Abstract
P53 is a transcription factor highly inducible by many stress signals such as DNA damage, oncogene activation, and nutrient deprivation. Cell-cycle arrest and apoptosis are the most prominent outcomes of p53 activation. Many studies showed that p53 cell-cycle and apoptosis functions are important for preventing tumor development. p53 also regulates many cellular processes including metabolism, antioxidant response, and DNA repair. Emerging evidence suggests that these noncanonical p53 activities may also have potent antitumor effects within certain context. This review focuses on the cell-cycle arrest and apoptosis functions of p53, their roles in tumor suppression, and the regulation of cell fate decision after p53 activation.
p53 most notably induces cell-cycle arrest and cell death, thereby blocking tumor progression. But p53 also regulates metabolism, antioxidant response, and DNA repair, which may block tumor initiation.
Cell-cycle arrest and apoptosis are the most noticeable biological outcomes of p53 activation in cell culture and animal experiments. The seminal finding of p53 as an inhibitor of oncogene-mediated transformation in foci formation is likely the result of its cell-cycle arrest or apoptosis activities (Finlay et al. 1989). The mammalian p53 DNA-binding domain has marginal thermostability, which facilitates the identification of temperature-sensitive mutants and provides a powerful tool for controlling p53 function. The ability of activated p53 to induce cell-cycle arrest in rat embryo fibroblasts and apoptosis in a leukemia cell line were discovered using temperature-sensitive p53–135V mutant (Martinez et al. 1991; Yonish-Rouach et al. 1991). Subsequent work using viral vector-mediated gene delivery and inducible expression validated these as the most prominent effects of p53 activation in cell culture.
Given the ability of p53 to induce both cell-cycle arrest and cell death, the regulation of cell fate decision has been the focus of numerous studies (Vousden and Prives 2009). This is a topic with significant clinical relevance because p53-mediated apoptosis in normal tissues are involved in chemotherapy toxicity, ischemia, and neurodegenerative diseases such as Alzheimer’s and Parkinson’s (Checler and Alves da Costa 2014). Induction of p53-mediated apoptosis in tumor cells is considered a desirable outcome of cancer therapy, whereas induction of cell-cycle arrest may interfere with drugs that target mitosis and reduce the efficacy of DNA-damaging drugs. Cell fate decision in response to p53 activation is determined at multiple levels, as discussed below.
MECHANISM OF CELL-CYCLE ARREST BY p53
Cell-cycle arrest by p53 is mainly mediated by the transcriptional activation of p21/WAF1 (el-Deiry et al. 1993; Harper et al. 1993). p53 binds to two sites 2.4 kb and 1.4 kb upstream of the p21 promoter. The 5′ site on the p21 promoter is one of the strongest p53-binding sites analyzed, with dissociation constant of ∼5 nm (Weinberg et al. 2005). The p21 mRNA is highly induced after p53 activation, and is the first p53 target gene isolated using an unbiased subtractive hybridization method (el-Deiry et al. 1993). p21 binds to cyclin E/Cdk2 and cyclin D/Cdk4 complexes to cause G1 arrest in the cell cycle (Harper et al. 1993). The inhibition of Cdk2 and Cdk4 by p21 blocks pRb phosphorylation, promotes pRb binding to E2F1, and promotes transcription silencing of E2F1 targets critical for DNA replication and cell-cycle progression. p21 also interacts with proliferating cell nuclear antigen (PCNA) and inhibits DNA replication in vitro, which may contribute to its cell-cycle arrest activity (Luo et al. 1995). p21-null mouse embryonic fibroblasts (MEFs) showed deficiency in cell-cycle arrest after DNA damage, indicating that it is a major mediator of cell-cycle arrest by p53 (Deng et al. 1995). However, p21-null cells are not completely defective for G1 arrest, suggesting that other p53 target genes also contribute to the growth arrest (Brugarolas et al. 1995).
p53 activation also arrests cells at the G2/M phases. Although p21 can also inhibit cyclin B/Cdc2 to inhibit cell-cycle progression through mitosis, other p53 target genes, such as 14-3-3σ, may participate in blocking G2/M transition (Martin-Caballero et al. 2001). p53 repression of the cdc25C promoter has also been shown to promote G2/M arrest after DNA damage (St Clair et al. 2004). Recent studies suggest that induction of microRNA mir34a contributes to growth arrest by p53, presumably by transcription silencing of multiple target genes (Chang et al. 2007; He et al. 2007; Tarasov et al. 2007). However, knockout of the mir34 gene family in mice did not affect p53-mediated arrest and apoptosis, suggesting that mir34 function in vivo is limited or redundant (Concepcion et al. 2012; Jain and Barton 2012).
CELL-CYCLE CHECKPOINT FUNCTIONS OF p53
In mammalian cells, p53-mediated arrest provides important cell-cycle checkpoint functions. DNA double-strand breaks by ionizing irradiation activates the ataxia telangiectasia mutated (ATM) kinase and inhibits MDM2 E3 ligase activity through phosphorylation, causing rapid accumulation of p53 and induction of p21 (Chen 2012). By arresting cells at the G1 phase and allowing time for the repair of potentially lethal double-strand breaks, p53 maintains chromosomal integrity and improves the survival of damaged cells. In addition to enforcing a cell-cycle checkpoint, p53 also regulates a group of genes involved in DNA recombination and repair (Gatz and Wiesmuller 2006). Furthermore, p53 regulates genes involved in heterochromatin formation to facilitate the timely repair of DNA strand breaks in constitutive heterochromatin regions. p53-null cells have deficiencies in the repair of double-strand breaks in heterochromatin and have reduced long-term viability after ionizing irradiation (Zheng et al. 2013).
p53 also has a postmitotic checkpoint function critical for preventing DNA re-replication when the mitotic spindle is disrupted. Treatment of normal mouse fibroblasts with mitotic spindle-destabilizing drugs, such as nocodazole and colcemid, causes them to exit mitosis and arrest at the G1 phase with 4N DNA content. In contrast, p53-deficient cells often initiate new rounds of DNA replication when cell division is blocked by a spindle poison, producing polyploid cells with 8N and higher DNA complement (Cross et al. 1995). As expected, p21 is also a critical mediator of this p53 postmitotic checkpoint function and is required for preserving chromosome stability (Lanni and Jacks 1998; Barboza et al. 2006). Absence of p53 increases chromosome instability, which was revealed by the rapid accumulation of aneuploid cells when fibroblasts from p53-null mice were cultured in vitro (Harvey et al. 1993). Although an absence of p53 per se is not sufficient to cause aneuploidy in vivo, cells without p53 are prone to accumulate abnormal chromosomes after oncogene activation (Duensing and Duensing 2005).
INDUCTION OF CELLULAR SENESCENCE BY p53
The cell-cycle arrest triggered by DNA damage is reversible after repair of DNA and down-regulation of p53. In contrast, excessive division of human fibroblasts results in p53-mediated irreversible arrest called replicative senescence. This is partly owing to chronic p53 activation triggered by telomere erosion and DNA damage signaling. Senescent cells have unique features such as large cell size, active autophagy, high lysosomal SA-β-gal activity, and secretion of proinflammatory cytokines (Campisi 2005). DNA-damaging treatment of tumor cells or activation of oncogenes in normal fibroblasts also induces cell-cycle arrest with features similar to replicative senescence, which is termed premature senescence. The ability of p53 to induce cell-cycle arrest is critical for senescence, because p21 knockout prevents senescence induction by p53 (Brown et al. 1997). In fact, sustained overexpression of p21 alone for 3–5 days is sufficient to induce senescence in tumor cells (Chang et al. 1999b). DNA damage also induces premature senescence in tumor cells, which can remain arrested for weeks after removal of the DNA-damaging drug (Chang et al. 1999a). Despite the general perception that senescence is irreversible, inactivation of p53 can cause cell-cycle reentry in senescent cells (Beausejour et al. 2003). Therefore, senescence is a unique state of cell-cycle arrest that is highly stable, but is not completely irreversible.
Several mechanisms may act cooperatively to maintain the stable cell-cycle arrest in senescent cells. Senescent cells have extensive formation of heterochromatin on E2F1 target genes (Narita et al. 2003). Many heterochromatin-inducing proteins can bind to methylated histones and promote the methylation of adjacent nucleosomes, a property well suited for maintaining self-sustaining gene repression (Bonasio et al. 2010). Therefore, when cells are subjected to persistent p21 expression and pRb activation, the level of heterochromatin marks on E2F1 target genes may exceed the threshold required for establishing a self-sustaining positive feedback. Furthermore, the expression of cytokines as part of the senescence-associated secretory phenotype (SASP) also has autocrine functions that reinforce senescence (Chien et al. 2011). The formation of stable heterochromatin in senescent cells may also play a role in generating persistent DNA damage signals, resulting in constitutive p53 activation (Rodier et al. 2011).
CELL FATE DECISION BETWEEN ARREST AND SENESCENCE
In cell culture models, induction of senescence often involves subjecting cells to chronic stresses such as telomere shortening, oncogene activation, or low-dose DNA-damaging drugs for 3 to 5 days. A certain amount of time may be needed to establish robust heterochromatin on cell-cycle genes and epigenetically reprogram senescence-related gene expression. Although the presence of p53 facilitates senescence induction by stress, p53 activation alone is not sufficient for inducing senescence in most conditions. In fact, it is somewhat disappointing from a therapeutic perspective that p53 activation by the MDM2 inhibitor Nutlin only induces reversible arrest in many tumor cells (Huang et al. 2009). Furthermore, Nutlin blocks senescence induction by ectopic p21 expression, suggesting that p53 has both pro- and antisenescence activities (Demidenko et al. 2010).
The functional status of several pathways and their cross talk with p53 may affect the decision between reversible arrest and senescence. In addition to p53, the activities of pRb, NF-κB, and mTOR are important for the induction of senescence. pRb promotes heterochromatin formation on E2F1 target genes, and NF-κB is required for proinflammatory cytokine expression in senescent cells. Activation of p53 can antagonize the functions of both pRb and NF-κB. For example, p53 has complex cross talk with the pRb pathway through MDM2-pRb binding (Sdek et al. 2005). Nutlin treatment causes down-regulation of pRb in a p53/MDM2-dependent fashion, which may limit formation of senescence-associated heterochromatin and increase reversibility (Du et al. 2009; Huang et al. 2009). p53 also inhibits NF-κB and mTOR, both required for the initiation of senescence (Feng et al. 2005; Dey et al. 2007). Inhibition of mTOR using rapamycin blocks senescence and keeps cells in a state of reversible quiescence after p21 overexpression (Demidenko et al. 2009). p53 also represses the expression of histone H3K9 methylase SUV39H1 and induces histone demethylase JMJD2d, which may prevent the establishment of stable gene silencing (Zheng et al. 2013). Therefore, cellular context and pathway cross talk may determine whether cells commit to irreversible cell-cycle arrest.
MECHANISM OF APOPTOSIS INDUCTION BY p53
On p53 activation, certain cell types (leukemia or transformed fibroblasts) undergo predominantly apoptosis instead of cell-cycle arrest. The apoptosis function of p53 is evolutionarily conserved in Drosophila and Caenorhabditis elegans (Derry et al. 2001; Sutcliffe and Brehm 2004). In response to DNA damage, the Drosophila proapoptotic genes Reaper, Hid, and Grim are transcriptionally induced by p53. These genes are sufficient to induce apoptosis by inhibiting the caspase inhibitor Diap1, causing the activation of initiator caspase Dronc and effector caspases DrICE and Dcp-1 (Sutcliffe and Brehm 2004; Xu et al. 2009). Unlike mammalian p53, Drosophila p53 does not induce cell-cycle arrest and does not activate the Drosophila p21 homolog Dacapo (Brodsky et al. 2004). Similarly, C. elegans p53 homolog Cep-1 causes apoptosis by inducing two BH3 domain proteins Egl-1 and Ced-13, but does not regulate the expression of p21 homolog cki-1 (Greiss et al. 2008). Presumably the p53 cell-cycle arrest function is not needed for these organisms because most cells in the adult body are postmitotic.
In mammalian cells, p53 induces a large number of genes involved in various steps of apoptosis signaling and execution (Riley et al. 2008). These include BH3 domain-only proapoptotic proteins (Puma, Noxa, Bad, Bax, Bak, p53AIP1); death receptors (Fas, Dr4, Killer/Dr5); and apoptosis execution factors (Apaf1, caspase 6, Bnip3L). Induction of Fas and Dr5 activates the extrinsic apoptosis pathway by promoting death receptor dimerization, recruitment and activation of procaspase 8, and activation of executioner caspase 3 and 7. Induction of the BH3-only proteins by p53 causes mitochondrial outer membrane permeabilization (MOMP), which is a key step in the intrinsic apoptosis pathway (Tait and Green 2010).
MOMP is determined by the balance of pro- and antiapoptotic Bcl2 family proteins. Central to this process, Bax and Bak are activated by interaction with tBid and Bim, insert into the outer mitochondrial membrane, and oligomerize to form pores to allow the release of cytochrome c and other proteins such as Smac and Omi from the intermembrane space. Cytochrome c binds to adenosine triphosphate (ATP) and Apaf1, promoting Apaf1 oligomerization to form the apoptosome complex. The apoptosome recruits and activates procaspase 9, which further activates executioner caspase 3 and 7 to induce cell death. Smac and Omi bind to and deactivate the IAP proteins that are inhibitors of caspases, therefore facilitating the commitment to apoptosis. The antiapoptotic proteins Bcl2, BclXL, and Mcl1 prevent the activation of Bax and Bak by sequestering Bim, whereas the derepressors, such as Puma and Noxa, promote apoptosis by releasing Bax, tBid, or Bak from inhibition by BclXL and Mcl1. Therefore, p53 induction of Puma, Bad, Bax, Noxa, Bak, and Apaf1 target multiple stages of the MOMP regulatory mechanism. The role of Puma has been tested in knockout mouse and is critical for p53-mediated apoptosis in thymus, spleen, and intestine after ionizing irradiation (Jeffers et al. 2003). Puma/Noxa double-null mice are more apoptosis resistant than Puma-null mice, suggesting that multiple p53 target genes act cooperatively (Michalak et al. 2008).
p53 FUNCTIONS AT THE MITOCHONDRIA
Several reports suggest that p53 also has transcription-independent apoptotic mechanisms by directly promoting MOMP at the mitochondria. A fraction of p53 protein can be detected at the mitochondria following DNA damage, preceding the loss of mitochondrial transmembrane potential and caspase activation (Marchenko et al. 2000; Mihara et al. 2003). Direct targeting of p53 to the mitochondria by fusion to a signal peptide was sufficient to induce apoptosis without activating transcription. Biochemically, p53 can bind to Bcl2 and BclXL, promoting the release and activation of Bax and Bak. The p53 core domain is involved in binding to BclXL via charge-mediated interaction (Petros et al. 2004; Sot et al. 2007). Other reports showed that the p53 core domain directly interacts with proapoptotic Bak, relieving Bak from inhibitory complexes with Mcl1 (Leu et al. 2004; Pietsch et al. 2008). These direct mitochondrial activities of p53 are inactivated by tumor-derived mutations in the core domain, suggesting that they require similar structural features of the core domain as for DNA binding (Tomita et al. 2006). Interestingly, a recent study suggests that p53 not only regulates mitochondria outer membrane permeability, it also induces permeabilization of the inner mitochondria membrane during oxidative stress (Vaseva et al. 2012). Mitochondrial-located p53 interacts with cyclophilin D and induces opening of the inner membrane permeability transition pore. Permeabilization of the mitochondria inner membrane dissipates the cross membrane potential, stopping oxidative phosphorylation and ATP production, resulting in necrosis.
Although p53 has transcription-independent MOMP-inducing activities, its significance relative to transcription remains unknown. The apoptosis defect of the Puma knockout mouse and the p53 transactivation domain mutant mouse suggests that transcription is indispensable for apoptosis induction (Jeffers et al. 2003; Brady et al. 2011). A study aimed at addressing this question showed that a subset of cytosolic p53 is sequestered in an inactive complex by BclXL. Stress activation of p53 induces Puma expression, which forms Puma-BclXL complex and liberates the cytosolic p53 to activate Bax in the cytoplasm (Chipuk et al. 2005). Therefore, the transcription-dependent and independent p53 activities may cooperate to induce apoptosis.
These findings suggest that p53 has evolved to function at the mitochondria in mammalian cells while still maintaining its role as a transcription factor in the nucleus. The structural flexibility of p53 may provide the basis for such functional diversification. The p53 core domain has poor thermostability and promiscuously interacts with multiple proteins, which is a feature of intrinsically disordered proteins (Friedler et al. 2005; Dunker et al. 2008). It is possible that at the higher body temperature of mammals p53 behaves similarly to intrinsically disordered proteins and gains new functions by interacting with BclXL or Bak without losing DNA-binding activity. The mitochondria permeabilization and cytochrome c–mediated mechanism of apoptosis is unique to vertebrates and absent in Drosophila and C. elegans (Oberst et al. 2008). Because p53 is not essential for cell survival, it may be able to evolve quickly and establish cross talk with the mitochondria cell death pathway in vertebrates.
REGULATION OF p53 APOPTOTIC FUNCTION BY POSTTRANSLATIONAL MODIFICATION
Induction of apoptosis in tumor cells is highly desirable during cancer chemotherapy, whereas p53-mediated apoptosis in normal tissues is an important cause of treatment side effects. Therefore, gaining the ability to fine-tune p53 apoptosis and cell-cycle arrest activities has significant therapeutic potential. Several mechanisms have been identified that alter p53 transcriptional activity toward cell-cycle and apoptosis target genes and shift the outcome of p53 activation.
Of the many posttranslational modifications on p53, phosphorylation and acetylation of certain residues have significant impact on cell fate decision. Phosphorylation of p53 S46 promotes the induction of proapoptotic target gene p53AIP1, thus favoring the induction of apoptosis (Oda et al. 2000). S46 is phosphorylated by the HIPK2 kinase after severe DNA damage (Puca et al. 2010). Several other kinases, including DYRK2, AMP-activated protein kinase (AMPK), p38, and protein kinase C (PKC)-δ, also promote S46 phosphorylation (Perfettini et al. 2005; Yoshida et al. 2006; Taira et al. 2007; Okoshi et al. 2008). Mice with heterozygous loss of HIPK2 and p53 develop more lymphomas after irradiation, suggesting that HIPK2 functionally cooperates with p53 (Mao et al. 2012). However, lymphomas developed in p53-null mice still undergo HIPK2 deletion, indicating that HIPK2 also has p53-independent mechanisms of tumor suppression. The physiological function of p53 S46 phosphorylation is still unclear, because mutation analysis of an equivalent site in the mouse has not been reported.
p53 is acetylated on multiple residues at the central DNA-binding domain and carboxy-terminal tail. Acetylation of K120 in the DNA-binding domain by acetylases Tip60 or hMOF specifically promotes apoptosis (Sykes et al. 2006; Tang et al. 2006). Mice expressing p53-K117R (equivalent to K120R in human p53) were defective for apoptosis without affecting the regulation of p21 (Li et al. 2012). The intuitive expectation that acetylation of p53 core domain regulates its target gene selectivity by affecting DNA binding appears to be incorrect. Instead, p53 acetylation regulates p53–MDM2 interaction and recruitment of MDM2 to DNA, suggesting a more complex mechanism that needs further investigation (Tang et al. 2008). The p53 carboxyl terminus is also modified by methylation on several arginine and lysine residues. The ΔCTD mouse that lacks the lysine-rich carboxyl terminus has increased p53 activity in some tissues (Hamard et al. 2013). Whether these modifications regulate cell fate decision between apoptosis and arrest is not clear.
REGULATION OF APOPTOSIS BY p53-BINDING PROTEINS
The ability of p53 to induce apoptosis target genes is regulated by the ASPP family proteins. ASPP1/2 proteins interact with the p53 core domain and stimulate p53 binding to the promoters of apoptotic genes (Samuels-Lev et al. 2001). The iASPP protein competes with ASPP using a similar p53-binding domain, and inhibits p53-mediated apoptosis (Bergamaschi et al. 2003). Interestingly, the p53-R72 polymorphic allele has lower affinity for iASPP binding, which correlates with stronger apoptosis activity than the P72 allele (Bergamaschi et al. 2006). Furthermore, phosphorylation of p53 S46 stimulates the recruitment of Pin1 prolyl isomerase and promotes the dissociation of iASPP to facilitate apoptosis (Mantovani et al. 2007). The cocrystal structure of p53BP2 (a fragment of ASPP2) with p53 core domain showed that ASPP2 occupies the DNA-binding surface of p53, which paradoxically suggests that ASPP2 should inactivate p53 (Gorina and Pavletich 1996; Samuels-Lev et al. 2001). Later work revealed that, in addition to binding the p53 core domain, ASPP also binds to the p53 proline-rich region (Bergamaschi et al. 2006). This multisite interaction may allow conformational switch in the complex to enhance p53 DNA binding. Antibodies that bind to the amino terminus of p53 activate core domain binding to DNA, suggesting that the amino terminus has autoinhibitory function (Cain et al. 2000). It is possible that in the ASPP-p53 complex, dynamic ASPP binding to the proline-rich region neutralizes the autoinhibition by the amino terminus and activates p53 DNA binding.
DIFFERENCES IN p53-BINDING SITES DETERMINE APOPTOSIS THRESHOLD
The fact that p53 modifications selectively alter apoptosis and cell-cycle gene expression suggests that intrinsic differences in these target promoters are important in setting the apoptosis threshold. In mammals, the p53-binding sites in cell-cycle target gene promoters are better conserved and have less deviation from the optimal sequence compared with apoptosis genes. This finding suggests that p53 regulation of cell-cycle genes has been consistently subjected to purifying selection through evolution, whereas apoptosis gene response to p53 is being selected by divergent requirements to suite survival in different environments (Horvath et al. 2007).
Biochemically, p53 targets involved in cell-cycle and DNA repair often have high-affinity binding sites, whereas the binding sites on apoptosis targets are more variable in affinity (Weinberg et al. 2005). p53 binding to the degenerate weak sites is more dependent on cooperative binding through tetramerization of the core domain (Schlereth et al. 2010). As such, at low p53 concentration, the occupancy on cell-cycle target sites will be higher than apoptosis genes, whereas a high p53 level is needed for significant binding and activation of apoptosis genes. This is consistent with the chromatin immunoprecipitation analysis of p53 occupancy on these genes (Kaeser and Iggo 2002). Reporter gene assays also showed that cell-cycle target promoters are generally more responsive to p53 compared with apoptosis gene promoters (Qian et al. 2002). Furthermore, cell-cycle target-binding sites can act alone without adjacent sequences, whereas additional promoter sequences in the apoptotic gene promoters are needed for strong p53 response, indicating that other inputs are required to trigger apoptosis (Qian et al. 2002). Corroborating these findings, experiments using inducible p53 showed that, in the absence of DNA damage, low-level p53 induced arrest, whereas high-level p53 induced apoptosis (Chen et al. 1996).
The difference in p53-binding affinity and involvement of adjacent promoter sequences may also alter the kinetics of transcriptional response. After p53 activation, the apoptosis targets tend to be induced with significantly slower kinetics compared with cell-cycle targets. For example, the induction of p21 and Gadd45 by p53 peaked 2–6 h after irradiation, whereas Bax induction peaked after 12–24 h (Purvis et al. 2012). The mRNA of p53AIP1 and Pig3 were also induced with 16- to 20-h delay compared with p21 after DNA damage (Oda et al. 2000; Szak et al. 2001). This feature may allow a cell-fate decision based on the duration of stress and p53 activation.
CELL TYPE AND ONCOGENE ACTIVITY DETERMINE APOPTOSIS SENSITIVITY
Highly proliferative cell types such as hematopoietic cells, germ cells, and intestinal crypt cells in the mouse are most susceptible to p53-mediated apoptosis. Apoptosis occurs whether p53 is activated by ionizing irradiation (thus accompanied by DNA damage signaling) or by eliminating MDM2 without additional stress (Ringshausen et al. 2006). The latter scenario underscores the dominance of cell-intrinsic properties in apoptosis sensitivity to p53 under physiological setting. This leads to the notion that p53 prevents tumor development by eliminating damaged cells that have high proliferation potential. Radiation-sensitive tissues in the mouse express high levels of p53 protein and mRNA compared with radioresistant tissues, suggesting that developmental and tissue-specific regulation of p53 promoter determines apoptosis sensitivity (MacCallum et al. 1996; Komarova et al. 1997). Ironically, much more attention has been directed to studying post-translational regulation of p53 stability because it is the major mechanism of p53 induction after stress in cell culture, whereas in normal tissues the p53 promoter may be a more important arbitrator of life and death.
In addition to the control of p53 expression by cell type and proliferation status, oncogenic signaling lowers the apoptotic threshold by direct transcription activation or repression of Bcl2 family proteins, several of which are targets of E2F1 (Croxton et al. 2002; Hershko and Ginsberg 2004). A hardwired connection between cell proliferation and apoptosis provides a safeguard against damaged, potentially malignant cells. The decisive role of oncogene activity was illustrated by the dramatic conversion of p53-mediated arrest to apoptosis after ectopic expression of E2F1 in mouse fibroblasts (Wu and Levine 1994).
THE ROLES OF ARREST AND APOPTOSIS IN TUMOR SUPPRESSION
Numerous studies have investigated the roles of cell-cycle arrest and apoptosis in tumor suppression by p53, as these are the most intuitive antitumor mechanisms. As we discuss experiments performed in different mouse models, it is noteworthy that the term “tumor suppression” was used to describe at least three different activities: (1) inhibition of spontaneous tumor development in normal mice, (2) delaying the progression of tumors in mice expressing activated oncogenes, and (3) inducing regression of tumors that are fully developed. Such classification will be helpful in reconciling some seemingly conflicting results.
The p53-null mice are prone to develop spontaneous thymic lymphomas. The ability of thymocytes to undergo apoptosis after irradiation or etoposide treatment was dependent on p53, suggesting that apoptosis is involved in suppressing thymic lymphomas (Clarke et al. 1993). Another early finding that implicated a role of p53-mediated apoptosis in tumor suppression was observed in the choroid plexus brain tumors induced by SV40 T antigen mutant defective for p53 binding. Unlike wild-type T antigen that induced rapid-growing tumors, the T antigen mutant without p53-binding domain induced slow-growing tumors with frequent apoptosis. Deletion of p53 in the mutant T antigen tumors resulted in rapid growth and reduced apoptosis, suggesting that p53 limits tumor expansion by inducing apoptosis (Symonds et al. 1994).
More recent studies attempted to use p53 mutants with selective functional defects to address the role of apoptosis in tumor suppression. Most p53 core domain mutations identified in human tumors completely abrogate both cell-cycle and apoptosis activities. However, several p53 mutants found in tumors or the germline of Li–Fraumeni patients have partial transcriptional activity. These mutants (e.g., R175P, E180K, and R181L) fail to activate apoptosis targets but retain p21 induction, suggesting an important role of apoptosis in blocking tumor development in the human setting (Rowan et al. 1996; Schlereth et al. 2010).
When the human-derived p53 mutations were introduced into the mouse germline, the p53-R172P (equivalent of human R175P) mutant mice were largely protected from spontaneous thymic lymphomas, suggesting that apoptosis was not needed for tumor suppression (Liu et al. 2004). However, p53-R172P only provided partial protection against Eµ-Myc-induced B-cell lymphoma, suggesting that apoptosis was important in this context (Post et al. 2010). The p53-E177R mutant (equivalent to human E180R) is deficient in cooperative DNA binding and does not induce apoptosis, but remains able to induce cell-cycle arrest, senescence, and regulate metabolic target genes. The p53-E177R mice were also quite well protected from spontaneous thymic lymphomas, and tumors that arose in other organs often remained benign, suggesting that apoptosis is not needed to suppress thymic lymphoma (Timofeev et al. 2013). Therefore, the pattern that emerges from these findings is that protection from spontaneous thymic tumor development does not require the full spectrum of p53 activities.
Genetic analysis using p53 missense mutants are not without caveats. Although certain p53 mutants may show severe defects in apoptosis without losing cell-cycle arrest, in reality all p53 activities are inevitably compromised to various degrees by the mutation. Therefore, it is unlikely that a clean separation of apoptosis and cell-cycle functions can be achieved using p53 point mutants. To this end, a different strategy showed that when apoptosis was blocked in the Eµ-Myc mouse model by expression of Bcl2, the selection pressure to inactivate p53 during lymphoma development was eliminated, arguing that in this model apoptosis by p53 is important for blocking tumor progression (Schmitt et al. 2002). Furthermore, Eµ-Myc-induced lymphomas were accelerated by knockout of Puma, suggesting that p53 blocks lymphoma development through apoptosis (Garrison et al. 2008).
p53-mediated premature senescence, which is a form of permanent cell-cycle arrest, is a potent antitumor mechanism in cells that are resistant to apoptosis. This effect was illustrated by blocking the apoptosis pathway in Myc-induced lymphoma with Bcl2. Treatment of established lymphomas with cyclophosphamide caused p53-dependent senescence and tumor regression (Schmitt et al. 2002). Restoration of normal p53 expression in hepatocellular carcinomas induced by activated H-rasV12 mutant caused tumor regression by senescence and immune clearance (Xue et al. 2007). Restoring p53 expression in sarcomas that developed in the absence of p53 also led to senescence and tumor regression (Ventura et al. 2007; Wang et al. 2011). The results suggest that p53-mediated cell-cycle arrest and senescence are effective mechanisms for inducing regression of established tumors.
p53-MEDIATED TUMOR SUPPRESSION WITHOUT APOPTOSIS AND ARREST
Despite the established dogma, recent studies produced surprising evidence that p53 can suppress spontaneous tumor development without inducing apoptosis and cell-cycle arrest. A study identified three p53 acetylation sites (K117, K161, and K162) that are important for the transactivation of classic p53 target genes such as p21 and Puma. The p533KR mice showed defects in both apoptosis and cell-cycle arrest, but were free of early-onset tumors. The p533KR mutant retains the ability to regulate target genes involved in energy metabolism and antioxidant response (Li et al. 2012). In a different mouse model, p5325,26 transactivation domain mutant with significant deficiency in regulating the canonical targets also retained tumor suppression function (Brady et al. 2011). Microarray analysis identified a group of genes that remained responsive to p5325,26. These genes are involved in signaling, DNA repair, and cytoskeletal function and may mediate the activity of p5325,26. Furthermore, p53 was able to block spontaneous tumor development in the p21/Puma/Noxa triple knockout mouse, showing that efficient cell-cycle arrest and apoptosis are not needed for preventing the development of spontaneous tumors (Valente et al. 2013).
SUPPRESSION OF TUMOR INITIATION AND PROGRESSION REQUIRE DIFFERENT p53 ACTIVITIES
The seemingly conflicting observations described above can be reconciled if the phenotype of tumor suppression is divided into two unique activities: (1) suppression of tumor initiation, and (2) suppression of tumor progression. The mouse models suggest that the noncanonical p53 activities in metabolism, antioxidant response, and DNA repair are sufficient to prevent the appearance of spontaneous thymic tumors (presumably a model of tumor initiation). In contrast, suppression of Myc-induced lymphomas (a model of tumor progression) requires p53 apoptosis or cell-cycle arrest functions.
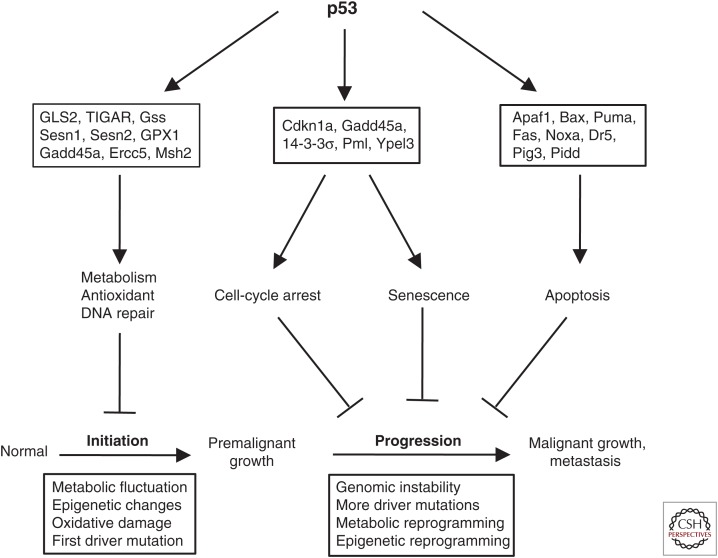
Although the exact initiating events in spontaneous tumor development are unknown, it is likely to include some of the following: (1) metabolic fluctuation from internal and external stress, (2) epigenetic reprogramming of gene expression caused by metabolic shift, (3) reactive oxygen species generation and DNA damage, and (4) activation of the first cancer-driving oncogene. The noncanonical p53 activities retained by p533KR or p5325,26 may be sufficient to regulate homeostasis and prevent the epigenetic/genetic changes that lead to abnormal proliferation, thus blocking the initiation of spontaneous tumors. For this phenotype, the apoptosis and arrest functions of p53 are dispensable because malignant cells never appeared in these animals. In contrast, tumor models expressing activated oncogenes have bypassed the early initiation steps and pose more severe challenges with rapidly growing tumors. The apoptosis and arrest functions of p53 may be necessary for stopping the rapid expansion of established tumors (Fig. 1).
Figure 1.
p53 induces different target genes to block different stages of tumor development. Metabolic/antioxidant/DNA repair genes reduce the chance of spontaneous tumor initiation by maintaining homeostasis, reducing mutation rates, blocking epigenetic changes, and promoting DNA damage repair. When tumors pass the initiation step, tumor progression is blocked by apoptosis/cell-cycle arrest/senescence activities.
Consistent with this hypothesis, p53 is able to prevent spontaneous thymic lymphoma in the p21/Puma/Noxa triple-null mice. In contrast, deletion of Puma alone in Myc-induced lymphomas accelerated tumor growth. Feeding p53-null mice with the antioxidant NAC was also sufficient to prevent the appearance of thymic lymphomas, consistent with the notion that p533KR blocks spontaneous thymic lymphoma through regulating antioxidant genes (Sablina et al. 2005; Li et al. 2012). Inhibition of aerobic glycolysis and other cancer-specific metabolic changes are promising strategies for cancer therapy. Whether the metabolic activity of p53 alone is able to induce regression of established tumors remains to be determined. However, the results from Eµ-Myc-induced lymphomas suggest that the entire spectrum of p53 activities may be needed to suppress such tumors.
THERAPEUTIC INDUCTION OF p53 APOPTOSIS AND ARREST FUNCTIONS
Animal studies showed that the apoptosis and senescence activities of p53 are potent inducers of tumor regression. A significant fraction of human tumors express wild-type p53 that is functionally compromised by binding to MDM2 or MDMX. The development of MDM2 inhibitors raised hope that stabilizing p53 in tumors may produce beneficial therapeutic effects. However, MDM2 inhibition predominantly led to reversible cell-cycle arrest in cell culture and a mouse colon tumor model (Tovar et al. 2006; Huang et al. 2009; Rigatti et al. 2012). To address this limitation, several studies showed that combining MDM2 inhibitor with DNA-damaging drugs resulted in more efficient apoptosis. A synthetic-lethal screen found that depletion of Met kinase or ATM cooperates with Nutlin to induce apoptosis (Sullivan et al. 2012). These findings suggest that it may be possible to identify new drug targets to selectively promote p53-mediated apoptosis.
An alternative strategy has been proposed to take advantage of the reversible cell-cycle arrest by MDM2 inhibitors. Blocking normal cells in G1 phase can protect them from drugs that target dividing cells (Carvajal et al. 2005; Kranz and Dobbelstein 2006). More than 50% of human tumors express mutant p53 and are not expected to respond to MDM2 inhibitors. For these patients, the MDM2 inhibitors may be useful for protecting normal tissues from chemotherapy toxicity by inducing arrest, while leaving their p53-defective tumors open to attack by the S/M phase-specific cancer drugs. However, for this strategy to be practical, the MDM2 inhibitors must display low toxicity to normal cells. The p53ERTAM mouse model showed that in the absence of MDM2, p53 is spontaneously activated and induces apoptosis in radio-sensitive organs (Ringshausen et al. 2006). Somatic knockout of MDM2 also causes apoptosis in both radio-sensitive and radio-insensitive tissues, resulting in organ damage and rapid death (Zhang et al. 2014). Clinical trials of MDM2 inhibitor RG7112 encountered toxicities consistent with the effects of p53 activation in the hematopoietic system, suggesting that the observations in mice may also be applicable to humans (Ray-Coquard et al. 2012). Clinical tests of additional classes of MDM2 inhibitors will be needed to determine the level of on-target toxicity in humans, and identify a safe therapeutic window.
Given the potential toxicity from MDM2 inhibition, targeting MDMX may provide a safer alternative. MDMX knockout in the mouse embryo causes growth arrest instead of apoptosis (Parant et al. 2001). Somatic MDMX knockout has less toxicity to many tissues, although apoptosis was still observed in certain cell types (Grier et al. 2006; Valentin-Vega et al. 2009). Reactivation of p53ERTAM in MDMX-null mice induces less tissue damage and does not cause rapid death, suggesting that MDMX inhibitors may have lower systemic toxicity (Garcia et al. 2011). Therefore, certain tumors that are addicted to MDMX, such as retinoblastomas or melanomas may be ideal targets for MDMX-specific inhibitors (Laurie et al. 2006; Gembarska et al. 2012). The current generation of MDM2 inhibitors does not specifically target MDMX (Khoo et al. 2014). The development of MDMX-specific inhibitor has not been successful, suggesting unexpected complexities of MDMX structure that require further study.
CONCLUDING REMARKS
The ability of p53 to induce cell-cycle arrest and apoptosis has significant antitumor potential that can be exploited for cancer treatment. However, cell-cycle arrest by p53 may also impede treatments with mitosis-targeting drugs, which may contribute to the absence of a general correlation between p53 mutation status and cancer-treatment response. Importantly, p53-mediated apoptosis and potential toxicity to normal organs demand significant attention as specific MDM2 inhibitors enter clinical trial. The future success of p53-targeted therapies will require better understanding of how p53 apoptosis and cell-cycle functions are regulated, developing the ability to selectively manipulate these responses and identifying the subsets of tumors that are most likely to respond to p53 activation with apoptosis or senescence.
The study of p53 tumor-suppressive mechanisms affirmed the important roles of cell-cycle arrest and apoptosis in preventing tumor progression and inducing regression of established tumors, but also suggested that the noncanonical p53 functions in metabolism and antioxidant response are sufficient for inhibiting spontaneous tumor initiation. Therefore, it will be important to further investigate the mechanisms of such “nontoxic” p53 activities, and whether the effects can be mimicked pharmacologically for the purpose of cancer prevention or treatment.
ACKNOWLEDGMENTS
The work of J.C. on p53 is supported by grants from the National Institutes of Health (CA141244, CA109636) and Florida Department of Health (4BB14). H. Lee Moffitt Cancer Center & Research Institute is supported by an NCI Comprehensive Cancer Center Support Grant (P30-CA076292).
Footnotes
Editors: Guillermina Lozano and Arnold J. Levine
Additional Perspectives on The p53 Protein available at www.perspectivesinmedicine.org
REFERENCES
- Barboza JA, Liu G, Ju Z, El-Naggar AK, Lozano G. 2006. p21 delays tumor onset by preservation of chromosomal stability. Proc Natl Acad Sci 103: 19842–19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, Campisi J. 2003. Reversal of human cellular senescence: Roles of the p53 and p16 pathways. EMBO J 22: 4212–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, O’Neil NJ, Trigiante G, Crook T, Hsieh JK, O’Connor DJ, Zhong S, Campargue I, Tomlinson ML, et al. 2003. iASPP oncoprotein is a key inhibitor of p53 conserved from worm to human. Nat Genet 33: 162–167. [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, Sullivan A, Zvelebil M, Breyssens H, Bisso A, Del Sal G, Syed N, Smith P, Gasco M, et al. 2006. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Genet 38: 1133–1141. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. 2010. Molecular signals of epigenetic states. Science 330: 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Kenzelmann Broz D, Basak S, Park EJ, McLaughlin ME, et al. 2011. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145: 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, Rubin GM. 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol 24: 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JP, Wei W, Sedivy JM. 1997. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science 277: 831–834. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377: 552–557. [DOI] [PubMed] [Google Scholar]
- Cain C, Miller S, Ahn J, Prives C. 2000. The N terminus of p53 regulates its dissociation from DNA. J Biol Chem 275: 39944–39953. [DOI] [PubMed] [Google Scholar]
- Campisi J. 2005. Senescent cells, tumor suppression, and organismal aging: Good citizens, bad neighbors. Cell 120: 513–522. [DOI] [PubMed] [Google Scholar]
- Carvajal D, Tovar C, Yang H, Vu BT, Heimbrook DC, Vassilev LT. 2005. Activation of p53 by MDM2 antagonists can protect proliferating cells from mitotic inhibitors. Cancer Res 65: 1918–1924. [DOI] [PubMed] [Google Scholar]
- Chang BD, Broude EV, Dokmanovic M, Zhu H, Ruth A, Xuan Y, Kandel ES, Lausch E, Christov K, Roninson IB. 1999a. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res 59: 3761–3767. [PubMed] [Google Scholar]
- Chang BD, Xuan Y, Broude EV, Zhu H, Schott B, Fang J, Roninson IB. 1999b. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene 18: 4808–4818. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26: 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checler F, Alves da Costa C. 2014. p53 in neurodegenerative diseases and brain cancers. Pharmacol Ther 142: 99–113. [DOI] [PubMed] [Google Scholar]
- Chen J. 2012. The roles of MDM2 and MDMX phosphorylation in stress signaling to p53. Genes Cancer 3: 274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ko LJ, Jayaraman L, Prives C. 1996. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev 10: 2438–2451. [DOI] [PubMed] [Google Scholar]
- Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, Premsrirut P, Luo W, Chicas A, Lee CS, et al. 2011. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev 25: 2125–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. 2005. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 309: 1732–1735. [DOI] [PubMed] [Google Scholar]
- Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, Wyllie AH. 1993. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 362: 849–852. [DOI] [PubMed] [Google Scholar]
- Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D’Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A. 2012. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet 8: e1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SM, Sanchez CA, Morgan CA, Schimke MK, Ramel S, Idzerda RL, Raskind WH, Reid BJ. 1995. A p53-dependent mouse spindle checkpoint. Science 267: 1353–1356. [DOI] [PubMed] [Google Scholar]
- Croxton R, Ma Y, Song L, Haura EB, Cress WD. 2002. Direct repression of the Mcl-1 promoter by E2F1. Oncogene 21: 1359–1369. [DOI] [PubMed] [Google Scholar]
- Demidenko ZN, Zubova SG, Bukreeva EI, Pospelov VA, Pospelova TV, Blagosklonny MV. 2009. Rapamycin decelerates cellular senescence. Cell Cycle 8: 1888–1895. [DOI] [PubMed] [Google Scholar]
- Demidenko ZN, Korotchkina LG, Gudkov AV, Blagosklonny MV. 2010. Paradoxical suppression of cellular senescence by p53. Proc Natl Acad Sci 107: 9660–9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82: 675–684. [DOI] [PubMed] [Google Scholar]
- Derry WB, Putzke AP, Rothman JH. 2001. Caenorhabditis elegans p53: Role in apoptosis, meiosis, and stress resistance. Science 294: 591–595. [DOI] [PubMed] [Google Scholar]
- Dey A, Wong ET, Bist P, Tergaonkar V, Lane DP. 2007. Nutlin-3 inhibits the NFκB pathway in a p53-dependent manner: Implications in lung cancer therapy. Cell Cycle 6: 2178–2185. [DOI] [PubMed] [Google Scholar]
- Du W, Wu J, Walsh EM, Zhang Y, Chen CY, Xiao ZX. 2009. Nutlin-3 affects expression and function of retinoblastoma protein: Role of retinoblastoma protein in cellular response to nutlin-3. J Biol Chem 284: 26315–26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duensing A, Duensing S. 2005. Guilt by association? p53 and the development of aneuploidy in cancer. Biochem Biophys Res Commun 331: 694–700. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. 2008. Function and structure of inherently disordered proteins. Curr Opin Struct Biol 18: 756–764. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817–825. [DOI] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S. 2005. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci 102: 8204–8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay CA, Hinds PW, Levine AJ. 1989. The p53 proto-oncogene can act as a suppressor of transformation. Cell 57: 1083–1093. [DOI] [PubMed] [Google Scholar]
- Friedler A, Veprintsev DB, Rutherford T, von Glos KI, Fersht AR. 2005. Binding of Rad51 and other peptide sequences to a promiscuous, highly electrostatic binding site in p53. J Biol Chem 280: 8051–8059. [DOI] [PubMed] [Google Scholar]
- Garcia D, Warr MR, Martins CP, Brown Swigart L, Passegue E, Evan GI. 2011. Validation of MdmX as a therapeutic target for reactivating p53 in tumors. Genes Dev 25: 1746–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison SP, Jeffers JR, Yang C, Nilsson JA, Hall MA, Rehg JE, Yue W, Yu J, Zhang L, Onciu M, et al. 2008. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol Cell Biol 28: 5391–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz SA, Wiesmuller L. 2006. p53 in recombination and repair. Cell Death Differ 13: 1003–1016. [DOI] [PubMed] [Google Scholar]
- Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, Zwolinska A, Haupt S, de Lange J, Yip D, et al. 2012. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med 18: 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorina S, Pavletich NP. 1996. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science 274: 1001–1005. [DOI] [PubMed] [Google Scholar]
- Greiss S, Schumacher B, Grandien K, Rothblatt J, Gartner A. 2008. Transcriptional profiling in C. elegans suggests DNA damage dependent apoptosis as an ancient function of the p53 family. BMC Genomics 9: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G. 2006. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol 26: 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamard PJ, Barthelery N, Hogstad B, Mungamuri SK, Tonnessen CA, Carvajal LA, Senturk E, Gillespie V, Aaronson SA, Merad M, et al. 2013. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev 27: 1868–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75: 805–816. [DOI] [PubMed] [Google Scholar]
- Harvey M, Sands AT, Weiss RS, Hegi ME, Wiseman RW, Pantazis P, Giovanella BC, Tainsky MA, Bradley A, Donehower LA. 1993. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene 8: 2457–2467. [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. 2007. A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko T, Ginsberg D. 2004. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem 279: 8627–8634. [DOI] [PubMed] [Google Scholar]
- Horvath MM, Wang X, Resnick MA, Bell DA. 2007. Divergent evolution of human p53 binding sites: Cell cycle versus apoptosis. PLoS Genet 3: e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Deo D, Xia M, Vassilev LT. 2009. Pharmacologic p53 activation blocks cell cycle progression but fails to induce senescence in epithelial cancer cells. Mol Cancer Res 7: 1497–1509. [DOI] [PubMed] [Google Scholar]
- Jain AK, Barton MC. 2012. Unmet expectations: miR-34 plays no role in p53-mediated tumor suppression in vivo. PLoS Genet 8: e1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al. 2003. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4: 321–328. [DOI] [PubMed] [Google Scholar]
- Kaeser MD, Iggo RD. 2002. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc Natl Acad Sci 99: 95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo KH, Verma CS, Lane DP. 2014. Drugging the p53 pathway: Understanding the route to clinical efficacy. Nat Rev Drug Discov 13: 217–236. [DOI] [PubMed] [Google Scholar]
- Komarova EA, Chernov MV, Franks R, Wang K, Armin G, Zelnick CR, Chin DM, Bacus SS, Stark GR, Gudkov AV. 1997. Transgenic mice with p53-responsive lacZ: p53 activity varies dramatically during normal development and determines radiation and drug sensitivity in vivo. EMBO J 16: 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz D, Dobbelstein M. 2006. Nongenotoxic p53 activation protects cells against S-phase-specific chemotherapy. Cancer Res 66: 10274–10280. [DOI] [PubMed] [Google Scholar]
- Lanni JS, Jacks T. 1998. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol 18: 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, et al. 2006. Inactivation of the p53 pathway in retinoblastoma. Nature 444: 61–66. [DOI] [PubMed] [Google Scholar]
- Leu JI, Dumont P, Hafey M, Murphy ME, George DL. 2004. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat Cell Biol 6: 443–450. [DOI] [PubMed] [Google Scholar]
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W. 2012. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149: 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Parant JM, Lang G, Chau P, Chavez-Reyes A, El-Naggar AK, Multani A, Chang S, Lozano G. 2004. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet 36: 63–68. [DOI] [PubMed] [Google Scholar]
- Luo Y, Hurwitz J, Massague J. 1995. Cell-cycle inhibition by independent CDK and PCNA binding domains in p21Cip1. Nature 375: 159–161. [DOI] [PubMed] [Google Scholar]
- MacCallum DE, Hupp TR, Midgley CA, Stuart D, Campbell SJ, Harper A, Walsh FS, Wright EG, Balmain A, Lane DP, et al. 1996. The p53 response to ionising radiation in adult and developing murine tissues. Oncogene 13: 2575–2587. [PubMed] [Google Scholar]
- Mantovani F, Tocco F, Girardini J, Smith P, Gasco M, Lu X, Crook T, Del Sal G. 2007. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat Struct Mol Biol 14: 912–920. [DOI] [PubMed] [Google Scholar]
- Mao JH, Wu D, Kim IJ, Kang HC, Wei G, Climent J, Kumar A, Pelorosso FG, DelRosario R, Huang EJ, et al. 2012. Hipk2 cooperates with p53 to suppress γ-ray radiation-induced mouse thymic lymphoma. Oncogene 31: 1176–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko ND, Zaika A, Moll UM. 2000. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem 275: 16202–16212. [DOI] [PubMed] [Google Scholar]
- Martin-Caballero J, Flores JM, Garcia-Palencia P, Serrano M. 2001. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res 61: 6234–6238. [PubMed] [Google Scholar]
- Martinez J, Georgoff I, Levine AJ. 1991. Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes Dev 5: 151–159. [DOI] [PubMed] [Google Scholar]
- Michalak EM, Villunger A, Adams JM, Strasser A. 2008. In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ 15: 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. 2003. p53 has a direct apoptogenic role at the mitochondria. Mol Cell 11: 577–590. [DOI] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716. [DOI] [PubMed] [Google Scholar]
- Oberst A, Bender C, Green DR. 2008. Living with death: The evolution of the mitochondrial pathway of apoptosis in animals. Cell Death Differ 15: 1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, et al. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102: 849–862. [DOI] [PubMed] [Google Scholar]
- Okoshi R, Ozaki T, Yamamoto H, Ando K, Koida N, Ono S, Koda T, Kamijo T, Nakagawara A, Kizaki H. 2008. Activation of AMP-activated protein kinase induces p53-dependent apoptotic cell death in response to energetic stress. J Biol Chem 283: 3979–3987. [DOI] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 29: 92–95. [DOI] [PubMed] [Google Scholar]
- Perfettini JL, Castedo M, Nardacci R, Ciccosanti F, Boya P, Roumier T, Larochette N, Piacentini M, Kroemer G. 2005. Essential role of p53 phosphorylation by p38 MAPK in apoptosis induction by the HIV-1 envelope. J Exp Med 201: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros AM, Gunasekera A, Xu N, Olejniczak ET, Fesik SW. 2004. Defining the p53 DNA-binding domain/Bcl-xL-binding interface using NMR. FEBS Lett 559: 171–174. [DOI] [PubMed] [Google Scholar]
- Pietsch EC, Perchiniak E, Canutescu AA, Wang G, Dunbrack RL, Murphy ME. 2008. Oligomerization of BAK by p53 utilizes conserved residues of the p53 DNA binding domain. J Biol Chem 283: 21294–21304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post SM, Quintas-Cardama A, Terzian T, Smith C, Eischen CM, Lozano G. 2010. p53-dependent senescence delays Eμ-myc-induced B-cell lymphomagenesis. Oncogene 29: 1260–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca R, Nardinocchi L, Givol D, D’Orazi G. 2010. Regulation of p53 activity by HIPK2: Molecular mechanisms and therapeutical implications in human cancer cells. Oncogene 29: 4378–4387. [DOI] [PubMed] [Google Scholar]
- Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. 2012. p53 dynamics control cell fate. Science 336: 1440–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Wang T, Naumovski L, Lopez CD, Brachmann RK. 2002. Groups of p53 target genes involved in specific p53 downstream effects cluster into different classes of DNA binding sites. Oncogene 21: 7901–7911. [DOI] [PubMed] [Google Scholar]
- Ray-Coquard I, Blay JY, Italiano A, Le Cesne A, Penel N, Zhi J, Heil F, Rueger R, Graves B, Ding M, et al. 2012. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: An exploratory proof-of-mechanism study. Lancet Oncol 13: 1133–1140. [DOI] [PubMed] [Google Scholar]
- Rigatti MJ, Verma R, Belinsky GS, Rosenberg DW, Giardina C. 2012. Pharmacological inhibition of Mdm2 triggers growth arrest and promotes DNA breakage in mouse colon tumors and human colon cancer cells. Mol Carcinog 51: 363–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. 2008. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412. [DOI] [PubMed] [Google Scholar]
- Ringshausen I, O’Shea CC, Finch AJ, Swigart LB, Evan GI. 2006. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell 10: 501–514. [DOI] [PubMed] [Google Scholar]
- Rodier F, Munoz DP, Teachenor R, Chu V, Le O, Bhaumik D, Coppe JP, Campeau E, Beausejour CM, Kim SH, et al. 2011. DNA-SCARS: Distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J Cell Sci 124: 68–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Ludwig RL, Haupt Y, Bates S, Lu X, Oren M, Vousden KH. 1996. Specific loss of apoptotic but not cell-cycle arrest function in a human tumor derived p53 mutant. EMBO J 15: 827–838. [PMC free article] [PubMed] [Google Scholar]
- Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. 2005. The antioxidant function of the p53 tumor suppressor. Nat Med 11: 1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels-Lev Y, O’Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X. 2001. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 8: 781–794. [DOI] [PubMed] [Google Scholar]
- Schlereth K, Beinoraviciute-Kellner R, Zeitlinger MK, Bretz AC, Sauer M, Charles JP, Vogiatzi F, Leich E, Samans B, Eilers M, et al. 2010. DNA binding cooperativity of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 38: 356–368. [DOI] [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, Lowe SW. 2002. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109: 335–346. [DOI] [PubMed] [Google Scholar]
- Sdek P, Ying H, Chang DL, Qiu W, Zheng H, Touitou R, Allday MJ, Xiao ZX. 2005. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol Cell 20: 699–708. [DOI] [PubMed] [Google Scholar]
- Sot B, Freund SM, Fersht AR. 2007. Comparative biophysical characterization of p53 with the pro-apoptotic BAK and the anti-apoptotic BCL-xL. J Biol Chem 282: 29193–29200. [DOI] [PubMed] [Google Scholar]
- St Clair S, Giono L, Varmeh-Ziaie S, Resnick-Silverman L, Liu WJ, Padi A, Dastidar J, DaCosta A, Mattia M, Manfredi JJ. 2004. DNA damage-induced downregulation of Cdc25C is mediated by p53 via two independent mechanisms: One involves direct binding to the cdc25C promoter. Mol Cell 16: 725–736. [DOI] [PubMed] [Google Scholar]
- Sullivan KD, Padilla-Just N, Henry RE, Porter CC, Kim J, Tentler JJ, Eckhardt SG, Tan AC, DeGregori J, Espinosa JM. 2012. ATM and MET kinases are synthetic lethal with nongenotoxic activation of p53. Nat Chem Biol 8: 646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JE, Brehm A. 2004. Of flies and men; p53, a tumour suppressor. FEBS Lett 567: 86–91. [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 24: 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds H, Krall L, Remington L, Saenz-Robles M, Lowe S, Jacks T, Van Dyke T. 1994. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell 78: 703–711. [DOI] [PubMed] [Google Scholar]
- Szak ST, Mays D, Pietenpol JA. 2001. Kinetics of p53 binding to promoter sites in vivo. Mol Cell Biol 21: 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K. 2007. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol Cell 25: 725–738. [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR. 2010. Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 11: 621–632. [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. 2006. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 24: 827–839. [DOI] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. 2008. Acetylation is indispensable for p53 activation. Cell 133: 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. 2007. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 6: 1586–1593. [DOI] [PubMed] [Google Scholar]
- Timofeev O, Schlereth K, Wanzel M, Braun A, Nieswandt B, Pagenstecher A, Rosenwald A, Elsasser HP, Stiewe T. 2013. p53 DNA binding cooperativity is essential for apoptosis and tumor suppression in vivo. Cell Rep 3: 1512–1525. [DOI] [PubMed] [Google Scholar]
- Tomita Y, Marchenko N, Erster S, Nemajerova A, Dehner A, Klein C, Pan H, Kessler H, Pancoska P, Moll UM. 2006. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J Biol Chem 281: 8600–8606. [DOI] [PubMed] [Google Scholar]
- Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, et al. 2006. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: Implications for therapy. Proc Natl Acad Sci 103: 1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente LJ, Gray DH, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL, Janic A, Strasser A. 2013. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep 3: 1339–1345. [DOI] [PubMed] [Google Scholar]
- Valentin-Vega YA, Box N, Terzian T, Lozano G. 2009. Mdm4 loss in the intestinal epithelium leads to compartmentalized cell death but no tissue abnormalities. Differentiation 77: 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. 2012. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 149: 1536–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. 2007. Restoration of p53 function leads to tumour regression in vivo. Nature 445: 661–665. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. 2009. Blinded by the light: The growing complexity of p53. Cell 137: 413–431. [DOI] [PubMed] [Google Scholar]
- Wang Y, Suh YA, Fuller MY, Jackson JG, Xiong S, Terzian T, Quintas-Cardama A, Bankson JA, El-Naggar AK, Lozano G. 2011. Restoring expression of wild-type p53 suppresses tumor growth but does not cause tumor regression in mice with a p53 missense mutation. J Clin Invest 121: 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR. 2005. Comparative binding of p53 to its promoter and DNA recognition elements. J Mol Biol 348: 589–596. [DOI] [PubMed] [Google Scholar]
- Wu X, Levine AJ. 1994. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci 91: 3602–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Woodfield SE, Lee TV, Fan Y, Antonio C, Bergmann A. 2009. Genetic control of programmed cell death (apoptosis) in Drosophila. Fly 3: 78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445: 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. 1991. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature 352: 345–347. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Liu H, Miki Y. 2006. Protein kinase C δ regulates Ser46 phosphorylation of p53 tumor suppressor in the apoptotic response to DNA damage. J Biol Chem 281: 5734–5740. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong S, Li Q, Hu S, Tashakori M, Van Pelt C, You MJ, Pageon L, Lozano G. 2014. Tissue-specific and age-dependent effects of global Mdm2 loss. J Pathol 233: 380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chen L, Pledger WJ, Fang J, Chen J. 2013. p53 promotes repair of heterochromatin DNA by regulating JMJD2b and SUV39H1 expression. Oncogene 33: 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]