoculated into YEP+2% raffinose at 25°C, grown to mid-log phase, and blocked in G1 by incubation (at O.D. ∼0.5) in fresh YEP+2% raffinose +7.5 nM α-factor at 25°C for 3 h. For induction, cells were pelleted and resuspended in YEP+2% galactose +7.5 nM α-factor at 25°C for 2 h, and cultures were released into S phase by centrifugation and resuspension (at O.D. ∼1.0) in fresh YEP+2% galactose +200 µg/mL Pronase E (Sigma-Aldrich, P5147) and sonicated to disperse cells. Early S phase analysis of replication was performed by including 0.2 M HU (Sigma-Aldrich, H8627) and BrdU at 400 µg/mL (Sigma-Aldrich, B5002) for 60 min at 25°C. For the time-course experiment, BrdU was used at 800 µg/mL. BrdU IP was carried out with anti-BrdU antibody (Invitrogen, 033900) at 1:1000. Bulk DNA content analysis was performed as described previously (Zhong et al. 2013). Immunoblot analysis of Fkh1 was performed with anti-Fkh1/2 antibody (Casey et al. 2008) at 1:1000. Anti-HA antibodies 12CA5 and 16B12 (Covance) were used at 1:1000.
qBrdU-seq
Genomic DNA isolation has been described previously (Viggiani et al. 2010). Genomic DNA was sheared to ∼300 bp average using a Covaris S2 instrument. One microgram sheared DNA was end-repaired and ligated with a barcoded, Illumina-compatible adapter as described (Dunham and Friesen 2013), using the NEBNext Ultra End Repair/dA-Tailing Module (E7442) and NEBNext Ultra Ligation Module (E7445). Adapter-ligated genomic DNA was purified and its concentration was determined by spectroscopy (Nanodrop). Equal amounts of multiple such barcoded DNA samples were pooled; 20 ng were set aside as “Input” and 1 µg of this pool was subject to BrdU-IP as described previously except that salmon sperm DNA was omitted (Viggiani et al. 2010). The IP and Input DNA samples were PCR-amplified separately with indexed Illumina-compatible primers. Amplified DNA was isolated using AMPure beads (Beckman Coulter, A63880) and validated and quantified on a Bioanalyzer (Agilent), and pooled. DNA sequencing (50 bp paired-end) was performed on a NextSeq instrument (Illumina) by the NextGen Sequencing Core of the USC Norris Cancer Center.
Analysis of qBrdU-seq data
Barcodes were split using the fastq-multx barcode splitter (http://code.google.com/p/ea-utils). Sequence libraries were aligned to S. cerevisiae genome release r.64 using Bowtie 2 (Langmead and Salzberg 2012). The first 5 bp were trimmed from the 5′ end to account for the barcode and allow for proper alignment. Aligned sequences were sorted and binned into 50-bp nonoverlapping bins (Li et al. 2009; Quinlan and Hall 2010), normalized by dividing each IP bin by its corresponding Input bin, yielding an IP:Input ratio, which was median-smoothed over a 500- or 2000-bp window for HU or time-course experiments, respectively. Experimental replicates were averaged and data were arbitrarily scaled, setting “1” for the maximum average WT signal by dividing all samples within a qBrdU-seq pool by the same value (i.e., the maximum average WT signal); in the time-course experiment, all time points were scaled to the maximum signal in WT among the time points. This optional scaling step was carried out only to facilitate comparisons between different qBrdU-seq experiments; it does not alter the quantitative comparison between pooled samples. BrdU peaks were called using MACS (P < 0.01) (Zhang et al. 2008). Origin peaks were subjected to Diffbind (using DESeq) analysis (adjusted P < 0.05) for calling of differential peak sizes (Anders and Huber 2010; Stark and Brown 2013), and called peaks were intersected among replicates. Only origin peaks identified within all three replicates of each strain were included in further analysis.
ChIP-chip
ChIP-chip was performed and analyzed as described previously with three independent replicates (Ostrow et al. 2014).
RNA-seq library preparation
One and one-half milliliters of culture were harvested, washed with TBS, pelleted, flash-frozen, and stored at −80°C. Total RNA was isolated using the MasterPure Yeast RNA Purification kit (Epicentre Biotechnologies, MPY03010). Poly(A) transcripts were isolated from 5 µg of total RNA using the NEB Poly(A) mRNA Magnetic Isolation kit (Epicentre, E7490S). First- and second-strand cDNA synthesis was carried out using the NEBNext FSS and SSS kits (NEB, E7525S and E6111S). cDNA was amplified by the standard Illumina protocol with inclusion of indexes for multiplexing. DNA sequencing (50 bp paired-end) was carried out on an Illumina HiSeq instrument by the FSU College of Medicine Translational Science Laboratory.
Analysis of RNA-seq data
To align reads and call differential expression of RNA transcripts, reads from two independent replicates, per condition, were first aligned using the TopHat2 sequence aligner (Kim et al. 2013) to S. cerevisiae genome release r.64 along with a known transcript file (.gtf format). Aligned reads were next subjected to the Cufflinks transcript assembly and differential expression pipeline including Cuffdiff to call differentially expressed transcripts (FDR ≤ 0.01) (Trapnell et al. 2010). Gene ontology analysis was performed with the GO term finder and GO slim mapper available at yeastgenome.org
Data access
The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE71052.
Acknowledgments
For helpful discussions and critical reading of the manuscript, we thank Drs. Joanna Haye, Simon Knott, and Zac Ostrow. For the Rnr3-OE plasmid, we thank Dr. Jeff Bachant (UC-Riverside). For assistance with high-throughput DNA sequence determination, we thank Selene Tyndale and Dr. Charles Nicolet of the NextGen Sequencing Core of the Norris Comprehensive Cancer Center at USC Keck School of Medicine, and Dr. Roger Mercer of the Translational Science Laboratory at Florida State University College of Medicine, and Dr. Michelle Arbeitman at Florida State University College of Medicine. This study was supported by National Institutes of Health (NIH) grant awards 2R01-GM065494 (to O.M.A.) and Cancer Center Support Grant P30CA014089 from the National Cancer Institute. J.M.P. received support from P50-HG002790.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.196857.115.
References
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM. 2013. Location, location, location: It's all in the timing for replication origins. Genes Dev 27: 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhoefer J, Rhind N. 2012. Replication timing and its emergence from stochastic processes. Trends Genet 28: 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- Casey L, Patterson EE, Muller U, Fox CA. 2008. Conversion of a replication origin to a silencer through a pathway shared by a Forkhead transcription factor and an S phase cyclin. Mol Biol Cell 19: 608–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabbert CD, Adjalley SH, Klaus B, Fritsch ES, Gupta I, Pelechano V, Steinmetz LM. 2015. A high-throughput ChIP-Seq for large-scale chromatin studies. Mol Syst Biol 11: 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A, Stillman B. 2007. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc Natl Acad Sci 104: 1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creager RL, Li Y, MacAlpine DM. 2015. SnapShot: origins of DNA replication. Cell 161: 418–418 e411. [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev 12: 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell 4: 983–993. [DOI] [PubMed] [Google Scholar]
- Dunham JP, Friesen ML. 2013. A cost-effective method for high-throughput construction of Illumina sequencing libraries. Cold Spring Harb Protoc 2013: 820–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BM, Brewer BJ, Reynolds AE, Fangman WL. 1991. A yeast origin of replication is activated late in S phase. Cell 65: 507–515. [DOI] [PubMed] [Google Scholar]
- Friedman KL, Brewer BJ, Fangman WL. 1997. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells 2: 667–678. [DOI] [PubMed] [Google Scholar]
- Futcher AB, Cox BS. 1984. Copy number and the stability of 2-μm circle-based artificial plasmids of Saccharomyces cerevisiae. J Bacteriol 157: 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. 2003. Global analysis of protein expression in yeast. Nature 425: 737–741. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2: 31–34. [DOI] [PubMed] [Google Scholar]
- Gilbert DM. 2010. Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet 11: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. 2012. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191: 33–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J, Masukata H. 2009. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat Cell Biol 11: 357–362. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavare S, Aparicio OM. 2012. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 148: 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Heck DJ, Nomura M, Horiuchi T. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev 12: 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11: 319–324. [DOI] [PubMed] [Google Scholar]
- Kwan EX, Foss EJ, Tsuchiyama S, Alvino GM, Kruglyak L, Kaeberlein M, Raghuraman MK, Brewer BJ, Kennedy BK, Bedalov A. 2013. A natural polymorphism in rDNA replication origins links origin activation with calorie restriction and lifespan. PLoS Genet 9: e1003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. 2010. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev 24: 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PC, Chretien L, Cote J, Kelly TJ, Forsburg SL. 2011. S. pombe replication protein Cdc18 (Cdc6) interacts with Swi6 (HP1) heterochromatin protein: region specific effects and replication timing in the centromere. Cell Cycle 10: 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke C, Klipp E, Lehrach H, Barberis M, Krobitsch S. 2013. Fkh1 and Fkh2 associate with Sir2 to control CLB2 transcription under normal and oxidative stress conditions. Front Physiol 4: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P. 2011. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J 30: 4805–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V, Hailey DW, Pot I, Givan SA, Hyland KM, Cagney G, Fields S, Davis TN, Hieter P. 2002. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev 16: 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Murakami H, Aiba H, Nakanishi M, Murakami-Tonami Y. 2010. Regulation of yeast forkhead transcription factors and FoxM1 by cyclin-dependent and polo-like kinases. Cell Cycle 9: 3233–3242. [DOI] [PubMed] [Google Scholar]
- Natsume T, Muller CA, Katou Y, Retkute R, Gierlinski M, Araki H, Blow JJ, Shirahige K, Nieduszynski CA, Tanaka TU. 2013. Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol Cell 50: 661–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrow AZ, Nellimoottil T, Knott SR, Fox CA, Tavare S, Aparicio OM. 2014. Fkh1 and Fkh2 bind multiple chromosomal elements in the S. cerevisiae genome with distinct specificities and cell cycle dynamics. PLoS One 9: e87647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PK, Kommajosyula N, Rosebrock A, Bensimon A, Leatherwood J, Bechhoefer J, Rhind N. 2008. The Hsk1(Cdc7) replication kinase regulates origin efficiency. Mol Biol Cell 19: 5550–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peace JM, Ter-Zakarian A, Aparicio OM. 2014. Rif1 regulates initiation timing of late replication origins throughout the S. cerevisiae genome. PLoS One 9: e98501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postnikoff SD, Malo ME, Wong B, Harkness TA. 2012. The yeast forkhead transcription factors fkh1 and fkh2 regulate lifespan and stress response together with the anaphase-promoting complex. PLoS Genet 8: e1002583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Brewer BJ, Fangman WL. 1997. Cell cycle–dependent establishment of a late replication program. Science 276: 806–809. [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. 2001. Replication dynamics of the yeast genome. Science 294: 115–121. [DOI] [PubMed] [Google Scholar]
- Rhind N, Gilbert DM. 2013. DNA replication timing. Cold Spring Harb Perspect Biol 5: a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Sharma K, Diffley JF. 1999. Activation of dormant origins of DNA replication in budding yeast. Genes Dev 13: 2360–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM. 2007. Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz DA, Orci L, Ravazzola M, Swaroop A, Kaiser CA. 1995. Human SEC13Rp functions in yeast and is located on transport vesicles budding from the endoplasmic reticulum. J Cell Biol 128: 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherriff JA, Kent NA, Mellor J. 2007. The Isw2 chromatin-remodeling ATPase cooperates with the Fkh2 transcription factor to repress transcription of the B-type cyclin gene CLB2. Mol Cell Biol 27: 2848–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith OK, Aladjem MI. 2014. Chromatin structure and replication origins: determinants of chromosome replication and nuclear organization. J Mol Biol 426: 3330–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark R, Brown R. 2013. DiffBind: differential binding analysis of ChIP-Seq peak data. bioconductor.org, University of Cambridge, Cancer Research UK–Cambridge Institute; http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf. [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. 2011. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol 21: 2055–2063. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veis J, Klug H, Koranda M, Ammerer G. 2007. Activation of the G2/M-specific gene CLB2 requires multiple cell cycle signals. Mol Cell Biol 27: 8364–8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viggiani CJ, Aparicio OM. 2006. New vectors for simplified construction of BrdU-Incorporating strains of Saccharomyces cerevisiae. Yeast 23: 1045–1051. [DOI] [PubMed] [Google Scholar]
- Viggiani CJ, Knott SR, Aparicio OM. 2010. Genome-wide analysis of DNA synthesis by BrdU immunoprecipitation on tiling microarrays (BrdU-IP-chip) in Saccharomyces cerevisiae. Cold Spring Harb Protoc 2010: pdb.prot5385. [DOI] [PubMed] [Google Scholar]
- Voth WP, Yu Y, Takahata S, Kretschmann KL, Lieb JD, Parker RL, Milash B, Stillman DJ. 2007. Forkhead proteins control the outcome of transcription factor binding by antiactivation. EMBO J 26: 4324–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Nurse P. 2009. Establishing the program of origin firing during S phase in fission yeast. Cell 136: 852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM. 2001. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294: 2357–2360. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Hori Y, Shinomiya T, Obuse C, Tsurimoto T, Yoshikawa H, Shirahige K. 1997. The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells 2: 655–665. [DOI] [PubMed] [Google Scholar]
- Yang SC, Rhind N, Bechhoefer J. 2010. Modeling genome-wide replication kinetics reveals a mechanism for regulation of replication timing. Mol Syst Biol 6: 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Bacal J, Desmarais D, Padioleau I, Tsaponina O, Chabes A, Pantesco V, Dubois E, Parrinello H, Skrzypczak M, et al. 2014. The histone deacetylases Sir2 and Rpd3 act on ribosomal DNA to control the replication program in budding yeast. Mol Cell 54: 691–697. [DOI] [PubMed] [Google Scholar]
- Zakian VA, Brewer BJ, Fangman WL. 1979. Replication of each copy of the yeast 2 micron DNA plasmid occurs during the S phase. Cell 17: 923–934. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yu Z, Fu X, Liang C. 2002. Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast. Cell 109: 849–860. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Nellimoottil T, Peace JM, Knott SR, Villwock SK, Yee JM, Jancuska JM, Rege S, Tecklenburg M, Sclafani RA, et al. 2013. The level of origin firing inversely affects the rate of replication fork progression. J Cell Biol 201: 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]